Abstract
Autophagy, self-eating, is a pivotal catabolic mechanism that ensures homeostasis and survival of the cell in the face of stressors as different as starvation, infection, or protein misfolding. The importance of the research in this field was recognized by the general public after the Nobel Prize for Physiology or Medicine was awarded in 2016 to Yoshinori Ohsumi for discoveries of the mechanisms of autophagy using yeast as a model organism. One of the seminal findings of Ohsumi was on the role ubiquitin-like proteins (UBLs)—Atg5, Atg12, and Atg8—play in the formation of the double-membrane vesicle autophagosome, which is the functional unit of autophagy. Subsequent work by several groups demonstrated that, like the founding member of the UBL family ubiquitin, these small but versatile protein and lipid modifiers interact with a plethora of proteins, which either directly regulate autophagosome formation, for example, components of the Atg1/ULK1 complex, or are involved in cargo recognition, for example, Atg19 and p62/SQSTM1. By tethering the cargo to the UBLs present on the forming autophagosome, the latter proteins were proposed to effectively act as selective autophagy receptors. The discovery of the selective autophagy receptors brought a breakthrough in the autophagy field, supplying the mechanistic underpinning for the formation of an autophagosome selectively around the cytosolic cargo, that is, a protein aggregate, a mitochondrion, or a cytosolic bacterium. In this historical overview, I highlight key steps that the research into selective autophagy has been taking over the past 20 years. I comment on their significance and discuss current challenges in developing more detailed knowledge of the mechanisms of selective autophagy. I will conclude by introducing the new directions that this dynamic research field is taking into its third decade.
Abbreviations: AIM, Atg8-interacting motif; Cvt, cytoplasm-to-vacuole targeting; ER, endoplasmic reticulum; FIR, FIP200-interacting region; KO, knockout; LIR, LC3-interacting region; LLPS, liquid–liquid phase separation; OMM, outer mitochondrial membrane; PAS, phagophore assembly site; PE, phosphatidylethanolamine; PI3P, phosphatidylinositol-3-phosphate; SAR, selective autophagy receptor; SLR, SQSTM1/p62-like receptor; UBL, ubiquitin-like protein
Keywords: Atg8, GABARAP, LC3, SAR, UBL
Graphical Abstract
Highlights
-
•
Historical overview of selective autophagy research over past 20 years
-
•
Discussion and modeling of selective autophagy pathway
-
•
Critical discussion on open questions
-
•
Indication of new directions in the field
Introduction
The discovery of the cellular process of self-eating, autophagy, was an important step in elucidating mechanisms of how the cell maintains its homeostasis in the face of different stress factors, including starvation, infection, and proteotoxic and oxidative stress (reviewed in Refs. [1], [2]). The major progress in the autophagy research field that has taken place over the past several decades, with its great impact on the modern understanding of human health and disease, was recognized by the general public latest in 2016 when the Nobel Prize in Physiology or Medicine was awarded to Yoshinori Ohsumi for his seminal work on basic autophagy mechanisms, primarily using yeast as a model organism. The key discovery made by Ohsumi at the University of Tokyo and later at the National Institute for Basic Biology in Okazaki, Japan [3], with independent contribution from Michael Thumm from the University of Stuttgart, Germany [4] and Daniel Klionsky from the University of California, USA [5], was the identification and characterization of the core autophagy (Atg) genes and proteins that govern the nucleation, initiation, and expansion of the autophagosome. This double-membrane vesicle is at the heart of the macroautophagy process: it ensures both physical sequestration of the autophagic cargo from the rest of the cytoplasm and its delivery into the vacuole of the yeast cell (lysosome in higher eukaryotes) where the degradation takes place.
The core Atg proteins found in yeast can be grouped into five multifunctional modules (reviewed in Refs. [6], [7]): (1) the Atg1 kinase complex, with Atg1, Atg13, Atg17, Atg29, and Atg31; (2) the class III phosphoinositide 3-kinase (PI3K) complex I, with Vps34, Vps15, Atg6, Atg14, and Atg38; (3) the phosphatidylinositol-3-phosphate (PI3P)-binding Atg2/Atg18 complex; (4) the ubiquitin-like Atg5–Atg12 and Atg8–phosphatidylethanolamine (PE) conjugation machineries, with Atg3, Atg4, Atg5, Atg7, Atg8, Atg10, Atg12, and Atg16; and (5) the only integral membrane protein Atg9. These Atg proteins assemble in a hierarchical fashion at the perivacuolar phagophore assembly site (PAS) described by Kuninori Suzuki and Yoshinori Ohsumi in 2001 [8], where the autophagosome is produced from a primordial membrane (known as phagophore or isolation membrane) in the yeast cell. Importantly, this core autophagy machinery is largely conserved in mammalian cells, with functional expansion of the ATG gene family and multiple sites of autophagosome formation in the cytosol being the hallmarks of the mammalian autophagy process [9].
The fact that autophagy can be selective, that is, autophagosomes can form around a particular cargo while excluding the rest of the cytoplasm, was suggested by the founding father of the autophagy research field Christian de Duve back in 1966 [10]; however, only in the last 20 years, advances in molecular and cell biology allowed deciphering the mechanisms behind this remarkable trait of the evolutionarily conserved degradation pathway. While all three main forms of autophagy, the chaperone-mediated autophagy, microautophagy (direct engulfment of cytosol by the lysosome or vacuole), and the macroautophagy, can be selective (with the chaperone-mediated autophagy being selective by definition, as it relies on the KFERQ-like sequence in the substrate proteins [11]), the explicit focus of this overview is on the autophagosome-dependent macroautophagy, which is hereafter referred to as simply “autophagy.” It is the main cellular mechanism that targets large structures, that is, protein aggregates, parts of the endoplasmic reticulum (ER) membrane, or entire organelles, such as mitochondria and peroxisomes, for degradation, thereby cleansing the cell of deleterious components and supplying metabolic intermediates, both critical for cellular homeostasis.
Peroxisomes [12] and mitochondria [13], [14] were the first large substrates shown to be degraded by autophagy selectively, so that specific terms “pexophagy” and “mitophagy” were coined to describe these respective degradation processes. Subsequently, discovery of every additional selective autophagy cargo prompted introduction of a new name, so that there is currently a growing number of pathways and corresponding terms recognized within the selective autophagy field (Table 1). For details, the reader is referred to a number of excellent reviews that cover the growing knowledge on these selective autophagy pathways [6], [15], [16], [17], [18], [19], [20].
Table 1.
Selective autophagy processes.
| Term | Process | References |
|---|---|---|
| Aggrephagy | Selective degradation of protein aggregates | [253] |
| Chlorophagy | Selective degradation of chloroplasts | [254] |
| ER-phagy or reticulophagy | Selective degradation of fragments of ER | [263], [264] |
| Ferritinophagy | Selective degradation of ferritin | [147] |
| Glycophagy | Selective degradation of glycogen | [146] |
| Granulophagy | Selective degradation of stress granules | [255] |
| Lipophagy | Selective degradation of lipid droplets | [256] |
| Lysophagy | Selective degradation of lysosomes | [257], [258] |
| Mitophagy | Selective degradation of mitochondria | [259] |
| Myelinophagy | Selective degradation of myelin | [260] |
| Nucleophagy | Selective degradation of parts of the nucleus | [261] |
| Pexophagy | Selective degradation of peroxisomes | [259] |
| Proteaphagy | Selective degradation of the proteasome | [262] |
| Ribophagy | Selective degradation of ribosomes | [77] |
| Virophagy | Selective degradation of viruses | [265] |
| Xenophagy | Selective degradation of bacteria and other intracellular pathogens | [266], [267] |
| Zymophagy | Selective degradation of zymogen granules | [268] |
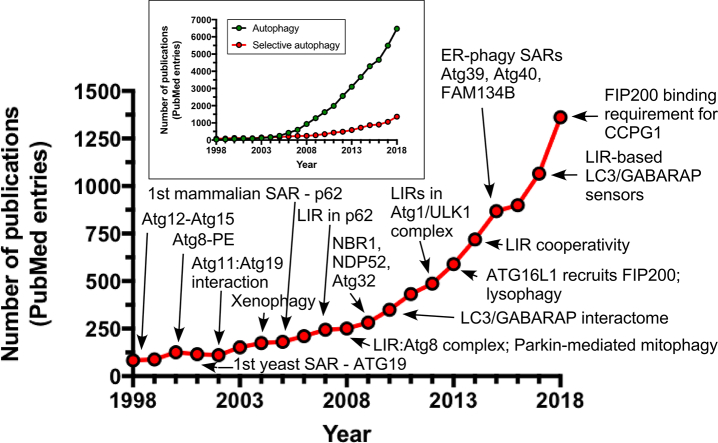
Our understanding of the molecular mechanisms of selective autophagy has recently been growing exponentially (Fig. 1). Today's immense progress in the field is indebted to two seminal discoveries: (1) identification and characterization of the autophagy-specific ubiquitin-like proteins (UBLs): Atg12, Atg5 [21], [22], and Atg8/LC3/GABARAPs [23], [24], [25]; and (2) the discovery of the LC3-interacting region [LIR; also known as Atg8-interacting motif (AIM), and LC3 recognition sequence, (LRS)] [26], [27], [28] within a set of selective autophagy receptor (SAR) proteins (Figs. 2 and 3). As I describe below, these two consecutive steps stimulated the massive international effort that has helped define the common principles behind the broad spectrum of the selective autophagy pathways (Table 1, Fig. 3).
Fig. 1.
Publication dynamics in the field of selective autophagy. Main graph: The numbers of publications per year are shown for the selective autophagy research field; selected discoveries that shaped the field are indicated. Inset graph: Comparison of the publication dynamics in the broader autophagy field versus that of selective autophagy. Note that in the PubMed database, the term “autophagy” was used to derive numbers of publications in the entire autophagy research field, while the combination of the following terms”selective autophagy“ OR “mitophagy” OR “aggrephagy” OR “xenophagy” OR “pexophagy” OR “ribophagy” OR “lipophagy” OR “zymophagy” OR “granulophagy” OR “nucleophagy” OR “glycophagy” OR “lysophagy” OR “ERphagy” OR “reticulophagy” OR “Cvt pathway” OR “ferritinophagy” was used to obtain the number of publications in the field of selective autophagy only. Abbreviations: Atg, autophagy-related; ER, endoplasmic reticulum; LIR, LC3 interacting region; SAR, selective autophagy receptor.
Fig. 2.
Key discoveries that have shaped the field of selective autophagy over the past 20 years. Note that not all relevant discoveries could be included due to space limitation. Abbreviations: Atg, autophagy related; Cvt, cytoplasm to vacuole targeting; ER, endoplasmic reticulum; IMM, inner mitochondrial membrane; LIR, LC3 interacting region; SAR, selective autophagy receptor; SLR, sequestosome-1-like receptor; UFIM, UFM1 interacting motif.
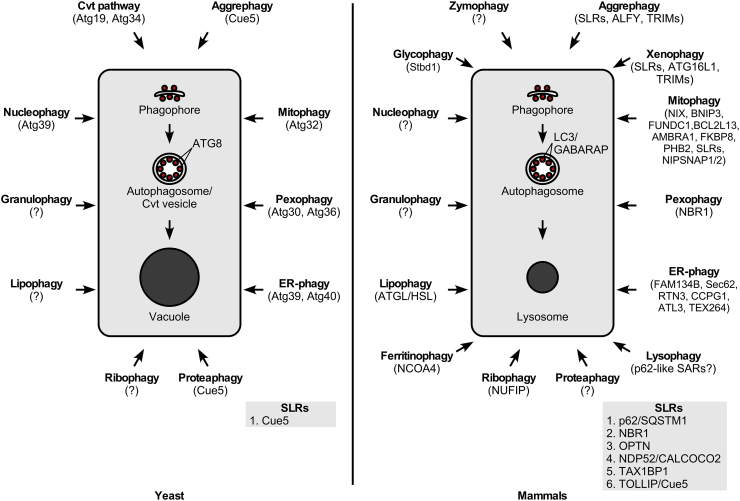
Fig. 3.
Main selective autophagy processes and their receptors. Main selective autophagy processes and the respective SARs in yeast versus mammalian cells are shown. Insets: Current lists of SLRs in yeast and mammals are provided. Abbreviations: Atg, autophagy related; Cvt, cytoplasm to vacuole targeting; SAR, selective autophagy receptor; SLR, sequestosome-1-like receptor.
Ab ovo—of the UBLs and the LIR
Ubiquitin, the small (76 amino acids) and highly conserved protein, is characterized by the globular β-grasp fold and its ability to become reversibly conjugated to other proteins following the three-step enzymatic cascade involving E1 (activating), E2 (conjugating), and E3 (ligating) enzymes (reviewed in Ref. [29]). The addition of monoubiquitin or polyubiquitin chains to a substrate protein creates a signal that is translated by ubiquitin-binding domain-containing proteins into the various biological functions of ubiquitin,that is, those associated with changes in stability, localization, or activity of the target protein. The signal can be terminated by the action of multiple deubiquitinating enzymes, which are also responsible for the proteolytic maturation of ubiquitin by exposing its conjugatable C-terminal Gly residue [30]. The key structural and functional features of ubiquitin are shared by the growing number of UBLs, such as NEDD8, FAT10, SUMO, UFM1, ISG15, Hub1/Ubl5, and URM1 (reviewed in Refs. [31], [32], [33]).
In 1998, the list of the known UBLs grew by two members when Ohsumi's group made the original discovery of a ubiquitin-like conjugation pathway required for autophagy in yeast. The two new UBLs, Atg12 and Atg5, were found to be conjugated to each other (Atg12 forms a stable conjugate with the Lys149 of Atg5), which depended on the activity of Atg7 and Atg10, and E1- and E2-like enzymes, respectively [21], [22]. In collaboration with Fuyuhiko Inagaki's group from the Hokkaido University in Japan, Ohsumi subsequently demonstrated that both Atg12 [34], [35] and Atg5 [36] display the characteristic ubiquitin fold, with Atg5 containing two ubiquitin-like domains linked by a central helix-rich domain.
In 2000, Ohsumi reported identification of the third autophagy-specific UBL called Atg8. Unlike any other known UBL, the exposed C-terminal Gly of Atg8 was conjugated to the amino group of a lipid, PE, enriched in the autophagosomal membrane. The Atg8–PE conjugation reaction is mediated by E1-like Atg7 and E2-like Atg3 enzymes [23], [24], while, as shown by Ohsumi in 2007, the stable Atg12–Atg5 conjugate, in complex with Atg16, acts as an E3 ligase [37]. Interestingly, the requirement for the formation of the covalent Atg12–Atg5 complex is waived in some organisms. As very recently shown by Noboru Mizushima's group from the University of Tokyo, apicomplexan parasites Plasmodium and Toxoplasma as well as the yeast Komagataella phaffii (previously known as Pichia pastoris) lack both the C-terminal Gly in Atg12 and the complete Atg10 protein required for Atg12–Atg5 conjugation, such that the non-covalent association between Atg12 and Atg5 ensures the functionality of the E3-like complex [38]. Importantly, as with ubiquitin, the Atg8–PE conjugation is reversed by the action of a deubiquitinating enzyme-like cysteine protease Atg4, also required for Atg8's proteolytic activation [24], [39], [40].
In the year of Atg8's discovery, Tamotsu Yoshimori, then at the National Institute for Basic Biology in Okazaki, characterized the mammalian homolog of Atg8 that he called LC3 (MAP1LC3B is the full name of the gene encoding LC3) [25]. Ever since, LC3 has been holding the title of the universal marker of autophagy based on the fact that it is associated with the autophagic membranes throughout the life cycle of an autophagosome: from the phagophore and autophagosome to the autolysosome—the product of fusion between the autophagosome and the lysosome [41], [42]. Today, the mammalian LC3/GABARAP family counts six members: LC3A (encoded by the MAP1LC3A gene), LC3B (in human cells encoded by two genes MAP1LC3B and MAP1LC3B2), LC3C (encoded by MAP1LC3C), GABARAP, GABARAPL1, and GABARAPL2/GATE-16 [43].
