Abstract
This study aims to model genetic, immunologic, metabolomics, and proteomic biomarkers for development of islet autoimmunity (IA) and progression to type 1 diabetes in a prospective high-risk cohort. We studied 67 children: 42 who developed IA (20 of 42 progressed to diabetes) and 25 control subjects matched for sex and age. Biomarkers were assessed at four time points: earliest available sample, just prior to IA, just after IA, and just prior to diabetes onset. Predictors of IA and progression to diabetes were identified across disparate sources using an integrative machine learning algorithm and optimization-based feature selection. Our integrative approach was predictive of IA (area under the receiver operating characteristic curve [AUC] 0.91) and progression to diabetes (AUC 0.92) based on standard cross-validation (CV). Among the strongest predictors of IA were change in serum ascorbate, 3-methyl-oxobutyrate, and the PTPN22 (rs2476601) polymorphism. Serum glucose, ADP fibrinogen, and mannose were among the strongest predictors of progression to diabetes. This proof-of-principle analysis is the first study to integrate large, diverse biomarker data sets into a limited number of features, highlighting differences in pathways leading to IA from those predicting progression to diabetes. Integrated models, if validated in independent populations, could provide novel clues concerning the pathways leading to IA and type 1 diabetes.
Introduction
Type 1 diabetes results from autoimmune destruction of insulin-producing pancreatic β-cells. Clinically apparent diabetes is typically preceded by a period of islet autoimmunity (IA), marked by appearance of autoantibodies against islet autoantigens (1). While there is consensus that chronic autoimmune destruction of β-cells is triggered by an interaction of environmental factor(s) with a relatively common genetic background, the specific cause remains elusive. Prospective cohort studies have reported a number of demographic, immune (2–4), genetic (5–10), metabolomic (11,12), and proteomic (13–15) predictors of IA and/or progression from IA to diabetes. Each analytic approach offers unique insights; however, single data stream analysis is unable to address the importance of technique-specific observations in the context of other analyses. Use of data fusion methods to integrate different data types can create models that are more complete and accurate than those derived from any individual source (16). Our objective was to provide proof of principle that machine learning Bayesian modeling of disparate biomarkers can yield useful integrated models for hypothesis generation. Applied to longitudinally collected biomarkers, such integrated models could provide novel clues concerning pathways leading to IA and/or diabetes. This integrative modeling approach could improve personalized prediction of progression through presymptomatic stages of type 1 diabetes.
Research Design and Methods
Study Participants
We performed a nested case-control study of children participating in the Diabetes Autoimmunity Study in the Young (DAISY) cohort. DAISY follows prospectively 2,547 children at increased risk for type 1 diabetes. The cohort consists of first-degree relatives of patients with type 1 diabetes (FDRs) and general population children with type 1 diabetes–susceptibility HLA DR-DQ genotypes identified by newborn screening (17,18), recruited between 1993 and 2004. Follow-up results are available through 29 September 2017. Written informed consent was obtained from subjects and parents. The Colorado Multiple Institutional Review Board approved all protocols.
Outcome Measures
Autoantibodies were tested at 9, 15, and 24 months and, if negative, annually thereafter; autoantibody-positive children were retested every 3–6 months. Radio-immunoassays for insulin (IAA), GAD (GADA), insulinoma-associated protein 2 (IA-2A), and/or zinc transporter 8 (ZnT8A) autoantibodies were conducted as previously described (19–23). Subjects were considered persistently islet autoantibody positive if they had two or more consecutive confirmed positive samples, not due to maternal islet autoantibody transfer, or had one confirmed positive sample and developed diabetes prior to next sample collection. Diabetes was diagnosed using American Diabetes Association criteria.
Selection of Subjects for Analyses
Sixty-seven children were selected in July of 2011 from the DAISY cohort for studies of metabolomic, proteomic, and immune predictors. Of those, 22 children developed diabetes (T1D group), 20 had developed persistent IA and were islet autoantibody positive at their last study visit (AbPos group), and 25 were control subjects (control group). Control subjects were frequency matched with subjects in the combined T1D and AbPos group on the HLA DR-DQ genotypes, age, sex, and FDR status. As of 29 September 2017, all control subjects have been negative for all islet autoantibodies. Of the AbPos group, four progressed to diabetes in subsequent years at median age 17.8 years. These individuals were retained in the AbPos group. Supplementary Fig. 1 describes subject selection. Supplementary Table 1 presents individual autoantibody histories of all case and control subjects at relevant time points.
Specimens for Analysis
When available, samples for each subject were analyzed for metabolomic, proteomic, and immune biomarkers at four time points: T1, earliest available sample prior to development of islet autoantibodies (typically age 9–15 months); T2, just prior to development of first autoantibody; T3, just after development of first autoantibody; and T4, just prior to diagnosis of diabetes or most recent sample for AbPos subjects at time of sample selection.
Of the subject with T1D, five were missing a T2 sample. Samples from control subjects were selected to frequency match storage time of samples from T1D and AbPos subjects combined.
Metabolomic Analysis
Global metabolic profiling combined two separate ultrahigh-performance liquid chromatography/tandem mass spectrometry (UHPLC/MS/MS2) injections, optimized for basic and acidic species, and gas chromatography/mass spectrometry (GS/MS) (Metabolon, Durham, NC). All serum samples were stored at −80°C ≤1 h after collection, never thawed until analyses, and processed essentially as described previously (24,25). Metabolites were identified by automated comparison of ion features in experimental samples with a reference library of chemical standard entries using software developed at Metabolon (26). A total of 382 named metabolites were included in this analysis. For statistical analyses and data display, any missing values were assumed to be below limits of detection, and these values were imputed with the compound minimum (minimum value imputation).
Proteomic Analysis
Relative abundance of 1,001 serum proteins were measured by aptamers (Somalogic, Boulder, CO) (27). Additionally, 49 peptides (representing 24 proteins) were measured by LC-MRM/MS in the laboratory of Drs. Thomas Metz and Qibin Zhang at Pacific Northwest National Laboratory as previously described (28).
