Summary
Recognition of non‐self structures on donor cells represents the main immunological barrier in solid organ transplantation. The human leukocyte antigens (HLA) are considered the most important non‐self (allo)antigens in transplantation. Long‐term graft attrition is mainly caused by the formation of alloreactive antibodies that are directed against non‐self structures (i.e., epitopes) on cell surface proteins. Recently published data provided evidence for a similar importance of non‐HLA mismatches between donors and recipients in acute rejection as well as long‐term kidney allograft survival. These data suggest a broader concept of immunological non‐self that goes beyond HLA incompatibility and expands the current concept of polymorphic non‐self epitopes on cell surface molecules from HLA to non‐HLA targets. Amino acid substitutions caused by single nucleotide variants in protein‐coding genes or complete loss of gene expression represent the basis for polymorphic residues in both HLA and non‐HLA molecules. To better understand these novel insights in non‐HLA alloimmunity, we will first review basic principles of the alloimmune response with a focus on the HLA epitope concept in donor‐specific antibody formation before discussing key publications on non‐HLA antibodies.
Keywords: donor‐specific antibodies, epitopes, genome‐wide incompatibility, non‐HLA Alloimmunity
Introduction
Recognition of non‐self structures on donor cells by the recipient’s adaptive immune system represents the main immunological barrier in solid organ transplantation. The human leukocyte antigens (HLA) encoded in the major histocompatibility complex (MHC) on the short arm of chromosome 6 are considered the most important alloantigens in transplantation 1. Quantification of serotype level HLA mismatch forms the basis of immunological graft allocation, and testing for anti‐HLA antibodies that are directed against the donor HLA types (HLA‐DSA) has been implemented into clinical routine 2. High‐resolution molecular typing of the MHC (in human HLA) region has identified an increasing number of HLA alleles over the last decade 3, 4.
In contrast to the MHC, so‐called minor histocompatibility antigens (mHA) include all proteins that are mismatched between donors and recipient and that are sufficiently antigenic to introduce a directed immune response against the non‐self antigen following transplantation 5. The importance of these mHAs in solid organ transplantation remains unresolved despite epidemiological data that suggested a significant contribution to long‐term graft survival. Single‐antigen approaches or a candidate set of previously identified mHA mismatches have not shown an impact on graft outcome following kidney transplantation 6. In HLA‐matched hematopoietic stem cell transplantation, however, the relevance of mHAs for graft‐versus‐host disease or graft‐versus‐leukemia effect is well established 7. On the genetic level, mHAs are caused by single nucleotide polymorphisms that result in an altered primary structure of proteins rendering them accessible to allorecognition. Alloantibodies against mHAs are mainly referred to as non‐HLA antibodies in the transplant literature.
More recently, an unexpectedly high number of genome‐wide genetic polymorphisms between unrelated individuals were identified with several thousand so‐called non‐synonymous genetic variants causing altered amino acid sequence in proteins 8. This high number of individual level genetic polymorphism opens a new approach for immunological non‐self definition.
Advances in the understanding of the pathophysiology of alloimmune graft injury have uncovered the molecular structures at the interaction site of alloantibodies and antigens: Polymorphic residues on cell surface molecules including both HLA (MHC) and non‐HLA antigens (mHA) represent B cell epitopes that are recognized by alloantibodies. Recent publications suggest that a quantitative approach that accounts for these polymorphic residues in both HLA molecules and non‐HLA transmembrane/extracellular proteins is strongly associated with the development of donor‐specific antibodies and reduced long‐term kidney allograft survival 9, 10.
Excellent reviews on the epitope concept in HLA alloimmunity and non‐HLA‐antibodies in kidney transplantation have been published before 11, 12, 13, 14, 15, 16. This review will focus on “non‐self” as it is seen by the antibody (i.e., B cell epitopes) and the emerging concept of genome‐wide genetic incompatibility as basis for a systematic approach to account for incompatibilities in mHAs. For a better understanding and to put these new data into context, we will sum up the current understanding of alloimmunity in general with a focus on indirect allorecognition. We will then revisit the concept of non‐self B cell epitopes in HLA molecules (e.g., eplet mismatch) as it pertains to HLA‐DSA formation before reviewing key publications on non‐HLA alloantibodies and discussing the general difficulties in differentiating non‐HLA alloreactivity from autoreactivity that is caused by loss of self‐tolerance 17.