In 2008, Klionsky, now at the University of Michigan in Ann Arbor, USA, demonstrated that Atg8 conjugation regulated the size of the autophagosome in the yeast Saccharomyces cerevisiae [44]. One potential explanation for the observed effect of the Atg8 proteins on the expansion of the autophagosomal membrane came from the experiments performed by the groups of Ohsumi [45] and Zvulun Elazar (at the Weizmann Institute of Science in Rehovot, Israel) [46], who showed that the α-helical domain in the N-terminus of Atg8/LC3/GABARAPs can mediate homotypic interactions, promoting tethering and hemifusion of lipid membranes in vitro.
In parallel to establishing the critical functions of the three UBLs in the autophagy pathway, by the early 2000s, strong evidence was mounting that some autophagic cargos are degraded in a highly selective manner, which required factors in addition to the core Atg machinery. Klionsky's laboratory pioneered the work on the selective autophagy-like process in yeast, the biosynthetic cytoplasm-to-vacuole targeting (Cvt) pathway, which is responsible for delivery of resident hydrolases, aminopeptidase I (Ape1) and α-mannosidase (Ams1), to the vacuole [47], [48]. (Necessary note: Although the Cvt pathway is in the strict sense of the term not a selective autophagy process, its principles are very similar to the ones employed by the selective autophagy. Therefore, for the purpose of this overview, it will be categorized as a selective autophagy-related process and its receptors Atg19 and Atg34 will be referred to as SARs.) In 1999, Klionsky showed that the Cvt pathway utilized components of the Atg machinery, as the Cvt and Atg mutants shared many of the genes (i.e., Atg1, Atg5, Atg7, Atg8, and Atg10). In fact the same set of genes was required for the selective degradation of peroxisomes—pexophagy [12]. However, in 2001, by studying yeast mutants that fail to maintain the Cvt pathway, Klionsky identified an additional protein that seemed to fit the definition of a Cvt cargo receptor protein: it interacted with the precursor form of Ape1 (prApe1, as also shown previously in a large-scale yeast two-hybrid screen performed by Jonathan Rothberg and colleagues at the University of Washington, USA [49]), and was delivered to the vacuole along with its cargo [50], [51]. Following studies performed in Klionsky's laboratory confirmed that the YOL082w, which was aptly re-named as Cvt19 and is now known as Atg19, directly binds the Cvt cargo and simultaneously two other proteins, which localize to the PAS in yeast cells [8]. One of the latter proteins was the Atg1 kinase-binding adaptor Atg11 (then known as Cvt9). Experimental evidence pointed at Atg11 being the key factor necessary for recruitment of the oligomeric prApe1–Atg19 complex to the PAS. Somewhat surprisingly, the other Atg19-interacting protein, besides prApe1 and Atg11, was the membrane-associated UBL Atg8. The significance of the Atg19:Atg8 interaction was therefore thought to be in recruiting some unidentified membrane-forming components by the cargo–receptor complex to the PAS [52], [53]. In 2010, Ohsumi's group reported identification of the second Cvt cargo receptor, Atg34 (Fig. 3), which is homologous to Atg19 and is required for efficient trafficking of Ams1 to the vacuole under starvation conditions [54], [55].
The rapid pace with which molecular mechanisms of selective autophagy had begun to unfold in the lower organism, enabled by the power of the yeast genetics, could not be kept up with by the studies in mammalian cells that lacked appropriate genetic tools at the turn of the millennium. In 2001, John Lemasters from the University of North Carolina, USA and Aviva Tolkovsky at the University of Cambridge, UK convincingly demonstrated that depolarized mitochondria were selectively removed from rat hepatocytes, neurons, and HeLa cells [56], [57], and several years later, in collaboration with Ohsumi, Lemasters reported formation of GFP-LC3 positive rings around mitochondria shortly before their depolarization [13]. However, it was the protein aggregation pathway that helped break the story of the selective autophagy machinery in mammalian cells.
Five years earlier, in 1996, Shin and colleagues from the Dana-Farber Cancer Institute in Boston, USA, had described molecular cloning of the p56lck SH2 ligand, a protein that interacted with the SH2 domain of the LCK kinase in a phospho-Tyr-independent manner [58]. This protein also bound ubiquitin in vivo [59]. At the same time, Tetsuro Ishii from the University of Tsukuba in Japan reported on a protein identical to the p56lck SH2 ligand, which they called A170, as the one induced by oxidative stress in macrophages [60]. This protein of 62 kDa was dubbed p62 and subsequently shown to play versatile roles as a signaling adaptor (reviewed in Refs. [61], [62]). It was also demonstrated that p62 was a common cargo-sequestering component of ubiquitin-positive protein aggregates, such as Mallory and Lewy bodies [63], [64], and hence its other name sequestosome-1 (SQSTM1). Subsequently, the group of Terje Johansen at the University of Tromso in Norway characterized the N-terminal Phox and Bem1p (PB1) domain of p62/SQSTM1 and demonstrated that it was responsible for p62/SQSTM1 oligomerization [65]. The PB1 domain oligomerization was also reported by the group of Roger Williams at the Medical Research Council Laboratory of Molecular Biology in Cambridge, UK [66].
Based on its oligomerization and ubiquitin-binding properties, Johansen asked whether p62/SQSTM1 was associated with autophagy of aggregated, ubiquitylated proteins in the mode similar to Atg19, which binds its oligomeric cargo prApe1. Indeed, in 2005, they could show for the first time that p62/SQSTM1 by itself formed protein aggregates, which were degraded in the lysosome. They could further demonstrate that ubiquitylated cargo recruited p62/SQSTM1 and that the oligomerization of p62/SQSTM1 was indeed required for efficient cargo degradation fulfilling the earlier definition of the receptor for autophagic degradation [67]. Building on this initial success, in 2007, the Johansen group reported on a direct interaction between p62/SQSTM1 and the members of the mammalian Atg8 family LC3/GABARAP proteins. Using the novel tandem fluorescence reporter mCherry-GFP fused with p62/SQSTM1, they unequivocally demonstrated delivery of the protein to the lysosome and mapped the short linear peptide found within the SAR as the determinant for its lysosomal trafficking [26]. This provided the first definition of a LIR, which was later found to be the defining feature of the SARs. Importantly, in the same year, the laboratory of Keiji Tanaka from the Tokyo Metropolitan Institute of Medical Science provided the first in vivo evidence for p62/SQSTM1's role in protein aggregation and aggrephagy in mouse tissues [68].
The initial effort in characterizing the SARs culminated in the elucidation of the structural characteristics of the AIM:Atg8 and LIR:LC3 interactions. In 2008, the groups of Masaaki Komatsu from the Juntendo University School of Medicine in Tokyo [27] and Fuyuhiko Inagaki from the Hokkaido University [28] published structural studies on the Atg19-derived AIM in complex with Atg8 and the p62/SQSTM1's LIR interacting with LC3B. Their key finding was that both, the C-terminal AIM of Atg19 and the intermediate LIR of p62/SQSTM1, form an intermolecular β-sheet with the β2 strand within the Atg8/LC3 core. This intermolecular structure is stabilized by (1) the hydrophobic interaction between the side chain of an invariant aromatic residue typical for a canonical LIR/AIM, which binds deeply within the hydrophobic pocket (HP) 1 of the Atg8/LC3, and (2) the binding between the second conserved, aliphatic residue of the LIR/AIM and the HP2 within the autophagy-specific UBLs. Subsequent studies also confirmed the importance of the acidic (or phosphorylated) groups immediately upstream of the core LIR sequence, which contribute a negative charge, further stabilizing the protein–protein interaction [69], [70], [71], as initially observed in experiments performed by Johansen [26].
Thus by 2008, 10 years after the discovery of Atg8, it was firmly established that the critical feature of a Cvt cargo receptor or an SAR is their ability to interact with (1) the substrate (prApe1 for Atg19 and ubiquitylated proteins for p62/SQSTM1) and (2) the autophagic machinery represented by Atg11 (in case of Atg19) and/or the UBL Atg8/LC3 (via the AIM/LIR motifs found in both the prototypical SARs, Atg19 and p62/SQSTM1). In addition, it became apparent that either the autophagic cargo itself (prApe1) or the cargo receptor (p62/SQSTM1) need to be oligomeric for them to be delivered into the vacuole/lysosome.
The “SAR Bomba”—The Explosive Growth of Knowledge on Selective Autophagy
The recognition of the first SARs in yeasts and mammals and understanding their common features greatly facilitated the search for new factors regulating selective autophagy (Fig. 3). In 2009, the group of Ivan Dikic at the Goethe University Frankfurt in Germany, together with the Johansen group and with contributions from the groups of Komatsu and Elazar, identified the p62/SQSTM1 homolog NBR1 as a ubiquitin- and LC3/GABARAP-binding protein and a major aggrephagy factor [72]. Noteworthy, p62/SQSTM1 and NBR1 interact with each other via their respective N-terminal PB1 domains [65], which plays an important role in both formation and clearance of protein aggregates. In 2010, Wade Harper's group at the Harvard Medical School in Boston, USA, published a comprehensive proteomic map of the autophagy pathway constructed with the help of an immunoprecipitation–mass spectrometry methodology. When using LC3/GABARAP proteins as baits, they identified p62/SQSTM1, NBR1, and several additional LC3/GABARAP-interacting proteins, which further expanded the mammalian autophagy landscape [73]. The realization that the mammalian genome encodes a number of proteins interacting with both LC3/GABARAP and the cargo additionally strengthened the SAR hypothesis and fueled further investigation into the selective autophagy machinery.
Ubiquitin-binding SARs
The fact that simultaneous binding to LC3/GABARAP and ubiquitin was the common denominator of the two mammalian SARs led Dikic and colleagues in 2009 to propose ubiquitin as a common “eat-me” signal for a range of cargos beyond protein aggregates [74]. Indeed, by the time, it had been known that ubiquitin was associated with cytosolic bacteria [75], peroxisomes [76], and ribosomes [77]. Subsequent studies substantiated this claim. Thus, in the same year, Felix Randow's group at the Medical Research Council Laboratory of Molecular Biology in Cambridge described another ubiquitin- and LC3-binding protein, NDP52 (also known as CALCOCO2), to be associated with ubiquitin-coated Salmonella enterica and required for the restriction of this pathogen [78]. Their later studies revealed that NDP52 preferentially interacts with the LC3C isoform via a novel type of LIR, which they called cLIR [79]. The NDP52 homolog TAX1BP1 was also found in 2015 by Folma Buss' group at the University of Cambridge to be associated with ubiquitylated S. typhimurium and required for xenophagy [80]. An additional ubiquitin-binding xenophagy receptor that was characterized in 2011 is optineurin (OPTN), whose role in restricting Salmonella was demonstrated by the Dikic group. An important discovery in that context was that phosphorylation of Ser177, upstream of the canonical LIR within OPTN, by TBK1 enhanced the affinity of the LIR to LC3B, thus modulating the OPTN:LC3 binding and overall xenophagy efficiency [70].
Active research in several laboratories is now addressing the nature of bacterial proteins that are ubiquitylated prior to their clearance by autophagy [81]. Candidate host E3 ligases induced in response to bacterial invasion have recently been identified and include RNF166 [82], LRSAM1 [83], LUBAC [84], ARIH1 [85], and SMURF1 [86]. On the other hand, results from Yoshimori's group at the Osaka University suggested that host proteins found on endosomal membranes, damaged during the bacterium's excursion into the cytosol, can also be ubiquitylated to facilitate xenophagy by recruiting ubiquitin-binding SARs, such as the mammalian ATG5-interacting protein ATG16L1, which is proposed to serve as an SAR in this instance [87].
Evidence for ubiquitin-driven mitophagy and the suspected role of the ubiquitin-binding SARs was first supplied in 2008 by Richard Youle's group at the National Institutes of Health in Bethesda, USA. Youle showed that the ubiquitin E3 ligase Parkin, mutated in familial cases of Parkinson's disease [88], is recruited to damaged mitochondria and mediates their degradation by mitophagy [89]. Subsequent efforts by the groups of Richard Youle [90], [91], Keiji Tanaka [92], and Noriyuki Matsuda [93] from the Tokyo Metropolitan Institute of Medical Science, as well as of Miratul Muqit from the University of Dundee, UK [94], and Nobutaka Hattori [95] and Yuzuru Imai [96] from the Juntendo University Graduate School of Medicine in Tokyo, revealed the signaling cascade in which the kinase PINK1, encoded by another Parkinson's disease-mutated gene [97], is stabilized in the outer mitochondrial membrane (OMM) of depolarized mitochondria where it phosphorylates both the UBL domain in Parkin and ubiquitin. Phospho-ubiquitin then interacts with Parkin releasing its autoinhibition and allowing ubiquitylation of OMM mitochondrial proteins (reviewed in [98]). The Harper group recently performed and published comprehensive analyses of the Parkin-dependent ubiquitinome of mitochondria [99], [100].
It would seem that all ubiquitin-binding SARs, that is, p62/SQSTM1, NBR1, OPTN, NDP52, TAX1BP1, and TOLLIP (Fig. 3), should be contributing to the autophagic clearance of ubiquitylated mitochondria. However, as shown by Youle's group in 2015, NDP52 and OPTN (but not p62/SQSTM1 or NBR1) may be the key receptors during Parkin-mediated mitophagy [101]. On the other hand, very recent data from Anne Simonsen's laboratory at the University of Oslo identify NIPSNAP1 and NIPSNAP2 proteins as major Parkin-dependent mitophagy factors which interact with LC3/GABARAP proteins as well as p62/SQSTM1, NBR1, NDP52, and TAX1BP1 (in addition to the non-ubiquitin-binding autophagy receptor ALFY/WDFY3) [102]. This suggests NIPSNAPs as potential hubs for ubiquitin-binding mitophagy SAR networks and might explain the SAR redundancy previously observed by Youle [101]. Importantly, Parkin-mediated ubiquitylation also mediates proteasome-dependent degradation of a number of OMM proteins [103], [104] and may be responsible for the rupture of the OMM allowing access of the autophagic machinery to the inner mitochondrial membrane [104].
Peroxisomes are single-membrane-bound organelles characterized by crystalline inclusions made from matrix proteins. They play important roles in β-oxidation of fatty acids and the detoxification of the poisonous hydrogen peroxide. The quantity of peroxisomes is regulated in part by autophagy, and the involvement of Atg and Cvt genes in this process in yeast cells undergoing changes in the source of nutrition was established by Klionsky in 1999 [12]. Eiki Kominami's group from the Juntendo University School of Medicine showed ATG gene dependence of pexophagy in mammalian cells in 2006 [105]. The important role of ubiquitylation as the driver of pexophagy was first demonstrated using mammalian cell models (ubiquitin-independent pexophagy was primarily studied in yeast and will be considered below). In 2008, the group of Jennifer Lippincott-Schwartz at the National Institutes of Health, USA, observed that fusion of GFP-monoubiquitin to the peroxisomal transporter PMP34 caused peroxisomal degradation [106]. Five years later, the group of Peter Kim from the Hospital for Sick Children in Toronto, Canada, in collaboration with Johansen's group, by manipulating NBR1 and p62/SQSTM1 levels, demonstrated the role of these ubiquitin-binding SARs in mammalian pexophagy. Interestingly, both the C-terminal ubiquitin-binding UBA domain and the intermediate membrane-binding α-helical J domain of NBR1, in addition to its LIR and coiled-coil domains, were required for NBR1's function as a pexophagy receptor [107]. The role of NBR1 in the degradation of peroxisomes was further corroborated by studies of PEX3-induced peroxisomal ubiquitylation and degradation [108]. The function of p62/SQSTM1 in pexophagy is less well understood: it may be increasing NBR1's efficiency [107] or more critically involved in special cases of pexophagy [109]. The candidate E3 ligase for ubiquitin-driven pexophagy is PEX2, which is induced upon amino acid starvation and ubiquitylates PEX5 and PMP70 on peroxisomal membranes [110].
Whether ubiquitin is a universal signal in other organisms and forms of selective autophagy is an active area of research. For a long time, the significance of ubiquitin-driven autophagy in yeast was not clear. However, in 2014, the work in the laboratory of Stefan Jentsch at the Max Planck Institute of Biochemistry in Martinsried, Germany, identified ubiquitin-binding CUE-domain protein Cue5, as the first aggrephagy SAR in yeasts: it interacts with both the ubiquitylated proteins and Atg8 and is required for clearance of aggregation-prone proteins. Interestingly, Cue5 is specific to aggrephagy, as neither pexophagy nor mitophagy or ribophagy was affected in yeast strains lacking Cue5 [111]. More recently, Cue5 was also proposed as a proteaphagy factor [112]. The specific function of the mammalian Cue5 homolog TOLLIP in autophagy is presently unclear.
