Abstract
In this review we critically summarize the evidence base and the progress to date regarding the genomic basis of periodontal disease and tooth morbidity (ie, dental caries and tooth loss), and discuss future applications and research directions in the context of precision oral health and care. Evidence for these oral/dental traits from genome‐wide association studies first emerged less than a decade ago. Basic and translational research activities in this domain are now under way by multiple groups around the world. Key departure points in the oral health genomics discourse are: (a) some heritable variation exists for periodontal and dental diseases; (b) the environmental component (eg, social determinants of health and behavioral risk factors) has a major influence on the population distribution but probably interacts with factors of innate susceptibility at the person‐level; (c) sizeable, multi‐ethnic, well‐characterized samples or cohorts with high‐quality measures on oral health outcomes and genomics information are required to make decisive discoveries; (d) challenges remain in the measurement of oral health and disease, with current periodontitis and dental caries traits capturing only a part of the health‐disease continuum, and are little or not informed by the underlying biology; (e) the substantial individual heterogeneity that exists in the clinical presentation and lifetime trajectory of oral disease can be identified and leveraged in a precision medicine framework or, if unappreciated, can hamper translational efforts. In this review we discuss how composite or biologically informed traits may offer improvements over clinically defined ones for the genomic interrogation of oral diseases. We demonstrate the utility of the results of genome‐wide association studies for the development and testing of a genetic risk score for severe periodontitis. We conclude that exciting opportunities lie ahead for improvements in the oral health of individual patients and populations via advances in our understanding of the genomic basis of oral health and disease. The pace of new discoveries and their equitable translation to practice will largely depend on investments in the education and training of the oral health care workforce, basic and population research, and sustained collaborative efforts..
Keywords: classification, dentistry, endophenotypes, genomics, precision, prediction
1. INTRODUCTION
The existence of innate susceptibility and heritability of oral health and disease traits is well‐established. In fact, scholarly work discussing possible hereditary components of dental caries was published as early as the 1920s.1 It was several decades later, and as a result of continued progress in the basic sciences, breakthroughs in the supporting technologies, and substantial investments in effort and resources, that dentistry began to enter the “genome era”. The first genome‐wide association studies of periodontitis and dental caries were published in 20102 and 2011,3 respectively. Nevertheless, other lines of research (including investigations among twins and families, and many candidate‐gene studies) have helped to build a solid case for the putative role of genetic factors in periodontal disease and dental caries. Excellent reviews and comprehensive summaries of the body of evidence supporting heritable components of oral disease have been published previously.4, 5, 6, 7, 8, 9
Genomic investigations (ie, aiming to study the genome, as opposed to genetics, traditionally focused on individual genes) have been successful in identifying important susceptibility loci for several complex disorders, including diabetes, obesity, Parkinson's disease, and several forms of cancer.10 Arguably, the major benefits of the genome‐wide association study approach include its agnostic nature (ie, hypothesis‐free as opposed to a candidate‐gene design) and coverage of a substantial proportion of the common variants found in the human genome (ie, producing information on millions of single nucleotide polymorphisms). From a methodologic standpoint, it is crucial to note that most reported associations from candidate‐gene studies fail to replicate in subsequent agnostic genome‐wide association study scans. There are several reasons behind this phenomenon, including the frequent low statistical power of genome‐wide association studies. However, the false‐positive ratio and the probable publication bias in the candidate‐gene literature are arguably more important issues. Ioannidis and colleagues,11 in a comprehensive quantitative analysis of candidate‐gene association replication in the genome‐wide association study era, found that ~1%‐5% of previously reported candidate‐gene associations were subsequently replicated by genome‐wide association studies. From an evidence‐based dentistry standpoint, this is important to acknowledge, as most oral health genomics evidence to date has been derived from candidate‐gene studies.12
Recently, its low cost has been added to the list of benefits of the genome‐wide association study methodology; high‐density genotyping can be undertaken for less than $100 per participant. The value and potential of genome‐wide association studies are now amplified by the increasing availability of whole genome sequence data, which can be used as reference panels for the imputation of additional (usually rare) markers that have not been directly genotyped,13 using two‐step14 or other imputation approaches. Moreover, the growth of publicly available data on the functional or regulatory role of single markers and genes offers additional opportunities for the annotation and functional interpretation of genome‐wide association study results.15, 16, 17, 18, 19
The efficient interrogation of the large and frequently multi‐omics data structures that accompany genome‐wide association study‐based research is certainly not straightforward, but parallel developments and advances in methodologic approaches and bioinformatics tools have made genome‐wide association studies increasingly accessible. Benefitting from this omics revolution, several genomics studies have been conducted in the oral health domain during the last decade, offering novel insights into the genomics of periodontal disease and tooth morbidity. The field is arguably in its early stages. However, several successes and noteworthy findings have marked the last decade and there is reasonable expectation that this progress will help to inform the realization of precision oral health and care, ultimately resulting in better individual clinical outcomes and improved population health.20, 21, 22
In the following section we provide an overview of research findings from genomic investigations of periodontal disease, dental caries, and several other related intermediate or composite traits. We highlight key points from each line of investigation, summarize where the field stands, and what future directions and opportunities lie ahead. As noted above, comprehensive reviews of periodontal genetics or genomics have been recently provided by Nibali et al,4 Schaefer,6 and Vieira and Albandar.7
2. GENOMICS OF TRADITIONAL CLINICAL DEFINITIONS OF ORAL AND DENTAL DISEASE
2.1. Periodontal disease
The genome‐wide association study of aggressive periodontitis reported by Schaefer et al2 marked the field's entry into the genome era. In that study, the investigators discovered and subsequently replicated the association of rs1537415, located in the glycosyltransferase gene (GLT6D1), with aggressive periodontitis. More recently, Sanders et al23 reported a significant association of a relatively rare TSNAX‐DISC1 noncoding RNA polymorphism (rs149133391) with chronic periodontitis among Hispanic/Latino people and subsequently replicated it among an independent sample of African‐Americans. Other studies24, 25, 26, 27, 28, 29 have implicated numerous loci without reaching genome‐wide statistical significance levels and/or demonstrating replication in an independent cohort. For example, a recent study among a small Italian population reported associations between EFCAB4B polymorphisms (rs242016 showing the strongest evidence of association) and localized periodontitis.24
Interestingly, several loci have been highlighted as showing suggestive evidence of association (with P values typically ranging between 5 × 10−6 and 5 × 10−8) in more than one (independent) genome‐wide association study and thus warrant attention. A prime example is SIGLEC5 (rs12461706), which was reported in a recent study of aggressive periodontitis30 and was the only locus that met genome‐wide statistical significance criteria in a large, consortium meta‐analysis of chronic periodontitis that combined clinical and self‐reported data.31 In a recent Korean study, Hong et al25reported TENM2 (ODZ2) as being putatively associated with chronic periodontitis, whereas an earlier genome‐wide association study by Divaris et al32 highlighted this locus for its association with Aggregatibacter actinomycetemcomitans subgingival colonization levels in a North American sample. Defensin alpha 1 and alpha 3 (DEFA1A3) polymorphisms (rs2978951 and rs2738058) have been reported by both a recent aggressive periodontitis study30 and an earlier chronic periodontitis study.29 NPY is another locus of interest. There was suggestive evidence of an association in a North American genome‐wide association study of chronic periodontitis27 and it was the locus with the strongest evidence of association in a German male‐only stratified sample of aggressive periodontitis.33 Additional genome areas with multiple, independent genome‐wide association level of evidence, albeit not reaching genome‐wide significance or formal genome‐wide replication, include ANRIL,34, 35, 36, 37, 38 CAMTA1/VAMP3,32, 38, 39 PF4/PPBP/CXCL5,40 NIN/CDKL1,27, 41 PLG,27, 30, 42, 43 VAMP8 (rs1561198),44 MTND1P5 (rs16870060), and LOC107984137/SHISA9 (rs729876).45 A gene‐centric and gene set enrichment re‐analysis of our group's single‐marker genome‐wide association study of chronic periodontitis27 and periodontal pathogen colonization32 has also been reported, including variable definitions of “gene boundaries”.46 Six genes showed genome‐wide evidence of association, four with severe chronic periodontitis (NIN, P = 1.6 × 10−7; ABHD12B, P = 3.6 × 10−7; WHAMM, P = 1.7 × 10−6; AP3B2, P = 2.2 × 10−6) and two with high periodontal pathogen colonization (red complex‐KCNK1, P = 3.4 × 10−7; Porphyromonas gingivalis‐DAB2IP, P = 1.0 × 10−6). The top‐ranked genes for moderate chronic periodontitis were HGD (P = 1.4 × 10−5), ZNF675 (P = 1.5 × 10−5), TNFRSF10C (P = 2.0 × 10−5), and EMR1 (P = 2.0 × 10−5). Loci containing NIN, EMR1, KCNK1, and DAB2IP had showed suggestive evidence of association in the earlier single nucleotide polymorphism‐based analysis,27 whereas WHAMM and AP2B2 emerged as novel candidates. Finally, a recent genome‐wide association study of lipopolysaccharide‐induced periodontitis in mice identified CXCR3 as a susceptibility locus for bone loss in that model.47
Other notable investigations have used genotyping arrays with exome content,48 whole‐exome sequencing,49, 50 RNA expression/multi‐omics profiling,51, 52 and several post hoc bioinformatics and machine learning classification approaches53, 54 to study the molecular and genomic basis of periodontitis. The study of gene expression and other more functional approaches (as opposed to genetic association studies) is a key complement to genome‐wide association studies. Of note, most signals and markers highlighted in genome‐wide association studies tend to be associated with regulatory areas of the genome.55 In a recent report, Kitagaki and colleagues49 used whole‐exome sequencing to identify GPR126 (lead marker: rs536714306) as a candidate genetic risk factor for aggressive periodontitis in a Japanese population. In another recent investigation, Sudo and colleagues50 used a two‐step approach in a family based study and used whole‐exome sequencing to identify novel mutations in the NOD2 gene, which is also associated with aggressive periodontitis.
