Abstract
Background
The temporal in vivo response of epithelial cells to a viral challenge and its association with viral clearance and clinical outcomes has been largely unexplored in asthma.
Objective
To determine gene expression profiles over time in nasal epithelial cells (NECs) challenged in vivo with rhinovirus‐16 (RV16) and compare to nasal symptoms and viral clearance.
Methods
Patients with stable mild to moderate asthma (n = 20) were challenged intranasally with RV16. Nasal brush samples for RNA sequencing were taken 7 days prior to infection and 3, 6 and 14 days post‐infection, and blood samples 4 days prior to infection and day 6 post‐infection. Viral load was measured in nasal lavage fluid at day 3, 6 and 14.
Results
Top differentially (>2.5‐fold increase) expressed gene sets in NECs post‐RV16 at days 3 and 6, compared with baseline, were interferon alpha and gamma response genes. Patients clearing the virus within 6 days (early resolvers) had a significantly increased interferon response at day 6, whereas those having cleared the virus by day 14 (late resolvers) had significantly increased responses at day 3, 6 and 14. Interestingly, patients not having cleared the virus by day 14 (non‐resolvers) had no enhanced interferon responses at any of these days. The daily Cold Symptom Scores (CSS) peaked at days 3 to 5 and correlated positively with interferon response genes at day 3 (R = 0.48), but not at other time‐points. Interferon response genes were also enhanced in blood at day 6 after RV16 challenge.
Conclusion and Clinical Relevance
This study shows that viral load and clearance varies markedly over time in mild to moderate asthma patients exposed to a fixed RV16 dose. The host's nasal interferon response to RV16 at day 3 is associated with upper respiratory tract symptoms. The temporal interferon response in nasal epithelium associates with viral clearance in the nasal compartment.
Keywords: asthma, epithelium, innate immunity, interferon response, nasal epithelium, rhinovirus, virus
1. INTRODUCTION
Asthma is a heterogeneous, inflammatory airway disease, in which respiratory viral infections can trigger worsening of symptoms. Rhinoviruses are the most common cause of virus‐induced worsening of asthma. There are no indications that asthma patients are more frequently infected than healthy controls, but they do develop more severe lower airway symptoms than healthy individuals,1, 2, 3 suggesting differences in the anti‐viral response.
Rhinoviruses predominantly infect and replicate in nasal and bronchial epithelial cells. Previous ex vivo studies have reported a deficient anti‐viral interferon production by bronchial epithelium 4, 5 and bronchoalveolar lavage cells 6 from asthma patients to rhinovirus 16 (RV16), which led to enhanced viral replication. In addition, in these and similar studies an exaggerated TH2 response to RV16 was found,7 which was paralleled by eosinophilic and neutrophilic inflammation.8 Other in vivo studies with asthma patients showed, as compared with baseline, pronounced local interferon responses to naturally occurring viral infections associated with exacerbations 9, 10 and also specifically to RV16 challenge.11
In a recent study with stable mild asthma patients, we showed a pronounced interferon response in bronchial epithelial cells after in vivo RV16 challenge, whereas we did not observe TH2‐mediator‐driven responses. This was confirmed (unpublished) retrospectively for 92 TH2‐associated genes, as provided by Ref.12 In addition, the host's interferon response, but not the viral load, correlated with both inflammatory and clinical parameters.13 In line herewith, others found that the viral load in nasal washes after naturally acquired rhinovirus 14 and experimental RV16 15 infections did not differ between individuals with and without asthma. In contrast, another study linked clinical severity of rhinovirus‐induced asthma exacerbations to the viral load in nasal lavage.3 The extent of control of asthma was offered as an explanation for these opposite findings.16 An alternative non‐exclusive hypothesis is that there are differences in timing in interferon responses amongst patients with asthma. For example, in a murine study of experimental asthma exacerbations we found that IL‐33, released early after viral exposure, inhibited the interferon response.17
The primary aim of this study was to examine changes in gene expression over time, particularly those of the interferon response genes, in epithelial cells upon in vivo RV16 challenge and compare those to viral replication and clinical outcomes. To that end, we subjected mild to moderate asthma patients, controlled by corticosteroids, to RV16 and sampled nasal epithelial cells to study gene expression over time, in relation to viral load and clinical symptoms. In addition, we collected blood to determine, if present, the systemic response to in vivo RV16 challenge and its relationship to the response measured in the nasal compartment.
2. METHODS
2.1. Subjects and design
The study set‐up (ClinicalTrials.gov: NCT01866306) is represented schematically in Figure 1. Twenty (out of 23) stable adult mild to moderate asthma patients on inhaled corticosteroids (≤500 µg/day fluticasone propionate or equivalent), all RV16‐seronegative (<1:4), were included in this analysis. The three excluded patients were either not infected (n = 1), infected with another virus at the time of inoculation (n = 1) or the sequence reads did not match with the non‐stranded sequenced samples during the stranded library preparation step (n = 1). Nasal lavage and brushed nasal epithelial cells (NECs) were collected 7 days prior to low dose rhinovirus16 (RV16UB) inoculation (100 TICD50) as baseline and at days 3, 6 and 14 after inoculation.18 RV16UB is a GMP RV16 stock prepared under auspices of U‐BIOPRED, where 100 TICD50 was found to be the lowest optimal dose for effective infections in healthy individuals and asthma patients (manuscript in preparation). Blood was obtained 4 days prior, as baseline, and at day 6 post‐RV16 exposure. Viral load was measured in nasal lavage fluid at days 3, 6 and 14. In addition, a PCR screening for respiratory viruses was performed in throat swabs 1 day before RV16 challenge to ensure no other viral infections. Fractional exhaled nitric oxide (FeNO) was measured, at the same days as nasal lavage was obtained. Cold Symptom Scores and FEV1% predicted (based on morning values) were measured every day from 7 days before until 14 days after RV16 challenge. The study was approved by the internal review boards of the participating centres, and written informed consent was obtained from all participating patients. Patient baseline characteristics are provided in Table 1. The inclusion and exclusion criteria for these patients are provided in the online data supplement.