Ubiquitin-independent SARs
The discovery of the LIR also prompted identification of those SARs that act independently of the ubiquitin signal by binding to the cargo directly. The first receptor of this kind, besides the Cvt cargo receptor Atg19, was the pexophagy-specific protein Atg30 identified by Suresh Subramani's group at the University of California in San Diego, USA, in 2008. The key features of Atg30, which is found in K. phaffii, are (1) its close association with the integral membrane peroxisomal proteins, peroxins Pex3 and Pex14, and (2) interaction with the components of the autophagic machinery: that is, Atg8, Atg11, and Atg17 [113]. The functional counterpart of Atg30 in S. cerevisiae is Atg36 reported by Ewald Hettema's group at the University of Sheffield, UK, 4 years later. Like Atg30, Atg36 interacts with Pex3 and Atg11, while no direct binding to Atg17 has been reported for Atg36 [114]. When mapping the LIRs in Atg30 and Atg36, the surprising finding was that the putative LIR sequences in these SARs had a very low affinity to Atg8 in their unphosphorylated state [115]. As with the LIR in OPTN, first when an upstream Ser was phosphorylated, Atg30 and Atg36 could bind Atg8 efficiently. The identity of the “LIR kinase” for the two pexophagy SARs remains elusive.
Mitophagy in yeast cells had been observed in response to growth in synthetic media with a carbon source other than glucose, with Uth1p [116] and Atg11 [117] initially shown to be specifically required for this process. In 2009, Klionsky's and Ohsumi's groups, using fluorescently labeled mitochondria, independently reported identification of the first yeast-specific mitophagy receptor Atg32 [118], [119]. The expression of this protein is induced during the respiratory growth of yeasts and, like Atg19, Atg32 binds both Atg11 and Atg8, obviating the need for any ubiquitin-binding SAR (which has not been discovered in yeast mitophagy to date). Interestingly, like with Atg30 and Atg36, phosphorylation of the Atg32 LIR by an unknown kinase is required to unleash the full-scale interaction between Atg8 and the LIR [115]. Of note, the binding between Atg11 and the SARs in yeast is also subject to phospho-regulation: casein kinase 2 (CK2) phosphorylates Atg32, as shown by the group of researchers led by Tamotake Kanki from the Niigata University, Japan, in 2013 [120] and Hrr25, a homolog of CK1δ, phosphorylates Atg19, Atg34, and Atg36 in their respective Atg11-binding regions, as demonstrated by Yoshinori Ohsumi and Hitoshi Nakatogawa from the Tokyo Institute of Technology in 2014 [121], [122]. On the other hand, as recently reported by Nakatogawa and Kanki, dephosphorylation of Atg32 by PP2A-like protein phosphatase Ppg1 inhibits mitophagy [123]. These data underscore the importance of the post-translational modifications in regulating selective autophagy.
The first mammalian SAR for ubiquitin- and Parkin-independent mitophagy was uncovered in 2010 by the groups of Ivan Dikic and Paul Ney from the St Jude Children's Hospital in Memphis, USA. They showed that NIX (also known as BNIP3L), which is induced by hypoxia [124] and during reticulocyte maturation [125], preferentially binds proteins from the GABARAP subfamily of the Atg8 family via its N-terminal LIR, which helps recruit the UBLs to depolarized mitochondria. In the reticulocyte maturation assay, which allows monitoring developmental mitophagy, a LIR-less, and hence LC3/GABARAP binding-deficient, version of NIX failed to mediate mitochondrial degradation efficiently [126]. These data helped explain, at least in part, the observed role of NIX in mitochondrial clearance in the erythroid lineage reported earlier [127]. Interestingly, BNIP3, the close NIX homolog with the highly conserved LIR, was also shown in 2012 by Asa Gustafsson's group at the University of California, USA, to be required for both mitophagy and ER-phagy [128]. The affinity of the LIR:LC3B interaction for both NIX and BNIP3 can be regulated by phosphorylation of an upstream Ser residue as demonstrated by the collaborative effort of Nathan Brady, Ivan Dikic, Volker Doetsch, and Vladimir Rogov in Germany [71], [129]. However, the kinase responsible for this has not been identified.
The search for mammalian mitophagy receptors is continuing, and the current list of LIR-containing mitochondrion-specific SARs (which do not depend on ubiquitin as a signal), besides the aforementioned NIX [126], BNIP3 [128] and NIPSNAP1/2 [102], includes FUNDC1, discovered in 2012 by Quan Chen's group at the Chinese Academy of Sciences [130]; BCL2L13, described in 2015 by Kinya Otsu's group from King's College London, UK [131]; AMBRA1, suggested in 2015 by Francesco Cecconi from the University of Rome Tor Vergata, Italy [132]; FKBP8, identified in 2017 by the groups of Terje Johansen and Vladimir Kirkin at Merck KGaA, Darmstadt, Germany [133]; and PHB2, published in 2017 by Beth Levine's group at the University of Texas Southwestern Medical Center in Dallas, USA [134]. Most of them, such as NIX, BNIP3, BCL2L13, FUNDC1, and FKBP8 (Fig. 3), are OMM proteins, with yet some (e.g., NIX, BNIP3, and FUNDC1) being inducibly expressed in response to hypoxia or as part of a developmental program. In contrast, PHB2 and NIPSNAPs are inner mitochondrial membrane and matrix proteins, respectively, which become accessible to cytosolic factors, LC3/GABARAPs and other SARs, upon mitochondrial membrane depolarization and rupture during PINK1/Parkin-mediated mitophagy [134]. Intriguingly, as shown by Valerian Kagan's group from the University of Pittsburgh in 2013, LC3/GABARAPs may directly interact with the inner mitochondrial membrane phospholipid cardiolipin, contributing yet another mechanism of mitophagy during mitochondrial membrane depolarization [135].
Selective degradation of the ER membrane (ER-phagy) had been documented during the unfolded protein response in yeast by the groups of Dan Klionsky [136] and Peter Walter [137] from the University of California, USA, in 2006. However, it had not been until 2015 that the first ER-phagy SARs were unearthed by Ohsumi and Nakatogawa, who identified Atg39 and Atg40 [138]. These are associated with the different parts of the yeast ER (Atg39 is localized to the perinuclear ER, whereas Atg40 is enriched on cortical and cytoplasmic ER) and, like most of the SARs in yeast, interact with both Atg8 and Atg11. The mammalian counterpart for Atg40, FAM134B, was described by Dikic and colleagues in 2015 and shown to be critical for homeostasis of sensory neurons [139]. Additional SARs specific to ER-phagy include Sec62, discovered in 2016 by the group led by Maurizio Molinari from the School of Life Sciences in Lausanne, Switzerland [140]; RTN3 reported in 2017 by the Dikic group [141]; CCPG1 published in 2018 by Simon Wilkinson from the University of Edinburgh, UK [142]; and, most recently, ATL3 described by Jianguo Chen's group at the Peking University in China [143] and TEX264 reported by the groups around Mizushima [144] and Harper [145]. These SARs (Fig. 3) are all integral ER membrane proteins that contain one or multiple LIRs in their cytosolic part, while CCPG1 seems to stand out in that, in addition to the LIR, it contains a novel protein–protein interaction module that binds the mammalian Atg11/Atg17-like protein FIP200 (also known as RB1CC1) via a linear FIP200-interacting region (FIR), which mediates recruitment of the Atg1/ULK1 kinase to the ER during the unfolded protein response [142].
Ubiquitin-independent SARs for selective autophagy pathways, other than the better-characterized aggrephagy, mitophagy, pexophagy, and ER-phagy, are being regularly identified based on their ability to interact both with the cargo and the Atg8/LC3/GABARAP proteins. Recent mammalian examples include the following: Stbd1 for glycophagy published by Peter Roach's group at the Indiana University School of Medicine, USA, in 2011 [146]; NCOA4 for the degradation of the iron-binding ferritin, independently discovered in 2014 by the groups of Alec Kimmelman at Dana-Farber Cancer Institute, Boston, USA [147], and Leon Murphy at the Novartis Institutes for Biomedical Research [148] ferritin was first described in 2011 as a bona fide autophagic cargo by the groups of Noboru Mizushima and Kazuhiro Iwai from the Osaka University in Japan [149]; ATGL and HSL for lipophagy identified in 2016 by Rajat Singh and colleagues from the Albert Einstein College of Medicine, Bronx, USA [150]; and NUFIP for ribophagy published in 2018 by the groups of Alessandro Ori from the Leibniz Institute on Aging-Fritz Lipmann Institute, Jena, Germany, and David Sabatini from the Massachusetts Institute of Technology, Cambridge, USA [151].
It needs to be added that there is a growing number of signaling proteins that have been shown to be selectively degraded by autophagy in the absence of an ostentatious link to ubiquitin- or SAR-dependent degradation. Recent examples include the following: DVL2 [152], β-catenin [153], FAS1 [154], Lamin B1 [155], paxillin [156], STING [157], CRY1 [158], and NCOR1 [159]. Most of these were shown to contain one or several LIR sequences, which mediate interactions with LC3/GABARAPs in a manner similar to the characterized SAR proteins. It remains to be investigated how such diverse proteins can all serve as efficient autophagy substrates.
Nature's blueprint of selective autophagy—hardwiring SARs with the ATG machinery
The molecular mechanism of autophagosome formation is one of the central questions in the autophagy field. Multiple studies performed since the original discovery of Atg genes in the mid-90s of the last century have led to the hierarchical model for the core Atg proteins, whose concerted action leads to the creation of the double-membrane vesicle at the PAS in response to starvation in yeast (reviewed in Refs. [9], [160]). Briefly, the apical Atg1 complex, consisting of Atg1 kinase and scaffolding proteins Atg13, Atg17, Atg29, and Atg31, is assembled at a perivacuolar PAS when the inhibitory TORC1 activity is low, for example, as a result of amino acid deprivation. Atg17 is the Atg1-binding scaffold protein that defines the site of the PAS during non-selective autophagy [161]. Once assembled and active, the Atg1 complex in turn activates the Vps34 system made of the lipid kinase Vps34 and the scaffolding proteins Vps15, Atg6, Atg14, and Atg38. The activation occurs via a poorly defined mechanism, likely involving direct phosphorylation of Vps34 complex subunits, such as Atg6 [162]. The net result of the Vps34 activity is the formation of the PI3P, which serves as a docking site for the lipid-binding Atg2/Atg18 complex. The Atg5–Atg12–Atg16 complex follows to the PAS, where it catalyzes the Atg8–PE conjugation and contributes to the membrane expansion. Atg9-containing vesicles shuttle between the Golgi and the PAS, contributing additional membrane to the nascent autophagosome. The selective autophagy-specific, Atg1-binding adaptor Atg11 [163], as well as its counterpart in non-selective autophagy Atg17 [164], also interact with Atg9, so that in the context of the selective and non-selective autophagy, respectively, the Atg1-binding scaffold proteins may at the same time be responsible for the recruitment of extra membrane via the Atg9 vesicles as shown by Ohsumi's group in 2012 [165]. In addition, in 2015, Ohsumi demonstrated that the Atg1-complex subunit Atg13 also interacts with Atg9, providing a feedforward mechanism for recruitment of the Atg9 vesicles [166]. Upon membrane closure, the autophagosome leaves the PAS and is able to fuse with the vacuole, releasing the inner vesicle with its contents into the lumen of the degradative compartment.
The general traits of this pathway seem to be well conserved in mammalian cells with two important differences: (1) widespread functional redundancy of the core ATG components in mammalian cells, with, for example, six mammalian LC3/GABARAP and four ATG4 proteins instead of one Atg8 and Atg4, respectively, in yeasts [167], and (2) simultaneous formation of autophagosomes at multiple locations (the concept of multiple PASs) at any given time during the autophagic response. The corresponding mammalian core ATG complexes are as follows (reviewed in Ref. [168]): (1) the ATG1/ULK1 kinase complex: ULK1, ULK2, ATG13, Atg17's homolog FIP200/RB1CC1, and ATG101; (2) the class III PI3K kinase complex: VPS34, VPS15, Atg6's homolog Beclin-1, and ATG14/Barkor; (3) the Atg2/Atg18-like complex: ATG2A, ATG2B, and Atg18's homologs WIPI1, WIPI2, WIPI3, and WIPI4; (4) the UBL conjugation machinery: ATG3, ATG4s (i.e., ATG4A, ATG4B, ATG4C, ATG4D), ATG5, ATG7, Atg8-like UBLs (i.e., LC3A, LC3B, LC3C, GABARAP, GABARAPL1, and GABARAPL2/GATE-16), ATG10, ATG12, and Atg16-like proteins ATG16L1 and ATG16L2; and (5) transmembrane ATG9A and ATG9B proteins homologous to the yeast Atg9.
De novo construction of the phagophore around the selective autophagy cargo shares many features with the non-selective autophagosome formation. However, it occurs in cells that are not experiencing starvation and thus possess active TORC1 complexes. So, the important questions in selective autophagy are as follows: How is Atg1/ULK1 recruited and activated at the surface of the cargo to set in motion the rest of the ATG machinery, and how is the observed close apposition of the growing autophagosomal membrane to the cargo achieved?
Lessons from yeast…
The first insights into the mechanics of the cargo-induced Atg1 recruitment were provided by the work of Klionsky using the Cvt pathway as a model. In 2001, he demonstrated the direct interaction between Atg1 and Atg11 [51] and, a year later, that the prApe1–Atg19 complex recruits Atg11 [61]. In 2005, he provided further evidence for Atg11 physically linking the Atg1 and prApe1–Atg19 complexes. The key features of Atg11, which allow it to perform its job as the adaptor between the cargo–receptor complex and the Atg machinery, are its pronounced ability to homo-oligomerize and simultaneously interact with both the cargo-bound Atg19 and the Atg1–Atg17 complex [169]. As all SARs identified in yeast to date (i.e., Atg19 [53], Atg30 [113], Atg32 [118], [119], Atg34 [54], Atg36 [114], Atg39 [138], and Atg40 [138]) bind Atg11, in addition to the cargo and Atg8 (Cue5 is the notable exception to the rule [111]), it can be assumed that Atg11 is the key adaptor/scaffold for SAR-, but not ubiquitin-, mediated selective autophagy in yeast [6]. However, it appears that at least in certain instances, for example, Atg30 in pexophagy [115], SARs may be able to recruit an additional Atg1-binding adaptor, such as Atg17 [170], [171].
In 2016, the critical role of Atg11 in selective autophagy was demonstrated by Claudine Kraft's group from the Max F. Perutz Laboratories at the University of Vienna, Austria, who tethered Atg11 directly to the cargo, prApe1 (Cvt pathway) or Pex3 (pexophagy), bypassing the requirement for the SARs, Atg19, or Atg36, respectively. This elegant approach revealed that the stable cargo–Atg11 complex was sufficient to induce selective autophagosome formation [172]. In addition, Kraft demonstrated that simple oligomerization of Atg1 was sufficient to activate Atg1 in situ, strongly corroborating the data on the direct oligomeric cargo-driven Atg1 activation from the laboratory of Vladimir Denic at the Harvard University in Cambridge, USA, published a year earlier [162].
The Atg11-centric model proposed by Kraft and colleagues may suggest that binding Atg8 by the yeast SARs may be optional. However, Atg11 may itself interact with Atg8, as proposed in 2014 by Richard Vierstra and colleagues from the University of Wisconsin, USA, for Atg11 in Arabidopsis [173]. Also, the discovery of the functional LIR/AIM in yeast Atg11 may have been precluded by the lack of a preceding phosphorylation event that would activate the Atg11:Atg8 interaction as shown for AIMs in the yeast SARs [115], [120], [121], [122].