Some of the limitations of genome‐wide association studies for periodontitis are related to the sample size of the study and the inherent complexity in defining the trait. A commonly cited limitation in the periodontal genome‐wide association study literature is the relatively modest sample sizes of individual studies compared with reports for other common complex diseases, ranging between a few hundred24, 28 and up to 10 000‐17 000 participants.23, 26 This has been somewhat overcome by the establishment of the Gene‐Lifestyle Interactions and Dental Endpoints consortium56 and a recent report of periodontitis and dental caries genome‐wide association studies among over half a million individuals.31 However, perhaps the greater limitation in studying this disease is the substantial variation in the clinical periodontal phenotypes and case definitions for periodontitis in the published reports of the genomics of periodontitis. Additionally, the recent 2017 world workshop on the classification of periodontal and peri‐implant diseases and conditions reclassified aggressive and chronic periodontitis into one entity (“periodontitis”);57 which is then further characterized based on a multidimensional system that includes stages (ie, severity classifiers) and grades (ie, information on biologic features, including progression rate and risk). No report has yet examined the implications of the new classification system on the discovery of susceptibility loci for the disease.
Additional sources of heterogeneity or potential bias in the reported literature are related to tooth loss, which is known to bias estimates of periodontitis in cross‐sectional studies. Cases of periodontitis may be unidentified because of tooth loss. In the extreme scenario of edentulism (ie, total tooth loss), periodontitis cannot be defined, with the possibility of missing severe or aggressive cases of periodontitis as a result of rapid tooth loss. Importantly, the risk and reasons for tooth loss may be population and study specific,58 possibly underlying the observed heterogeneity between genome‐wide association studies of periodontitis. 31 Classification systems that capture and operationalize patterns of tooth loss have been developed. Although they are useful vehicles for the harmonization of different periodontal cohorts and samples,59, 60 they have yet to be interrogated in the context of genome‐wide association studies.
Key points: The number of genetic loci associated with periodontal disease obtained from genome‐wide association studies of aggressive and chronic periodontitis is increasing. Few loci, mainly identified for aggressive and severe forms of the disease, have met genome‐wide statistical significance criteria and have been replicated in independent investigations. Most genome‐wide association studies of periodontitis have been based on moderate or small sample sizes and there is substantial heterogeneity in their studied populations, the methods used, and the results reported.
2.2. Dental caries
The first genome‐wide association study for dental caries, published in 2011, was carried out for “childhood caries”, namely dental caries lesions manifested in the primary dentition.3 A subsequent report comprising five independent cohorts61 investigated dental caries in the permanent dentition and was published 1 year later. None of these investigations detected significant genome‐wide signals, although several loci had suggestive evidence of an association or had emerged from stratified analyses. More recent genome‐wide association studies were conducted for adult dental caries62 and early childhood caries63 and, similar to the previous studies, reported no significant loci but there were several suggestive, plausible ones (eg, NAMPT and BMP7 for adult dental caries). A recent consortium meta‐analysis of childhood caries64 reported two significant genome‐wide loci (ALLC, rs1594318 and NEDD9, rs7738851) as well as heterogeneity and low heritability (1%) in the measured trait, compared with individual studies or previously published estimates.65
Although not the traditional clinical definition, dental caries patterns (ie, groups of tooth surfaces, such as pits and fissures) and subtypes have been used, with slightly more noteworthy outcomes in the context of genome‐wide association studies. For example, significant genome‐wide signals were reported for LYZL2 (rs399593) and AJAP1 (rs3896439) for subtypes (ie, dental caries patterns) of adult dental caries,66 and for KPNA4 (rs17236529) for pit‐and‐fissure dental caries lesions in the primary dentition.67 A similar genome‐wide association study investigating clusters of pit‐and‐fissure and smooth surface dental caries in the primary dentition did not identify any significant signals.68 Additional loci not meeting genome‐wide significance criteria but with multiple lines of supporting evidence requiring some attention, include MMP16 (rs2046315 and rs10429371),69 PKD2 (rs17013735, rs11938025, rs2725270) and SIBLING (rs2725233),70 MPPED2 and ACTN2, 71 and TRAV4.72 Although additional insights have been gained by gene set enrichment re‐analyses of a dental caries genome‐wide association study reported by Wang et al73, the recent, large‐scale consortium meta‐analysis that included clinical and self‐reported data (including denture use) among half a million individuals, discovered 47 novel loci for adult dental caries.31
As is the case with periodontitis, there are substantial variations and methodologic areas for improvement in the reported genome‐wide association studies of dental caries, especially with clinical phenotype ascertainment. One such example pertains to the childhood disease domain, where primary teeth begin to shed after the age of 6 years, leading to the loss of potentially disease‐informative surfaces between the ages of 6 and 12 years (which is considered the age range of primary dentition caries). Additionally, dental restorative work (eg, fillings, crowns, dentures, and extractions) is known to inflate measures of dental caries burden compared with what would be measured among “untreated” individuals.74 Finally, and similar to studies of periodontitis, tooth loss and its cause (ie, dental caries, periodontitis, orthodontic reasons, trauma, or congenitally missing) may not always be discernable and will probably vary between populations and study samples.58
Key points: Genome‐wide association evidence of dental caries is still limited, but a few promising and plausible candidates with corroborating evidence have been reported. A recent, large‐scale, consortium meta‐analysis identified many loci associated with dental caries, providing a rich resource of candidates to be followed‐up in subsequent investigations. Nevertheless, sizeable genotyped cohorts with high‐quality clinical data, including detailed phenotypes of dental caries, are warranted.
3. GENOMICS OF COMPOSITE, INTERMEDIATE, AND BIOLOGICALLY INFORMED TRAITS OF ORAL HEALTH AND DISEASE
Clinical definitions of disease typically capture a subset of the disease process or its expression. Interestingly, case definitions and diagnostic criteria for both periodontitis and dental caries have evolved over time, mostly as a result of changes in their population prevalence and, to some degree, improvements in our understanding of the disease process.75, 76, 77, 78 For example, most genome‐wide association study evidence for periodontitis has been generated by studies that used clinical measures of probing depth and attachment loss, whereas varying thresholds and criteria have been applied to derive person‐level dental caries experience indices. These measures are very popular and understandable by both scientific and clinical audiences. However, they arguably fail to capture the biologic aspects of periodontitis and dental caries. Of note, both diseases are essentially of dysbiotic‐microbial nature and share common risk factors.79 In spite of these similarities, periodontitis is characterized by an aberrant inflammatory response to commensal and dysbiotic subgingival microbial communities, whereas dental caries is a sustained tooth surface‐supragingival biofilm dysbiosis that leads to progressive demineralization of the dental hard tissues. Of note, the measurement of both diseases is undermined to some degree by tooth loss, especially in cross‐sectional studies, where reasons for tooth loss may be unknown or unclear. In this section we outline the genomic investigations of composite (ie, comprising more than one disease), intermediate (ie, part of the disease process, typically including inflammation and microbiome), and biologically informed (ie, clinical traits enriched with information on biologic parameters, typically inflammation and microbiome). We demonstrate the potential benefits of these traits over conventional clinical taxonomies.
3.1. Tooth morbidity
Tooth loss is the most common type of oral impairment and disability, with 79% of American adults aged 50 years and older having lost one or more teeth and 11% being edentulous.80 Apart from the obvious functional, biologic, and psychosocial consequences, edentulism (partial and complete) is associated with substantial rehabilitation costs and affects quality of life. In terms of etiology, tooth loss is attributed predominantly to the two most common oral diseases, caries and periodontitis. Accurate estimates of the individual contributions of caries and periodontitis to tooth loss are lacking and are probably heterogeneous across populations and between study samples;58 these proportions are also probably affected by numerous factors, including age, diet, smoking, dental care, and others. Nevertheless, the two diseases share a common etiologic basis: they are associated with pathogenic shifts in the oral microbiome and their pathogenesis entails complex interactions of highly organized intra‐oral biofilms with host immunity and protective factors. As reviewed earlier, genome‐wide evidence regarding genetic risk loci in caries and periodontitis has emerged. However, little attention has been given to the examination of the joint effects of these diseases on the dentition. To address this knowledge gap, our group proposed a composite “tooth morbidity” index [the sum of decayed, missing due to all causes (ie, “total”), and filled surfaces; DMTFS], which captures the cumulative dental effects of caries and periodontitis (Figure 1) and has been interrogated in a genome‐wide association study context.81 Conceptually, this analysis can be considered analogous to a genome‐wide association study of all‐cause mortality.82
Figure 1.