Figure 1.
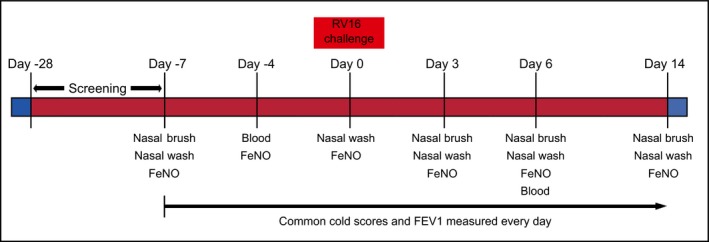
An overview of the study design. Screening was performed 28 d before experimental rhinovirus infection. Nasal washes, brushes and blood were collected on indicated days. Cold Symptom Scores and FEV1 were measured every d from 7 d before RV16 until 14 d after RV16 administration to asthma patients
Table 1.
Characteristics of moderate asthma patients enrolled in the study
| Patient characteristics | ||||
|---|---|---|---|---|
| Age (y)[Link] | 33.8 (22.1‐53.1) | |||
| Sex ratio (M:F) | 1:1 | |||
| Baseline | Day 3 | Day 6 | Day 14 | |
| Subjects (n)[Link] | 20 | 20 | 19 | 19 |
| FEV1 pre‐bronchodilator (L)[Link] | 3.33 (0.72) | 3.32 (0.76) | 3.33 (0.74) | 3.27 (0.73) |
| FEV1 pre‐bronchodilator (% pred)[Link] | 90.22 (10.79) | 89.81 (11.52) | 90.91 (11.36) | 88.68 (11.81) |
| Cold Symptom Scores[Link] | 0.25 (0.53) | 5.6 (2.76) | 4.15 (3.15) | 1.2 (3.09) |
| FeNO (ppb)[Link] | 35.17 (25.72) | 32.92 (19.2) | 32.9 (23.96) | 33.97 (25.9) |
Mean (range).
Arithmetic mean (SD).
1 asthma patient dropped out for days 6 and 14.
2.2. Study procedures
Cold Symptom Scores (CSS) were assessed using the Jackson Cold Symptom Score questionnaire, recording daily sneezing, nasal discharge, stuffy nose, sore throat, cough, chest pain, fever, chills and headache.19 The FEV1% predicted was measured in the morning according to the recommendations by the European Respiratory Society.20 The drop in FEV1 was determined by the maximal change in FEV1% predicted after RV16 challenge at days 3, 6 and 14 compared with FEV1% predicted before RV16 (average of 7 measurements taken every day from 7 days before until 1 day before RV16). Nasal brushings and collection of nasal lavage were performed by standard procedures.21 FeNO was measured by a chemiluminescence analyzer (NIOX MINO®).22 Rhinovirus in nasal lavage was quantified by PCR.23 The lower limit of detection for viral titre was 500.
2.3. RNA isolation and sequencing
The nasal brushings consisted predominantly of nasal epithelial cells, verified by the transcriptome of nasal brushes that did not express specific markers for eosinophils (eosinophilic cationic protein), neutrophils (myeloperoxidase) and macrophages (CD206), which are predominant in nasal airways. Two brushings at each time‐point were pooled by centrifugation at 1000 g (standard table centrifuge) for 5 minutes at 4°C. The pellet was dissolved in 1 mL of TRIzol® and stored at −80°C. Once all samples were obtained, they were thawed to room temperature and 200 µL of chloroform was added, followed by inverting 10 times. After incubating at room temperature for 5 minutes, the samples were centrifuged at 3500 rpm for 10 minutes at 4°C. The RNA was purified from the obtained aqueous phase, using protocol 4.C. of SV 96 total RNA isolation system (Promega). The RNA from blood samples was isolated by standard PAXgene® RNA isolation kit. All samples with RIN (RNA integrity number) score of >6 assessed by bioanalyzer and/or that passed the quality control with FASTQ and alignment quality analysis with Omicsoft (Qiagen®) were used for analysis. In total, 78 out of 80 nasal brush samples and 38 out of 40 blood samples were sequenced using Illumina HiSeq 2500 sequencer. The detailed methods on cDNA preparation, RNA sequencing and analysis are provided in the online data supplement. The data sets will be submitted to Gene Expression Omnibus website within 6 months after publication.
2.4. Statistical analyses
The read counts were loaded into the edgeR for TMM (trimmed mean of M‐values) normalization and voom function in limma to remove any variations in sample quality. This was followed by linear modelling and determining the differentially expressed genes in limma. The comparisons were pair‐wise for the respective time‐point to the baseline gene expression, and an adjusted P value of <.05 was considered statistically significant. The Z scores used for correlation with viral load and clinical parameters were calculated by subtracting the mean interferon response gene expression of all patients from the gene expression of the individual patient and divided by the standard deviation, sum of the scores for all genes in the gene set and divided the result by the square‐root of the total number of genes.24 For Ingenuity Pathway Analysis (IPA), log2 fold change and P value adjusted were used as inputs. For all correlations, Pearson's correlation coefficients with two‐tailed analysis were used. These statistical analyses were performed using GraphPad Prism 7.