Importantly, several reports strongly supported the idea that, next to Atg11, recruitment of Atg8 by SARs, is playing a leading role in cargo-induced activation of the Atg1 complex in yeast. In 2012, three groups independently described identification of functional AIMs/LIRs in Atg1/ULK1 complexes: Yoshinori Ohsumi's group [174] and the one led by Matthias Peter from the ETH Zuerich, Switzerland [175] identified the AIM in yeast Atg1, whereas Peter's [175] and Johansen's [176] groups found the LIR in the mammalian homologs ULK1 and ULK2. The AIM in Atg1 was required for Atg1 localization to the autophagosome and its transport into the vacuole, where the Atg1–Atg13 complex is degraded. Defects in the Atg1:Atg8 interaction caused faults in both Cvt and autophagy pathways [174], [175]. Of significant interest was the finding that at least in the human ULK1/2 complex, also ATG13 and FIP200 contain functional LIR motifs, as demonstrated by Johansen [176], and, in the case of ATG13, studied in detail by the groups of Tamotsu Yoshimori and Soichi Wakatsuki from the Institute of Materials Structure Science in Tsukuba, Japan [177]. Thus, much tighter association between the Atg1/ULK1–Atg13/ATG13–Atg17/FIP200 complex and Atg8/LC3/GABARAPs can be expected.
Given the relatively low affinity of the binding between Atg8/LC3/GABARAPs and AIMs/LIRs (in the range of 0.1–100 μM [20]), as already mentioned, the oligomerization of cargo/receptor/scaffolding proteins or the presence of multiple LIR sequences in proteins is required for stronger interaction. In 2014, using biochemical methods, Sascha Martens' group from the Max F. Perutz Laboratories identified multiple AIMs in Atg19 that could be revealed upon the binding of Atg19 to its oligomeric cargo, prApe1 [178]. Importantly, the multiple AIMs in Atg19 also ensure its tight apposition with Atg8-coated membranes in a fully reconstituted, cell-free system as shown by the same group in 2016 [179].
… and mammalian cells
The mechanism of the selective autophagosome formation in mammalian cells is much more convoluted and correspondingly less understood, in part, because of the increasing complexity of the core ATG and SAR protein systems and, in part, because of the lack of a powerful in vivo model system, like the biosynthetic Cvt pathway in yeast. SARs for aggrephagy/pexophagy, mitophagy, and ER-phagy are among the best studied in the mammalian cell. They are either oligomeric by the nature of their domain organization (e.g., p62/SQSTM1 oligomerizes via its PB1 domain [67]) or may be exposed in a concentrated way on the surface of the target organelle (the surmised feature of the membrane-bound SARs, such as NIX [126] and FAM134B [139]). Historically, the first defining feature of the mammalian SARs, besides their cargo-binding or self-oligomerizing domains, was the LIR [20], [180], which ensures interaction of the mammalian SARs with the members of the LC3/GABARAP family. One possibility is that LC3/GABARAP's recruitment can be required for bringing in the ULK1/2–ATG13–FIP200 [174] [175], [176] and VPS34–VPS15–Beclin-1–ATG14 [181] complexes via the LIRs present therein. In addition to the already mentioned pieces of evidence, in 2015, the laboratory of Sharon Tooze at The Francis Crick Institute in London, UK, demonstrated that activation of ULK1 on starvation-induced phagophores depends on the functional GABARAP as well as the LIR within the ULK1 [182]. More recently, Johansen, in collaboration with Tooze, also described functional LIRs in all key components of the class III PI3K kinase complex: VPS34, Beclin-1, and ATG14. Significantly, mutation of the LIR within the autophagy-specific subunit ATG14 prevented its phosphorylation by ULK1, mediated by direct binding of ATG13 and ATG14 (as previously demonstrated by Daniel Voytas and Do-Hyung Kim's groups at the University of Minnesota, Minneapolis, USA [183]), and impaired mitophagy [181]. This indeed suggests a much broader role for the mammalian ATG8 proteins in the recruitment of the core autophagy machinery.
ATG16L1, which is a mammalian homolog of the yeast Atg16 and the scaffolding component of the ATG5–ATG12:ATG16L1 E3 ligase complex, acts as an unusual SAR in that it binds its cargo, ubiquitin [87], but interacts with LC3/GABARAPs only indirectly, via the E2 enzyme ATG3, which is recruited by ATG12 to the phagophore [37]. However, it does interact directly with FIP200, as demonstrated in 2013 independently by the groups of Yoshimori [87] and Mizushima [184], as well as Michael Overholtzer and Xuejun Jiang at the Memorial Sloan Kettering Cancer Center in New York, USA [185].
FIP200 was proposed by Mizushima' in 2008 to be the homolog for the yeast Atg11 and Atg17 proteins that are missing in the mammalian system [186]. In 2014, the Tooze laboratory showed that the PI3P-binding protein WIPI2 at the phagophore recruits the ATG5–ATG12–ATG16L1 complex by directly binding ATG16L1, thus further amplifying both the ULK1 activation and LC3/GABARAP–PE conjugation. Of interest, FIP200 was not required for this process [187]. Coupled with the observation that the ER-phagy SAR CCPG1 has both LIR and FIR motifs and is thus capable of recruiting simultaneously LC3/GABARAP and FIP200 to the site of ER-phagy [142], the requirement for cargo-mediated recruitment of both LC3/GABARAP–PE and ULK1/2-activating systems may be the new defining feature of the mammalian selective autophagy.
Cooperativity between SARs may be the strategy for mammalian cells to enhance the cargo-induced recruitment of LC3/GABARAP and FIP200 (and hence ULK1/2, and possibly VPS34 complexes) to the SAR–cargo complex. p62/SQSTM1 interacts directly with NBR1 [65], so that dual p62/SQSTM1 and NBR1 enlistment can increase the local concentration of LIR motifs. Furthermore, as shown in 2014 by Ronggui Hu and colleagues from the Chinese Academy of Sciences in Shanghai, p62/SQSTM1 can recruit ubiquitylated OPTN [188]. Consequently, it can be envisaged that all six known p62/SQSTM1-like receptors (SLRs), p62/SQSTM1, NBR1, NDP52, TAX1BP1, OPTN and TOLLIP, might form higher-order molecular scaffolds made of the ubiquitylated cargo and SARs [189]. Intriguingly, the founding member of the SLRs, p62/SQSTM1, may also lead in the process of selective autophagosome formation by binding FIP200 directly, as now shown by Marten's group [190], thus recruiting both LC3/GABARAPs and FIP200 to ubiquitylated cargo, similar to CCPG1 in the process of ER-phagy [142]. Of note, also other SLRs may interact with FIP200, as now also demonstrated for NDP52 by two recent reports contributed by the groups of Felix Randow, Giampietro Schiavo (The University College London, UK), and Richard Youle [191], [192], suggesting that the parallel binding to LC3/GABARAP and FIP200 may indeed be a defining paradigm also for mammalian SARs, similar to the Atg8/Atg11-binding SARs found in yeast.
Members of the tripartite motif (TRIM) family containing an N-terminal RING domain, a B box, and a coiled-coil domain, were shown in 2014 by the group of Vojo Deretic at the University of New Mexico Health Sciences Center, Albuquerque, USA, to act as possible organizers of selective autophagy signaling platforms [193]. One of the members, TRIM5α, interacts with a plethora of ATG proteins, including ULK1, Beclin-1, and GABARAP, acting as a conduit for ULK1 and VPS34 activation. As TRIMs also directly interact with various cargo (e.g., TRIM5α binds retroviral capsids [194]), they are proposed to act as bona fide SARs for special types of selective autophagy, such as virophagy [193] and aggrephagy [195] (Fig. 3).
Curiously, recent work suggested that the best-studied SAR p62/SQSTM1 may be dispensable for some forms of selective autophagy. In 2015, Youle's group published the results of their reconstitution experiments in HeLa cells lacking five SLRs (p62/SQSTM2, NBR1, OPTN, NDP52, and TAX1BP1), showing that NDP52 and OPTN (to a lesser extent TAX1BP1, but not p62/SQSTM1 or NBR1) were recruited in a PINK1- and pS65 ubiquitin-dependent manner to depolarized mitochondria and able to rescue Parkin-mediated mitophagy [101]. Furthermore, at the beginning of this year, Michael Lazarou's laboratory at the Monash University in Melbourne, Australia, refined Youle's model of mitophagy showing that the LIR motifs of NDP52 and OPTN were dispensable for ubiquitin-driven recruitment of the SLRs but required for the amplification of the mitophagy by enlisting the ULK1/2 complex via the LC3/GABARAP and stimulating additional rounds of LC3/GABARAP lipidation and ULK1/2 complex recruitment at the surface of mitochondria [196]. These results highlight the complexity and potential redundancy of the selective autophagy components in the mammalian cell.
Digesting issues with the selective autophagosome formation
The tremendous progress achieved in the past two decades, primed by the elucidation of the role of UBLs and identification of SARs, has allowed drafting the first model (Fig. 4) of how an autophagosome can form around a cargo selectively [6], [20]. Many laboratories around the globe have recently been working toward refining this concept by providing experimental evidence in order to explain cargo-initiated seeding of the core ATG machinery. However, unexpected findings raised important issues, some of which are addressed below.
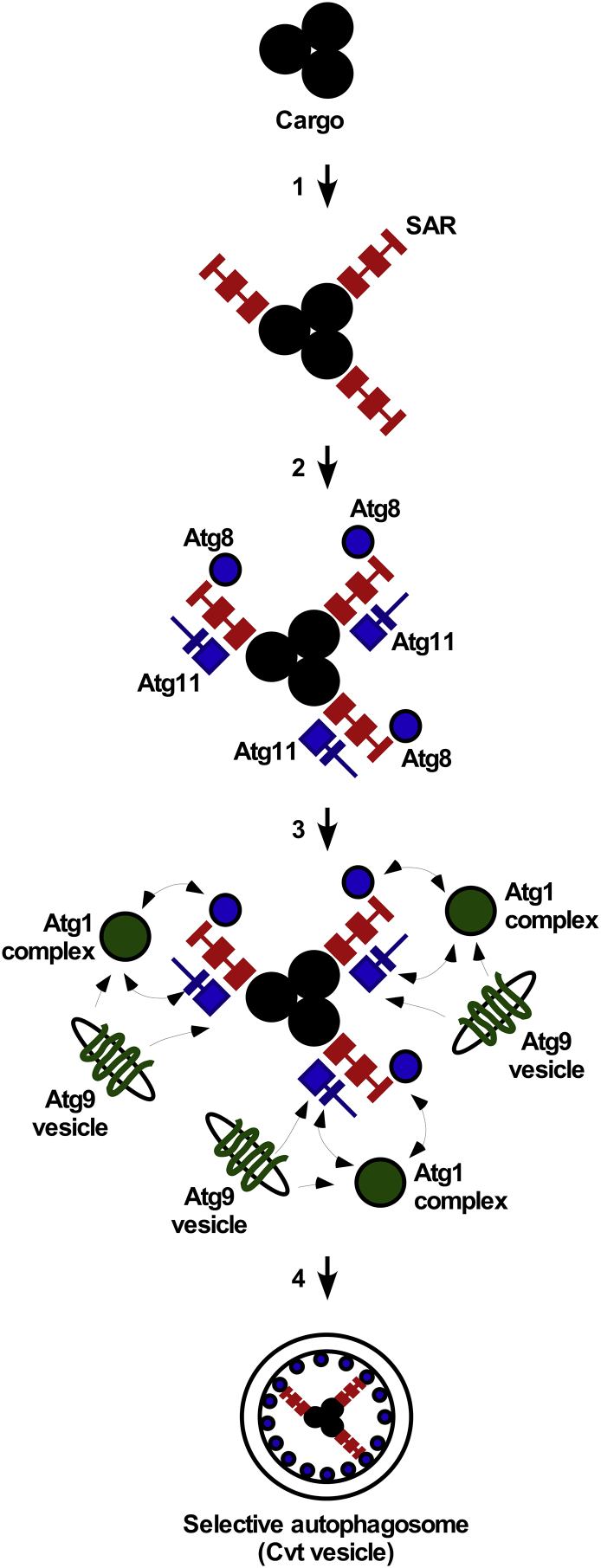
Fig. 4.
A simplified model of selective autophagosome formation. Based on studies using yeast as a model organism, the following steps of selective autophagosome formation can be discerned: (1) Oligomeric cargo is labeled by SARs. Oligomerization of the SARs (not shown) can further contribute to the cargo/receptor complex formation. (2) SARs recruit the Atg1- and Atg9-binding adaptor Atg11 as well as Atg8, which may or may not be conjugated to PE (not shown) at this step. The SAR:Atg11 and SAR:Atg8 interactions can be regulated by post-translational phosphorylation (not shown). (3) Atg11 and Atg8 recruit the components of the Atg1 kinase complex responsible for the activation of the catalytic cascade that leads to the production of the selective autophagosome in situ (4). In addition, both Atg11 and components of the Atg1 complex interact with the transmembrane protein Atg9 anchored in the Atg9 vesicles, regulating the growth of the lipid membrane. Multiple components of the system, such as Atg11 and the Atg1 complex, are oligomeric (not shown), which further contributes to the formation of the selective autophagosome (4). Abbreviations: Atg, autophagy-related, Cvt, cytoplasm to vacuole targeting; SAR, selective autophagy receptor.
Are Atg8/LC3/GABARAPs strictly necessary for autophagosome formation?
Perhaps, one of the most puzzling questions is the one about the role of the Atg8/LC3/GABARAP family of autophagy-specific UBLs in the autophagosome formation. In 2016, Lazarou's group, while performing reconstitution experiments in HeLa cells lacking six members of the LC3/GABARAP family (LC3A, LC3B, LC3C, GABARAP, GABARAPL1, and GABARAPL2), demonstrated that the UBLs were dispensable for the sequestration of mitochondria in selective autophagosomes during Parkin-dependent mitophagy. However, they observed that the autophagosome–lysosome fusion was defective in the hexa-knockout (KO) HeLa cells [197]. There is currently no good explanation for the redundancy of LC3/GABARAPs in this form of mitophagy; however, one could speculate that the remaining MAP1LC3B2 gene, which also encodes LC3B in human cells and not disrupted in hexa-KO HeLa cells, could have provided some compensation for the LC3/GABARAP function in membrane expansion. In addition, the massive ubiquitylation of the OMM proteins caused by Parkin overexpression might have overridden the dependence on LC3/GABARAP proteins by recruiting ULK1/2-ATG13-FIP200 complexes via ATG16L1 directly. Also, other SARs might bring the ULK1/2 complex and the ubiquitylated mitochondria together, via their FIRs, leading to the membrane expansion in an LC3/GABARAP-independent fashion. Indeed, simple tethering of a FIP200-binding peptide derived from ATG16L1 to mitochondria is sufficient to drive mitophagy [191].
Other reports also indicated the apparent redundancy of LC3/GABARAP–PE conjugation machinery for some forms of selective autophagy. In 2017, Murphy's group described that ferritinophagy, selective delivery of ferritin to the lysosomes via NCOA4, was dependent on ATG9A, FIP200, and VPS34 but could proceed normally in cells lacking the E1 (ATG7), E2 (ATG7), and E3 (ATG16L1) elements required for the LC3/GABARAP–PE conjugation. Instead, they observed that TAX1BP1 was directly binding NCOA4, driving its degradation in ATG7-deficient cells [198]. That SLRs, including NBR1 and TAX1BP1, can be degraded by a lysosome-dependent pathway in ATG7 KO cells was also demonstrated by the Denic group [199]. The results from the groups of Johansen and Harald Stenmark, from the Oslo University Hospital, suggest that components of the endosomal sorting complex required for transport (ESCRT)-III complex may be responsible for endosome-mediated delivery of p62/SQSTM1, NBR1, TAX1BP1, NDP52, and also NCOA4, into the lysosome and their rapid degradation during the first hours of amino acid starvation [200]. This can explain the ATG7- and LC3/GABARAP lipidation-independent degradation of SARs observed in the earlier study [198]. However, much more work is required to understand the interplay between the endosomal and autophagy membrane trafficking systems in cargo degradation, as several components, including subunits of the VPS34 kinase complex, for example, Beclin-1 [201], are shared between the two lysosome trafficking pathways.
Is Atg5 an overlooked selective autophagy factor?