A brief summary of the current understanding of alloimmunity
HLA matching as key determinant of graft patency
Early in the history of transplantation, the importance of HLA mismatch as predominant determinant of histocompatibility was identified 18. Donor and recipient matching on HLA serotype level became feasible through large national and international organ sharing efforts providing a large pool of donors. Epidemiological data confirmed a reduction of acute rejection episodes and longer kidney allograft survival in better matched individuals 19.
However, the importance of HLA matching on kidney graft survival was reduced by the introduction of highly effective immunosuppressive regimen in combination with induction therapy that largely interferes with T cell activation 20. This led to a significant reduction in acute rejection episodes and improved one‐year renal allograft survival. Nevertheless, recent analyses confirmed that HLA serotype level mismatch remains associated with both acute rejection episodes and graft survival in large cohorts 21.
Direct allorecognition
Together with the graft donor antigen‐presenting cells (APCs) are introduced into the recipient. In direct allorecognition intact non‐self HLA on these donor APCs interact with the T cell receptors (TCR) on recipient T cells. (Fig. 1a) 22. Allorecognition in this way results in a strong immune response due to the high precursor frequency of directly alloreactive T cells ranging between 1% and 10% of all circulating T cells 23, 24. Direct allorecognition is predominantly important for acute T cell‐mediated rejection early after transplantation. The specific properties of direct allorecognition depending on TCR interaction with non‐self HLA explain the importance of the HLA antigen mismatch in the setting of acute rejection. However, it is not fully understood if the non‐self HLA molecule itself serves as the ligand for host alloreactive T cells or if a different peptide repertoire that is presented in the peptide‐binding groove of the non‐self HLA molecule is decisive. In the latter case, one must keep in mind that differences in peptide restriction of HLA molecules can give rise to a vastly different donor peptide repertoire.
Figure 1.
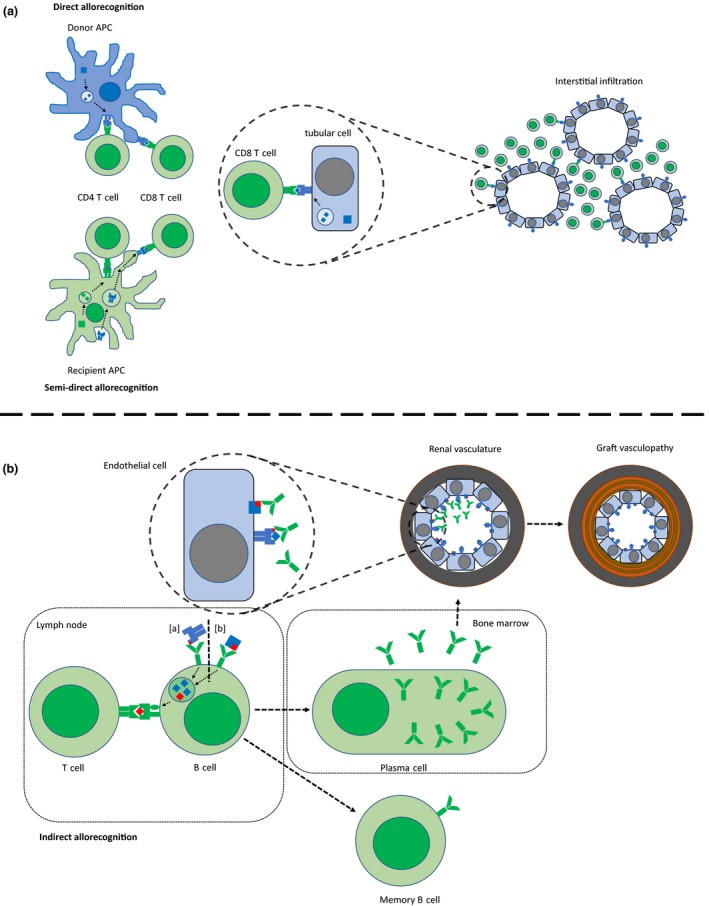
Direct and indirect allorecognition. (a) Directly alloreactive recipient T cells recognize intact donor HLA on donor APCs. The semi‐direct pathway enables direct allorecognition beyond the early post‐transplant period when recipient APCs acquire donor HLA and present it on the cell surface. Cytotoxic CD8+ T cells recognize donor HLA on graft tubular cells. The predominant histopathological finding of acute cellular rejection is tissue infiltration by T cells. (b) Indirectly alloreactive CD4+ recipient T cells recognize non‐self peptides (from [a] HLA or [b] non‐HLA antigens) presented in the peptide‐binding groove of self HLA class II molecules on the recipient’s B cells once the antigen has been internalized following binding of the antigen by the B cell receptor. T cell help is required for B cell differentiation. Plasma cells in the bone marrow produce alloantibodies against HLA and non‐HLA antigens that cause endothelial cell injury (microvascular inflammation) leading to acute humoral rejection and chronic allograft injury.