3. RESULTS
3.1. Interferon response gene expression is enhanced at day 3 and remains high at day 6 after RV16 challenge in nasal epithelium from asthma patients
To determine nasal epithelial gene expression upon in vivo RV16 challenge, we collected nasal epithelial brushings from 20 asthma patients at 7 days pre‐challenge (baseline) and 3, 6 and 14 days post‐challenge. Compared with baseline, 871 genes were differentially expressed at day 3 (Table S1), 4199 genes at day 6 (Table S2) and only 101 genes at day 14 (Table S3), indicating that changes induced by RV16 in nasal epithelial cells were mostly back to baseline by day 14. As displayed in the Venn diagram, 352 of the differentially expressed genes are unique for day 3, 3610 for day 6 and 9 for day 14 (Figure S1A). Gene set enrichment analysis (GSEA) revealed that, compared with baseline, the top differentially regulated set of genes at days 3 and 6 were the interferon gamma and alpha response gene sets (Figure 2A). At day 3, the other differentially expressed gene sets within the top 10 belong to inflammatory response, apoptosis and signalling pathways involving TNF‐α, JAK‐STAT3, STAT2, complement and p53 pathway. Most of these genes in these sets overlap with the interferon response gene sets. At day 6, the other differentially expressed gene sets are MYC and E2F targets, MTORC1 signalling, glycolysis, oxidative phosphorylation and apoptosis that all relate to cell metabolism. No specific gene sets were differentially expressed at day 14. Heat maps of genes induced by both interferon alpha and gamma (Figure 2B) or specifically by interferon alpha (Figure S1B) or interferon gamma (Figure S1C) showed similar patterns. The corresponding viral loads are shown for convenience and discussed further below. The Z scores of the common interferon response genes were significantly higher at days 3, 6 and 14 after RV16 challenge, compared with baseline, with a trend wise decrease at day 14 (Figure 2C). To visualize regulators of differentially expressed genes in a pathway, and their localizations and functions, IPA analysis was used. It also displayed both type I and type II interferon signalling as the most up‐regulated pathway at days 3 and 6 after RV16 exposure in nasal epithelium of asthma patients (Figure 2D).
Figure 2.
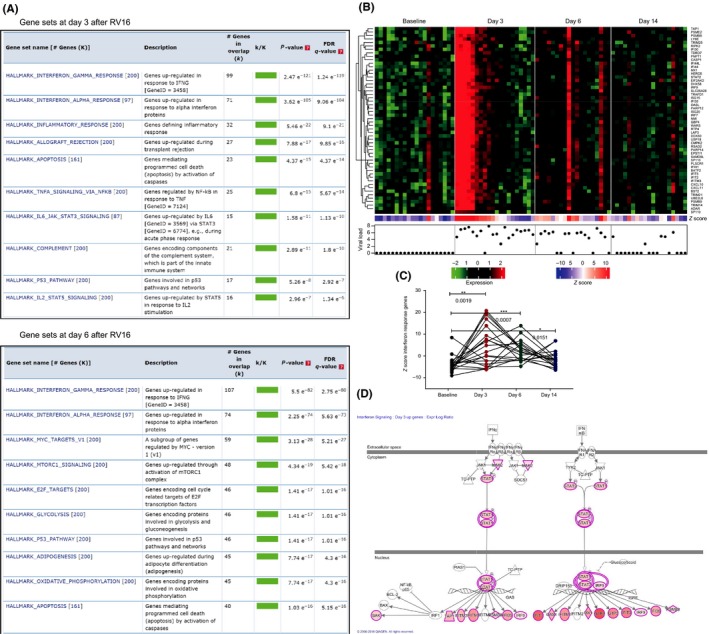
Interferon alpha and gamma response gene sets are the most differentially expressed sets, relative to baseline, in nasal epithelium at days 3 and 6. A, Gene set enrichment analysis of all differentially expressed genes showed that interferon gamma and alpha response gene sets were up‐regulated at days 3 and 6 after RV16 in nasal epithelial cells of asthma patients. B, Heat maps of interferon response genes common for both interferon alpha and gamma were enhanced at day 3 (n = 20), day 6 (n = 19) and day 14 (n = 19) after RV16 challenge compared with baseline (7 d before RV16; n = 20). The heat maps for respective patients are shown in the same order for each day. The viral load represented as log10, measured in nasal brushes, is provided below the heat maps for respective days and corresponding asthma patients. C, Z scores of interferon response genes were plotted per patient and were significantly increased at day 3, day 6 and day 14 after RV16 compared with baseline (7 d before RV16); *P < .05, **P < .01, ***P < .001, paired t tests. D, Ingenuity pathway analysis of all differentially expressed genes revealed that interferon signalling pathway is up‐regulated after RV16 at days 3 and 6 after RV16, but not at day 14. The coloured nodes represent differentially expressed genes, where the higher the intensity of the node colour, the higher the gene expression is. The genes in white nodes are not differentially expressed
3.2. Up‐regulation of interferon response genes relates to viral clearance in asthma patients
The course of the infection varies based on the viral titres in nasal lavage, as seen in Figure 3A. Given the time‐points available in the present study design, asthma patients that cleared the virus by day 7 were denoted as early resolvers (red); those clearing virus by day 14 as late resolvers (green); and those with persisting virus at day 14 as non‐resolvers (blue) (Figure 3A). The Z scores of interferon response genes were significantly increased at days 3, 6 and 14 for late resolvers and at day 6 for early resolvers, compared with baseline of respective groups. Importantly, there was no significant increase in interferon response genes from baseline at any of the days for the non‐resolvers. However, non‐resolvers had high baseline expression (P = .03) of interferon response genes compared with late resolvers (Figure 3B). IPA analysis showed that these differences between early, late and non‐resolvers could not be explained by variation in type I and type II interferons or anti‐viral genes (data not shown). There were no other differentially expressed gene sets post‐RV16, like the apoptosis genes (Figure S2A), that could explain differentiation between early, late and non‐resolvers (Figure S2B). To analyse whether patients with high viral load also have enhanced interferon response gene expression, we correlated these two parameters for the respective days. The viral load did not correlate with the Z score of interferon response genes at days 3 and 6. At day 14, there was a positive correlation of interferon response genes and viral load (P = .01; R2 = 0.32), but this correlation was skewed by one patient with a high viral load at day 14 (Figure 3C). The early, late and non‐resolvers did not significantly differ in any of the patient characteristics listed in supplementary Table S4.