Another plausible reason for the unexpected findings in ATG7 [198] and also ATG3 [202] KO cells, which apparently support LC3/GABARAP lipidation-independent autophagosome formation, is the possibility that at least some functions of the Atg8/LC3/GABARAP proteins, albeit less efficiently, might be taken over by the other autophagy-specific UBL Atg5. In 2012, groups of Martens and Kraft, using purified Atg proteins and giant unilamellar vesicles (GUVs), demonstrated that Atg5 could bind lipid membranes [203]. In 2014, Thomas Wollert's laboratory from the Max Planck Institute of Biochemistry in Martinsried also showed that the Atg12–Atg5–Atg16 complex has the ability to remain associated with the lipid membrane after the Atg8–PE conjugate formation. In 2018, the group led by John Brumell from the Hospital for Sick Children in Toronto demonstrated the importance of the ATG12–ATG5–ATG16L1 complex for the repair of damaged membranes. This was independent of the ESCRT complex, typically involved in this process, and not connected to the known role of ATG12–ATG5–ATG16L1 in autophagy [204]; however, this finding did further implicate the ATG12–ATG5–ATG16L1 complex in lipid membrane remodeling.
If Atg5 is associated with autophagic membranes, are there SARs that can specifically interact with this UBL? In 2010, groups of Anne Simonsen and Ai Yamamoto from the Columbia University in New York, USA, described the ATG5- and PI3P-interacting protein ALFY/WDFY3 that is required for efficient clearance of the mutated huntingtin protein [205]. This suggested the existence of ATG5-binding SARs, although in 2014 Simonsen showed that ALFY also has a functional LIR and thus binds LC3/GABARAPs [206]. In 2016, Martens' laboratory published the curious finding that the AIMs of the yeast SARs, Atg19 and Atg34, as well as the LIRs in the human SARs p62/SQSTM1, NDP52, and OPTN interact with ATG5 [207], suggesting that the LIR-containing SARs might also recognize membrane-localized Atg5/ATG5. The Atg8:AIM and Atg5:AIM interactions are mutually exclusive, so that it is unclear how the complex interplay between the cargo receptors and the UBLs takes place in vivo. Clearly, the role of ATG5 in selective autophagy, as an additional membrane-associated UBL, deserves thorough investigation.
What is the identity of the mammalian Atg11?
Atg11 is the core Atg factor in the cargo-dependent, selective autophagosome formation in yeast. It oligomerizes and binds both the SAR–cargo complex (e.g., Atg19–prApe1) and the Atg1 complex, thereby bridging the cargo with the key kinase activity required for the autophagosome formation (reviewed in Ref. [208]). In addition, Atg11 may interact with autophagic membranes via Atg9 [163] and Atg8 [173]. FIP200 has been proposed to act as an Atg17-like factor tethering ULK1/2 complex to the site of autophagosome formation in mammalian cells [186]. However, a growing body of evidence strongly points also at the Atg11-like functions of FIP200 in selective autophagy. An increasing number of mammalian SARs, such as ATG16L1 [87], [184], [185], CCPG1 [142], p62/SQSTM1 [190], and NDP52 [191], [192], have now been shown to interact with FIP200 directly. Interestingly, the FIR of p62/SQSTM1 encompasses its LIR and shows mutually exclusive binding with FIP200 and LC3B, respectively [190]. This raises interesting questions regarding the sequence of events in the recruitment of ULK1 complex via either LC3/GABARAPs or FIP200.
Intriguingly, the large protein huntingtin, notoriously mutated in Huntington's disease, also exhibits an Atg11-like sequence and is able to interact with both ULK1 and GABARAPs, as revealed in 2014 by a group of researchers led by Joan Steffan from the University of California, USA [209]. In addition, huntingtin directly binds p62/SQSTM1, as shown in 2015 by Anna Maria Cuervo's group from the Albert Einstein College of Medicine and Sheng Zhang's group from The University of Texas Medical School at Houston, USA [210], which increases its similarity with FIP200. Undoubtedly, further studies into huntingtin will follow revealing its role in selective autophagy, especially in the context of neurodegeneration.
In the light of the growing evidence for the role of mammalian Atg17/Atg11-like proteins in selective autophagy, the previously somewhat LIR-centric discovery of SARs will need to adopt the growing knowledge of FIP200-binding motifs and FIRs. More broadly, researchers working in the field of selective autophagy will need to consider the role of interactions between the SARs and the core ATG proteins as well as between SARs and lipid membranes to gain more complete understanding of the process of the selective autophagosome formation.
Why are not all LIR-containing proteins targeted to the selective autophagosome?
Definition of the AIM/LIR [176], [180] has helped identify a plethora of proteins involved in the process of autophagy either as part of the selective autophagy machinery (SAR and ATG proteins) or as autophagic cargo. To aid discovery of new LIR-containing proteins, groups of Ioannis Nezis from the University of Warwick, UK, and Vasilis Promponas from the University of Cyprus created a web-based resource in 2014, which they called iLIR. It can be used for predicting LIRs in eukaryotic proteins [211], [212]. A number of proteins have since been found to contain a LIR(s) and also be degraded by autophagy: for example, DVL2 [152], β-catenin [153], FAS1 [154], Lamin B1 [155], paxillin [156], STING [157], CRY1 [158], and NCOR1 [159], while others seem to contain a LIR(s) but perform a scaffolding/regulatory function and not subject to lysosomal degradation: for example, TP53INP2 [213], FYCO1 [214], TBC1D5 [215], TBC1D25 [216], TECPR2 [217], PLEKHM1 [218], Ankyrin-3 [219], PCM1 [220], and BRUCE [221]. This poses the question: Why do some LIR-containing proteins serve as substrates for autophagy, while others do not seem to be “visible” to the degradation machinery? One suggestion is that recruitment to the outer membrane of the autophagosome, rather than inside the autophagosome, can be the reason for the escape of the protein from the autophagosome before it is fused with the lysosome. What determines the topology of the recruited LIR-containing protein is, however, unclear.
The other possibility is that, in order to be an efficient autophagic cargo, the protein needs to create higher-order oligomers and be able to recruit a critical number of Atg8/LC3/GABARAP and/or Atg11/FIP200 proteins, which in turn can enlist and activate the Atg1/ULK1 and/or Vps34 complexes. Recently, several groups started to explore whether the biophysical model of biomolecular condensation underpinned by multivalent macromolecular interactions (reviewed in Ref. [222]) could explain how high-order cargo:SAR complexes are formed in the cytosol prior to being engulfed by the autophagic membrane. As shown by Martens [223], as well as Pilong Li and Li Yu from the Tsinghua University, Beijing, China [224], recombinant p62/SQSTM1 undergoes liquid–liquid phase separation (LLPS) and forms biomolecular clusters (condensates) in the presence of polyubiquitin chains (model cargo) in vitro. This phase separation activity of p62/SQSTM1 is enhanced by mutations that increase its affinity toward the ubiquitin (i.e., S403E in the UBA domain). As the PB1 domain of p62/SQSTM1 was required for the phase separation, it can be concluded that p62/SQSTM1 oligomerization is another key driver for this process. Importantly, the LLPS could be observed in cells, where p62/SQSTM1 bodies were shown to be liquid-like, dynamic, and governed by the same principles as those identified using the recombinant proteins [224]. In addition, the presented p62/SQSTM1 LLPS model was compatible with p62/SQSTM1's interactions with NBR1 (which enhanced p62/SQSTM1:ubiquitin clustering) and LC3B (which reduced the number and the size of p62/SQSTM1:ubiquitin clusters) [223].
The LLPS principles may govern cargo:receptor complex assembly observed in other model systems for selective autophagy. Thus, investigators around Nobuo Noda from the Institute of Microbial Chemistry in Tokyo have demonstrated that the Cvt pathway-specific Atg19:prApe1 binding competes with the self-association of the prApe1 propeptide, thus regulating the size of the dodecameric prApe1 substrate and permitting its interaction with the Atg8 via the AIMs harbored in Atg19 [225]. Hong Zhang and colleagues from the Chinese Academy of Sciences in Beijing showed that the size, composition, and stability of the P granules found in Caenorhabditis elegans embryos, which are a classical example of LLPS, are regulated by the C. elegans-specific SAR SEPA-1, which promotes LLPS of the P granule components PGL-1 and PGL-3 [226] and is reminiscent of the coalescence between p62/SQSTM1 and polyubiquitin chains. Of interest is also the finding that mTOR activity modulates the LLPS of the P granules suggesting a role of this autophagy-modulating kinase also in some forms of selective autophagy [226].
The role of other core ATG proteins, especially Atg11, in LLPS has not been analyzed to date. However, it can be expected that ATG proteins will modulate the dynamics of the SAR:cargo LLPS and eventually define whether a cargo will become an optimal substrate for autophagy.
How is the assembly of the cargo–SAR–ATG complex regulated?
The dynamic assembly of the cargo-SAR complex and their LLPS may be spontaneous in situations where there is instantaneous self-oligomerization of the cargo and/or the SARs. However, it can be envisaged that in order to respond to an abrupt fluctuation in the stoichiometry of the cargo–SAR complex (e.g., rapid increase in the cargo not matched by availability of SARs), the affinity of the interaction between the cargo and the receptor might require adjustment. Phosphorylation of Ser403 within the UBA domain of p62/SQSTM1 increases its affinity to ubiquitin chains as demonstrated by the group of Nobuyuki Nukina at the RIKEN Brain Science Institute in Wako, Japan, in 2011 [227]. Deretic, in collaboration with the Johansen group, showed in 2012 that TBK1 phosphorylates this residue in response to mycobacterial infection [228]. Furthermore, in 2017, Ronggui Hu's group, in collaboration with an international team of scientists, demonstrated how monoubiquitylation of p62/SQSTM1, mediated by the p62-interacting E2 enzymes, UBE2D2 and UBE2D3, activates its ubiquitin-binding UBA domain, thus also increasing the affinity of the p62/SQSTM1:ubiquitin interaction [229]. It is also likely that the oligomerization of p62/SQSTM1 is regulated, as it is critical for the tight binding between the SAR and ubiquitin as well as the SAR and LC3 [230]. In 2015, the group led by Kwon at the Seoul National University, Korea, showed that arginylation of several substrate proteins activates the intermediate ZZ domain in p62/SQSTM1 and promotes its oligomerization and aggregation, while increasing the interaction with LC3 and stimulating autophagy [231].
Once the cargo–receptor complex is formed, a further round of regulation is at play. As mentioned previously, several kinases have been found to phosphorylate the Atg11-interacting sequences within yeast SARs and increase the affinity of the Atg11:SAR interaction [120], [121], [122]. Similarly, phosphorylation is likely to regulate the SAR:FIP200 interaction in mammalian cells, as now shown for both p62/SQSTM1 [190] and NDP52 [191]. On the other hand, phospho-regulation of the affinity of the AIM:Atg8 or LIR:LC3/GABARAP interactions is another well-recognized step in activation of some SARs, with the prominent examples including OPTN and Atg30/Atg36, as discussed above.
Phosphorylation of LC3, similar to phospho-ubiquitin, has been observed [232], [233], and this too will affect the LIR:LC3 affinity, potentially regulating selective autophagy. Acetylation has been shown to regulate LC3 nuclear-cytoplasmic shuttling [234] but may equally regulate the affinity of the binding between LC3 and the LIRs within SARs.
Conclusions and Perspectives
The countless discoveries of the past 20 years have brought to light the growing complexity of selective autophagy pathways. In vivo studies, mostly in yeasts, and in vitro experiments, using purified proteins and artificial lipid membranes, greatly facilitated reconstruction and modeling of the steps involved in the selective autophagosome formation (summarized in an extremely simplified form in Fig. 4). As the autophagosome machinery is well conserved across the phyla, it can be expected that selective autophagy mechanisms will also be underpinned by some common principles. The preservation of the Atg8:SAR system is now well documented in model organisms other than mammals and yeasts: NBR1 in Arabidopsis is the functional counterpart for both p62/SQSTM1 and NBR1 [235], while Ref (2)P is the p62/SQSTM1 homolog in Drosophila [236]. Also, SEPA-1 and SQST-1 are two SARs in C. elegans [237], [238]. Similarly, conservation of the Atg11:SAR interaction can be expected. It will therefore be possible to probe into the selective autophagosome formation mechanisms in multicellular model organisms with lower complexity of the selective autophagy components. Once the basic components and regulatory loops have been identified, it will be important to confirm the findings in human cells that will eventually lead to medically relevant break-throughs in understanding of selective autophagy processes and their exploitation in finding cures for human diseases.
Selective autophagy is implicated in neurodegeneration, as different protein aggregates accumulate in the brains of patients with Parkinson's, Huntingtin's, Alzheimer's, and other proteinopathies (reviewed in Ref. [239]). It is highly desirable to induce selective degradation of protein aggregates or their intermediates, which has been shown to lead to amelioration of symptoms of neurodegeneration in model organisms (reviewed in Ref. [240]). Here, the recent data using ATG16L1-derived FIR peptides to induce mitophagy are very encouraging [191]. Similarly, in infectious diseases, one could envision great utility for drugs that would both selectively eliminate viruses and bacteria and enhance presentation of the pathogen-derived antigens by stimulating targeted autophagosome formation. Also, the role of selective autophagy in cancer is understood only poorly but is thought to be important in the context of cancer cell-specific protein aggregates or feeding the tumor antigen presenting pathways. It would therefore be extremely important to gain opportunities to modulate selective autophagy in the pathological states.
Exploitation of the UBL:LIR interaction could represent the entry point for therapies targeting selective autophagy. Peptides or small molecules interfering with the formation of the intermolecular β-sheet, by having affinities to the LC3/GABARAP proteins higher than naturally occurring LIRs, could lead to potent inhibition of selective autophagosome formation. Ultra-high-affinity, extended LIR peptides (e.g., with a lower nanomolar affinity to GABARAP) derived from ankyrins have recently been described by Chao Wang and Mingjie Zhang groups from the Hong Kong University of Science and Technology [241]. These are able to inhibit selective autophagy in C. elegans, providing an in vivo proof of principle for the LIR peptide-based approach. With activation of selective autophagy being probably more desirable, especially in the context of neurodegenerative and infectious diseases, it is anticipated that better understanding of the mechanisms of selective autophagy will allow to design strategies to seed a selective autophagosome on or at the surface of a cargo. There are now tool compounds available that may unlock the SAR function. XIE62–1004 is one of them discovered by the group of Xiang-Qun Xie (The University of Pittsburg, USA) and Yong Tae Kwon's and Bo Yeon Kim's groups at the Seoul National University in Korea in 2017 [242]. It mimics protein arginylation and was shown to cause conformational changes in p62/SQSTM1's ZZ domain, thus stimulating its binding to LC3 and autophagic degradation, as shown by the groups of Yong Tae Kwon and Tatiana Kutateladze from the University of Colorado School of Medicine, USA, in 2018 [243].
The knowledge of how AIM/LIRs recognize Atg8/LC3/GABARAPs also helps design new tools to monitor the process of autophagy. In 2017, the groups of Ivan Dikic as well as Deok-Jin Jang and Jin-A Lee from the Kyungpook National University National University and Hannam University, respectively, in Korea, developed LIR-based sensors based on GFP-LIR fusions, which hold the promise of delivering specific reagents for different LC3/GABARAP isoforms, which is not always possible using isoform-specific antibodies [244], [245]. Interestingly, novel LC3/GABARAP interaction sequences are being discovered, such as the 3D LIR in Atg12 published by Wollert's group in 2014 [246]; the dual UFM1-Interacting Motif (UFIM)/LIR in the UFM1-specific E1 enzyme UBA5 identified in 2016 by Kirkin, Komatsu, and Rogov's groups [247]; the α-helical LIR in Trim5α described by the groups of Jonathan Stoye from the Francis Crick Institute and David Goldstone, the University of Auckland, New Zeland [248]; and, most recently, the ubiquitin-interacting motif (UIM)-like Atg8 binding sequence that utilizes an alternative hydrophobic patch, as elegantly demonstrated by the Vierstra lab [249]. It is, however, important to realize that Atg8/LC3/GABARAP proteins play roles in processes other than autophagy, such as LC3-associated phagocytosis (LAP) [250], V-ATPase-dependent endolysosomal LC3 lipidation [251], and some others (reviewed in Ref. [252]); so that additional markers for autophagy (e.g., other autophagosome assembly and flux markers) will have to be analyzed when developing and using tools and drugs exploiting and targeting the Atg8:SAR interactions.
In conclusion, with the massively accumulating knowledge of the past two decades, we are now well positioned to start exploring and exploiting the mechanics of making the selective autophagosome around various cargos, which holds great promise for novel approaches to medicine and biotechnology.