Immune Markers
Cytokines were measured using a Human Custom Cytokine 9-Plex assay (Meso Scale Discovery, Rockville, MD) and included interferon (IFN)-α2a, interleukin (IL)-6, IL-17, IL-1β, interferon γ–induced protein (IP)-10, monocyte chemotactic protein (MCP)-1, IFN-γ, IL-1α, and IL-1ra (4).
Genotyping
All 106 non-HLA SNPs available for these subjects were included. Locus and reference for genotyping are shown in Supplementary Table 2. As genetic data were derived from several analyses, some genes were represented by more than one SNP and the rs2476601 SNP for PTPN22 was present in two genetic feature sets from separate analyses (Supplementary Table 2).
Metadata
Individual metadata included in the model consisted of age at sampling, sex, Hispanic ethnicity, and FDR status (classified as mother with type 1 diabetes, other FDR with type 1 diabetes, or no FDR) Additionally, subjects were classified by four HLA risk categories based on typing for HLA class II alleles as previously described (29).
Statistics and Machine Learning
SAS, version 9.4 (SAS Institute, Cary, NC), was used to analyze descriptive data for groups. Integrative machine learning, based on the set of demographic characteristics, gene variants, cytokines, proteins, and metabolites, was used to evaluate whether predictive models can separate future cases from control subjects, as well as identify the primary features that distinguish the groups. Two disease stages were modeled. 1) Development of IA: transition from T1 to T2 among combined AbPos and T1D subjects versus control subjects. 2) Progression to diabetes: transition from T3 to T4 among T1D versus AbPos subjects. Transition between time points is represented in analysis by determining log fold difference of cytokine, protein, or metabolite between time points. Integrative machine learning was performed using probability-based integration of multiple data streams via the Posterior Probability Product (P3) (16,30). Feature selection was performed in the context of the integrated model using a Repeated Optimization for Feature Interpretation (ROFI) approach (31). The ROFI-P3 algorithm for this analysis is described in detail in Supplementary Fig. 2. It has several key characteristics amenable to identifying and evaluating important features of diabetes progression across disparate data sources, which include allowing each data set to be modeled with the optimal machine learning algorithm and features to be assigned importance metrics through repeated analyses.
The first step requires selection of the machine learning method to model each data set (e.g., metabolomic, proteomic, etc.). We evaluated seven machine learning classification methods with all features in each data set where a feature is the ratio of the measurements between time points for a case or control subject, including random forest (RF), logistic regression (LR), k-nearest neighbors (KNN), linear discriminant analysis (LDA), support vector machine (SVM) with a radial basis function (RBF) kernel, SVM with a linear (LIN) kernel, and naive Bayes (NB). Supplementary Fig. 3 shows average area under the receiver operating characteristic (ROC) curve (AUC) based on fivefold CV repeated 100 times. The only requirement of this step is that a machine learning classifier can output the posterior probability, defined as the probability that class i
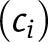 is observed given the data for subject s of data set j
is observed given the data for subject s of data set j
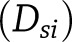 ; P(ci|Dsj). The posterior probabilities are generated using the standard functions in the R programming language for each machine learning algorithm.
; P(ci|Dsj). The posterior probabilities are generated using the standard functions in the R programming language for each machine learning algorithm.
Integration via the P3 approach is a naïve product-based integration as the product of the posterior probability of each sample as related to each data sets:
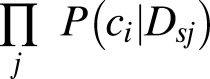 |
(16,30). These integrated probabilities can be used to compute the accuracy of the integrated model using a standard AUC. Feature selection is performed on the integrated model to assure that features selected are those that work best in combination across disparate sources. Selection utilizes a statistical optimization algorithm, such as simulated annealing, which is not affected by the order of features in the data set and allows the algorithm to move out of local minima by updating the solution at each iteration based on the current feature state and sampling in proportion to including or excluding the variable of interest. Thus, for each feature change proposal, this is based on looking at the difference of the accuracy of the current state (AUCCurrent) and an updated solution (AUCUpdated). The updated solution is selected in proportion to:
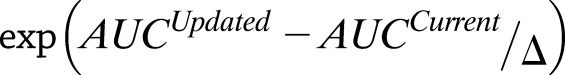 |
based on a uniform distribution between 0 and 1, where Δ = 0.25 for this analysis. For each run of the algorithm, we perform 100 random changes of individual features and keep or discard the change based on this exponential difference between AUC values. After each 100 proposals and potential updates, we determine whether the solution has converged based on the difference between the AUC prior to the 100 feature evaluations and the current solution. If this value is <δ, which was set to 1E-4 for this analysis, it is determined that the solution has converged (31).
Within ROFI-P3, the AUC is computed based on fivefold CV for every feature evaluation. We repeat the algorithm in conjunction with CV for 100 repetitions, each of which yields a single feature set solution. We use the 100 repetitions to obtain a feature ensemble solution, which gives the likelihood that the feature would be selected for inclusion in the model. This is represented as the percentage of times that a specific feature was selected to be in the model. This also has the additional benefit of yielding robust measures of uncertainty on our classification accuracy metrics.
To evaluate the performance of the ROFI-P3 method versus established optimization methods, we compared ROFI-P3 with standard recursive feature elimination (RFE) approaches. RFE is a method used extensively in biology in combination with various machine learning algorithms, such as LDA and SVM (16,30,32,33). RFE is readily available in most statistical programming languages and is simple to implement. It is a greedy algorithm that sequentially eliminates the feature that yields the maximum AUC.
Data and Resource Availability
The data sets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Results
Characteristics of the study subjects are presented in Table 1. Ages at visits ranged from 6 months to almost 21 years old with similar ages at time points T1, T2, T3, and T4 (Supplementary Table 3). Demographic (metadata), genetic, immune, metabolic and proteomic biomarkers were analyzed across these four distinct time points.
Table 1.