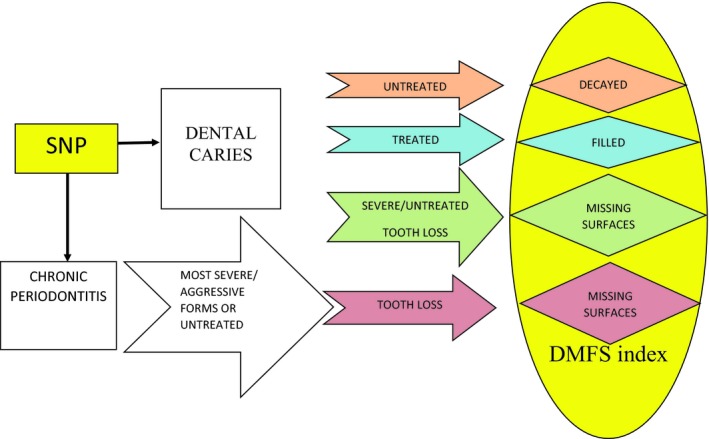
Theorized pathways contributing to the tooth morbidity (DMFS) index, emanating from dental caries and periodontitis. SNP, single nucleotide polymorphism
The genome‐wide association study was carried out on a sample of approximately 4500 European‐American participants (mean age = 62 years) of the Atherosclerosis Risk in Communities study.83 Genotyping was carried out using the Affymetrix 6.0 platform and imputation to 2.5 million markers was based on HapMap II‐CEU.27, 81 Dental examination data were used to construct the composite, tooth morbidity index (DMTFS), comprising decayed surfaces, missing surfaces (total, due to all causes), and filled surfaces (excluding third molars and range: 0‐128). The association between single nucleotide polymorphisms (minor allele frequency ≥ 5%) and DMTFS was estimated using linear regression models assuming additive genetic effects. Models were adjusted for age, sex, study examination center, and population stratification, and a conventional multiple‐testing correction (P < 5 × 10−8) was applied. Exploratory analyses included additional adjustment for smoking, diabetes, body mass index, and chronic periodontitis, and stratification by periodontitis diagnosis, ie, healthy/mild periodontitis, moderate periodontitis, and severe periodontitis.
The mean DMTFS of the sample was 69 (standard deviation = 26). Four loci showed genome‐wide statistically significant evidence of an association with tooth morbidity (Table 1): PMAIP1 (rs11664212; b = 3.65, P = 1.5 × 10−10), SPC25 (rs477309; b = 4.23, P = 2.7 × 10−9), MC4R (rs752720; b = 3.74, P = 3.1 × 10−9), and MPP7 (rs1262024; b = 5.80, P = 3.7 × 10−8). There was no evidence of heterogeneity (ie, effect measure modification) in periodontitis diagnosis‐stratified analyses. Of note, these associations persisted after adjustment for periodontitis diagnosis, smoking, diabetes, and body mass index (Table 2). These loci, with the exception of MPP7, also showed significant associations with the number of remaining natural teeth, but not with periodontitis diagnosis (Table 3), suggesting that dental caries is probably the driver of this association signal in this study population. Of these loci, PMAIP1/MC4R (Figure 2) is of particular interest, as it was subsequently reported with a significant genome‐wide signal for dental caries in the recent meta‐analysis of the Gene‐Lifestyle Interactions and Dental Endpoints consortium, which included over half a million individuals.31
Table 1.
Genome‐wide association analysis results of tooth morbidity (number of decayed, missing, or filled tooth surfaces) among the dental Atherosclerosis Risk in Communities study participants, overall and stratified by chronic periodontitis diagnosis. The table presents the lead single nucleotide polymorphism (ie, the marker with the lowest P value) for each of the four loci that met genome‐wide significance criteria (P < 5 × 10−8 and minor allele frequency ≥5%) in the study sample
| Chromsome | Locus | SNP | MAFa | Risk allele | Entire sample (n = 4398) | Chronic periodontitis diagnosisb | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Healthy (n = 1785) | Mild/moderate (n = 1862) | Severe (n = 751) | ||||||||||||||
| Beta | SE | P | Beta | SE | P | Beta | SE | P | Beta | SE | P | |||||
| 18 | PMAIP1 | rs11664212 | [G] 0.35 | [G] | 3.65 | 0.57 | 1.5 × 10−10 | 4.09 | 0.92 | 8.8 × 10−6 | 3.58 | 0.83 | 1.7 × 10−5 | 3.14 | 1.44 | 2.8 × 10−2 |
| 2 | SPC25 | rs477309 | [T] 0.14 | [C] | 4.23 | 0.71 | 2.7 × 10−9 | 5.61 | 1.14 | 8.1 × 10−7 | 3.03 | 1.08 | 4.8 × 10−3 | 4.08 | 1.71 | 1.6 × 10−2 |
| 18 | MC4R | rs752720 | [T] 0.46 | [C] | 3.74 | 0.63 | 3.1 × 10−9 | 3.69 | 1.00 | 2.2 × 10−4 | 3.90 | 0.95 | 4.0 × 10−5 | 3.79 | 1.59 | 1.6 × 10−2 |
| 10 | MPP7 | rs1262024 | [A] 0.08 | [A] | 5.80 | 1.05 | 3.7 × 10−8 | 6.18 | 1.64 | 1.6 × 10−4 | 4.01 | 1.63 | 1.3 × 10−2 | 8.50 | 2.58 | 9.2 × 10−4 |
Abbreviations: MAF, minor allele frequency; SE, standard error; SNP, single nucleotide polymorphism.
Based on HapMap II‐CEU.
All homogeneity X2 (test of between chronic periodontitis diagnosis strata heterogeneity) P > 0.2.
Table 2.
Genome‐wide association analysis results of tooth morbidity (number of decayed, missing, or filled tooth surfaces) among the dental Atherosclerosis Risk in Communities study participants, “unadjusted” and adjusted for smoking and diabetic status, body mass index, and periodontitis diagnosis. The table presents the lead single nucleotide polymorphism (ie, marker with the lowest P value) for each of the four loci that met genome‐wide significance criteria (P < 5 × 10−8 and minor allele frequency ≥5%) in the entire study sample
| Chromosome | Locus | SNP | MAFa | Risk allele | “Unadjusted” (n = 4398) | Fully adjusted modelsb | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chronic periodontitis diagnosis | Smoking, diabetes, body mass index | Smoking, diabetes, body mass index, and chronic periodontitis diagnosis | ||||||||||||||
| Beta | SE | P | Beta | SE | P | Beta | SE | P | Beta | SE | P | |||||
| 18 | PMAIP1 | rs11664212 | [G] 0.35 | [G] | 3.65 | 0.57 | 1.5 × 10−10 | 3.66 | 0.57 | 1.2 × 10−10 | 3.48 | 0.56 | 6.9 × 10−10 | 3.47 | 0.56 | 7.8 × 10−10 |
| 2 | SPC25 | rs477309 | [T] 0.14 | [C] | 4.23 | 0.71 | 2.7 × 10−9 | 4.25 | 0.71 | 2.3 × 10−9 | 4.29 | 0.70 | 1.0 × 10−9 | 4.22 | 0.70 | 2.0 × 10−9 |
| 18 | MC4R | rs752720 | [T] 0.46 | [C] | 3.74 | 0.63 | 3.1 × 10−9 | 3.75 | 0.63 | 2.8 × 10−9 | 3.48 | 0.63 | 2.7 × 10−8 | 3.50 | 0.63 | 2.2 × 10−8 |
| 10 | MPP7 | rs1262024 | [A] 0.08 | [A] | 5.80 | 1.05 | 3.7 × 10−8 | 5.81 | 1.05 | 3.5 × 10−8 | 5.96 | 1.04 | 1.1 × 10−8 | 5.81 | 1.04 | 2.6 × 10−8 |
Abbreviations: MAF, minor allele frequency; SE, standard error; SNP, single nucleotide polymorphism.
Based on HapMap II‐CEU.
All changes in estimate after adjustment <10%.
Table 3.
Association results of the four loci that were prioritized from the tooth morbidity genome‐wide association study with edentulousness and chronic periodontitis traits among the Atherosclerosis Risk in Communities study participants. P values were based on logistic regression models for the edentulous and chronic periodontitis traits, and a linear regression model for the number of remaining natural teeth
| Chromosome | Locus | SNP | Edentulous vs dentatea (n = 8103) | Number of natural teethb (0‐32) (n = 5538) | Moderate chronic periodontitis vs healthyc | Severe chronic periodontitis vs healthyc |
|---|---|---|---|---|---|---|
| 18 | PMAIP1 | rs11664212 | 0.18 | 0.00049 | 0.21 | 0.22 |
| 2 | SPC25 | rs477309 | 0.97 | 0.0087 | 0.26 | 0.06 |
| 18 | MC4R | rs752720 | 0.22 | 0.00082 | 0.53 | 0.17 |
| 10 | MPP7 | rs1262024 | 0.34 | 0.24 | 0.10 | 0.09 |
Abbreviation: SNP, single nucleotide polymorphism.
Based on dental screening results among the Atherosclerosis Risk in Communities study participants.
Based on a combination of dental screening and complete dental examinations among the Atherosclerosis Risk in Communities study participants.
Based on comprehensive periodontal examinations among the dental Atherosclerosis Risk in Communities study participants.