Figure 3.
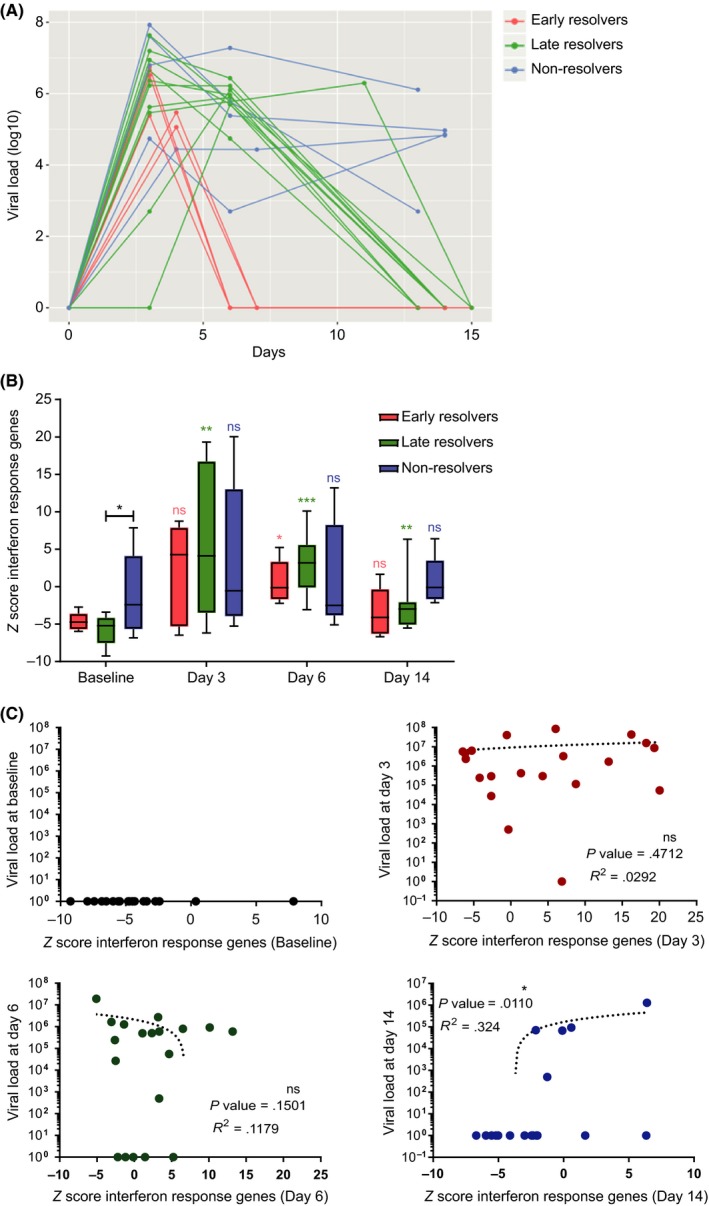
Interferon response gene expression in nasal epithelium from asthma patients correlates with clearance of RV16. A, Asthma patients separated in three groups based on viral clearance. In early resolvers (red), virus was cleared by day 6, and in late resolvers (green), virus was cleared by day 14. In the non‐resolvers (blue), virus was not yet cleared at day 14. B, The Z scores of common interferon response genes for early (n = 5), late (n = 10) and non‐resolvers (n = 5); *P < .05, **P < .01, ***P < .001, paired t tests (all comparisons of the respective groups to their baseline at different days). At baseline, non‐resolvers had higher interferon response gene expression compared with late resolvers; *P < .05, unpaired t test. C, Viral load in nasal lavage did not correlate with Z scores of interferon response gene sets at any of the days in nasal epithelial cells from asthma patients (Pearson's correlation coefficient)
3.3. The RV16‐induced local interferon response is also reflected in blood
We examined whether exposure to RV16 resulted in a systemic response and, if so, how this compared to the response in nasal epithelium. Blood was collected 4 days before and 6 days after RV16 challenge. Compared with baseline, 674 genes were differentially expressed in blood at day 6 after RV16 (see Table S5). Similar to nasal epithelial cells, GSEA analysis showed that interferon alpha and gamma are the most differentially expressed gene sets in blood at day 6 (Figure 4A). The heat map shows the individual differentially expressed interferon response genes at day 6 after RV16 compared with baseline (Figure 4B). The Z scores obtained from the heat maps showed a highly significant up‐regulation of interferon response genes after RV16 exposure (Figure 4C). Interestingly, the Z score of interferon response genes at day 6 in nasal epithelium and in blood correlated significantly (P = .03; R2 = 0.23), thus reflecting a similar degree of up‐regulation of interferon response genes to RV16 (Figure 4D). The highest scoring network with an IPA score of 36 was associated with the defence response to virus. The key components in the network are IFI27, RSAD2, IFI44L, IFIT1, CMPK2, IFI44, HERC5, OAS3, MX1 and EPSTI1 (Figure 4E).
Figure 4.