Characteristics of the study participants
| Control | AbPos | T1D | P | |
|---|---|---|---|---|
| N | 25 | 20 | 22 | |
| HLA DR group, n | 0.55 | |||
| 4/4, 4/3, or 4/X and DQB1*03:02 | 17 | 15 | 17 | |
| 3/3 or 3/X | 5 | 3 | 3 | |
| Other | 3 | 2 | 2 | |
| FDR, n (%) | 9 (36) | 11 (55) | 15 (68) | 0.08 |
| Female sex, n (%) | 13 (52) | 8 (40) | 10 (45) | 0.72 |
| NHW ethnicity, n (%) | 20 (80) | 15 (75) | 21 (95) | 0.17 |
| Age (years) at development of IA, median (IQR) | 7.4 (5.4, 9.9) | 5.2 (2.9, 7.9) | 0.06 | |
| Age (years) at development of diabetes, median (IQR) | 11.0 (9.4, 13.7) | — |
All comparisons by χ2 except age at IA, which was compared using Wilcoxon rank sum test. IQR, interquartile range; NHW, non-Hispanic white; X, neither HLA DR4 nor DR3.
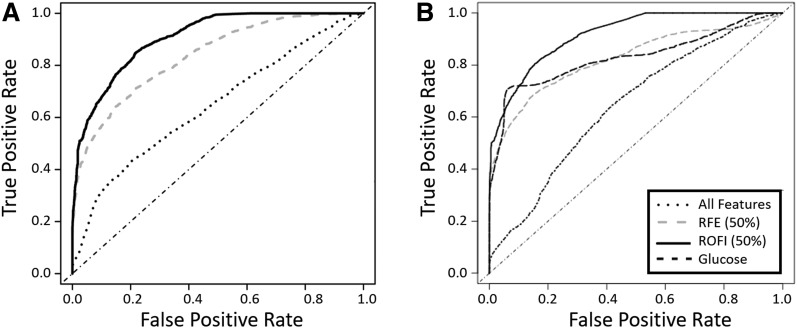
First, we determined the optimal machine learning algorithm for each of the data types (Supplementary Fig. 3). Next, we analyzed the ability of ROFI-P3 to predict the changes leading up to two stages of type 1 diabetes: 1) seroconversion and 2) progression to diabetes. ROFI-P3 was performed, and for each repetition features were selected. Each feature is represented as the percentage of times it is selected as part of the model during 100 repetitions. For comparison, we also ran RFE for multiple repetitions, each time permuting the features, since the order of the features has a direct relationship with those selected. This allowed us also to represent RFE features as the percentage of times they were selected. To evaluate how well the methods work, a feature selection threshold is selected and an ROC curve was generated only on this reduced model using fivefold CV to build and test the model independently and to minimize overfitting. Fig. 1A and B show the results of these comparisons at a 50% frequency selection, i.e., features selected at least 50% of the time for both ROFI-P3 and RFE, as well as if no feature selection is performed. Various thresholds were evaluated, and the ROFI-P3 integrated feature selection approach was consistently more accurate than RFE. This demonstrated a clear advantage over both the RFE selection and simple combination of all features for prediction of development of IA (AUC 0.91 vs. 0.84 and 0.64, respectively, P < 0.0001) and progression (AUC 0.92 vs. 0.82 and 0.64, respectively, P < 0.0001) at a 50% feature selection threshold. We further evaluated the method in the context of the classification of specific individuals rather than a global metric of classification. If we select a defined reasonable false positive rate of 10% for the development of IA (Fig. 1A), we would correctly classify 67.6% of those who developed IA with ROFI-P3. The percentages drop to 56.5% and 31.5% for the RFE selection and simple combination approaches, respectively. The prediction is slightly better for the progression end point; ROFI-P3 correctly identifies 71.6% compared with 61.4% for RFE and 18.3% for simple combination (Fig. 1B). The top features selected by ROFI-P3 at a 50% frequency or higher for analysis of development of IA (Supplementary Fig. 4A) and progression to diabetes (Supplementary Fig. 4B) included features from five of the six data sets.
Figure 1.
ROC curves. A: Comparing development of IA in control group vs. combined AbPos and T1D groups at transition from earliest time point (T1) to preseroconversion (T2). B: Comparing progression to T1D—transition from postseroconversion (T3) to before T1D diagnosis (T4)—in AbPos vs. T1D groups. Dotted line, prediction based on all features; gray dashed line (RFE), prediction based on features selected 50% of the time or more using recursive feature elimination; solid black line (ROFI), prediction based on features selected at least 50% of the time using ROFI-P3 algorithm; black dashed line (in panel B only), prediction based on glucose change from T3 to T4.
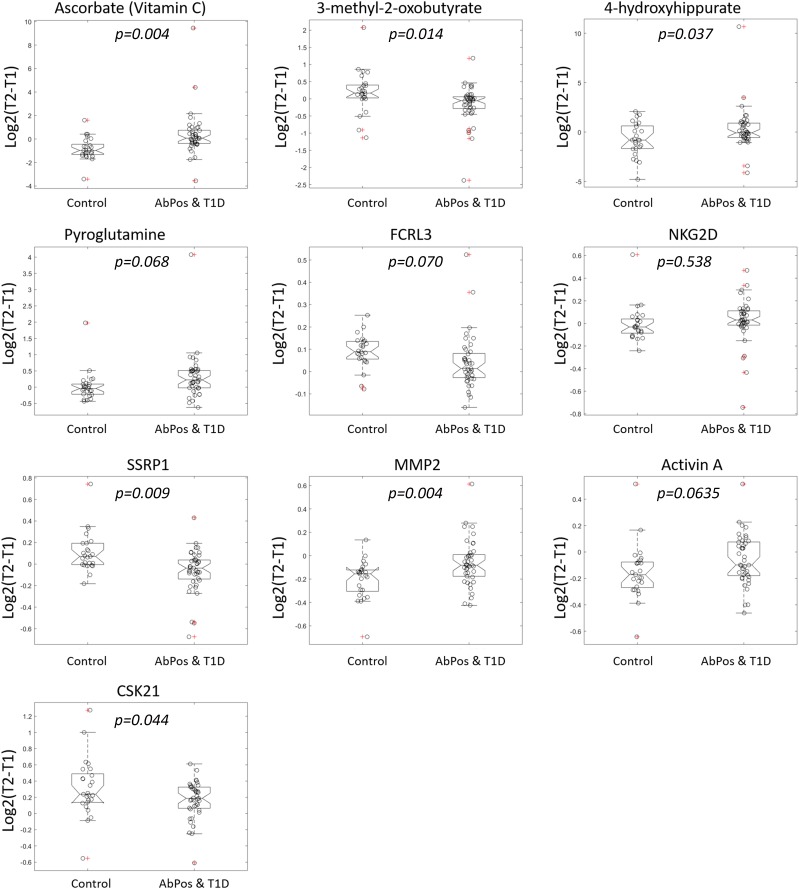
The top features selected by ROFI-P3 as predictors of IA are shown in Table 2. Percent selected is a measure of the number of times a particular feature was selected in the 100 iterations of the algorithm, an indication of the importance of that feature in the prediction of outcome. Supplementary Table 4 shows all 76 features selected at 50% frequency or higher. Further detail regarding these features is shown in Supplementary Tables 5–8. Box plots of the log fold change in abundance in these top metabolites, proteins, and peptides from T1 to T2 for IA and control subjects are shown in Fig. 2.