Acknowledgments
I thank Terje Johansen, The University of Tromso, Norway, and Rajesh Chopra, The Institute of Cancer Research London, UK, for critical reading of this manuscript. The work in the laboratory of V.K. is supported by the CRUK program grant C2739/A22897 and by a Marie Skłodowska-Curie ETN grant under the European Union's Horizon 2020 Research and Innovation Program (Grant Agreement No 765912). I apologize to the authors of many other important discoveries in the field of selective autophagy whose work could not be cited here due to space limitation.
References
- 1.Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24:9–23. doi: 10.1038/cr.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky D.J. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 3.Tsukada M., Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 4.Thumm M., Egner R., Koch B., Schlumpberger M., Straub M., Veenhuis M. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 5.Harding T.M., Morano K.A., Scott S.V., Klionsky D.J. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farre J.C., Subramani S. Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat Rev Mol Cell Biol. 2016;17:537–552. doi: 10.1038/nrm.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizushima N., Yoshimori T., Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 10.De Duve C., Wattiaux R. Functions of lysosomes. Annu. Rev. Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- 11.Cuervo A.M., Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 2014;24:92–104. doi: 10.1038/cr.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hutchins M.U., Veenhuis M., Klionsky D.J. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J. Cell Sci. 1999;112:4079–4087. doi: 10.1242/jcs.112.22.4079. Pt 22. [DOI] [PubMed] [Google Scholar]
- 13.Kim IR-E S., Mizushima N., Ohsumi Y., Lemasters J.J. Autophagic degradation of mitochondria in GFP-LC3 transgenic mouse hepatocytes after nutrient deprivation. Hepatology. 2004;40(Suppl. 1):291A. [Google Scholar]
- 14.Kim I., Rodriguez-Enriquez S., Lemasters J.J. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gatica D., Lahiri V., Klionsky D.J. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 2018;20:233–242. doi: 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaffagnini G., Martens S. Mechanisms of selective autophagy. J. Mol. Biol. 2016;428:1714–1724. doi: 10.1016/j.jmb.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stolz A., Ernst A., Dikic I. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 2014;16:495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 18.Anding A.L., Baehrecke E.H. Cleaning house: selective autophagy of organelles. Dev. Cell. 2017;41:10–22. doi: 10.1016/j.devcel.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khaminets A., Behl C., Dikic I. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 2016;26:6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 20.Rogov V., Dotsch V., Johansen T., Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 21.Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M.D. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 22.Mizushima N., Sugita H., Yoshimori T., Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J. Biol. Chem. 1998;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- 23.Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 24.Kirisako T., Ichimura Y., Okada H., Kabeya Y., Mizushima N., Yoshimori T. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 27.Ichimura Y., Kumanomidou T., Sou Y.S., Mizushima T., Ezaki J., Ueno T. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 28.Noda N.N., Kumeta H., Nakatogawa H., Satoo K., Adachi W., Ishii J. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 29.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 30.Nijman S.M., Luna-Vargas M.P., Velds A., Brummelkamp T.R., Dirac A.M., Sixma T.K. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Kirkin V., Dikic I. Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr. Opin. Cell Biol. 2007;19:199–205. doi: 10.1016/j.ceb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Taherbhoy A.M., Schulman B.A., Kaiser S.E. Ubiquitin-like modifiers. Essays Biochem. 2012;52:51–63. doi: 10.1042/bse0520051. [DOI] [PubMed] [Google Scholar]
- 33.van der Veen A.G., Ploegh H.L. Ubiquitin-like proteins. Annu. Rev. Biochem. 2012;81:323–357. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- 34.Hanada T., Ohsumi Y. Structure–function relationship of Atg12, a ubiquitin-like modifier essential for autophagy. Autophagy. 2005;1:110–118. doi: 10.4161/auto.1.2.1858. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki N.N., Yoshimoto K., Fujioka Y., Ohsumi Y., Inagaki F. The crystal structure of plant ATG12 and its biological implication in autophagy. Autophagy. 2005;1:119–126. doi: 10.4161/auto.1.2.1859. [DOI] [PubMed] [Google Scholar]
- 36.Matsushita M., Suzuki N.N., Obara K., Fujioka Y., Ohsumi Y., Inagaki F. Structure of Atg5.Atg16, a complex essential for autophagy. J. Biol. Chem. 2007;282:6763–6772. doi: 10.1074/jbc.M609876200. [DOI] [PubMed] [Google Scholar]
- 37.Hanada T., Noda N.N., Satomi Y., Ichimura Y., Fujioka Y., Takao T. The Atg12–Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 38.Pang Y., Yamamoto H., Sakamoto H., Oku M., Mutungi J.K., Sahani M.H. Evolution from covalent conjugation to non-covalent interaction in the ubiquitin-like ATG12 system. Nat. Struct. Mol. Biol. 2019;26:289–296. doi: 10.1038/s41594-019-0204-3. [DOI] [PubMed] [Google Scholar]
- 39.Lang T., Schaeffeler E., Bernreuther D., Bredschneider M., Wolf D.H., Thumm M. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 1998;17:3597–3607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J., Huang W.P., Klionsky D.J. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshii S.R., Mizushima N. Monitoring and measuring autophagy. Int. J. Mol. Sci. 2017;18 doi: 10.3390/ijms18091865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shpilka T., Weidberg H., Pietrokovski S., Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 2011;12:226. doi: 10.1186/gb-2011-12-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie Z., Nair U., Klionsky D.J. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakatogawa H., Ichimura Y., Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 46.Weidberg H., Shpilka T., Shvets E., Abada A., Shimron F., Elazar Z. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev. Cell. 2011;20:444–454. doi: 10.1016/j.devcel.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 47.Lynch-Day M.A., Klionsky D.J. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584:1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klionsky D.J., Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 49.Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 50.Scott S.V., Guan J., Hutchins M.U., Kim J., Klionsky D.J. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol. Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim J., Kamada Y., Stromhaug P.E., Guan J., Hefner-Gravink A., Baba M. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J., Huang W.P., Stromhaug P.E., Klionsky D.J. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shintani T., Huang W.P., Stromhaug P.E., Klionsky D.J. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki K., Kondo C., Morimoto M., Ohsumi Y. Selective transport of alpha-mannosidase by autophagic pathways: identification of a novel receptor, Atg34p. J. Biol. Chem. 2010;285:30019–30025. doi: 10.1074/jbc.M110.143511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe Y., Noda N.N., Kumeta H., Suzuki K., Ohsumi Y., Inagaki F. Selective transport of alpha-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34. J. Biol. Chem. 2010;285:30026–30033. doi: 10.1074/jbc.M110.143545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elmore S.P., Qian T., Grissom S.F., Lemasters J.J. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 57.Xue L., Fletcher G.C., Tolkovsky A.M. Mitochondria are selectively eliminated from eukaryotic cells after blockade of caspases during apoptosis. Curr. Biol. 2001;11:361–365. doi: 10.1016/s0960-9822(01)00100-2. [DOI] [PubMed] [Google Scholar]
- 58.Joung I., Strominger J.L., Shin J. Molecular cloning of a phosphotyrosine-independent ligand of the p56lck SH2 domain. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5991–5995. doi: 10.1073/pnas.93.12.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vadlamudi R.K., Joung I., Strominger J.L., Shin J. p62, a phosphotyrosine-independent ligand of the SH2 domain of p56lck, belongs to a new class of ubiquitin-binding proteins. J. Biol. Chem. 1996;271:20235–20237. doi: 10.1074/jbc.271.34.20235. [DOI] [PubMed] [Google Scholar]
- 60.Ishii T., Yanagawa T., Kawane T., Yuki K., Seita J., Yoshida H. Murine peritoneal macrophages induce a novel 60-kDa protein with structural similarity to a tyrosine kinase p56lck-associated protein in response to oxidative stress. Biochem. Biophys. Res. Commun. 1996;226:456–460. doi: 10.1006/bbrc.1996.1377. [DOI] [PubMed] [Google Scholar]
- 61.Sanchez-Martin P., Saito T., Komatsu M. p62/SQSTM1: ‘Jack of all trades’ in health and cancer. FEBS J. 2019;286:8–23. doi: 10.1111/febs.14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moscat J., Karin M., Diaz-Meco M.T. p62 in Cancer: signaling adaptor beyond autophagy. Cell. 2016;167:606–609. doi: 10.1016/j.cell.2016.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zatloukal K., Stumptner C., Fuchsbichler A., Heid H., Schnoelzer M., Kenner L. p62 is a common component of cytoplasmic inclusions in protein aggregation diseases. Am. J. Pathol. 2002;160:255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuusisto E., Salminen A., Alafuzoff I. Ubiquitin-binding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. Neuroreport. 2001;12:2085–2090. doi: 10.1097/00001756-200107200-00009. [DOI] [PubMed] [Google Scholar]
- 65.Lamark T., Perander M., Outzen H., Kristiansen K., Overvatn A., Michaelsen E. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 2003;278:34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- 66.Wilson M.I., Gill D.J., Perisic O., Quinn M.T., Williams R.L. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol. Cell. 2003;12:39–50. doi: 10.1016/s1097-2765(03)00246-6. [DOI] [PubMed] [Google Scholar]
- 67.Bjorkoy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Komatsu M., Waguri S., Koike M., Sou Y.S., Ueno T., Hara T. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 69.Rogov V.V., Suzuki H., Fiskin E., Wild P., Kniss A., Rozenknop A. Structural basis for phosphorylation-triggered autophagic clearance of Salmonella. Biochem. J. 2013;454:459–466. doi: 10.1042/BJ20121907. [DOI] [PubMed] [Google Scholar]
- 70.Wild P., Farhan H., McEwan D.G., Wagner S., Rogov V.V., Brady N.R. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu Y., Massen S., Terenzio M., Lang V., Chen-Lindner S., Eils R. Modulation of serines 17 and 24 in the LC3-interacting region of Bnip3 determines pro-survival mitophagy versus apoptosis. J. Biol. Chem. 2013;288:1099–1113. doi: 10.1074/jbc.M112.399345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kirkin V., Lamark T., Sou Y.S., Bjorkoy G., Nunn J.L., Bruun J.A. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 73.Behrends C., Sowa M.E., Gygi S.P., Harper J.W. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kirkin V., McEwan D.G., Novak I., Dikic I. A role for ubiquitin in selective autophagy. Mol. Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 75.Perrin A.J., Jiang X., Birmingham C.L., So N.S., Brumell J.H. Recognition of bacteria in the cytosol of mammalian cells by the ubiquitin system. Curr. Biol. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 76.Platta H.W., Erdmann R. Peroxisomal dynamics. Trends Cell Biol. 2007;17:474–484. doi: 10.1016/j.tcb.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 77.Kraft C., Deplazes A., Sohrmann M., Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 78.Thurston T.L., Ryzhakov G., Bloor S., von Muhlinen N., Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 79.von Muhlinen N., Akutsu M., Ravenhill B.J., Foeglein A., Bloor S., Rutherford T.J. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol. Cell. 2012;48:329–342. doi: 10.1016/j.molcel.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tumbarello D.A., Manna P.T., Allen M., Bycroft M., Arden S.D., Kendrick-Jones J. The autophagy receptor TAX1BP1 and the molecular motor myosin VI are required for clearance of Salmonella typhimurium by autophagy. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Herhaus L., Dikic I. Regulation of Salmonella–host cell interactions via the ubiquitin system. Int J Med Microbiol. 2018;308(1):176–184. doi: 10.1016/j.ijmm.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 82.Heath R.J., Goel G., Baxt L.A., Rush J.S., Mohanan V., Paulus G.L.C. RNF166 determines recruitment of adaptor proteins during antibacterial autophagy. Cell Rep. 2016;17:2183–2194. doi: 10.1016/j.celrep.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huett A., Heath R.J., Begun J., Sassi S.O., Baxt L.A., Vyas J.M. The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella typhimurium. Cell Host Microbe. 2012;12:778–790. doi: 10.1016/j.chom.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Noad J., von der Malsburg A., Pathe C., Michel M.A., Komander D., Randow F. LUBAC-synthesized linear ubiquitin chains restrict cytosol-invading bacteria by activating autophagy and NF-kappaB. Nat. Microbiol. 2017;2 doi: 10.1038/nmicrobiol.2017.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Polajnar M., Dietz M.S., Heilemann M., Behrends C. Expanding the host cell ubiquitylation machinery targeting cytosolic Salmonella. EMBO Rep. 2017;18:1572–1585. doi: 10.15252/embr.201643851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Franco L.H., Nair V.R., Scharn C.R., Xavier R.J., Torrealba J.R., Shiloh M.U. The ubiquitin ligase Smurf1 functions in selective autophagy of Mycobacterium tuberculosis and anti-tuberculous host defense. Cell Host Microbe. 2017;22:421–423. doi: 10.1016/j.chom.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fujita N., Morita E., Itoh T., Tanaka A., Nakaoka M., Osada Y. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J. Cell Biol. 2013;203:115–128. doi: 10.1083/jcb.201304188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 89.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kane L.A., Lazarou M., Fogel A.I., Li Y., Yamano K., Sarraf S.A. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 94.Kazlauskaite A., Kondapalli C., Gourlay R., Campbell D.G., Ritorto M.S., Hofmann K. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shiba-Fukushima K., Imai Y., Yoshida S., Ishihama Y., Kanao T., Sato S. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci. Rep. 2012;2:1002. doi: 10.1038/srep01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shiba-Fukushima K., Arano T., Matsumoto G., Inoshita T., Yoshida S., Ishihama Y. Phosphorylation of mitochondrial polyubiquitin by PINK1 promotes Parkin mitochondrial tethering. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 98.Matsuda N. Phospho-ubiquitin: upending the PINK–Parkin–ubiquitin cascade. J. Biochem. 2016;159:379–385. doi: 10.1093/jb/mvv125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ordureau A., Paulo J.A., Zhang W., Ahfeldt T., Zhang J., Cohn E.F. Dynamics of PARKIN-dependent mitochondrial ubiquitylation in induced neurons and model systems revealed by digital snapshot proteomics. Mol. Cell. 2018;70:211–27 e8. doi: 10.1016/j.molcel.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sarraf S.A., Raman M., Guarani-Pereira V., Sowa M.E., Huttlin E.L., Gygi S.P. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lazarou M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Princely Abudu Y., Pankiv S., Mathai B.J., Hakon Lystad A., Bindesboll C., Brenne H.B. NIPSNAP1 and NIPSNAP2 act as “eat me” signals for mitophagy. Dev. Cell. 2019 doi: 10.1016/j.devcel.2019.03.013. pii: S1534-5807(19)30224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tanaka A., Cleland M.M., Xu S., Narendra D.P., Suen D.F., Karbowski M. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yoshii S.R., Kishi C., Ishihara N., Mizushima N. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J. Biol. Chem. 2011;286:19630–19640. doi: 10.1074/jbc.M110.209338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Iwata J., Ezaki J., Komatsu M., Yokota S., Ueno T., Tanida I. Excess peroxisomes are degraded by autophagic machinery in mammals. J. Biol. Chem. 2006;281:4035–4041. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- 106.Kim P.K., Hailey D.W., Mullen R.T., Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Deosaran E., Larsen K.B., Hua R., Sargent G., Wang Y., Kim S. NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci. 2013;126:939–952. doi: 10.1242/jcs.114819. [DOI] [PubMed] [Google Scholar]