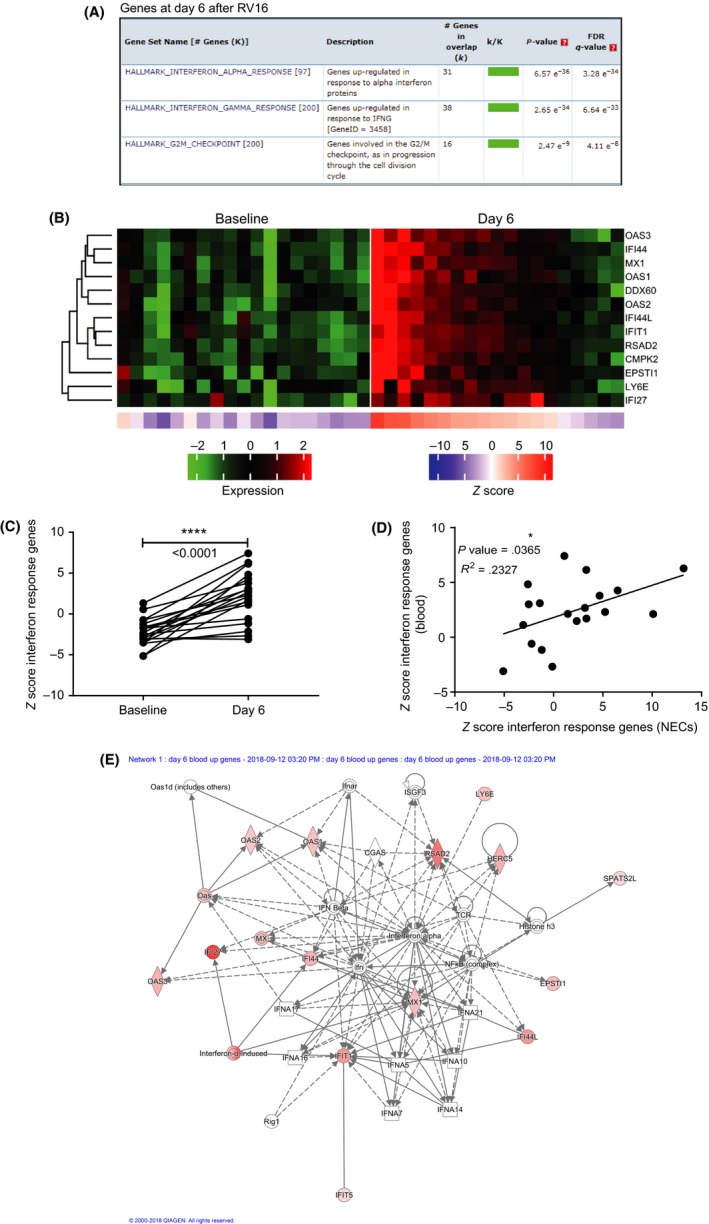
Systemic interferon response gene expression in blood is increased at day 6, as in the nasal compartment. A, In blood, gene set enrichment analysis showed that interferon response gene sets (both alpha and gamma) are up‐regulated at day 6. B, Heat maps of interferon response for respective patients (same order before and after RV16) are enhanced at day 6 after RV16 (n = 19). C, Z scores of blood interferon response genes of individual patients significantly increased 6 days after RV16 compared with baseline (day = −4); ****P < .0001, paired t tests. D, The Z scores of interferon response genes at day 6 after RV16 in blood and nasal epithelium correlated significantly (Pearson's correlation analysis). E, Gene network identification by IPA representing the most influential interferon‐related genes. Genes are connected by solid lines (direct interaction) and dotted lines (indirect interaction)
3.4. Cold Symptom Scores (CSS) and interferon response genes correlate at the peak of upper airway symptoms, but not to FEV1
Previously, we found that the interferon response genes in bronchial epithelial cells upon RV16 exposure correlated with lower airway symptoms. Now, we assessed whether the enhanced interferon response genes in NECs correlated with clinical parameters of the upper airways. CSS were measured every day from 7 days before until 14 days after RV16 administration. The CSS were highest at days 3 to 5 (manuscript in preparation). The Z scores of interferon response genes in nasal epithelium and the CSS, both at day 3 after RV16, correlated significantly (P = .02; R2 = 0.24). This correlation was not observed at days 6, 14 or at baseline (Figure 5A). The Z scores of interferon response genes in blood at day 6 also correlated positively with the CSS at day 6 (Figure S3). The drop in FEV1% predicted from baseline (averaged FEV1% predicted values of 7 days before RV16) was low as expected for patients on corticosteroids and, as a group, did not significantly differ between days 1‐14 after RV16 (manuscript in preparation). To determine whether the nasal interferon responses are reflected in the bronchial compartment, we correlated enhanced interferon response genes and the drop in FEV1. There was no correlation of interferon response genes in nasal epithelium and the RV16‐induced percental drop in FEV1 at days 3, 6 and 14 (Figure 5B). Moreover, the Z scores of interferon response at day 3 did not correlate with the maximum drop in FEV1% predicted over 14 days after RV16 challenge (Figure 5C). Interestingly, unlike for the Z scores of interferon response genes, the viral load did not correlate with the CSS at either days 3, 6, 14 or at baseline Figure S4A). The CSS measured post‐RV16 (day of maximum increase) did not differ between early, late and non‐resolvers (Figure S4B). The FeNO levels were not affected in these asthma patients after RV16 at days 3, 6 and 14 compared with baseline (day 0 and 7 days before; manuscript in preparation).
Figure 5.