Figure 2.
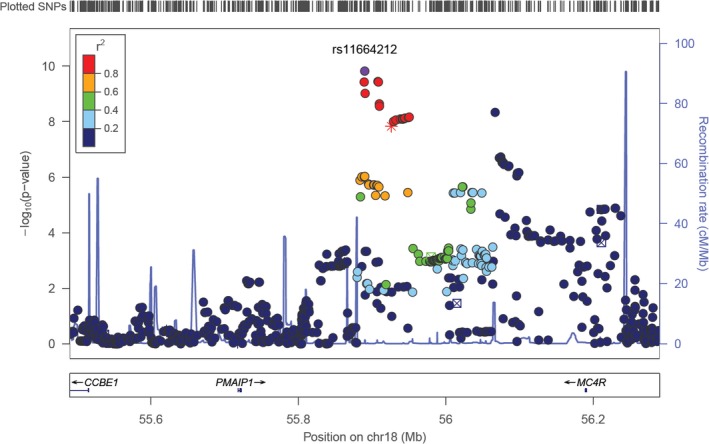
Regional association (Locus Zoom) plot of the PMAIP1/MC4R locus, which showed two independent association signals (rs11664212, minor allele frequency = 0.35, P = 1.5 × 10−10 and rs752720, minor allele frequency = 0.46, 3.1 × 10−9) in the genome‐wide association study of tooth morbidity in the dental Atherosclerosis Risk in Communities study. SNP, single nucleotide polymorphism
Key points: Composite measures of oral disease offer promising and perhaps efficient alternatives to conventional and individual measures of dental caries and periodontitis. A genome‐wide association study of tooth morbidity among a moderately sized sample of European‐Americans identified four significant genome‐wide signals; one of these loci (PMAIP1/MC4R) was subsequently replicated in a consortium meta‐analysis.
3.2. The oral microbiome and inflammatory mediators
Understanding the genomic basis of clinical traits and directly observable health and disease end points is of natural interest to both clinicians and investigators. In the context of common complex diseases, such as periodontitis and tooth morbidity, it is expected that several loci contribute to disease development.84 Different loci and environmental factors are also probably involved, driving the disease incidence among different population subgroups. Theoretically, the presumably weak association signals of these loci should be detectable in clinical traits if large sample sizes are available. Another, alternative and complementary, approach is the interrogation of “biologic proximate” disease traits that are more likely to demonstrate a direct biologic connection with genetic loci. This approach has been successfully implemented in psychiatric and neurologic traits,85 where the observed traits can be very heterogeneous; these intermediate traits are often referred to as “endophenotypes”. This approach is conceptually analogous to the genetic interrogation of serum lipids (the endophenotypes) in the context of genetic studies of cardiovascular disease (the clinical end point).86
The composition and the function of the oral microbiome in states of oral health and disease87 are obvious target endophenotypes for both periodontitis and dental caries/tooth morbidity. To date, no exploration of the genomic basis of the supragingival (ie, dental caries‐related) microbiome composition has been undertaken; however, some evidence in the context of periodontitis exists. This line of investigation has been termed “infectogenomics” by Nibali and colleagues,88, 89, 90 and several plausible candidates associated with subgingival pathogen colonization have emerged from candidate‐gene studies (not reviewed here). The only genome‐wide association study of periodontal pathogen colonization, carried out by our group in 2012,32 did not detect any significant genome‐wide association signals in single marker (single nucleotide polymorphism) analyses. However, two loci showed evidence of a statistically significant genome‐wide association in subsequent gene‐centric re‐analyses (KCNK1, P = 3.4 × 10−7 for high “red complex” colonization and DAB2IP, P = 1.0 × 10−6 for Porphyromonas gingivalis high colonization).45 Moreover, three single nucleotide polymorphisms initially prioritized (P < 5 × 10−6) by our genome‐wide association study of periodontitis (rs2521634; NPY locus)27 and periodontal pathogen colonization (rs10010758; TBC1D1 locus and rs10043775; FBXO30 locus)32 were subsequently found to be associated with periodontal pathogen colonization by a recent independent study by Cavalla et al.91 Specifically, the NPY locus was associated with Tannerella forsythia, Actinomyces gerencseriae, Fusobacterium periodonticum, and Prevotella nigrescens colonization, the TBC1D1 locus with P. gingivalis, and the FBXO30 locus with Prevotella intermedia, after adjustment for multiple testing. Of note, both the genome‐wide association study and the subsequent candidate‐polymorphism study were carried out using DNA‐DNA checkerboard for the characterization of the subgingival microbiota. One can reasonably anticipate improvements in this line of host genomics investigation via the utilization of microbial community‐wide assessments (eg, whole genome sequencing shotgun or metagenomics), as well as more “functional” approaches (eg, RNA sequencing or metatranscriptomics).
Several lines of investigation have examined the host response and inflammation in the context of periodontitis and the discovery of their genomic basis is of great interest. From a clinical standpoint, a severe gingival inflammation index has been interrogated in the genome‐wide association study context.92 Our group identified a significant genome‐wide association signal in the ASIC2 (formerly known as ACCN1) locus (lead marker: rs11652874; P = 3.9 × 10−8). However, this finding has not been replicated or mechanistically confirmed and should be treated with caution. From a more biologic standpoint, gingival crevicular fluid is an easily obtainable and highly informative biofluid that has been used as a marker of periodontal tissue inflammation and, thus, an endophenotype for periodontitis. The gingival crevicular fluid is a serum transudate that is modified by the local host response to the subgingival microbiome and it can serve as a biomarker of the microbial activation of the host's immune response. To date, gingival crevicular fluid interleukin‐1beta expression is the only periodontitis‐specific inflammatory endophenotype for which evidence of a genome‐wide association exists.93 Importantly, interleukin‐1beta has been established as a robust marker for severe inflammation, bone loss, and periodontal disease progression, and is known to be strongly genetically controlled. In brief, in a recent comprehensive genome‐wide association study and mechanistic follow‐up investigation, our group recently reported that variants in the IL37 locus (lead marker: rs3811046) strongly controlled gingival crevicular fluid interleukin‐1beta expression (P = 3.3 × 10−22) and were also associated with 10‐year incident tooth loss and aggressive periodontitis assessed in an independent cohort. This investigation also showed a previously undetected heterogeneity in the genetic control of gingival crevicular fluid interleukin‐1beta expression. Specifically, we found that the IL37 locus predominantly controlled (lead marker: rs3811046; P = 7.2 × 10−20) the high‐end of the distribution (eg, “top 10%” or profoundly hyper‐inflammatory vs bottom 50%) as opposed to the IL1B locus (lead marker: rs16944), which predominantly controlled (P = 3.2 × 10−8) mild elevations in interleukin‐1beta expression (eg, 50th‐75th percentile vs bottom 50%). This is a key finding in the search for the elements of the “hyper‐inflammatory trait”, as IL37, a member of the interleukin‐1 family of cytokines, is now being recognized as a natural suppressor of inflammatory and immune responses.94
Key points: Endophenotypes of oral diseases, mainly measures of the microbiome and the host response, are primary candidates for genomics studies in the context of periodontitis and dental caries/tooth morbidity. Although not always directly linked with a clinically measurable end point, these lines of investigation can uncover important biologic pathways that may otherwise be hard to detect via the study of heterogeneous clinical traits. Some genome‐wide evidence exists to support the genomic basis of both the microbiome and the host response in the context of oral health and disease.
3.3. Biologically informed, complex traits
A logical extension of the genomic interrogation of dental and periodontal endophenotypes is the combination of these biologic intermediates with clinical measures of health and disease, to create “biologically informed” complex traits. Therefore, our group recently combined clinical (ie, periodontal) and biologic (ie, subgingival periodontal pathogen colonization and gingival crevicular fluid interleukin‐1beta expression) data, using a principal components approach, to create six periodontal complex traits.95 This methodology has been used previously in genome‐wide association studies of complex facial morphology96 and bone traits.97
The six periodontal complex traits showed distinct and identifiable microbial and inflammatory profiles (eg, periodontal complex trait‐1 was characterized by a uniformly high pathogen load; periodontal complex trait‐3 showed high inflammatory and A. actinomycetemcomitans loading; and periodontal complex trait‐5 was dominated by P. gingivalis). Using these biologically informed, complex periodontal traits, we reported evidence of a genome‐wide association for 12 novel loci, specifically: periodontal complex trait‐1: CLEC19A, TRA, GGTA2P, TM9SF2, IFI16, and RBMS3; periodontal complex trait‐3: C1QTNF7 and TSNARE; periodontal complex trait‐4: HPVC1; and periodontal complex trait‐5: SLC15A4, PKP2, and SNRPN. Some additional follow‐up evidence on the role of IFI16/AIM2 variants in periodontitis and their association with inflammatory and microbiologic parameters has since been reported by Marchesan et al.98
The biologic enrichment of these complex traits as well as the endophenotypes reviewed in the previous sections has obvious advantages in the search for associated genomic loci, as the traits are more proximal to biologic processes that are controlled by the genome. At the same time, the study of these traits comes with several limitations. A major limitation is related to the impossibility or difficulty in replicating genome‐wide association signals for these traits, as, thus far, they tend to be unique to the study samples they are created in (ie, the Atherosclerosis Risk in Communities study is the only one to have analyzed gingival crevicular fluid‐interleukin‐1beta expression and the subgingival microbiome in a clinical periodontitis cohort). Other limitations, also relevant to the traditional clinical disease definitions, are related to measurement, ie, differences under which studies are carried out and measurements are taken, and the known influence of tooth loss on periodontitis ascertainment.