Indirect allorecognition
In contrast, alloantibody formation requires the full cascade of immune activation including T cell help through indirect allorecognition 22. In indirect allorecognition, TCR recognize allopeptides presented by self‐MHC class II molecules on professional antigen‐presenting cells (APCs), for example, B cells, macrophages, dendritic cells, or monocytes. Indirect allorecognition includes not only peptide fragments from alloreactive HLA molecules but virtually all polymorphic and mismatched proteins. Alloantigens that are shed from the transplanted organ are internalized by recipient APCs and processed in the same way as exogenous antigens before being presented as peptides in self‐MHC class II molecules to CD4+ T cells (Fig. 1b). The frequency of indirectly reactive T cells is lower compared to directly alloreactive ones, and they presumably are of importance later after transplantation.
Focus on humoral alloimmune responses
In contrast to the significant reduction in acute rejection episodes and improved one‐year graft survival in kidney transplantation, current immunosuppressive strategies turned out to be less effective in improving long‐term graft survival. Approximately half of the transplanted kidneys are lost over a period between 10 and 15 years 25. After the first year, the annual attrition rate of transplanted kidneys is about three to five percent per year and this number remained stable over the last 20 years 26. An ongoing alloimmune response in the graft leading to chronic inflammation and fibrosis has been proposed as main driver for late allograft loss. 27. This response is mainly mediated by the development of donor‐specific antibodies directed against polymorphic donor antigens as mediators of chronic graft rejection 28, 29, 30, 31, 32.
HLA as B cell epitope
Complexity of the HLA system
Through HLA sequencing, over 15 000 different HLA alleles have been identified to date. HLA nomenclature of serotypes was historically based on serological reactivity patterns and allele groups which do not reflect differences in immunogenicity of mismatches in the transplant setting 3. High‐resolution HLA genotypes provide the amino acid sequences of the different HLA alleles and allow for the identification of mismatches on a molecular level 11. Not all identified amino acid polymorphisms show the same level of immunogenicity and have a different impact on the immune response in the recipient. Some amino acid substitutions change the physico‐chemical properties of the protein (e.g., charge, hydrophobicity), and others may have only minor impact on the molecular structure.
B cell epitopes
HLA‐DSA bind to polymorphic residues on the surface of the HLA molecule. Each HLA molecule contains multiple of these antibody/B cell epitopes that define a unique pattern of non‐self surface structures. Individual epitopes are shared among different HLA alleles (Fig. 2) 11. The number of different non‐self epitopes shows high variability within the same level of antigen mismatch (e.g., HLA‐A, HLA‐B, HLA‐DR mismatch of 3 can represent an epitope mismatch of 10‐50) 16. The complexity that is introduced by the increasing number of genetically distinct HLA alleles that are recognized makes complete matching on the HLA antigen level impossible. The focus on a limited number of defined polymorphic residues proven to be targets of HLA antibodies may help to reduce complexity and improve matching of kidney allograft donors and recipients to prevent both (i) development of de novo donor specific antibodies (dnDSA) and (ii) identify acceptable mismatches in patients with already established donor specific antibody (DSA) 33.
Figure 2.