Table 2.
The top 16 predictors for development of IA
| Selected (%) | Source | Feature | Function/description |
|---|---|---|---|
| 100 | Metabolite | Ascorbate (vitamin C) | Antioxidant and coenzyme |
| 100 | Metadata | Age (years) | Age at T1 |
| 98 | Metadata | First-degree relative status | Grouped by mother with type 1 diabetes, other FDR (sibling or father), or no FDR |
| 94 | Metabolite | 3-methyl-2-oxobutyrate | Branched-chain organic acid; precursor to leucine and valine synthesis |
| 93 | Protein | FCRL3 (Fc receptor-like protein 3) | Promotes TLR9-induced B-cell proliferation, activation, and survival |
| 91 | Metabolite | 4-hydroxyhippurate | Microbial end product derived from polyphenol metabolism by the microflora in the intestine |
| 90 | Metadata | Hispanic | Self-report of Hispanic ethnicity |
| 90 | Protein | NKG2D type II integral membrane protein/KLRK1 (Killer cell lectin-like receptor subfamily K member 1) | Stimulatory and costimulatory innate immune response on activated killer cells; involved in immunosurveillance of virus-infected cells |
| 89 | SNP | rs2476601 (PTPN22) | Autoimmunity gene; negative regulator of T-cell receptor signaling |
| 89 | Protein | SSRP1 (Structure Specific Recognition Protein 1)/FACT (Facilitates Chromatin Transcription) complex subunit | The FACT complex plays a role in mRNA elongation, DNA replication, and DNA repair |
| 89 | Metabolite | Pyroglutamine | Glutamine and glutathione metabolism |
| 87 | Protein | MMP-2 | Metalloproteinase involved in diverse functions including angiogenesis, tissue repair, and inflammation |
| 86 | Protein | Activin A | Member of TGF-β superfamily of cytokines; plays role in regulation of tissue homeostasis, organ development, inflammation, cell proliferation, and apoptosis |
| 85 | SNP | rs2476601 (PTPN22) | Autoimmunity gene; negative regulator of T-cell receptor signaling |
| 84 | Protein | CSK21 (Casein kinase II subunit alpha) | Regulates various cellular processes including cell cycle progression, apoptosis, and transcription as well as response to viral infections |
| 84 | SNP | rs3087243 (CTLA4) | Autoimmunity gene; negative regulator of T-cell responses |
Analysis by ROFI-P3 comparing control subjects with pooled antibody-positive subjects and subjects with type 1 diabetes. Ranking by selection frequency for metabolites and proteins (fold change from T1 to T2) or SNPs (risk allele count). Proteins: www.genecards.org and www.uniprot.org. SNPs: www.SNPedia.com. Metabolite: www.hmdb.ca.
Figure 2.
The top 10 protein, peptide, and metabolite predictors for development of IA. For each analyte, the box plots show log fold change from time 1 (T1) to time 2 (T2) for case and control subjects with individual values noted by circles. The value of log2(T2 − T1) is positive with increasing trajectory and negative with decreasing trajectory.
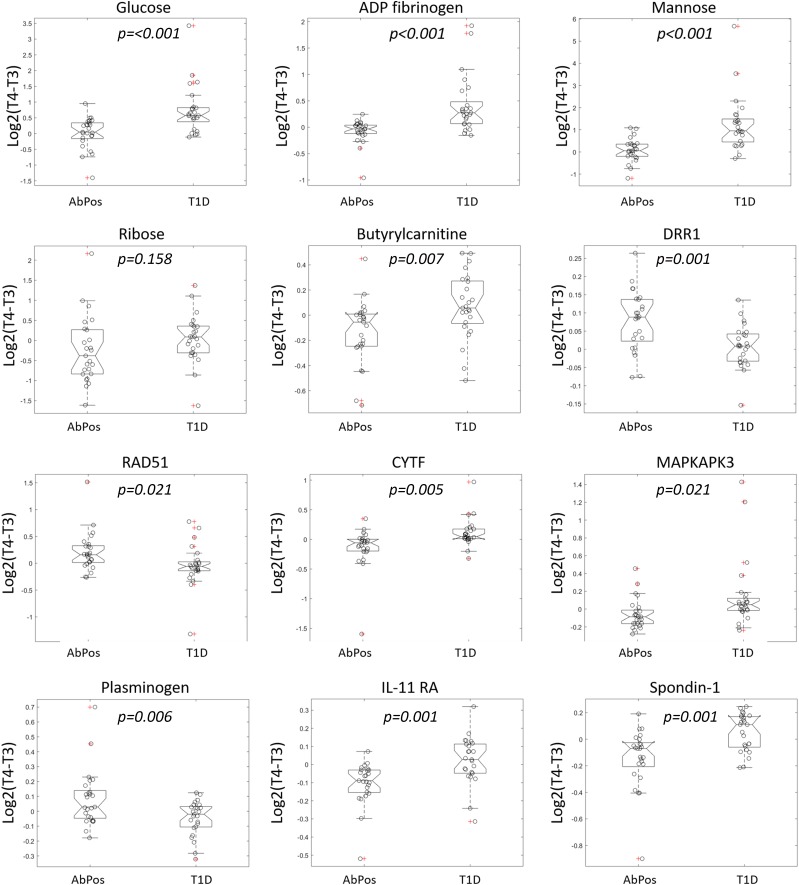
The top features selected by ROFI-P3 as predictors of progression from IA to diabetes are shown in Table 3, while all 83 features selected at 50% frequency or higher are shown in Supplementary Table 9. Characteristics of features including genotypes or direction of change in abundance from T3 to T4 are further described in Supplementary Tables 10–13. Figure 3 shows the box plots for control and AbPos groups of the log fold change of each feature from T3 to T4.