Key points: The study of biologically informed, complex traits (ie, combining clinical, microbial, and host‐response information) has been the most productive approach to date for detecting genome‐wide signals and promising candidates for further mechanistic studies in periodontitis. However, these complex traits are virtually impossible to replicate in independent samples and have been generated from relatively small sample sizes. For these reasons, these findings should be treated with caution.
3.4. “Precision” periodontal traits
To address the limitations imposed by the influence of tooth loss on disease measurement, the need to facilitate clinical data harmonization across studies and the opportunity to capitalize on all available (eg, tooth‐level) clinical information, our group recently embarked upon a novel, latent class analysis99 approach to derive a new classification system for periodontitis.59, 60, 100 In brief, the approach is analogous to an unsupervised clustering procedure, where individuals and teeth are placed within mutually exclusive categories – periodontal profile classes and tooth profile classes, respectively. Our group identified seven distinct periodontal profile classes and seven distinct tooth profile classes that aid in patient stratification,59 are predictive of periodontitis progression and tooth loss,60 and are arguably better‐suited for precision oral health applications than current conventional disease classifications.100, 101 A genome‐wide association study of the periodontal profile class classification has not yet been undertaken. Nevertheless, the new classification has resulted in phenotypes that are familiar to clinicians who recognize patterns of missing teeth, areas of recession, diminished periodontal support, and other aspects of the dentition. Similar efforts, albeit not based on latent class analysis, have been undertaken in the dental caries domain: Shaffer and colleagues102 reported the use of principal component and factor analysis approaches to derive heritable and nonheritable patterns of dental caries (ie, total disease burden, pit‐and‐fissure, and smooth surface decay) in the permanent dentition.
Key points: The identification and operationalization of classes of patients or disease variance vectors are aligned with the notion of precision oral health and offer advantages over traditional disease taxonomies. Some evidence exists in the dental caries domains that certain disease subtypes may be more heritable than others. Additional research in the periodontitis domain is needed to understand whether the periodontal profile class/tooth profile class classification can lead to novel genomics insights or discoveries.
4. POTENTIAL UTILITY OF GENOME‐WIDE ASSOCIATION STUDY FINDINGS FOR THE CONSTRUCTION OF A PERIODONTITIS GENETIC RISK SCORE
Our genome‐wide association study of periodontitis27 (as defined by the Centers for Disease Control and Prevention/American Academy of Periodontology classification criteria) demonstrated that considerable proportions of the phenotypic variance can be explained by genome‐wide association study single nucleotide polymorphisms. This proportion can be interpreted as heritability and was 0.22 (standard error = 0.19) for severe periodontitis. Interestingly, the heritable variance increased to 0.52 (standard error = 0.35) when a genome × smoking interaction term was considered. Naturally, once risk loci and specific risk markers have been discovered and confirmed for a disease or condition, questions regarding their predictive ability, and ultimately utility, emerge. However, virtually no work has been carried out in the oral health domain to examine the utility and feasibility of using genome‐wide association study findings in the construction of informative periodontitis or dental caries “predictive” and risk models. Admittedly, risk assessment and outcome prediction using genome‐wide association study single nucleotide polymorphisms are categorically challenging for complex diseases,103 which tend to be polygenic and with a strong environmental or behavioral risk component. Additionally, the oral health field has not yet reached a consensus on validated and replicated genetic risk markers for dental caries and periodontitis. In spite of this, our group sought to demonstrate the potential utility of genome‐wide association study findings for the development and evaluation of periodontitis “predictive” models using sets of demographic, behavioral, and genetic factors prioritized from agnostic genome‐wide association study scans.
We used linkage disequilibrium pruning to identify 658 independent genomic loci that had a minor allele frequency >5%, an imputation quality score >0.8, and were associated at P < 0.001 with severe periodontitis.104 A logistic regression model for severe chronic periodontitis prediction was saturated after the inclusion of the top 287 markers; area under the curve = 0.998, subjects correctly classified = 98.5%, positive predictive value = 98.1%. In contrast, a model for moderate chronic periodontitis required almost double the number of prioritized genome markers to reach saturation (n = 481). Models including fewer numbers of markers also showed reasonable performance. For example, for the top 40 loci (Table 4): area under the curve = 0.866, subjects correctly classified = 81.5%, positive predictive value = 73.5%; for the top 100 loci: area under the curve = 0.929, subjects correctly classified = 87.7%, positive predictive value = 82.8%. The classification performance of all of these models clearly outperformed models wherein simulated, uninformative single nucleotide polymorphisms were used instead of those of the genome‐wide association study.
Table 4.
The 40 most strongly associated (lowest P value in the discovery gemome‐wide association study of chronic periodontitis in the dental Atherosclerosis Risk in Communities study) loci with severe periodontitis, after linkage disequilibrium pruning. The top single nucleotide polymorphism in each locus is retained, with minor allele frequency >5% and imputation quality score >0.8
| No. | Chromosome | Single nucleotide polymorphism rs id | Minor allele frequency | Effect allele |
|---|---|---|---|---|
| 1 | 14 | rs12883458 | 0.104 | C |
| 2 | 3 | rs11925054 | 0.134 | G |
| 3 | 7 | rs2521634 | 0.246 | G |
| 4 | 1 | rs12073917 | 0.128 | G |
| 5 | 15 | rs2890313 | 0.217 | C |
| 6 | 7 | rs2106737 | 0.067 | C |
| 7 | 20 | rs271972 | 0.481 | A |
| 8 | 21 | rs7281463 | 0.414 | C |
| 9 | 5 | rs16889923 | 0.128 | A |
| 10 | 1 | rs11577771 | 0.127 | C |
| 11 | 7 | rs1688605 | 0.122 | G |
| 12 | 11 | rs10765844 | 0.478 | G |
| 13 | 4 | rs1534582 | 0.213 | G |
| 14 | 4 | rs17006135 | 0.071 | C |
| 15 | 11 | rs10790919 | 0.212 | A |
| 16 | 21 | rs2410204 | 0.406 | A |
| 17 | 7 | rs258920 | 0.145 | A |
| 18 | 20 | rs6029598 | 0.127 | C |
| 19 | 6 | rs7772901 | 0.213 | A |
| 20 | 17 | rs2871289 | 0.377 | C |
| 21 | 6 | rs12175557 | 0.077 | A |
| 22 | 6 | rs9791329 | 0.302 | A |
| 23 | 11 | rs1398282 | 0.451 | C |
| 24 | 6 | rs4485988 | 0.054 | C |
| 25 | 7 | rs4527765 | 0.139 | A |
| 26 | 2 | rs11695297 | 0.372 | A |
| 27 | 10 | rs4751326 | 0.324 | C |
| 28 | 4 | rs6814571 | 0.469 | C |
| 29 | 6 | rs2305089 | 0.485 | C |
| 30 | 12 | rs10771435 | 0.460 | C |
| 31 | 5 | rs10043322 | 0.290 | C |
| 32 | 18 | rs4800313 | 0.213 | T |
| 33 | 12 | rs12314141 | 0.110 | A |
| 34 | 7 | rs1541363 | 0.112 | T |
| 35 | 14 | rs8008037 | 0.331 | A |
| 36 | 11 | rs12364480 | 0.213 | T |
| 37 | 9 | rs373637 | 0.429 | C |
| 38 | 15 | rs4238336 | 0.231 | C |
| 39 | 16 | rs4270178 | 0.500 | A |
| 40 | 12 | rs10444531 | 0.236 | T |
We further examined the distribution and properties of the summary score of the “top 40” set of markers, ie, the genetic risk score‐40 (Figure 3, top panel). Its theoretical range is 0‐80; in our sample it was normally distributed with median = 37.1, mean = 37.1 (standard deviation = 3.9), range = 24.4‐51.7; there was no difference between sexes and no association with the age of the participants. As expected, the score was strongly positively associated with the model‐predicted probability of a participant having severe periodontitis (Figure 3, bottom panel). When categorized in deciles, the score also correlated well with periodontitis diagnoses in the entire dental Atherosclerosis Risk in Communities study sample (Figure 4).
Figure 3.