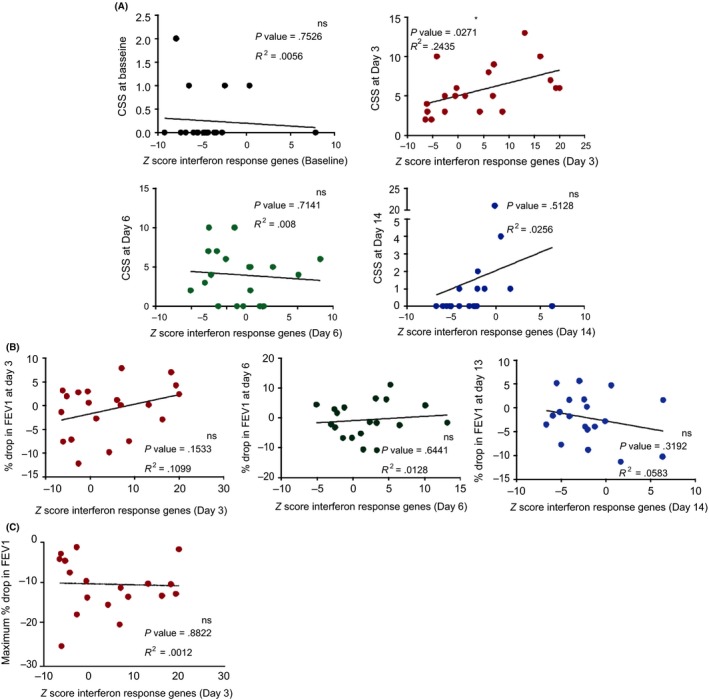
Interferon response gene expression in nasal epithelium at day 3 correlates with peak of upper airway symptoms. A, Cold Symptom Score (CSS) correlated with Z scores of interferon response genes only at day 3 (red) at which there was the highest expression of interferon response genes. While at baseline (black), day 6 (green) and day 13 (blue), there was no correlation observed between CSS and interferon response genes at their respective time‐points (Pearson's correlation coefficient). B, The Z scores of interferon response genes did not correlate with the drop in % FEV1 measured at day 3 (n = 20), day 6 (n = 19) and day 13 (n = 19) and C, also the maximum drop in % FEV1 over 14 d after RV16 (n = 20). All correlations were performed using Pearson's correlation coefficient
4. DISCUSSION
In this study, we depicted sequential changes over 3, 6 and 14 days in the transcriptome of nasal epithelial cells from asthma patients in response to an in vivo viral challenge, which allowed us to recognize differential responses that could be linked to the course of the infection. As also various clinical parameters were measured, it enabled to link virus‐induced responses to clinical outcomes. Nasal epithelial cells from mild to moderate asthma patients upon in vivo RV16 challenge displayed, compared with baseline, enhanced interferon response gene expression at day 3, remained high at day 6 post‐infection and returned to near‐baseline at day 14. Interestingly, the increased interferon responses in nasal epithelium at day 3, but not the viral load, correlated well with symptom scores at day 3, corresponding to the peak of symptom scores. Asthma patients with a relatively high interferon response gene expression at baseline, were unable to mobilize an elevated interferon response at days 3 and 6 compared with baseline and did not clear the virus by day 14. In contrast, those who cleared the virus all displayed a temporal increased interferon response. RNA sequencing of blood, 6 days after RV16 exposure, showed similar up‐regulation of interferon response genes compared with nasal brushings. These data suggest that enhanced interferon responses at days 3 and 6 after in vivo RV16 challenge are associated with viral clearance and higher cold symptom scores.
In our previous in vivo challenge study with RV16 in mild asthma patients,13 we found, compared with baseline, an enhanced interferon response in bronchial epithelium 6 days post‐infection. In both studies, the extent of the initial interferon response correlated with clinical symptoms, whereas the viral load did not correlate with clinical symptoms. This is in line with another study 25 that shows that children with upper respiratory rhinovirus infection had higher levels of interferons in nasal washes and enhanced levels linked with more wheezing, but no difference in viral titres. The viral load in any given compartment is the sum of viral replication and clearance of the virus. Viral replication in bronchial epithelium is the major driver of interferon production in asthma patients.26 Viral load in the bronchial compartment correlated with the local interferon response, but this was not the case in the nasal compartment where the virus was administered. The maximal viral load in the nasal compartment varied about 4 logs between patients, which indicates that despite a fixed infectious dose, the number of viral particles generated by replication varies considerably between patients. As we have previously shown that IL‐33 inhibits interferon responses,17 we also searched for IL‐33 response genes 27 in NECs at days 3, 6 and 14 post RV16 but found no differences (data not shown). For the bronchial compartment, the correlation between viral load and interferon response suggests that the viral load is more representative of viral replication. This also implies that the bronchial presence of IL‐33 and the variation in time taken by the virus to reach the bronchial compartment 17, 28 have little impact on the interferon response in the bronchial compartment, at least at days 3 and 6 post‐infection. The interferon response over time, but not the viral load, did correlate with the course of the infection, which is in line with the anti‐viral activity of interferons. In the nasal compartment, viral clearance was independent of IL‐33 response genes as no differences were found between early, late and non‐resolvers groups (data not shown). The association between viral clearance and interferon response genes is very specific as other differentially expressed gene sets, for example the apoptosis genes or the metabolic genes at day 6, were not different among the viral clearance groups.
In the current study, we found a marked interferon response gene expression in nasal epithelial cells despite the use of inhaled corticosteroids (ICS) by these asthma patients. Such maintenance treatment was inevitable, because it is unethical to withdraw ICS in moderate asthmatics in this study set‐up. However, we do not know whether ICS affects the nasal compartment and, if so, to what extent. Previously though, a rhinovirus challenge in asthma patients on ICS showed enhanced levels of CXCL‐10 in sputum and nasal lavage,29 and of CXCL‐10 and CXCL‐11 in upper airway secretions at day 4 after RV16.11 In line with the current findings, CXCL‐10 and CXCL‐11 were the genes in the nasal epithelium with the highest fold change after RV16 at days 3 and 6. Previous studies have shown that oral corticosteroids increase RV16 titres at day 3 after RV16 challenge 30 and this was considered to be due to a decreased anti‐viral response by corticosteroids. We, however, show that within this patient group, which is homogeneous in ICS use, the interferon response gene expression varies between patients and determines the viral clearance. The source of interferons in asthma patients after in vivo RV16 challenge has not yet been determined. This also applies to the systemic interferon response, which also is relevant to potential spill‐over effects on the bronchial compartment.