Table 3.
The top 16 predictors of progression from IA to diabetes
| Selected (%) | Source | Feature | Function/description |
|---|---|---|---|
| 100 | Metabolite | Glucose | Carbohydrate metabolism |
| 100 | Metadata | Age (years) | Age at T3 |
| 100 | Metabolite | ADP fibrinogen | Coagulation |
| 100 | Protein | DRR1 (downregulated in renal cell carcinoma 1)/actin-associated protein FAM107A | Regulation of cytoskeleton organization and cell growth |
| 99 | Metabolite | Mannose | Carbohydrate metabolism |
| 98 | Protein | RAD51 (DNA repair protein RAD51 homolog 1) | Response to DNA damage; DNA repair |
| 98 | Protein | CYTF (cystatin-F) | Inhibits cathepsin L; may play a role in immune regulation |
| 97 | Protein | MAPKAPK3 (MAP kinase–activated protein kinase 3) | Stress-activated serine/threonine protein kinase |
| 92 | SNP | MHC (rs3117103) | SLE-associated genetic variant on chromosome 6 |
| 91 | SNP | From GWAS for T1D (rs7221109) | Type 1 diabetes–associated genetic variant on chromosome 17 |
| 90 | Protein | Plasminogen | Dissolves fibrin in blood clots; plays a role in inflammation and tissue remodeling |
| 89 | Metabolite | Ribose | Carbohydrate metabolism |
| 89 | Protein | IL-11 RA (interleukin-11 receptor subunit α) | Development and proliferation of mesenchymal cells |
| 89 | SNP | HLA DQB1 8.1 (rs2157678) | Type 1 diabetes–associated genetic variant; associated with HLA DR3-B8-A1 |
| 87 | Metabolite | Butyrylcarnitine | Fatty acid ester |
| 86 | Protein | Spondin-1 | Cell adhesion protein |
Analysis by ROFI-P3 comparing islet autoantibody–positive subjects who progressed to diabetes with those who did not progress. Ranking by selection frequency for metabolites and proteins (fold change from T3 to T4) or SNPs (risk allele count). Proteins: www.genecards.org and www.uniprot.org. SNPs: www.SNPedia.com. Metabolite: www.hmdb.ca. GWAS, genome-wide association studies; SLE, systemic lupus erythematosus.
Figure 3.
The top 12 protein, peptide, and metabolite predictors for progression to diabetes. For each analyte, the box plots show log fold change from time 3 (T3) to time 4 (T4) for case and control subjects with individual values noted by circles. The value of log2(T4 − T3) is positive with increasing trajectory and negative with decreasing trajectory.
Of note, the metabolite glucose is the top selected feature, as could be expected during the progression to T1D. To evaluate the value of adding additional features to glucose, we ran a logistic regression using glucose alone as the predictor for progression to T1D compared with that of the selected ensemble. Change in glucose alone was able to classify a majority of case subjects, but the AUC for the ROFI-P3 (0.91) was significantly higher than that for glucose alone (0.83, P < 0.00001) (Fig. 1B). As expected for the end point of development of IA, the AUC of glucose is not predictive: 0.48.
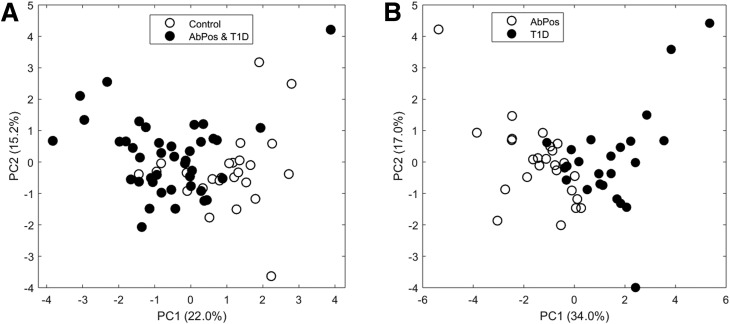
Overfitting is often an issue with machine learning, especially when sample sizes are only large enough to allow CV. To evaluate whether the top features could separate the groups with an unsupervised approach, principal component analysis (PCA) was utilized on only the top qualitative omics features in Tables 1 and 2. From the PCA plot, the first component can visually separate the two groups for both predictors of IA (Fig. 4A) and progression from IA to diabetes (Fig. 4B) without prior knowledge of the groups. This demonstrates that although there may be some overfitting, the methodology in general is identifying features that can discriminate the groups of interest.
Figure 4.
PCA of (A) predictors of IA based on top 4 metabolites and 6 proteins and (B) progression from IA to diabetes on top 5 metabolites and 7 proteins. Open circles represent control subjects, and closed circles represent combined AbPos and T1D groups. PC1, principal component 1; PC2, principal component 2.
Discussion
Identification of causative factors in the development of IA and type 1 diabetes has been elusive. Recent observations regarding the role of vitamin D in risk of IA has underlined the importance of understanding environmental exposures in the context of genetic background (34). Thus, analysis that integrates multiple data streams has the potential to identify unique ensembles of pathogenic features. This proof-of-concept analysis represents the first integration of disparate omics data sets for the prediction of IA and type 1 diabetes. The ROFI-P3 approach solves the feature selection process through hundreds of iterations, resulting in a probability measure for each individual feature. This allows the reduction of large data sets to a smaller, more informative set of features as well as a robust measure of feature-level uncertainty. The biomarker panels identified in this analysis represent an individualized prediction algorithm based on a set of disparate features (e.g., metabolites, proteins in combination with genetics, and standard risk factors) selected in at least 50% of the iterations. These models predicted development of IA and progression to diabetes with an AUC of 0.91 and 0.92, respectively. It should be noted that as the analysis incorporated change in protein, metabolite, or cytokine over time, selected features represent features whose change, not absolute value, is associated with outcome.