- 108.Yamashita S., Abe K., Tatemichi Y., Fujiki Y. The membrane peroxin PEX3 induces peroxisome–ubiquitination-linked pexophagy. Autophagy. 2014;10:1549–1564. doi: 10.4161/auto.29329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang J., Tripathi D.N., Jing J., Alexander A., Kim J., Powell R.T. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol. 2015;17:1259–1269. doi: 10.1038/ncb3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sargent G., van Zutphen T., Shatseva T., Zhang L., Di Giovanni V., Bandsma R. PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation. J. Cell Biol. 2016;214:677–690. doi: 10.1083/jcb.201511034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lu K., Psakhye I., Jentsch S. Autophagic clearance of polyQ proteins mediated by ubiquitin–Atg8 adaptors of the conserved CUET protein family. Cell. 2014;158:549–563. doi: 10.1016/j.cell.2014.05.048. [DOI] [PubMed] [Google Scholar]
- 112.Marshall R.S., McLoughlin F., Vierstra R.D. Autophagic turnover of inactive 26S proteasomes in yeast is directed by the ubiquitin receptor Cue5 and the Hsp42 chaperone. Cell Rep. 2016;16:1717–1732. doi: 10.1016/j.celrep.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 113.Farre J.C., Manjithaya R., Mathewson R.D., Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Motley A.M., Nuttall J.M., Hettema E.H. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 2012;31:2852–2868. doi: 10.1038/emboj.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Farre J.C., Burkenroad A., Burnett S.F., Subramani S. Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep. 2013;14:441–449. doi: 10.1038/embor.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kissova I., Deffieu M., Manon S., Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. J. Biol. Chem. 2004;279:39068–39074. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- 117.Kanki T., Klionsky D.J. Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 2008;283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kanki T., Wang K., Cao Y., Baba M., Klionsky D.J. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Okamoto K., Kondo-Okamoto N., Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 120.Kanki T., Kurihara Y., Jin X., Goda T., Ono Y., Aihara M. Casein kinase 2 is essential for mitophagy. EMBO Rep. 2013;14:788–794. doi: 10.1038/embor.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tanaka C., Tan L.J., Mochida K., Kirisako H., Koizumi M., Asai E. Hrr25 triggers selective autophagy-related pathways by phosphorylating receptor proteins. J. Cell Biol. 2014;207:91–105. doi: 10.1083/jcb.201402128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pfaffenwimmer T., Reiter W., Brach T., Nogellova V., Papinski D., Schuschnig M. Hrr25 kinase promotes selective autophagy by phosphorylating the cargo receptor Atg19. EMBO Rep. 2014;15:862–870. doi: 10.15252/embr.201438932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Furukawa K., Fukuda T., Yamashita S.I., Saigusa T., Kurihara Y., Yoshida Y. The PP2A-like protein phosphatase Ppg1 and the far complex cooperatively counteract CK2-mediated phosphorylation of Atg32 to inhibit mitophagy. Cell Rep. 2018;23:3579–3590. doi: 10.1016/j.celrep.2018.05.064. [DOI] [PubMed] [Google Scholar]
- 124.Fei P., Wang W., Kim S.H., Wang S., Burns T.F., Sax J.K. Bnip3L is induced by p53 under hypoxia, and its knockdown promotes tumor growth. Cancer Cell. 2004;6:597–609. doi: 10.1016/j.ccr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 125.Schweers R.L., Zhang J., Randall M.S., Loyd M.R., Li W., Dorsey F.C. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Novak I., Kirkin V., McEwan D.G., Zhang J., Wild P., Rozenknop A. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sandoval H., Thiagarajan P., Dasgupta S.K., Schumacher A., Prchal J.T., Chen M. Essential role for nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hanna R.A., Quinsay M.N., Orogo A.M., Giang K., Rikka S., Gustafsson A.B. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J. Biol. Chem. 2012;287:19094–19104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rogov V.V., Suzuki H., Marinkovic M., Lang V., Kato R., Kawasaki M. Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins. Sci. Rep. 2017;7:1131. doi: 10.1038/s41598-017-01258-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu L., Feng D., Chen G., Chen M., Zheng Q., Song P. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 131.Otsu K., Murakawa T., Yamaguchi O. BCL2L13 is a mammalian homolog of the yeast mitophagy receptor Atg32. Autophagy. 2015;11:1932–1933. doi: 10.1080/15548627.2015.1084459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Strappazzon F., Nazio F., Corrado M., Cianfanelli V., Romagnoli A., Fimia G.M. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2015;22:419–432. doi: 10.1038/cdd.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bhujabal Z., Birgisdottir A.B., Sjottem E., Brenne H.B., Overvatn A., Habisov S. FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Rep. 2017;18:947–961. doi: 10.15252/embr.201643147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wei Y., Chiang W.C., Sumpter R., Jr., Mishra P., Levine B. Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell. 2017;168:224–38 e10. doi: 10.1016/j.cell.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chu C.T., Ji J., Dagda R.K., Jiang J.F., Tyurina Y.Y., Kapralov A.A. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 2013;15:1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yorimitsu T., Nair U., Yang Z., Klionsky D.J. Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bernales S., McDonald K.L., Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mochida K., Oikawa Y., Kimura Y., Kirisako H., Hirano H., Ohsumi Y. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522:359–362. doi: 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- 139.Khaminets A., Heinrich T., Mari M., Grumati P., Huebner A.K., Akutsu M. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 140.Fumagalli F., Noack J., Bergmann T.J., Cebollero E., Pisoni G.B., Fasana E. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat. Cell Biol. 2016;18:1173–1184. doi: 10.1038/ncb3423. [DOI] [PubMed] [Google Scholar]
- 141.Grumati P., Morozzi G., Holper S., Mari M., Harwardt M.I., Yan R. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife. 2017;6 doi: 10.7554/eLife.25555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Smith M.D., Harley M.E., Kemp A.J., Wills J., Lee M., Arends M. CCPG1 is a non-canonical autophagy cargo receptor essential for ER-phagy and pancreatic ER proteostasis. Dev. Cell. 2018;44:217–32 e11. doi: 10.1016/j.devcel.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen Q., Xiao Y., Chai P., Zheng P., Teng J., Chen J. ATL3 is a tubular ER-phagy receptor for GABARAP-mediated selective autophagy. Curr. Biol. 2019;29(5):846–855. doi: 10.1016/j.cub.2019.01.041. [DOI] [PubMed] [Google Scholar]
- 144.Chino H., Hatta T., Natsume T., Mizushima N. Intrinsically disordered protein TEX264 mediates ER-phagy. Mol. Cell. 2019 doi: 10.1016/j.molcel.2019.03.033. pii: S1097-2765(19)30257-6. [DOI] [PubMed] [Google Scholar]
- 145.An H., Ordureau A., Paulo J.A., Shoemaker C.J., Denic V., Harper J.W. TEX264 is an endoplasmic reticulum-resident ATG8-interacting protein critical for ER remodeling during nutrient stress. Mol. Cell. 2019 doi: 10.1016/j.molcel.2019.03.034. S1097-2765(19)30258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jiang S., Wells C.D., Roach P.J. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem. Biophys. Res. Commun. 2011;413:420–425. doi: 10.1016/j.bbrc.2011.08.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mancias J.D., Wang X., Gygi S.P., Harper J.W., Kimmelman A.C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dowdle W.E., Nyfeler B., Nagel J., Elling R.A., Liu S., Triantafellow E. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat. Cell Biol. 2014;16:1069–1079. doi: 10.1038/ncb3053. [DOI] [PubMed] [Google Scholar]
- 149.Asano T., Komatsu M., Yamaguchi-Iwai Y., Ishikawa F., Mizushima N., Iwai K. Distinct mechanisms of ferritin delivery to lysosomes in iron-depleted and iron-replete cells. Mol. Cell. Biol. 2011;31:2040–2052. doi: 10.1128/MCB.01437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Martinez-Lopez N., Garcia-Macia M., Sahu S., Athonvarangkul D., Liebling E., Merlo P. Autophagy in the CNS and periphery coordinate lipophagy and lipolysis in the brown adipose tissue and liver. Cell Metab. 2016;23:113–127. doi: 10.1016/j.cmet.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wyant G.A., Abu-Remaileh M., Frenkel E.M., Laqtom N.N., Dharamdasani V., Lewis C.A. NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science. 2018;360:751–758. doi: 10.1126/science.aar2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gao C., Cao W., Bao L., Zuo W., Xie G., Cai T. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat. Cell Biol. 2010;12:781–790. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
- 153.Petherick K.J., Williams A.C., Lane J.D., Ordonez-Moran P., Huelsken J., Collard T.J. Autolysosomal beta-catenin degradation regulates Wnt–autophagy–p62 crosstalk. EMBO J. 2013;32:1903–1916. doi: 10.1038/emboj.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Shpilka T., Welter E., Borovsky N., Amar N., Shimron F., Peleg Y. Fatty acid synthase is preferentially degraded by autophagy upon nitrogen starvation in yeast. Proc. Natl. Acad. Sci. U. S. A. 2015;112:1434–1439. doi: 10.1073/pnas.1409476112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dou Z., Xu C., Donahue G., Shimi T., Pan J.A., Zhu J. Autophagy mediates degradation of nuclear lamina. Nature. 2015;527:105–109. doi: 10.1038/nature15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sharifi M.N., Mowers E.E., Drake L.E., Collier C., Chen H., Zamora M. Autophagy promotes focal adhesion disassembly and cell motility of metastatic tumor cells through the direct interaction of Paxillin with LC3. Cell Rep. 2016;15:1660–1672. doi: 10.1016/j.celrep.2016.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liu D., Wu H., Wang C., Li Y., Tian H., Siraj S. STING directly activates autophagy to tune the innate immune response. Cell Death Differ. 2018 doi: 10.1038/s41418-018-0251-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Toledo M., Batista-Gonzalez A., Merheb E., Aoun M.L., Tarabra E., Feng D. Autophagy regulates the liver clock and glucose metabolism by degrading CRY1. Cell Metab. 2018;28:268–81 e4. doi: 10.1016/j.cmet.2018.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Saito T., Kuma A., Sugiura Y., Ichimura Y., Obata M., Kitamura H. Autophagy regulates lipid metabolism through selective turnover of NCoR1. Nat. Commun. 2019;10:1567. doi: 10.1038/s41467-019-08829-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shibutani S.T., Yoshimori T. A current perspective of autophagosome biogenesis. Cell Res. 2014;24:58–68. doi: 10.1038/cr.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Suzuki K., Kubota Y., Sekito T., Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 162.Kamber R.A., Shoemaker C.J., Denic V. Receptor-bound targets of selective autophagy use a scaffold protein to activate the Atg1 kinase. Mol. Cell. 2015;59:372–381. doi: 10.1016/j.molcel.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.He C., Song H., Yorimitsu T., Monastyrska I., Yen W.L., Legakis J.E. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J. Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sekito T., Kawamata T., Ichikawa R., Suzuki K., Ohsumi Y. Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells. 2009;14:525–538. doi: 10.1111/j.1365-2443.2009.01299.x. [DOI] [PubMed] [Google Scholar]
- 165.Yamamoto H., Kakuta S., Watanabe T.M., Kitamura A., Sekito T., Kondo-Kakuta C. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J. Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Suzuki S.W., Yamamoto H., Oikawa Y., Kondo-Kakuta C., Kimura Y., Hirano H. Atg13 HORMA domain recruits Atg9 vesicles during autophagosome formation. Proc. Natl. Acad. Sci. U. S. A. 2015;112:3350–3355. doi: 10.1073/pnas.1421092112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Slobodkin M.R., Elazar Z. The Atg8 family: multifunctional ubiquitin-like key regulators of autophagy. Essays Biochem. 2013;55:51–64. doi: 10.1042/bse0550051. [DOI] [PubMed] [Google Scholar]
- 168.Bento C.F., Renna M., Ghislat G., Puri C., Ashkenazi A., Vicinanza M. Mammalian autophagy: how does it work? Annu. Rev. Biochem. 2016;85:685–713. doi: 10.1146/annurev-biochem-060815-014556. [DOI] [PubMed] [Google Scholar]
- 169.Yorimitsu T., Klionsky D.J. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol. Biol. Cell. 2005;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cheong H., Yorimitsu T., Reggiori F., Legakis J.E., Wang C.W., Klionsky D.J. Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cheong H., Nair U., Geng J., Klionsky D.J. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 2008;19:668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Torggler R., Papinski D., Brach T., Bas L., Schuschnig M., Pfaffenwimmer T. Two independent pathways within selective autophagy converge to activate Atg1 kinase at the vacuole. Mol. Cell. 2016;64:221–235. doi: 10.1016/j.molcel.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 173.Li F., Chung T., Vierstra R.D. AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell. 2014;26:788–807. doi: 10.1105/tpc.113.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Nakatogawa H., Ohbayashi S., Sakoh-Nakatogawa M., Kakuta S., Suzuki S.W., Kirisako H. The autophagy-related protein kinase Atg1 interacts with the ubiquitin-like protein Atg8 via the Atg8 family interacting motif to facilitate autophagosome formation. J. Biol. Chem. 2012;287:28503–28507. doi: 10.1074/jbc.C112.387514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kraft C., Kijanska M., Kalie E., Siergiejuk E., Lee S.S., Semplicio G. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 2012;31:3691–3703. doi: 10.1038/emboj.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Alemu E.A., Lamark T., Torgersen K.M., Birgisdottir A.B., Larsen K.B., Jain A. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J. Biol. Chem. 2012;287:39275–39290. doi: 10.1074/jbc.M112.378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Suzuki H., Tabata K., Morita E., Kawasaki M., Kato R., Dobson R.C. Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism. Structure. 2014;22:47–58. doi: 10.1016/j.str.2013.09.023. [DOI] [PubMed] [Google Scholar]
- 178.Sawa-Makarska J., Abert C., Romanov J., Zens B., Ibiricu I., Martens S. Cargo binding to Atg19 unmasks additional Atg8 binding sites to mediate membrane-cargo apposition during selective autophagy. Nat. Cell Biol. 2014;16:425–433. doi: 10.1038/ncb2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Abert C., Kontaxis G., Martens S. Accessory interaction motifs in the Atg19 cargo receptor enable strong binding to the clustered ubiquitin-related Atg8 protein. J. Biol. Chem. 2016;291:18799–18808. doi: 10.1074/jbc.M116.736892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Birgisdottir A.B., Lamark T., Johansen T. The LIR motif—crucial for selective autophagy. J. Cell Sci. 2013;126:3237–3247. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- 181.Birgisdottir A.B., Mouilleron S., Bhujabal Z., Wirth M., Sjottem E., Evjen G. Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs. Autophagy. 2019:1–23. doi: 10.1080/15548627.2019.1581009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Joachim J., Jefferies H.B., Razi M., Frith D., Snijders A.P., Chakravarty P. Activation of ULK kinase and autophagy by GABARAP trafficking from the centrosome is regulated by WAC and GM130. Mol. Cell. 2015;60:899–913. doi: 10.1016/j.molcel.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Park J.M., Jung C.H., Seo M., Otto N.M., Grunwald D., Kim K.H. The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14. Autophagy. 2016;12:547–564. doi: 10.1080/15548627.2016.1140293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nishimura T., Kaizuka T., Cadwell K., Sahani M.H., Saitoh T., Akira S. FIP200 regulates targeting of Atg16L1 to the isolation membrane. EMBO Rep. 2013;14:284–291. doi: 10.1038/embor.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gammoh N., Florey O., Overholtzer M., Jiang X. Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and -independent autophagy. Nat. Struct. Mol. Biol. 2013;20:144–149. doi: 10.1038/nsmb.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J.L. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Dooley H.C., Razi M., Polson H.E., Girardin S.E., Wilson M.I., Tooze S.A. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol. Cell. 2014;55:238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Liu Z., Chen P., Gao H., Gu Y., Yang J., Peng H. Ubiquitylation of autophagy receptor optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell. 2014;26:106–120. doi: 10.1016/j.ccr.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Conway O., Kirkin V. Love laughs at locksmiths: ubiquitylation of p62 unlocks its autophagy receptor potential. Cell Res. 2017;27:595–597. doi: 10.1038/cr.2017.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Turco E., Witt M., Abert C., Bock-Bierbaum T., Su M.Y., Trapannone R. FIP200 claw domain binding to p62 promotes autophagosome formation at ubiquitin condensates. Mol. Cell. 2019;74(2):330–346. doi: 10.1016/j.molcel.2019.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Vargas J.N.S., Wang C., Bunker E., Hao L., Maric D., Schiavo G. Spatiotemporal control of ULK1 activation by NDP52 and TBK1 during selective autophagy. Mol. Cell. 2019;74(2):347–362. doi: 10.1016/j.molcel.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ravenhill B.J., Boyle K.B., von Muhlinen N., Ellison C.J., Masson G.R., Otten E.G. The cargo receptor NDP52 initiates selective autophagy by recruiting the ULK complex to cytosol-invading bacteria. Mol. Cell. 2019;74:320–9 e6. doi: 10.1016/j.molcel.2019.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mandell M.A., Jain A., Arko-Mensah J., Chauhan S., Kimura T., Dinkins C. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev. Cell. 2014;30:394–409. doi: 10.1016/j.devcel.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Stremlau M., Perron M., Lee M., Li Y., Song B., Javanbakht H. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Jena K.K., Kolapalli S.P., Mehto S., Nath P., Das B., Sahoo P.K. TRIM16 controls assembly and degradation of protein aggregates by modulating the p62-NRF2 axis and autophagy. EMBO J. 2018;37 doi: 10.15252/embj.201798358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Padman B.S., Nguyen T.N., Uoselis L., Skulsuppaisarn M., Nguyen L.K., Lazarou M. LC3/GABARAPs drive ubiquitin-independent recruitment of optineurin and NDP52 to amplify mitophagy. Nat. Commun. 2019;10:408. doi: 10.1038/s41467-019-08335-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nguyen T.N., Padman B.S., Usher J., Oorschot V., Ramm G., Lazarou M. Atg8 family LC3/GABARAP proteins are crucial for autophagosome–lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J. Cell Biol. 2016;215:857–874. doi: 10.1083/jcb.201607039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Goodwin J.M., Dowdle W.E., DeJesus R., Wang Z., Bergman P., Kobylarz M. Autophagy-independent lysosomal targeting regulated by ULK1/2–FIP200 and ATG9. Cell Rep. 2017;20:2341–2356. doi: 10.1016/j.celrep.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Shoemaker C.J., Huang Q.T., Weir N.R., Polyakov N., Denic V. A CRISPR screening approach for identifying novel autophagy-related factors and cytoplasm-to-lysosome trafficking routes. bioRxiv. 2017 [Google Scholar]