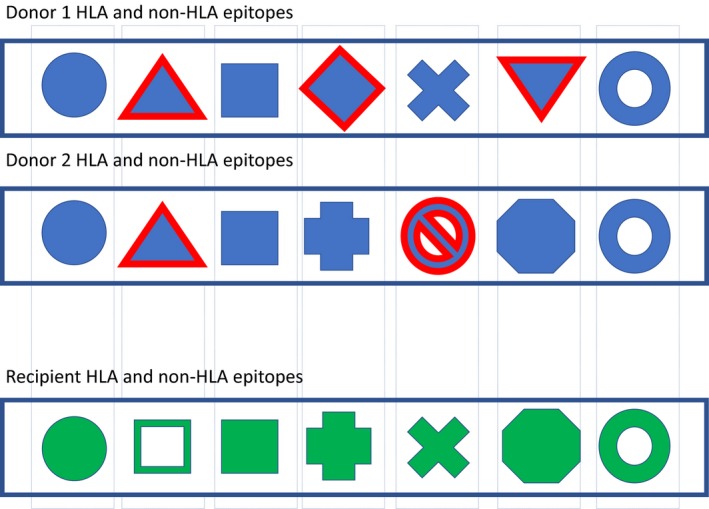
Each individual carries a unique set of polymorphic residues on HLA molecules as well as other endothelial antigens (e.g., non‐synonymous single‐nucleotide polymorphisms in transmembrane proteins, loss of function variants) representing B cell epitopes. Each donor–recipient pair is mismatched for different epitopes. A quantitative approach suggests that a higher number of mismatches predispose for alloantibody development.
The first characterization of these epitopes was performed computationally (based on calculated amino acid mismatch and later developed into the eplet concept) as well as experimentally (Terasaki epitopes) 34, 35 with both approaches showing a high correlation 36. Cross‐reactivity of antibodies against different HLA alleles was already observed in the days of serological characterization of HLA antigens: Different HLA alleles exhibiting shared antigenic determinants were combined into CREGs (Cross REactive Groups). Nowadays, these can be better characterized by individual epitopes 37.
Eplets
A quantitative concept of B cell epitope mismatch was developed to assess immunological risk 38. These scores show superiority in terms of prediction of development of dnDSA and antibody‐mediated rejection (ABMR) compared to the HLA antigen mismatch. The idea was first introduced by Duquesnoy et al. 39 who determined triplets based on amino acid polymorphism in the HLA molecules that were mismatched between donors and recipients. This was further developed into the eplet concept including linear and conformational non‐self residues on the surface of the HLA molecule based on longer sequences and stereochemical 3‐D modeling of crystalized antibody/antigen complexes 35, 40. Eplets represent the central part of an epitope and consist of clusters of a few amino acids that are close to each other (stretching over 3–3.5 angstrom [0.3–0.35nm]) and are located on the antibody‐accessible site of the HLA molecule. They represent the smallest functional unit of the antibody–antigen binding site (epitope) and determine the antibody specificity through interaction with the central complementarity‐determining region of the antibody paratope, whereas the epitope covers the complete antigen/antibody interface and has a size of 15 angstrom (1.5 nm). Besides the eplet representing the amino acids that determine antibody specificity (functional epitope), the entire B cell epitope also includes other (either polymorphic or non‐polymorphic) residues that determine affinity (binding strength) but not specificity of the antigen/antibody interaction (structural epitope) (Fig. 3a).
Figure 3.