To predict development of IA, several metadata features were included, which serves to adjust the analysis for these factors. Among the most highly selected features were all five metadata elements: age, FDR status, Hispanic ethnicity, HLA risk group, and sex, indicating that these categories were important in conjunction with other features in the prediction of IA.
Two highly selected features were genetic markers associated with development of IA: PTPN22 (rs2476601) and CTLA4 (rs3087243 and rs231775) (5,35). Both PTPN22 (rs2476601) and CTLA4 (rs3087243 and rs231775) were selected twice in this analysis, as they were included in the feature set comprising data from multiple separate genetic analyses. The observation that the same SNPs were selected twice provides additional evidence of robustness of this analytical approach.
Many of the most frequently selected features were metabolites. The highest selected feature was ascorbate (vitamin C), an important antioxidant. Ascorbate was present at lower relative abundance in participants who developed IA at the earliest time point (T1) relative to control subjects and rose over time (Fig. 2), while control subjects started with a higher level and then showed a downward trend in ascorbate levels. Discrepant trajectories between these two groups were significantly associated with IA outcome (Supplementary Table 6). Other metabolites whose change over time predicted outcome included 3-methyl-oxobutyrate (α-ketoisovaleric acid), a degradation product from valine as well as a precursor to valine for leucine synthesis. These branched-chain amino acids are known to predict development of insulin resistance. They play an intriguing role in promoting lymphocyte growth and proliferation as well as cytotoxic T- lymphocyte activity and have been previously identified as elevated prior to seroconversion (36). 4-hydroxyhippuric acid is a microbial end product produced through polyphenol metabolism by intestinal microflora (37), and serum levels are affected by altered gut permeability in mice (38). Pyroglutamic acid is a derivative of l-glutamic acid, formed nonenzymatically from glutamate, glutamine, and γ-glutamylated peptides. Elevated blood levels of pyroglutamine may indicate problems in antioxidant glutathione metabolism (39). This metabolite increased during progression to IA in case subjects but decreased in control subjects.
Finally, among the most frequent features selected for development of IA were multiple proteins involved in immunity and inflammation: FCRL3 (Fc receptor-like protein 3), KLRK1, MMP-2, and activin A. Also selected were SSRP1, a protein involved in DNA repair, and CSK21, which plays a role in apoptosis and response to viral infections.
Of the five elements of metadata included in analysis for progression from IA to diabetes, only age and FDR status were among the highest selected features, indicating that these features were important in conjunction with the constellation of other highly selected features.
Interestingly, top-selected SNPs associated with development of IA were different from those associated with progression to diabetes. Of these, rs2157678 (HLA DQB1 8.1) is associated with the ancestral HLA DR3-B8-A1 haplotype (40).
In analysis of progression from IA to diabetes, selected metabolomic features included multiple carbohydrates: glucose, mannose, and ribose (Table 3 and Fig. 3). All three metabolites increased in abundance in children progressing to diabetes but decreased from T3 to T4 in the control group (Supplementary Table 11). Additionally, butyrylcarnitine, an acylcarnitine, was noted to increase from T3 to T4 in case subjects but decrease in control subjects. This could be explained by an overall increase in lipolysis secondary to progressive insulinopenia as one approaches clinical diabetes.
Among the top proteomic features in progression from IA to diabetes was cystatin-F, a protein that modulates natural killer and T cell cytotoxity and RAD51, which plays a role in DNA repair. Plasminogen, a protease important for lysis blood clots, also plays a role in activating the complement system. Proteins involved in cell adhesion and growth (DRR1, IL-11 RA, and spondin-1) were also among the most frequently selected features.
In summary, we demonstrated that the ROFI-P3 algorithm can identify and evaluate known and novel predictors of development of IA and progression to diabetes across disparate data sources. Importantly, in children with high-risk HLA genotypes, changes in relative abundance of certain proteins and metabolites as well as genetic markers predicted development of IA, and a distinct constellation of features predicted progression of persistent IA to diabetes. Seroconversion was associated with altered antioxidant profile, a finding that has been noted in humans (36) and NOD mice (41). Additionally, there are indications of altered gut permeability, another proposed pathogenic mechanism (42). In contrast, progression from IA to diabetes was associated with altered sugars and acyl carnitines, indicating a potential switch to alternate metabolic pathways as relative insulin deficiency becomes more prominent.
The goal of this study was to develop a robust statistical machine learning model that predicts development of IA and progression from IA to diabetes. The major advantage of this study is the prospective characterization of developing autoimmunity over a prolonged period of time, with repeat longitudinal measurements of biomarkers. The DAISY cohort has >20 years of follow-up. While a peak incidence of IA has been observed within the first 2 years of life (3,43), new seroconversion has been observed well into adolescence and beyond (44). In addition, data from multiple sources (clinical data, genetics, metabolomics, and proteomics) are available to be integrated in a machine learning framework. Limitations of this study include the relatively small numbers of subjects in each group. Larger cohort size could allow additional analysis, including examination of whether age at seroconversion or specific endotypes play a role in the features selected. A further limitation of the study is the potential bias to individuals with later seroconversion. Both IA and T1D groups (Table 1) were older than the reported median seroconversion age of 2.3 years in The Environmental Determinants of Diabetes in the Young (TEDDY) study at 7 years of follow-up (45). In contrast, studies such as DAISY (46) and BABYDIAB (43), with longer follow-up beyond the early peak in autoimmunity, observe ongoing seroconversion into later childhood. Thus, selection of participants included individuals with seroconversion at older ages. Further, attention to requisite sample availability may have biased against individuals with early seroconversion who often have exceedingly rapid progression. This may impact generalizability to such rapidly progressing individuals.