There are a few limitations of this study. The use of ICS could have attenuated particularly the clinical parameters of the lower airways,31 but apparently not those of the nasal compartment. Thus, the use of ICS could have a bias in the correlation of interferon response gene expression in nasal epithelial cells with lower airway symptoms. Although we differentiated inflammatory and immune cells from nasal lavages (data not shown), the very high standard deviations of ±46.13 and ±20.57 for baseline eosinophil and neutrophil relative counts precluded a comparison with the interferon response gene expression. Thus, it is not clear whether the correlation between the interferon response and inflammation in the bronchial compartment also applies to the nasal compartment. Given that the current findings and conclusions do resemble those obtained in the bronchial compartment in our previous study, we would argue that there is similarity between both compartments. We cannot exclude, however, that local mediators like IL‐33 and anti‐inflammatory effects of ICS impose additional effects in the bronchial compartment.
In conclusion, our study reveals that viral replication and load vary considerably over time after challenge with a fixed infectious dose of RV16 in asthmatic subjects on ICS. The interferon response gene expression is heterogeneous but prominent in nasal epithelium after RV16. Our data further indicate that the increment of interferon response at days 3 and 6 compared with baseline, but not the interferon response at day 3 itself, relates to viral clearance. The interferon response at day 3, however, relates to symptoms. These findings may explain why IFN‐β therapy was not effective in all asthma patients 32 and opens a window of opportunity for tailored IFN‐β therapy. It would be of interest to further delineate the potential contributions of the other differentially expressed gene sets to the course of the infection.
CONFLICT OF INTEREST
The research leading to these results has received support from the Innovative Medicines Initiative (IMI) Joint Undertaking, under grant agreement no. 115010, resources for which are composed of financial contribution from the European Union's Seventh Framework Programme (FP7/2007‐2013) and kind contributions from companies in the European Federation of Pharmaceutical Industries and Associations (EFPIA) (http://www.imi.europa.eu). The authors declare no other conflict of interests.
AUTHOR'S CONTRIBUTION
AR analysed the data and prepared the manuscript. MC is the lead data analyst for this study. M.vd P. carried out the PN218 study. SY analysed clinical data and contributed to experimental set‐up definitions. AA, BT, LC, AB, MR, I.d.L., DS, JMJ, PJS and NK devised the study and contributed to the experimental set‐up definition (L.C is currently affiliated with Celgene, New Jersey, USA. M.R is currently affiliated with Galapagos NV, Mechelen, Belgium). RL devised the study, contributed to the experimental set‐up and prepared the manuscript. All authors approved the final version of the manuscript. M.C, SY, A.A, B.T, L.C, A.B, M.R and I.d.L. are/were employees of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc, Kenilworth, NJ, USA, or Merck Sharp & Dohme (MSD) Belgium, all of whom may own/hold stock/stock options in Merck & Co., Inc, Kenilworth, NJ, USA.
Supporting information
ACKNOWLEDGEMENTS
The authors would like to acknowledge Dr Jan Koster from the Department of Oncogenomics, Amsterdam University Medical Center for crosschecking datasets in R2 platform. We are also thankful to Dr Brendon P. Scicluna and Hina Naz Khan (Center for Experimental and Molecular Medicine, Amsterdam University Medical Center) for providing access to IPA platform. The authors are very grateful to all patients for their participation in the present studies; without their commitment and suggestions, this study would not have been possible.
Ravi A, Chang M, van de Pol M, et al; on behalf of U‐BIOPRED Study Group . Rhinovirus‐16 induced temporal interferon responses in nasal epithelium links with viral clearance and symptoms. Clin Exp Allergy. 2019;49:1587–1597. 10.1111/cea.13481
Ravi and Chang both authors contributed equally.
Contributor Information
Abilash Ravi, Email: a.ravi@amc.nl.
René Lutter, Email: r.lutter@amsterdamumc.nl.
REFERENCES
- 1. van der Sluijs KF, van de Pol MA, Kulik W, et al. Systemic tryptophan and kynurenine catabolite levels relate to severity of rhinovirus‐induced asthma exacerbation: a prospective study with a parallel‐group design. Thorax. 2013;68(12):1122‐1130. [DOI] [PubMed] [Google Scholar]
- 2. Corne JM, Marshall C, Smith S, et al. Frequency, severity, and duration of rhinovirus infections in asthmatic and non‐asthmatic individuals: a longitudinal cohort study. Lancet. 2002;359(9309):831‐834. [DOI] [PubMed] [Google Scholar]
- 3. Message SD, Laza‐Stanca V, Mallia P, et al. Rhinovirus‐induced lower respiratory illness is increased in asthma and related to virus load and Th1/2 cytokine and IL‐10 production. Proc Natl Acad Sci USA. 2008;105(36):13562‐13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wark PAB, Johnston SL, Bucchieri F, et al. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J Exp Med. 2005;201(6):937‐947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Edwards MR, Regamey N, Vareille M, et al. Impaired innate interferon induction in severe therapy resistant atopic asthmatic children. Mucosal Immunol. 2013;6(4):797‐806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Contoli M, Message SD, Laza‐Stanca V, et al. Role of deficient type III interferon‐lambda production in asthma exacerbations. Nat Med. 2006;12(9):1023‐1026. [DOI] [PubMed] [Google Scholar]