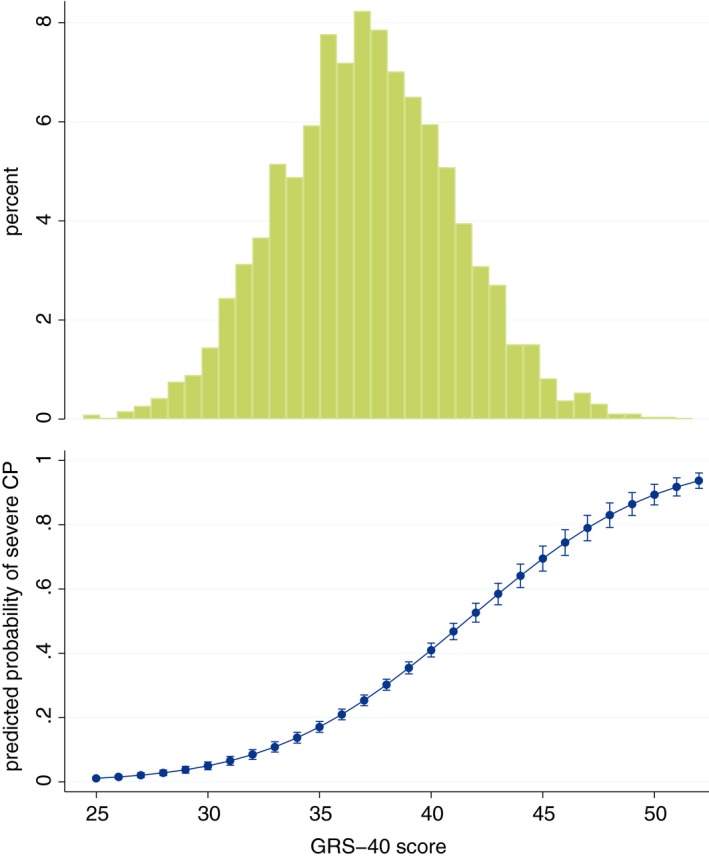
Distribution of the genetic risk score (GRS)‐40 index (top panel), together with the predicted probability (and 95% confidence intervals) of severe periodontitis (vs healthy/mild, according to the Centers for Disease Control and Prevention/American Academy of Periodontology classification) in the dental Atherosclerosis Risk in Communities study sample (bottom panel), derived from the genome‐wide association study of chronic periodontitis (CP)
Figure 4.
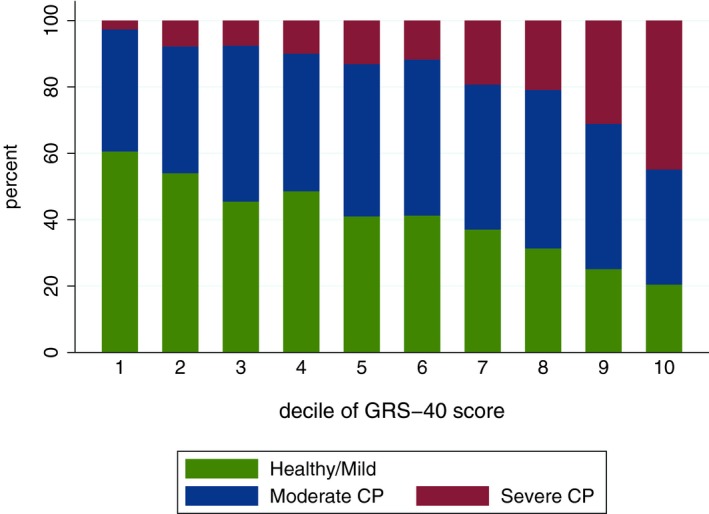
Distribution of periodontitis diagnoses (healthy/mild, moderate and severe chronic periodontitis, according to the Centers for Disease Control and Prevention/American Academy of Periodontology classification criteria) according to decile‐categories of the genetic risk score (GRS)‐40 index, derived from the genome‐wide association study of chronic periodontitis (CP)
We caution that the genetic risk score‐40 is by no means a validated, predictive, genetic score for periodontitis propensity, for several reasons. First, the genetic markers used were discovered in only one cohort and do not represent validated or replicated risk indicators. The markers and the risk score itself must be examined for replication in independent cohorts. Second, the discovery genome‐wide association study was conducted among a sample of middle‐age European‐Americans and for a specific clinically defined taxonomy of chronic periodontitis; both of these features severely limit the transferability of these findings to other populations and settings. Third, factors other than genomics are known to play important roles in the development of oral disease (eg, behavioral risk factors, socioeconomic environment, access to oral health care, to name a few). Therefore, although these indices (upon validation and replication) may provide useful information that is true “on average” and at the population level, they will not always work at the individual level. A more comprehensive approach, taking into consideration individual susceptibility, environment, and behaviors, and operationalizing disease subtypes aligned with the notion of precision medicine,20, 21, 101 will probably be effective. In spite of these limitations, the work presented here demonstrates that once a consensus set of validated and replicated genomic markers for periodontitis, dental caries, or tooth morbidity is developed, the genetic association signals can be efficiently combined to create aids for oral health and disease risk assessment and individual susceptibility estimation. In the future, this approach can ultimately be informative for screening, prevention, therapy, and the overall management of oral health and disease.
Key points: Genomics association information gained from genome‐wide association studies can be efficiently combined in risk models and indices that are predictive of health and disease case statuses. We are far from reaching a consensus with regard to specific genomic risk markers that are validated and replicated for their association with oral disease. However, once these markers have been discovered, validated, and replicated, genetic risk scores can be developed and will be useful in the context of precision oral health. This approach will probably be informative for the more severe disease categories, where genomics presumably plays a greater role.
5. SUMMARY AND CONCLUSIONS
Various groups have identified few loci associated with aggressive and severe forms of periodontal disease. However, most genome‐wide association studies of periodontitis have been based on moderate or small sample sizes and there is substantial heterogeneity in the studied populations, the methods used, and the results reported. In respect to dental caries, genome‐wide association evidence is still limited, but a recent, large‐scale, consortium meta‐analysis has identified many loci associated with dental caries, providing a rich resource of candidates to be followed‐up in subsequent investigations. Nevertheless, sizeable genotyped cohorts with high‐quality clinical data including detailed phenotypes on dental caries are warranted.
Biologically informed composite traits of oral diseases, including the microbiome and the host response, offer alternatives to conventional classifications of diseases that have traditionally measured the history of disease experience. This allows the identification of different biologic endophenotypes that may have overlapping clinical presentations. It has been the most productive approach to date to detect genome‐wide significant association signals and promising candidates for further mechanistic investigations in periodontitis. However, these complex traits have their own limitations and should be interpreted with caution.
The application of precision medicine in oral health has introduced the opportunity to use data‐driven and biologically informed phenotypes in the management of oral health and disease. On the research side, we expect that this will also facilitate clinical data harmonization across studies and will offer ample opportunities to maximize the usable clinical information.
We conclude that exciting opportunities lie ahead to improve the oral health of individual patients and populations via advances in our understanding of the genomic basis of oral health and disease. The pace of new discoveries and their equitable translation to practice will largely depend on investments in the education and training of the oral health care workforce, basic and population research, and sustained collaborative efforts.
ACKNOWLEDGEMENT
Cary Agler and Kimon Divaris are supported by NIH/NIDCR grant U01DE025046.
Morelli T, Agler CS, Divaris K. Genomics of periodontal disease and tooth morbidity. Periodontol 2000. 2020;82:143–156. 10.1111/prd.12320
REFERENCES
- 1. Kappes LO. Factors in the decay of teeth. Am J Dis Child. 1928;36(2):268‐276. [Google Scholar]
- 2. Schaefer AS, Richter GM, Nothnagel M, et al. A genome‐wide association study identifies GLT6D1 as a susceptibility locus for periodontitis. Hum Mol Genet. 2010;19(3):553‐562. [DOI] [PubMed] [Google Scholar]
- 3. Shaffer JR, Wang X, Feingold E, et al. Genome‐wide association scan for childhood caries implicates novel genes. J Dent Res. 2011;90(12):1457‐1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nibali L, Di Iorio A, Tu YK, Vieira AR. Host genetics role in the pathogenesis of periodontal disease and caries. J Clin Periodontol. 2017;44(Suppl 18):S52‐S78. [DOI] [PubMed] [Google Scholar]
- 5. Loos BG, Papantonopoulos G, Jepsen S, Laine ML. What is the contribution of genetics to periodontal risk? Dent Clin North Am. 2015;59(4):761‐780. [DOI] [PubMed] [Google Scholar]
- 6. Schaefer AS. Genetics of periodontitis: discovery, biology, and clinical impact. Periodontol 2000. 2018;78(1):162‐173. [DOI] [PubMed] [Google Scholar]
- 7. Vieira AR, Albandar JM. Role of genetic factors in the pathogenesis of aggressive periodontitis. Periodontol 2000. 2014;65(1):92‐106. [DOI] [PubMed] [Google Scholar]
- 8. Laine ML, Jepsen S, Loos BG. Progress in the identification of genetic factors in periodontitis. Curr Oral Health Rep. 2014;1(4):272-278. [Google Scholar]
- 9. Masumoto R, Kitagaki J, Fujihara C, et al. Identification of genetic risk factors of aggressive periodontitis using genome wide association studies in association with those of chronic periodontitis. J Periodontal Res. 2019;54(3):199-206. 10.1111/jre.12620 [DOI] [PubMed] [Google Scholar]
- 10. Beckmann JS, Antonarakis SE. Lessons from the genome‐wide association studies for complex multifactorial disorders and traits In: Speicher MR, Motulsky AG, Antonarakis SE, eds. Vogel and Motulsky's Human Genetics. Berlin: Springer; 2010:287-297. [Google Scholar]
- 11. Ioannidis JP, Tarone R, McLaughlin JK. The false‐positive to false‐negative ratio in epidemiologic studies. Epidemiology. 2011;22(4):450‐456. [DOI] [PubMed] [Google Scholar]
- 12. de Coo A, Quintela I, Blanco J, Diz P, Carracedo Á. Assessment of genotyping tools applied in genetic susceptibility studies of periodontal disease: a systematic review. Arch Oral Biol. 2018;92:38‐50. [DOI] [PubMed] [Google Scholar]
- 13. McCarthy S, Das S, Kretzschmar W, et al. Haplotype Reference Consortium. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279‐1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu YJ, Li Y, Auer PL, Lin DY. Integrative analysis of sequencing and array genotype data for discovering disease associations with rare mutations. Proc Natl Acad Sci USA. 2015;112(4):1019‐1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. ENCODE Project Consortium . An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rivas MA, Pirinen M, Conrad DF, et al. Human genomics. Effect of predicted protein‐truncating genetic variants on the human transcriptome. Science. 2015;348(6235):666‐669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248‐249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gamazon ER, Zhang W, Konkashbaev A, et al. SCAN: SNP and copy number annotation. Bioinformatics. 2010;26(2):259‐262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haeussler M, Zweig AS, Tyner C, et al. The UCSC genome browser database: 2019 update. Nucleic Acids Res. 2019;47(D1):D853-D858. 10.1093/nar/gky1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Divaris K. Fundamentals of precision medicine. Compend Contin Educ Dent. 2017;38(Suppl 8):30‐32. [PMC free article] [PubMed] [Google Scholar]