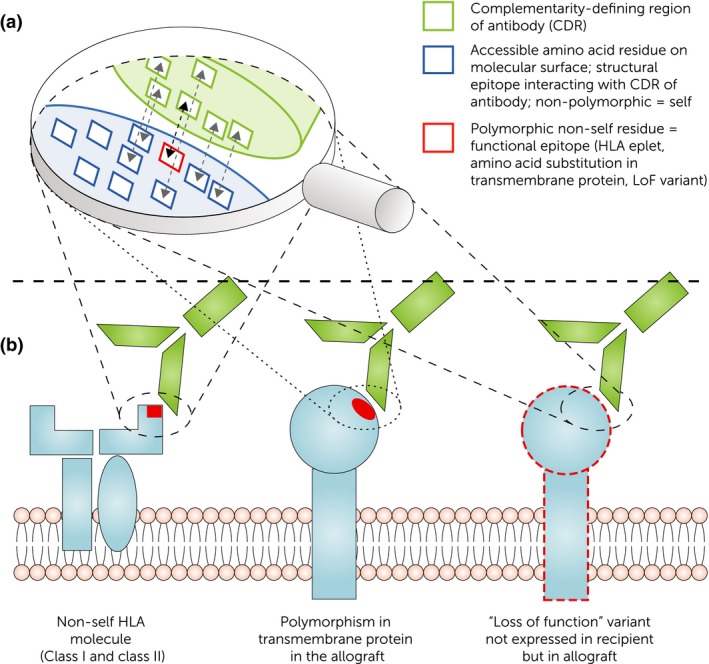
B cell epitopes recognized by donor‐specific antibodies on alloantigens. (a) Interaction of complementarity‐determining region (CDR) of alloantibody with the HLA and non‐HLA molecules on endothelial cells. Next to the functional epitope responsible for specificity (e.g., eplet, amino acid substitution in transmembrane protein, loss of function variant carrying a non‐self epitope), the structural epitope covers non‐polymorphic (“self”) residues important for binding strength/affinity. (b) Concept of non‐self epitopes on HLA and non‐HLA antigens as binding site for donor‐specific alloantibodies (DSA). HLA‐DSA binding to polymorphic residue on HLA molecule. Non‐HLA‐DSA recognizing non‐self residues on polymorphic transmembrane proteins as well as non‐self residues on loss of function variants (i.e., recipient has complete loss of gene expression of a specific allele but the donor carries at least one functioning copy).
The eplet mismatch between donors and recipients can be calculated using the HLAMatchmaker algorithm (http://www.epitopes.net). An increasing number of clinical data shows that kidney transplant recipients with a high eplet mismatch are at greater risk for the development of dnDSA 9. For the HLA‐DR and HLA‐DQ locus, cut‐off levels for eplet mismatch have been proposed based on data from a single‐center cohort from Canada (>9 and >16 for DR and DQ, respectively). However, development of dnDSA does not exclusively occur in individuals with high eplet mismatch. With respect to hard endpoints, eplet mismatch is also associated with ABMR and long‐term kidney allograft survival and shows superiority compared to HLA serotype mismatch 41. Lately, a refinement of the eplet mismatch was proposed that calculates the eplet score for each HLA class II molecule individually showing even better correlation with the development of dnDSA 42. This further supports the concept that the level of polymorphic residues on each HLA molecule defines the risk for antibody formation. However, there is no causal relationship between the number of eplet mismatches and the occurrence of DSA. A higher number of mismatches merely increase the chance that one of these mismatches is immunogenic and stimulates an alloimmune response. Thus, antibodies can also develop against a donor organ with only a single HLA eplet mismatch.
The eplet approach also helped to define acceptable antigen mismatches as allocation criterion for highly sensitized individuals compared to previous allocation solely based on HLA antigen identity with the donor. It was integrated in the Eurotransplant Acceptable Mismatch program to identify suitable kidney transplant donors and helped to decrease waiting time while showing graft survival comparable to non‐sensitized individuals 43.
Different approaches to define B cell epitopes
There are alternative approaches to define potential B cell epitopes on HLA molecules including physico‐chemical properties (hydrophobicity, electrical charge etc.) and the amino acid mismatch score that were both investigated by a group from Cambridge 44. Both models were association with the development of dnDSA. A recent analysis comparing eplet mismatch and amino acid mismatch as well as the electrostatic mismatch score and hydrophobicity mismatch score showed high correlation between all approaches 45. However, to date there are not enough data to finally conclude superiority of one score over the other. Nevertheless, the majority of published clinical data on epitope mismatch uses the eplet mismatch calculated by the HLAMatchmaker.
Prediction of indirect allorecognition
Another approach to quantify immunogenic polymorphism in the HLA region focuses on the indirect allorecognition pathway. For IgG antibody formation, T cell help is required through indirect recognition. If a non‐self HLA antigen contains a B cell epitope, but lacks the accompanying linked T cell epitope, no IgG DSA formation can occur.