- 7. Jackson DJ,Makrinioti H, Rana BM, et al. IL‐33‐dependent type 2 inflammation during rhinovirus‐induced asthma exacerbations in vivo. Am J Respir Crit Care Med. 2014;190(12):1373‐1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Toussaint M, Jackson DJ, Swieboda D, et al. Host DNA released by NETosis promotes rhinovirus‐induced type‐2 allergic asthma exacerbation. Nat Med. 2017;23(6):681‐691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schwantes EA, Manthei DM, Denlinger LC, et al. Interferon gene expression in sputum cells correlates with the asthma index score during virus‐induced exacerbations. Clin Exp Allergy. 2014;44(6):813‐821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Swanson CL, Babineau D, Whalen E, et al. An exaggerated type i interferon antiviral response is associated with exacerbations in pediatric asthma. J Allergy Clin Immunol. 2018;141(2):p. Ab116. [Google Scholar]
- 11. Hansel TT, Tunstall T, Trujillo‐Torralbo M, et al. A comprehensive evaluation of nasal and bronchial cytokines and chemokines following experimental rhinovirus infection in allergic asthma: increased interferons (IFN‐gamma and IFN‐lambda) and type 2 Inflammation (IL‐5 and IL‐13). EBioMedicine. 2017;19:128‐138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peters MC, Ringel L, Dyjack N, et al. A transcriptomic method to determine airway immune dysfunction in T2‐high and T2‐low asthma. Am J Respir Crit Care Med. 2019;199(4):465‐477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ravi A, Koster J, Dijkhuis A, et al. Interferon‐induced epithelial response to rhinovirus‐16 in asthma relates to inflammation and FEV<sub>1</sub>. J Allergy Clin Immunol. 2019;143(1):442‐447.e10. [DOI] [PubMed] [Google Scholar]
- 14. Kennedy JL, Shaker M, McMeen V, et al. Comparison of viral load in individuals with and without asthma during infections with rhinovirus. Am J Respir Crit Care Med. 2014;189(5):532‐539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DeMore JP, Weisshaar EH, Vrtis RF, et al. Similar colds in subjects with allergic asthma and nonatopic subjects after inoculation with rhinovirus‐16. J Allergy Clin Immunol. 2009;124(2):pp. 245–52, 252 e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jackson DJ, Trujillo‐Torralbo M‐B, del‐Rosario J, et al. The influence of asthma control on the severity of virus‐induced asthma exacerbations. J Allergy Clin Immunol. 2015;136(2):pp. 497–500.e3. [DOI] [PubMed] [Google Scholar]
- 17. Ravanetti L, Dijkhuis A, Dekker T, et al. IL‐33 drives influenza‐induced asthma exacerbations by halting innate and adaptive anti‐viral immunity. J Allergy Clin Immunol. 2019;143(4):1355–1370. e16. [DOI] [PubMed] [Google Scholar]
- 18. Gwaltney JM Jr,Hendley O, Hayden FG, et al. Updated recommendations for safety‐testing of viral inocula used in volunteer experiments on rhinovirus colds. Prog Med Virol. 1992;39:256‐263. [PubMed] [Google Scholar]
- 19. Jackson GG, Dowling HF, Spiesman IG, Boand AV. Transmission of the common cold to volunteers under controlled conditions. I. The common cold as a clinical entity. AMA Arch Intern Med. 1958;101(2):267‐278. [DOI] [PubMed] [Google Scholar]
- 20. Miller MR, Hankinson J, Brusasco V, et al. Standardisation of spirometry. Eur Respir J. 2005;26(2):319‐338. [DOI] [PubMed] [Google Scholar]
- 21. Thavagnanam S, Parker JC, McBrien ME, Skibinski G, Shields MD, Heaney LG. Nasal epithelial cells can act as a physiological surrogate for paediatric asthma studies. PLoS One. 2014;9(1):e85802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rijssenbeek‐Nouwens LH, Fieten KB, Bron AO, Hashimoto S, Bel EH, Weersink EJ. High‐altitude treatment in atopic and nonatopic patients with severe asthma. Eur Respir J. 2012;40(6):1374‐1380. [DOI] [PubMed] [Google Scholar]
- 23. Jansen RR, Wieringa J, Koekkoek SM, et al. Frequent detection of respiratory viruses without symptoms: toward defining clinically relevant cutoff values. J Clin Microbiol. 2011;49(7):2631‐2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee E, Chuang H‐Y, Kim J‐W, Ideker T, Lee D. Inferring pathway activity toward precise disease classification. PLoS Comput Biol. 2008;4(11):e1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miller EK, Hernandez JZ, Wimmenauer V, et al. A mechanistic role for type III IFN‐lambda1 in asthma exacerbations mediated by human rhinoviruses. Am J Respir Crit Care Med. 2012;185(5):508‐516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Patel DA, You Y, Huang G, et al. Interferon response and respiratory virus control are preserved in bronchial epithelial cells in asthma. J Allergy Clin Immunol. 2014;134(6):pp. 1402‐1412.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nagarkar DR, Ramirez‐Carrozzi V, Choy DF, et al. IL‐13 mediates IL‐33‐dependent mast cell and type 2 innate lymphoid cell effects on bronchial epithelial cells. J Allergy Clin Immunol. 2015;136(1):202‐205. [DOI] [PubMed] [Google Scholar]
- 28. Werder RB, Zhang V, Lynch JP, et al. Chronic IL‐33 expression predisposes to virus‐induced asthma exacerbations by increasing type 2 inflammation and dampening antiviral immunity. J Allergy Clin Immunol. 2018;141(5):p. 1607‐1619.e9. [DOI] [PubMed] [Google Scholar]
- 29. Adura PT, Reed E, Macintyre J, et al. Experimental rhinovirus 16 infection in moderate asthmatics on inhaled corticosteroids. Eur Respir J. 2014;43(4):1186‐1189. [DOI] [PubMed] [Google Scholar]
- 30. Gustafson L, Proud D, Hendley J, Hayden F, Gwaltneyjr J. Oral prednisone therapy in experimental rhinovirus infections. J Allergy Clin Immunol. 1996;97(4):1009‐1014. [DOI] [PubMed] [Google Scholar]
- 31. Lange P, Scharling H, Ulrik CS, Vestbo J. Inhaled corticosteroids and decline of lung function in community residents with asthma. Thorax. 2006;61(2):100‐104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Djukanovic R, Harrison T, Johnston SL, et al. The effect of inhaled IFN‐beta on worsening of asthma symptoms caused by viral infections. A randomized trial. Am J Respir Crit Care Med. 2014;190(2):145‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