- 21. Kornman KS. Contemporary approaches for identifying individual risk for periodontitis. Periodontol 2000. 2018;78(1):12‐29. [DOI] [PubMed] [Google Scholar]
- 22. Kornman KS, Polverini PJ. Clinical application of genetics to guide prevention and treatment of oral diseases. Clin Genet. 2014;86(1):44‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sanders AE, Sofer T, Wong Q, et al. Chronic periodontitis genome‐wide association study in the Hispanic Community Health Study/Study of Latinos. J Dent Res. 2017;96(1):64‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bevilacqua L, Navarra CO, Pirastu N, Lenarda RD, Gasparini P, Robino A. A genome‐wide association study identifies an association between variants in EFCAB4B gene and periodontal disease in an Italian isolated population. J Periodontal Res. 2018;53(6):992‐998. [DOI] [PubMed] [Google Scholar]
- 25. Hong KW, Shin MS, Ahn YB, Lee HJ, Kim HD. Genomewide association study on chronic periodontitis in Korean population: results from the Yangpyeong health cohort. J Clin Periodontol. 2015;42(8):703‐710. [DOI] [PubMed] [Google Scholar]
- 26. Shimizu S, Momozawa Y, Takahashi A, et al. A genome‐wide association study of periodontitis in a Japanese population. J Dent Res. 2015;94(4):555‐561. [DOI] [PubMed] [Google Scholar]
- 27. Divaris K, Monda KL, North KE, et al. Exploring the genetic basis of chronic periodontitis: a genome‐wide association study. Hum Mol Genet. 2013;22(11):2312‐2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Feng P, Wang X, Casado PL, et al. Genome wide association scan for chronic periodontitis implicates novel locus. BMC Oral Health. 2014;14:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Teumer A, Holtfreter B, Völker U, et al. Genome‐wide association study of chronic periodontitis in a general German population. J Clin Periodontol. 2013;40(11):977‐985. [DOI] [PubMed] [Google Scholar]
- 30. Munz M, Willenborg C, Richter GM, et al. A genome‐wide association study identifies nucleotide variants at SIGLEC5 and DEFA1A3 as risk loci for periodontitis. Hum Mol Genet. 2018;27(5):941‐942. [DOI] [PubMed] [Google Scholar]
- 31. Shungin D, Haworth S, Divaris K, et al. Genome-wide analysis of dental caries and periodontitis combining clinical and self-reported data. Nat Commun. 2019;10(1):2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Divaris K, Monda KL, North KE, et al. Genome‐wide association study of periodontal pathogen colonization. J Dent Res. 2012;91(7 Suppl):21S‐28S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Freitag‐Wolf S, Dommisch H, Graetz C, et al. Genome‐wide exploration identifies sex‐specific genetic effects of alleles upstream NPY to increase the risk of severe periodontitis in men. J Clin Periodontol. 2014;41(12):1115‐1121. [DOI] [PubMed] [Google Scholar]
- 34. Schaefer AS, Bochenek G, Manke T, et al. Validation of reported genetic risk factors for periodontitis in a large‐scale replication study. J Clin Periodontol. 2013;40(6):563‐572. [DOI] [PubMed] [Google Scholar]
- 35. Schaefer AS, Richter GM, Groessner‐Schreiber B, et al. Identification of a shared genetic susceptibility locus for coronary heart disease and periodontitis. PLoS Genet. 2009;5(2):e1000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hubberten M, Bochenek G, Chen H, et al. Linear isoforms of the long noncoding RNA CDKN2B‐AS1 regulate the c‐myc‐enhancer binding factor RBMS1. Eur J Hum Genet. 2019;27(1):80‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aarabi G, Zeller T, Seedorf H, et al. Genetic susceptibility contributing to periodontal and cardiovascular disease. J Dent Res. 2017;96(6):610‐617. [DOI] [PubMed] [Google Scholar]
- 38. Ernst FD, Uhr K, Teumer A, et al. Replication of the association of chromosomal region 9p21.3 with generalized aggressive periodontitis (gAgP) using an independent case‐control cohort. BMC Med Genet. 2010;11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bochenek G, Häsler R, El Mokhtari NE, et al. The large non‐coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Hum Mol Genet. 2013;22(22):4516‐4527. [DOI] [PubMed] [Google Scholar]
- 40. Shusterman A, Munz M, Richter G, et al. The PF4/PPBP/CXCL5 gene cluster is associated with periodontitis. J Dent Res. 2017;96(8):945‐952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shaffer JR, Polk DE, Wang X, et al. Genome‐wide association study of periodontal health measured by probing depth in adults ages 18‐49 years. G3. 2014;4(2):307‐314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schaefer AS, Bochenek G, Jochens A, et al. Genetic evidence for plasminogen as a shared genetic risk factor of coronary artery disease and periodontitis. Circ Cardiovasc Genet. 2015;8(1):159‐167. [DOI] [PubMed] [Google Scholar]
- 43. Munz M, Chen H, Jockel‐Schneider Y, et al. A haplotype block downstream of plasminogen is associated with chronic and aggressive periodontitis. J Clin Periodontol. 2017;44(10):962‐970. [DOI] [PubMed] [Google Scholar]
- 44. Munz M, Richter GM, Loos BG, et al. Meta‐analysis of genome‐wide association studies of aggressive and chronic periodontitis identifies two novel risk loci. Eur J Hum Genet. 2019;27(1):102‐113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. . Munz M , Richter GM, Loos BG, et al. Genome‐wide association meta‐analysis of coronary artery disease and periodontitis reveals a novel shared risk locus. Sci Rep. 2018;8(1):13678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rhodin K, Divaris K, North KE, et al. Chronic periodontitis genome‐wide association studies: gene‐centric and gene set enrichment analyses. J Dent Res. 2014;93(9):882‐890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hiyari S, Green E, Pan C, et al. Genome‐wide association study identifies Cxcl family members as partial mediators of LPS‐induced periodontitis. J Bone Miner Res. 2018;33(8):1450‐1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kasbohm E, Holtfreter B, Völker U, et al. Exome variant analysis of chronic periodontitis in 2 large cohort studies. J Dent Res. 2017;96(1):73‐80. [DOI] [PubMed] [Google Scholar]
- 49. Kitagaki J, Miyauchi S, Asano Y, et al. A putative association of a single nucleotide polymorphism in GPR126 with aggressive periodontitis in a Japanese population. PLoS One. 2016;11(8):e0160765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. . Sudo T, Okada Y, Ozaki K, et al. Association of NOD2 mutations with aggressive periodontitis. J Dent Res. 2017;96(10):1100‐1105. [DOI] [PubMed] [Google Scholar]
- 51. Kebschull M, Papapanou PN. Exploring genome‐wide expression profiles using machine learning techniques In: Seymour G, Cullinan M, Heng N, eds. Oral Biology. Methods in Molecular Biology, Vol. 1537. New York, NY: Humana Press; 2017:347‐364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kebschull M, Hülsmann C, Hoffmann P, Papapanou PN. Genome‐wide analysis of periodontal and peri‐implant cells and tissues In: Seymour G, Cullinan M, Heng N, eds. Oral Biology. Methods in Molecular Biology, Vol. 1537. New York, NY: Humana Press; 2017:307‐326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kebschull M, Demmer RT, Grün B, Guarnieri P, Pavlidis P, Papapanou PN. Gingival tissue transcriptomes identify distinct periodontitis phenotypes. J Dent Res. 2014;93(5):459‐468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sawle AD, Kebschull M, Demmer RT, Papapanou PN. Identification of master regulator genes in human periodontitis. J Dent Res. 2016;95(9):1010‐1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Razzouk S. Regulatory elements and genetic variations in periodontal diseases. Arch Oral Biol. 2016;72:106‐115. [DOI] [PubMed] [Google Scholar]
- 56. Shungin D, Cornelis MC, Divaris K, et al. Using genetics to test the causal relationship of total adiposity and periodontitis: Mendelian randomization analyses in the Gene‐Lifestyle Interactions and Dental Endpoints (GLIDE) Consortium. Int J Epidemiol. 2015;44(2):638‐650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Papapanou PN, Sanz M, Buduneli N, et al. Periodontitis: consensus report of workgroup 2 of the 2017 world workshop on the classification of periodontal and peri‐implant diseases and conditions. J Periodontol. 2018;89(Suppl 1):S173‐S182. [DOI] [PubMed] [Google Scholar]