In contrast to B cell epitopes that represent polymorphic structures on pathogens or cell surface molecules interacting with specific antibodies, T cell epitopes are only recognized by antigen‐specific T cells when presented in a self HLA molecule by antigen‐presenting cells. This concept has been integrated into the PIRCHE‐II score that estimates the number of indirectly recognizable T cell epitopes based on the donor HLA type and restriction of recipient HLA class II peptide expression 46. Currently, however, it is impossible to estimate which of the presentable peptides will eventually induce a T cell response. Nevertheless, evaluation of the PIRCHE‐II score in two large kidney transplant cohorts confirmed its association with dnDSA occurrence and long‐term kidney allograft survival independent of eplet mismatch 41, 47.
Missing pieces in the theory of chronic antibody‐mediated rejection
A recent study by Stegall et al. 48 showed that in late protocol biopsies the predominant histopathological lesion was arterial hyalinosis rather than features of chronic rejection. Transcriptome analysis from kidney biopsy samples suggested that hyalinosis reflects calcineurin inhibitor exposure and therefore adherence to adequate levels of immunosuppression 49. The importance of under‐immunosuppression in chronic rejection was further emphasized by a study from Wiebe et al. 50 that showed that nonadherent patients with a high eplet mismatch are at the highest risk for development of HLA‐DSA. DQ eplet mismatch was also highly predictive for development of dnDSA in the CTOT‐09 study on tacrolimus withdrawal 51.
Epitope mismatch and indirect allorecognition represent two concepts that allow us to estimate the likelihood for dnDSA formation as risk factor for ABMR and allograft loss. More recently, the histopathological phenotype of ABMR in absence of HLA‐DSA has drawn more attention. It remains unclear if this entity has the same impact on graft survival as DSA‐positive ABMR. A recent analysis by Senev et al. found better long‐term kidney allograft survival in patients with HLA‐DSA‐negative ABMR compared to patients with detectable HLA‐DSA 52. The detection of donor‐reactive memory B cells without currently detectable antibody levels further identifies patients at risk 53. Other explanations for DSA‐negative ABMR include not yet characterized non‐HLA antibodies.
Non‐HLA antibody response
Evidence for non‐HLA alloimmunity
In clinical transplantation, the focus has been so far on HLA‐related alloimmunity. However, occurrence of ABMR following kidney transplantation from HLA identical siblings suggested the importance of non‐HLA antigens in alloimmunity 54, 55. In a retrospective analysis of UNOS registry data, Terasaki et al. 56 deduced that 38% of kidney allograft losses could be due to non‐HLA‐related immunological factors compared to only 18% that were due to HLA mismatches. Further evidence for the importance of non‐HLA alloimmune responses was provided by Opelz et al. when they analyzed the impact of pre‐transplant panel reactive antibody (PRA) levels on the long‐term outcome in kidney transplant recipients from sibling donors that were fully matched at the HLA‐A, HLA‐B, and HLA‐DR locus. They estimated the likelihood of incompatibilities in other HLA loci (especially DQ and DP) to be <3% in HLA‐A‐, HLA‐B‐ and HLA‐DR‐matched siblings. Most interestingly, the effect of PRA only became apparent after the first post‐transplant year. The authors therefore concluded that non‐HLA immunity has a much stronger role in clinical transplantation than previously suspected and that non‐HLA alloimmunity is mainly associated with chronic allograft loss 57.
Definition of minor histocompatibility antigens
The term minor histocompatibility antigens is commonly used to refer to non‐HLA antigens. From an immunological point of view, these are defined as any non‐MHC encoded polymorphic protein that is sufficiently antigenic to induce an immune response through indirect allorecognition by a T cell when transplanted into an individual with absent or altered gene expression. mHAs have been extensively studied in HLA identical hematopoietic stem cell transplantation and are associated with graft‐versus‐host disease and graft‐versus‐leukemia effect, but less in solid organ transplantation 7. The genetic basis for these mHAs is non‐synonymous single‐nucleotide polymorphism (nsSNP) giving rise to polymorphic proteins 5. Examples for polymorphic proteins that act as mHAs in solid organ transplantation include Y chromosomal encoded proteins (HY antigens) in gender‐mismatched transplantation and autosomal mHAs like the MHC Class I Polypeptide‐Related Sequence A (MICA) 58, 59. For the development of an alloantibody response, indirect allorecognition by CD4+ T cells is required.