- 200.Mejlvang J., Olsvik H., Svenning S., Bruun J.A., Abudu Y.P., Larsen K.B. Starvation induces rapid degradation of selective autophagy receptors by endosomal microautophagy. J. Cell Biol. 2018;217(10):3640–3655. doi: 10.1083/jcb.201711002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Funderburk S.F., Wang Q.J., Yue Z. The Beclin 1–VPS34 complex—at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tsuboyama K., Koyama-Honda I., Sakamaki Y., Koike M., Morishita H., Mizushima N. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 2016;354:1036–1041. doi: 10.1126/science.aaf6136. [DOI] [PubMed] [Google Scholar]
- 203.Romanov J., Walczak M., Ibiricu I., Schuchner S., Ogris E., Kraft C. Mechanism and functions of membrane binding by the Atg5–Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31:4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Tan J.M.J., Mellouk N., Osborne S.E., Ammendolia D.A., Dyer D.N., Li R. An ATG16L1-dependent pathway promotes plasma membrane repair and limits Listeria monocytogenes cell-to-cell spread. Nat. Microbiol. 2018;3:1472–1485. doi: 10.1038/s41564-018-0293-5. [DOI] [PubMed] [Google Scholar]
- 205.Filimonenko M., Isakson P., Finley K.D., Anderson M., Jeong H., Melia T.J. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol. Cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lystad A.H., Ichimura Y., Takagi K., Yang Y., Pankiv S., Kanegae Y. Structural determinants in GABARAP required for the selective binding and recruitment of ALFY to LC3B-positive structures. EMBO Rep. 2014;15:557–565. doi: 10.1002/embr.201338003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Fracchiolla D., Sawa-Makarska J., Zens B., Ruiter A., Zaffagnini G., Brezovich A. Mechanism of cargo-directed Atg8 conjugation during selective autophagy. Elife. 2016;5 doi: 10.7554/eLife.18544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Papinski D., Kraft C. Regulation of autophagy by signaling through the Atg1/ULK1 complex. J. Mol. Biol. 2016;428:1725–1741. doi: 10.1016/j.jmb.2016.03.030. [DOI] [PubMed] [Google Scholar]
- 209.Ochaba J., Lukacsovich T., Csikos G., Zheng S., Margulis J., Salazar L. Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc. Natl. Acad. Sci. U. S. A. 2014;111:16889–16894. doi: 10.1073/pnas.1420103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Rui Y.N., Xu Z., Patel B., Chen Z., Chen D., Tito A. Huntingtin functions as a scaffold for selective macroautophagy. Nat. Cell Biol. 2015;17:262–275. doi: 10.1038/ncb3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Jacomin A.C., Samavedam S., Promponas V., Nezis I.P. iLIR database: a web resource for LIR motif-containing proteins in eukaryotes. Autophagy. 2016;12:1945–1953. doi: 10.1080/15548627.2016.1207016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kalvari I., Tsompanis S., Mulakkal N.C., Osgood R., Johansen T., Nezis I.P. iLIR: a web resource for prediction of Atg8-family interacting proteins. Autophagy. 2014;10:913–925. doi: 10.4161/auto.28260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Nowak J., Archange C., Tardivel-Lacombe J., Pontarotti P., Pebusque M.J., Vaccaro M.I. The TP53INP2 protein is required for autophagy in mammalian cells. Mol. Biol. Cell. 2009;20:870–881. doi: 10.1091/mbc.E08-07-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Pankiv S., Alemu E.A., Brech A., Bruun J.A., Lamark T., Overvatn A. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 2010;188:253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Popovic D., Akutsu M., Novak I., Harper J.W., Behrends C., Dikic I. Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol. Cell. Biol. 2012;32:1733–1744. doi: 10.1128/MCB.06717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Itoh T., Kanno E., Uemura T., Waguri S., Fukuda M. OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J. Cell Biol. 2011;192:839–853. doi: 10.1083/jcb.201008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Stadel D., Millarte V., Tillmann K.D., Huber J., Tamin-Yecheskel B.C., Akutsu M. TECPR2 cooperates with LC3C to regulate COPII-dependent ER export. Mol. Cell. 2015;60:89–104. doi: 10.1016/j.molcel.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 218.McEwan D.G., Popovic D., Gubas A., Terawaki S., Suzuki H., Stadel D. PLEKHM1 regulates autophagosome–lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell. 2015;57:39–54. doi: 10.1016/j.molcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 219.Tseng W.C., Jenkins P.M., Tanaka M., Mooney R., Bennett V. Giant ankyrin-G stabilizes somatodendritic GABAergic synapses through opposing endocytosis of GABAA receptors. Proc. Natl. Acad. Sci. U. S. A. 2015;112:1214–1219. doi: 10.1073/pnas.1417989112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Joachim J., Razi M., Judith D., Wirth M., Calamita E., Encheva V. Centriolar satellites control GABARAP ubiquitination and GABARAP-mediated autophagy. Curr. Biol. 2017;27:2123–36 e7. doi: 10.1016/j.cub.2017.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ebner P., Poetsch I., Deszcz L., Hoffmann T., Zuber J., Ikeda F. The IAP family member BRUCE regulates autophagosome–lysosome fusion. Nat. Commun. 2018;9:599. doi: 10.1038/s41467-018-02823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zaffagnini G., Savova A., Danieli A., Romanov J., Tremel S., Ebner M. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 2018;37 doi: 10.15252/embj.201798308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sun D., Wu R., Zheng J., Li P., Yu L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 2018;28:405–415. doi: 10.1038/s41422-018-0017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Yamasaki A., Watanabe Y., Adachi W., Suzuki K., Matoba K., Kirisako H. Structural basis for receptor-mediated selective autophagy of aminopeptidase I aggregates. Cell Rep. 2016;16:19–27. doi: 10.1016/j.celrep.2016.05.066. [DOI] [PubMed] [Google Scholar]
- 226.Zhang G., Wang Z., Du Z., Zhang H. mTOR regulates phase separation of PGL granules to modulate their autophagic degradation. Cell. 2018;174:1492–506 e22. doi: 10.1016/j.cell.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 227.Matsumoto G., Wada K., Okuno M., Kurosawa M., Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 228.Pilli M., Arko-Mensah J., Ponpuak M., Roberts E., Master S., Mandell M.A. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223–234. doi: 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Peng H., Yang J., Li G., You Q., Han W., Li T. Ubiquitylation of p62/sequestosome1 activates its autophagy receptor function and controls selective autophagy upon ubiquitin stress. Cell Res. 2017;27:657–674. doi: 10.1038/cr.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Wurzer B., Zaffagnini G., Fracchiolla D., Turco E., Abert C., Romanov J. Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. Elife. 2015;4 doi: 10.7554/eLife.08941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Cha-Molstad H., Sung K.S., Hwang J., Kim K.A., Yu J.E., Yoo Y.D. Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat. Cell Biol. 2015;17:917–929. doi: 10.1038/ncb3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Cherra S.J., 3rd, Kulich S.M., Uechi G., Balasubramani M., Mountzouris J., Day B.W. Regulation of the autophagy protein LC3 by phosphorylation. J. Cell Biol. 2010;190:533–539. doi: 10.1083/jcb.201002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Wilkinson D.S., Jariwala J.S., Anderson E., Mitra K., Meisenhelder J., Chang J.T. Phosphorylation of LC3 by the Hippo kinases STK3/STK4 is essential for autophagy. Mol. Cell. 2015;57:55–68. doi: 10.1016/j.molcel.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Huang R., Xu Y., Wan W., Shou X., Qian J., You Z. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell. 2015;57:456–466. doi: 10.1016/j.molcel.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 235.Svenning S., Lamark T., Krause K., Johansen T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy. 2011;7:993–1010. doi: 10.4161/auto.7.9.16389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Nezis I.P., Simonsen A., Sagona A.P., Finley K., Gaumer S., Contamine D. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J. Cell Biol. 2008;180:1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Lin L., Yang P., Huang X., Zhang H., Lu Q., Zhang H. The scaffold protein EPG-7 links cargo–receptor complexes with the autophagic assembly machinery. J. Cell Biol. 2013;201:113–129. doi: 10.1083/jcb.201209098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zhang Y., Yan L., Zhou Z., Yang P., Tian E., Zhang K. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 2009;136:308–321. doi: 10.1016/j.cell.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 239.Soto C., Pritzkow S. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci. 2018;21:1332–1340. doi: 10.1038/s41593-018-0235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Rubinsztein D.C., Bento C.F., Deretic V. Therapeutic targeting of autophagy in neurodegenerative and infectious diseases. J. Exp. Med. 2015;212:979–990. doi: 10.1084/jem.20150956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Li J., Zhu R., Chen K., Zheng H., Zhao H., Yuan C. Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins. Nat. Chem. Biol. 2018;14:778–787. doi: 10.1038/s41589-018-0082-8. [DOI] [PubMed] [Google Scholar]
- 242.Cha-Molstad H., Yu J.E., Feng Z., Lee S.H., Kim J.G., Yang P. p62/SQSTM1/Sequestosome-1 is an N-recognin of the N-end rule pathway which modulates autophagosome biogenesis. Nat. Commun. 2017;8:102. doi: 10.1038/s41467-017-00085-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhang Y., Mun S.R., Linares J.F., Ahn J., Towers C.G., Ji C.H. ZZ-dependent regulation of p62/SQSTM1 in autophagy. Nat. Commun. 2018;9:4373. doi: 10.1038/s41467-018-06878-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Stolz A., Putyrski M., Kutle I., Huber J., Wang C., Major V. Fluorescence-based ATG8 sensors monitor localization and function of LC3/GABARAP proteins. EMBO J. 2017;36:549–564. doi: 10.15252/embj.201695063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Lee Y.K., Jun Y.W., Choi H.E., Huh Y.H., Kaang B.K., Jang D.J. Development of LC3/GABARAP sensors containing a LIR and a hydrophobic domain to monitor autophagy. EMBO J. 2017;36:1100–1116. doi: 10.15252/embj.201696315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Kaufmann A., Beier V., Franquelim H.G., Wollert T. Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell. 2014;156:469–481. doi: 10.1016/j.cell.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 247.Habisov S., Huber J., Ichimura Y., Akutsu M., Rogova N., Loehr F. Structural and functional analysis of a novel interaction motif within UFM1-activating enzyme 5 (UBA5) required for binding to ubiquitin-like proteins and ufmylation. J. Biol. Chem. 2016;291:9025–9041. doi: 10.1074/jbc.M116.715474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Keown J.R., Black M.M., Ferron A., Yap M., Barnett M.J., Pearce F.G. A helical LC3-interacting region mediates the interaction between the retroviral restriction factor Trim5alpha and mammalian autophagy-related ATG8 proteins. J. Biol. Chem. 2018;293:18378–18386. doi: 10.1074/jbc.RA118.004202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Marshall R.S., Hua Z., Mali S., McLoughlin F., Vierstra R.D. ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell. 2019;177:766–81 e24. doi: 10.1016/j.cell.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Heckmann B.L., Boada-Romero E., Cunha L.D., Magne J., Green D.R. LC3-associated phagocytosis and inflammation. J. Mol. Biol. 2017;429:3561–3576. doi: 10.1016/j.jmb.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Florey O., Gammoh N., Kim S.E., Jiang X., Overholtzer M. V-ATPase and osmotic imbalances activate endolysosomal LC3 lipidation. Autophagy. 2015;11:88–99. doi: 10.4161/15548627.2014.984277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Subramani S., Malhotra V. Non-autophagic roles of autophagy-related proteins. EMBO Rep. 2013;14:143–151. doi: 10.1038/embor.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Overbye A., Fengsrud M., Seglen P.O. Proteomic analysis of membrane-associated proteins from rat liver autophagosomes. Autophagy. 2007;3:300–322. doi: 10.4161/auto.3910. [DOI] [PubMed] [Google Scholar]
- 254.Seay M., Hayward A.P., Tsao J., Dinesh-Kumar S.P. Something old, something new: plant innate immunity and autophagy. Curr. Top. Microbiol. Immunol. 2009;335:287–306. doi: 10.1007/978-3-642-00302-8_14. [DOI] [PubMed] [Google Scholar]
- 255.Buchan J.R., Kolaitis R.M., Taylor J.P., Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Hung Y.H., Chen L.M., Yang J.Y., Yang W.Y. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat. Commun. 2013;4:2111. doi: 10.1038/ncomms3111. [DOI] [PubMed] [Google Scholar]
- 258.Maejima I., Takahashi A., Omori H., Kimura T., Takabatake Y., Saitoh T. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 2013;32:2336–2347. doi: 10.1038/emboj.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Scott S.V., Klionsky D.J. Delivery of proteins and organelles to the vacuole from the cytoplasm. Curr. Opin. Cell Biol. 1998;10:523–529. doi: 10.1016/s0955-0674(98)80068-9. [DOI] [PubMed] [Google Scholar]
- 260.Gomez-Sanchez J.A., Carty L., Iruarrizaga-Lejarreta M., Palomo-Irigoyen M., Varela-Rey M., Griffith M. Schwann cell autophagy, myelinophagy, initiates myelin clearance from injured nerves. J. Cell Biol. 2015;210:153–168. doi: 10.1083/jcb.201503019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Park Y.E., Hayashi Y.K., Bonne G., Arimura T., Noguchi S., Nonaka I. Autophagic degradation of nuclear components in mammalian cells. Autophagy. 2009;5:795–804. doi: 10.4161/auto.8901. [DOI] [PubMed] [Google Scholar]
- 262.Marshall R.S., Li F., Gemperline D.C., Book A.J., Vierstra R.D. Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol. Cell. 2015;58:1053–1066. doi: 10.1016/j.molcel.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Bernales S., Schuck S., Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. 2007;3:285–287. doi: 10.4161/auto.3930. [DOI] [PubMed] [Google Scholar]
- 264.Maiuri M.C., Criollo A., Tasdemir E., Vicencio J.M., Tajeddine N., Hickman J.A. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L) Autophagy. 2007;3:374–376. doi: 10.4161/auto.4237. [DOI] [PubMed] [Google Scholar]
- 265.Sumpter R., Jr., Sirasanagandla S., Fernandez A.F., Wei Y., Dong X., Franco L. Fanconi anemia proteins function in mitophagy and immunity. Cell. 2016;165:867–881. doi: 10.1016/j.cell.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 267.Nakagawa I., Amano A., Mizushima N., Yamamoto A., Yamaguchi H., Kamimoto T. Autophagy defends cells against invading group A streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 268.Grasso D., Ropolo A., Lo Re A., Boggio V., Molejon M.I., Iovanna J.L. Zymophagy, a novel selective autophagy pathway mediated by VMP1–USP9x–p62, prevents pancreatic cell death. J. Biol. Chem. 2011;286:8308–8324. doi: 10.1074/jbc.M110.197301. [DOI] [PMC free article] [PubMed] [Google Scholar]