- 58. Haworth S, Shungin D, Kwak SY, et al. Tooth loss is a complex measure of oral disease: determinants and methodological considerations. Community Dent Oral Epidemiol. 2018;46(6):555‐562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Morelli T, Moss KL, Beck J, et al. Derivation and validation of the periodontal and tooth profile classification system for patient stratification. J Periodontol. 2017;88(2):153‐165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Morelli T, Moss KL, Preisser JS, et al. Periodontal profile classes predict periodontal disease progression and tooth loss. J Periodontol. 2018;89(2):148‐156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang X, Shaffer JR, Zeng Z, et al. Genome‐wide association scan of dental caries in the permanent dentition. BMC Oral Health. 2012;12:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Morrison J, Laurie CC, Marazita ML, et al. Genome‐wide association study of dental caries in the Hispanic Communities Health Study/Study of Latinos (HCHS/SOL). Hum Mol Genet. 2016;25(4):807‐816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ballantine JL, Carlson JC, Ferreira Zandoná AG, et al. Exploring the genomic basis of early childhood caries: a pilot study. Int J Paediatr Dent. 2018;28(2):217‐225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Haworth S, Shungin D, van der Tas JT, et al. Consortium‐based genome‐wide meta‐analysis for childhood dental caries traits. Hum Mol Genet. 2018;27(17):3113‐3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Divaris K. Precision dentistry in early childhood: the central role of genomics. Dent Clin North Am. 2017;61(3):619‐625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shaffer JR, Feingold E, Wang X, et al. GWAS of dental caries patterns in the permanent dentition. J Dent Res. 2013;92(1):38‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zeng Z, Feingold E, Wang X, et al. Genome‐wide association study of primary dentition pit‐and‐fissure and smooth surface caries. Caries Res. 2014;48(4):330‐338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zeng Z, Shaffer JR, Wang X, et al. Genome‐wide association studies of pit‐and‐fissure‐ and smooth‐surface caries in permanent dentition. J Dent Res. 2013;92(5):432‐437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lewis DD, Shaffer JR, Feingold E, et al. Genetic association of MMP10, MMP14, and MMP16 with dental caries. Int J Dent. 2017;2017:8465125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Eckert S, Feingold E, Cooper M, et al. Variants on chromosome 4q21 near PKD2 and SIBLINGs are associated with dental caries. J Hum Genet. 2017;62(4):491‐496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Stanley BO, Feingold E, Cooper M, et al. Genetic association of MPPED2 and ACTN2 with dental caries. J Dent Res. 2014;93(7):626‐632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Briseño‐Ruiz J, Shimizu T, Deeley K, et al. Role of TRAV locus in low caries experience. Hum Genet. 2013;132(9):1015‐1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang Q, Jia P, Cuenco KT, et al. Association signals unveiled by a comprehensive gene set enrichment analysis of dental caries genome‐wide association studies. PLoS One. 2013;8(8):e72653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. . Divaris K. Predicting dental caries outcomes in children: a “risky” concept. J Dent Res. 2016;95(3):248‐254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. . Page RC , Eke PI. Case definitions for use in population‐based surveillance of periodontitis. J Periodontol. 2007;78(7 Suppl):1387‐1399. [DOI] [PubMed] [Google Scholar]
- 76. Eke PI, Page RC, Wei L, Thornton‐Evans G, Genco RJ. Update of the case definitions for population‐based surveillance of periodontitis. J Periodontol. 2012;83(12):1449‐1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pitts N. “ICDAS”–an international system for caries detection and assessment being developed to facilitate caries epidemiology, research and appropriate clinical management. Community Dent Health. 2004;21(3):193‐198. [PubMed] [Google Scholar]
- 78. Pitts NB, Ekstrand KR, ICDAS Foundation . International Caries Detection and Assessment System (ICDAS) and its International Caries Classification and Management System (ICCMS) ‐ methods for staging of the caries process and enabling dentists to manage caries. Community Dent Oral Epidemiol. 2013;41(1):e41‐e52. [DOI] [PubMed] [Google Scholar]
- 79. Chapple IL, Bouchard P, Cagetti MG, et al. Interaction of lifestyle, behaviour or systemic diseases with dental caries and periodontal diseases: consensus report of group 2 of the joint EFP/ORCA workshop on the boundaries between caries and periodontal diseases. J Clin Periodontol. 2017;44(Suppl 18):S39‐S51. [DOI] [PubMed] [Google Scholar]
- 80. Dye BA, Weatherspoon DJ, Lopez Mitnik G. Tooth loss among older adults according to poverty status in the United States from 1999 through 2004 and 2009 through 2014. J Am Dent Assoc. 2019;150(1):9‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Divaris K, North KE, Slade GD, et al. Genome‐wide association study of tooth morbidity. J Dent Res. 2014;92(Spec Iss B):190702(IADR/AMER). [Google Scholar]
- 82. Walter S, Atzmon G, Demerath EW, et al. A genome‐wide association study of aging. Neurobiol Aging. 2011;32(11):2109.e15‐2109.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. The ARIC Investigators . The Atherosclerosis Risk in Communities (ARIC) study: design and objectives. Am J Epidemiol. 1989;129(4):687‐702. [PubMed] [Google Scholar]
- 84. Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747‐753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cannon TD, Keller MC. Endophenotypes in the genetic analyses of mental disorders. Annu Rev Clin Psychol. 2006;2:267‐290. [DOI] [PubMed] [Google Scholar]
- 86. Hoffmann TJ, Theusch E, Haldar T, et al. A large electronic‐health‐record‐based genome‐wide study of serum lipids. Nat Genet. 2018;50(3):401‐413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2013;69(1):137‐143. [DOI] [PubMed] [Google Scholar]
- 88. Nibali L, Di Iorio A, Onabolu O, Lin GH. Periodontal infectogenomics: systematic review of associations between host genetic variants and subgingival microbial detection. J Clin Periodontol. 2016;43(11):889‐900. [DOI] [PubMed] [Google Scholar]
- 89. Nibali L, Donos N, Henderson B. Periodontal infectogenomics. J Med Microb. 2009;58:1269‐1274. [DOI] [PubMed] [Google Scholar]
- 90. Nibali L, Tonetti MS, Ready D, et al. Interleukin‐6 polymorphisms are associated with pathogenic bacteria in subjects with periodontitis. J Periodontol. 2008;79:677‐683. [DOI] [PubMed] [Google Scholar]
- 91. Cavalla F, Biguetti CC, Melchiades JL, et al. Genetic association with subgingival bacterial colonization in chronic periodontitis. Genes. 2018;9(6):271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhang S, Divaris K, Moss K, et al. The novel ASIC2 locus is associated with severe gingival inflammation. JDR Clin Trans Res. 2016;1(2):163‐170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Offenbacher S, Jiao Y, Kim SJ, et al. GWAS for interleukin‐1β levels in gingival crevicular fluid identifies IL37 variants in periodontal inflammation. Nat Commun. 2018;9(1):3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cavalli G, Dinarello CA. Suppression of inflammation and acquired immunity by IL‐37. Immunol Rev. 2018;281(1):179‐190. [DOI] [PubMed] [Google Scholar]
- 95. Offenbacher S, Divaris K, Barros SP, et al. Genome‐wide association study of biologically informed periodontal complex traits offers novel insights into the genetic basis of periodontal disease. Hum Mol Genet. 2016;25(10):2113‐2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Liu F, van der Lijn F, Schurmann C, et al. A genome‐wide association study identifies five loci influencing facial morphology in Europeans. PLoS Genet. 2012;8(9):e1002932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Karasik D, Cupples LA, Hannan MT, Kiel DP. Genome screen for a combined bone phenotype using principal component analysis: the Framingham study. Bone. 2004;34(3):547‐556. [DOI] [PubMed] [Google Scholar]
- 98. Marchesan JT, Jiao Y, Moss K, et al. Common polymorphisms in IFI16 and AIM2 genes are associated with periodontal disease. J Periodontol. 2017;88(7):663‐672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lanza ST, Rhoades BL. Latent class analysis: an alternative perspective on subgroup analysis in prevention and treatment. Prev Sci. 2013;14(2):157‐168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Beck JD, Moss KL, Morelli T, Offenbacher S. In search of appropriate measures of periodontal status: the periodontal profile phenotype (P3) system. J Periodontol. 2018;89(2):166‐175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Beck JD, Philips K, Moss K, Divaris K, Morelli T, Offenbacher S. Advances in precision oral health. Periodontol 2000. 2020;82(1):266‐283. [DOI] [PubMed] [Google Scholar]
- 102. Shaffer JR, Feingold E, Wang X, et al. Heritable patterns of tooth decay in the permanent dentition: principal components and factor analyses. BMC Oral Health. 2012;12:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet. 2013;14(7):507‐515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Divaris K, North KE, Slade GD, et al. Genome‐wide association‐based chronic periodontitis risk modeling: development and initial evaluation. J Dent Res. 2015;94(Spec Iss A):0440(IADR/AADR). [Google Scholar]