Concept of Anti‐endothelial cell antibodies
Antibody‐mediated graft injury mostly affects the endothelial surface of the graft as the endothelial layer is the barrier between recipient circulation (including immune cells/proteins) and allograft vascular tissue 60. It remains to be finally resolved, if these antigens mediate graft injury or are secondary effects following tissue damage. Besides HLA, most endothelial antigens in transplantation are unknown. The concept of anti‐endothelial cell antibodies (AECA) was introduced in the late 1970s and focuses on the site of antibody/antigen interaction rather than antigen specificity 61, 62. Characterization of target antigens is still ongoing including proteomics approaches. Mechanisms of endothelial cell injury have to be resolved and include complement‐mediated cell injury as well as endothelial cell activation 13. In contrast to HLA, individual non‐HLA antigens are often not highly expressed on the cell surface. Flow cytometry‐based endothelial cell crossmatch tests, however, showed that AECA are associated with adverse outcomes following kidney transplantation (higher rate of rejection and lower 1‐year graft survival) 63, 64. There is a high heterogeneity of endothelial cells used in these analyses including donor‐derived progenitor cells as well as third‐party umbilical cells. The use of donor‐derived cells allows for detection of antibody responses against polymorphic proteins that are mismatched between the donor and the recipient.
G‐protein‐coupled receptors
Antibodies against angiotensin II type 1 receptor (AT1R) have been identified as endothelial cell antibodies in patients with acute rejection 65. AT1R is expressed on the endothelium and represents a G‐protein‐coupled receptor such that antibodies interacting with the receptor activate signaling pathways 66. A recent study reported an HLA‐DSA‐independent effect on allograft rejection with a distinct histopathological (endothelial activation) and clinical phenotype (hypertension) 67. The endothelin receptor type A (ETAR) represents another G‐protein‐coupled receptor that has been identified as endothelial antigen. Interestingly, not all transplant recipients with AT1R and ETAR antibodies experience allograft dysfunction and a high variability in antibody titers was observed between studies 66.
Both AT1R and ETAR do not harbor polymorphisms in protein coding regions that result in an altered amino acid sequence or changed protein structure. Single nucleotide polymorphisms that were identified in both AT1R and ETAR were mainly associated with expression levels 68, 69. Antibodies that activate G‐protein‐coupled receptors were also detected in patients without transplantation and showed an association with cardiovascular disease, preeclampsia, hypertension, aging, autoimmune diseases, and cancer 66. AT1R and ETAR are therefore mainly considered autoantibodies rather than alloantibodies directed against non‐self residues. For a more detailed discussion of G‐protein‐coupled receptors in transplantation, the reader is referred to a recent review by Philogene et al. 66.
HY antigens
Proteins encoded on the Y chromosome have a 90% sequence similarity with their X chromosomal homologs. As example, the protein RSP4Y on the Y chromosome (40S ribosomal protein S4, Y isoform 1) differs from the RSP4X variant on the X chromosome by 19 amino acid substitutions based on single nucleotide polymorphisms 70. Following the identification of HY‐specific alloimmune T cells in gender‐mismatched transplantation, the development of HY alloantibodies was later observed in individuals with acute allograft injury 59. On an epidemiological level, transplantation of male donor kidneys into female recipients was associated with a negative impact on long‐term graft function in a retrospective cohort study in 200 000 kidney transplant recipients 71.
MICA
MICA antigens are highly polymorphic and have been extensively studied in kidney transplantation. Alloantibodies against mismatched MICA antigens were associated with acute rejection episodes and reduced 1‐year graft survival in initial publications, but a retrospective analysis in a large European cohort of 779 kidney transplant recipients did not show an independent association with four‐year death‐censored graft survival 58, 72, 73. The understanding of pathophysiology of MICA is incomplete, and testing has not been implemented in clinical routine.
Loss of self‐tolerance
In patients with chronic ABMR, a broad reactivity against a unique set of antigenic targets was observed 74. Graft injury creates an inflammatory milieu that causes breakdown of B cell tolerance and sets stage for the development of antibodies against intracellular and kidney tissue antigens through posttranslational modifications 75. More recently, the role of polyreactive natural antibodies in allograft rejection has been identified 76. The broad spectrum of reactivity profiles could be caused by polyreactive natural antibodies rather than a multitude of monospecific antibodies 77.