Building predictive models via machine learning is an emerging strategy for identification of predictive biomarkers in type 1 diabetes and other diseases; however, challenges remain in the integration of large and diverse data sets. Machine learning strategies that incorporate feature selection allow identification of biomarkers that perform well in combination. This not only selects the most predictive features from among many but also may lend insight into important biological mechanisms. Although P3 and ROFI have both been used previously to study omics data, this is the first combination for feature selection in an integrative fashion. The feature sets identified using the ROFI-P3 strategy perform well in prediction of both IA and type 1 diabetes outcome. Further, identification of distinct panels of predictors underlines differences between processes leading to development of IA from pathways involved in progression to diabetes. The associated measure of probability adds further information for interpreting the utility of various biomarkers and could help researchers in identifying the best candidates to focus limited resources on validation. Further studies will determine whether these selected features can be validated in independent populations to predict progression to IA or type 1 diabetes.
Supplementary Material
Article Information
Funding. This work was supported by JDRF (grants 17-2013-535, 11-2010-206, and 5-ECR-2017-388-A-N) and the National Institutes of Health (grants R01 DK32493, DK32083, DK049654, K12 DK094712, and P30 DK57516).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. B.I.F., B.-J.W.-R., and M.R. contributed to the design of the study, analyzed data, contributed to the discussion, and wrote the manuscript. L.M.B. and S.M.R. analyzed data, contributed to the discussion, and reviewed and edited the manuscript. K.W., A.K.S., and J.M.N. researched data, contributed to discussion, and reviewed and edited the manuscript. M.R. is principal investigator of DAISY. All authors approved the final version of the manuscript. M.R. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 77th Scientific Sessions of the American Diabetes Association, San Diego, CA, 9–13 June 2017.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db18-1263/-/DC1.
References
- 1.Insel RA, Dunne JL, Atkinson MA, et al. Staging presymptomatic type 1 diabetes: a scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diabetes Care 2015;38:1964–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker JM, Barriga KJ, Yu L, et al.; Diabetes Autoimmunity Study in the Young . Prediction of autoantibody positivity and progression to type 1 diabetes: Diabetes Autoimmunity Study in the Young (DAISY). J Clin Endocrinol Metab 2004;89:3896–3902 [DOI] [PubMed] [Google Scholar]
- 3.Ilonen J, Hammais A, Laine A-P, et al. Patterns of β-cell autoantibody appearance and genetic associations during the first years of life. Diabetes 2013;62:3636–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waugh K, Snell-Bergeon J, Michels A, et al. Increased inflammation is associated with islet autoimmunity and type 1 diabetes in the Diabetes Autoimmunity Study in the Young (DAISY). PLoS One 2017;12:e0174840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hermann R, Laine AP, Veijola R, et al. The effect of HLA class II, insulin and CTLA4 gene regions on the development of humoral beta cell autoimmunity. Diabetologia 2005;48:1766–1775 [DOI] [PubMed] [Google Scholar]
- 6.Steck AK, Wong R, Wagner B, et al. Effects of non-HLA gene polymorphisms on development of islet autoimmunity and type 1 diabetes in a population with high-risk HLA-DR,DQ genotypes. Diabetes 2012;61:753–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Törn C, Hadley D, Lee H-S, et al.; TEDDY Study Group . Role of type 1 diabetes–associated SNPs on risk of autoantibody positivity in the TEDDY study. Diabetes 2015;64:1818–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winkler C, Krumsiek J, Buettner F, et al. Feature ranking of type 1 diabetes susceptibility genes improves prediction of type 1 diabetes. Diabetologia 2014;57:2521–2529 [DOI] [PubMed] [Google Scholar]
- 9.Krischer JP, Liu X, Lernmark Å, et al.; TEDDY Study Group . The influence of type 1 diabetes genetic susceptibility regions, age, sex, and family history on the progression from multiple autoantibodies to type 1 diabetes: a TEDDY study report. Diabetes 2017;66:3122–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frohnert BI, Laimighofer M, Krumsiek J, et al. Prediction of type 1 diabetes using a genetic risk model in the Diabetes Autoimmunity Study in the Young. Pediatr Diabetes 2018;19:277–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orešič M. Metabolomics in the studies of islet autoimmunity and type 1 diabetes. Rev Diabet Stud 2012;9:236–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pflueger M, Seppänen-Laakso T, Suortti T, et al. Age- and islet autoimmunity–associated differences in amino acid and lipid metabolites in children at risk for type 1 diabetes. Diabetes 2011;60:2740–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moulder R, Bhosale SD, Erkkilä T, et al. Serum proteomes distinguish children developing type 1 diabetes in a cohort with HLA-conferred susceptibility. Diabetes 2015;64:2265–2278 [DOI] [PubMed] [Google Scholar]
- 14.von Toerne C, Laimighofer M, Achenbach P, et al. Peptide serum markers in islet autoantibody-positive children. Diabetologia 2017;60:287–295 [DOI] [PubMed] [Google Scholar]
- 15.Liu C-W, Bramer L, Webb-Robertson B-J, Waugh K, Rewers MJ, Zhang Q. Temporal expression profiling of plasma proteins reveals oxidative stress in early stages of type 1 diabetes progression. J Proteomics 2018;172:100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beagley N, Stratton KG, Webb-Robertson B-JM. VIBE 2.0: visual integration for bayesian evaluation. Bioinformatics 2010;26:280–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rewers M, Bugawan TL, Norris JM, et al. Newborn screening for HLA markers associated with IDDM: diabetes autoimmunity study in the young (DAISY). Diabetologia 1996;39:807–812 [DOI] [PubMed] [Google Scholar]
- 18.Norris JM, Yin X, Lamb MM, et al. Omega-3 polyunsaturated fatty acid intake and islet autoimmunity in children at increased risk for type 1 diabetes. JAMA 2007;298:1420–1428 [DOI] [PubMed] [Google Scholar]
- 19.Gianani R, Rabin DU, Verge CF, et al. ICA512 autoantibody radioassay. Diabetes 1995;44:1340–1344 [DOI] [PubMed] [Google Scholar]
- 20.Grubin CE, Daniels T, Toivola B, et al. A novel radioligand binding assay to determine diagnostic accuracy of isoform-specific glutamic acid decarboxylase antibodies in childhood IDDM. Diabetologia 1994;37:344–350 [DOI] [PubMed] [Google Scholar]
- 21.Yu L, Robles DT, Abiru N, et al. Early expression of antiinsulin autoantibodies of humans and the NOD mouse: evidence for early determination of subsequent diabetes. Proc Natl Acad Sci U S A 2000;97:1701–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonifacio E, Yu L, Williams AK, et al. Harmonization of glutamic acid decarboxylase and islet antigen-2 autoantibody assays for national institute of diabetes and digestive and kidney diseases consortia. J Clin Endocrinol Metab 2010;95:3360–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wenzlau JM, Juhl K, Yu L, et al. The cation efflux transporter ZnT8 (Slc30A8) is a major autoantigen in human type 1 diabetes. Proc Natl Acad Sci U S A 2007;104:17040–17045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohta T, Masutomi N, Tsutsui N, et al. Untargeted metabolomic profiling as an evaluative tool of fenofibrate-induced toxicology in Fischer 344 male rats. Toxicol Pathol 2009;37:521–535 [DOI] [PubMed] [Google Scholar]
- 25.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem 2009;81:6656–6667 [DOI] [PubMed] [Google Scholar]
- 26.Dehaven CD, Evans AM, Dai H, Lawton KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform 2010;2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lollo B, Steele F, Gold L. Beyond antibodies: new affinity reagents to unlock the proteome. Proteomics 2014;14:638–644 [DOI] [PubMed] [Google Scholar]
- 28.Zhang Q, Fillmore TL, Schepmoes AA, et al. Serum proteomics reveals systemic dysregulation of innate immunity in type 1 diabetes. J Exp Med 2013;210:191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steck AK, Dong F, Wong R, et al. Improving prediction of type 1 diabetes by testing non-HLA genetic variants in addition to HLA markers. Pediatr Diabetes 2014;15:355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webb-Robertson BJ, McCue LA, Beagley N, et al. A Bayesian integration model of high-throughput proteomics and metabolomics data for improved early detection of microbial infections. Pac Symp Biocomput 2009:451–463 [PMC free article] [PubMed] [Google Scholar]
- 31.Webb-Robertson BJM, Bramer LM, Reehl SM, et al. ROFI - the use of Repeated Optimization for Feature Interpretation. In 2016 International Conference on Computational Science and Computational Intelligence (CSCI). 2016. p. 29–33. [Google Scholar]
- 32.Webb-Robertson B-J, Kreuzer H, Hart G, et al. Bayesian integration of isotope ratio for geographic sourcing of castor beans. J Biomed Biotechnol 2012;2012:450967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rolandsson O, Hägg E, Nilsson M, Hallmans G, Mincheva-Nilsson L, Lernmark A. Prediction of diabetes with body mass index, oral glucose tolerance test and islet cell autoantibodies in a regional population. J Intern Med 2001;249:279–288 [DOI] [PubMed] [Google Scholar]
- 34.Norris JM, Lee H-S, Frederiksen B, et al.; TEDDY Study Group . Plasma 25-Hydroxyvitamin D concentration and risk of islet autoimmunity. Diabetes 2018;67:146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steck AK, Liu SY, McFann K, et al. Association of the PTPN22/LYP gene with type 1 diabetes. Pediatr Diabetes 2006;7:274–278 [DOI] [PubMed] [Google Scholar]
- 36.Orešič M, Simell S, Sysi-Aho M, et al. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J Exp Med 2008;205:2975–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobs DM, Spiesser L, Garnier M, et al. SPE-NMR metabolite sub-profiling of urine. Anal Bioanal Chem 2012;404:2349–2361 [DOI] [PubMed] [Google Scholar]
- 38.Orrego-Lagarón N, Martínez-Huélamo M, Vallverdú-Queralt A, Lamuela-Raventos RM, Escribano-Ferrer E. High gastrointestinal permeability and local metabolism of naringenin: influence of antibiotic treatment on absorption and metabolism. Br J Nutr 2015;114:169–180 [DOI] [PubMed] [Google Scholar]
- 39.Emmett M. Acetaminophen toxicity and 5-oxoproline (pyroglutamic acid): a tale of two cycles, one an ATP-depleting futile cycle and the other a useful cycle. Clin J Am Soc Nephrol 2014;9:191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baschal EE, Aly TA, Jasinski JM, et al.; Type 1 Diabetes Genetics Consortium . The frequent and conserved DR3-B8-A1 extended haplotype confers less diabetes risk than other DR3 haplotypes. Diabetes Obes Metab 2009;11(Suppl. 1):25–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brosche T, Platt D. The biological significance of plasmalogens in defense against oxidative damage. Exp Gerontol 1998;33:363–369 [DOI] [PubMed] [Google Scholar]
- 42.Vaarala O, Atkinson MA, Neu J. The “perfect storm” for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes 2008;57:2555–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ziegler A-G, Bonifacio E; BABYDIAB-BABYDIET Study Group . Age-related islet autoantibody incidence in offspring of patients with type 1 diabetes. Diabetologia 2012;55:1937–1943 [DOI] [PubMed] [Google Scholar]
- 44.Yu L, Rewers M, Gianani R, et al. Antiislet autoantibodies usually develop sequentially rather than simultaneously. J Clin Endocrinol Metab 1996;81:4264–4267 [DOI] [PubMed] [Google Scholar]
- 45.Vehik K, Lynch KF, Schatz DA, et al.; TEDDY Study Group . Reversion of β-cell autoimmunity changes risk of type 1 diabetes: TEDDY study. Diabetes Care 2016;39:1535–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frohnert BI, Ide L, Dong F, et al. Late-onset islet autoimmunity in childhood: the Diabetes Autoimmunity Study in the Young (DAISY). Diabetologia 2017;60:998–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.