Current approach to identify endothelial cell antigens
Protein arrays that allow for simultaneous screening of antibody responses against up to 9 000 different human proteins have been used to identify non‐HLA antigens that are expressed on the vascular endothelium 78, 79, 80. Overall, these studies revealed a broad variety of antigens associated with rejection and chronic allograft injury. Most recently, Delville et al. 81 identified 38 patients with acute microvascular rejection in the absence of anti‐HLA antibodies in a French nationwide cohort of kidney transplant recipients. Microvascular inflammation serves as surrogate for an antibody‐mediated injury in the graft. Using a combination of transcriptomic and proteomic techniques, they were able to identify new endothelial targets for non‐HLA antibodies. Interestingly, these antigens showed only little redundancy among individuals. Previously proposed anti‐endothelial cell antibodies against AT1R, endothelin‐1 type A or natural antibodies did not increase in patients with acute microvascular rejection. In the light of recent data on genetic polymorphisms, it remains unclear if the observed antibody reactivity in these studies was directed against self‐epitopes rather than polymorphic residues of the targeted proteins that could not be differentiated based on the assays used to determine antibody reactivity.
Genome‐wide non‐HLA genetic mismatch
The HapMap Project and 1 000 Genomes Project have uncovered an unexpected large genetic diversity in humans 8, 82. Each individual carries on average over 9 000 nsSNPs causing a significant level of protein polymorphism between individuals on the amino acid level 83. In addition, the human genome also contains genetic variants that result in a complete loss of function (LoF) in protein‐coding genes. When both alleles are affected, this results in a complete loss of gene expression. An individual genome contains on average 100 of these LoF variants with about 20 affecting both alleles 84.
Systematic approach to define genome‐wide incompatibility
Identification of genome‐wide genetic variants in both donors and recipients allows for a systematic approach to identify individual level non‐HLA mismatch. This includes nsSNP that cause alteration in the amino acid sequence of proteins as well as complete loss of gene expression in the recipient (LoF variants) 17. In the field of hematopoietic stem cell transplantation, novel mHAs were successfully identified using similar approaches 85. The aforementioned HY antigens and MICA serve as examples for nsSNP‐based protein polymorphism in solid organ transplantation.
Especially, polymorphisms in immune‐accessible transmembrane proteins represent plausible mHAs when the donor carries an allele that is not present in the recipient (representing a non‐self/allo‐epitope). Non‐self peptides can be presented by the recipients professional APCs/B cells to indirectly alloreactive T cells 17. The high number of potential genome‐wide mismatches suggests that each donor and recipient pair caries a unique set of mismatched genetic variations. Mesnard et al. 86 were the first to quantify genome‐wide mismatches outside of the HLA region and developed the allogenomics mismatch score (AMS). Exome sequencing was performed in 53 living donor and recipient pairs, and the number of predicted amino acid mismatches in transmembrane proteins was calculated. The AMS showed a statistically significant effect on estimated glomerular filtration rate (eGFR) at one to three years after transplantation. The effect was independent of HLA mismatch in the HLA‐A, HLA‐B, and HLA‐DR loci. Subsequently, Pineda et al. identified non‐HLA mismatches based on exome sequencing data from 28 kidney allograft donor and recipient pairs. In their analysis, the non‐HLA mismatch was significantly higher in patients with biopsy‐proven ABMR compared to patients without rejection. They identified a set of 123 variants that were associated with the risk for ABMR 87.
Antibody responses against genetically defined non‐HLA antigens
More recently, Reindl‐Schwaighofer et al. calculated the genome‐wide mismatch in immune‐accessible transmembrane and secreted proteins between kidney allograft donor and recipient pairs (excluding genetic variants in the HLA region on chromosome 6) in a prospective transplant cohort of 477 patients using a customized genotyping array: A median of 1892 mismatches in non‐synonymous genetic variants that result in protein polymorphisms were identified per donor and recipient pair 10. The degree of these non‐HLA mismatches was independently associated with graft loss in a multivariable model adjusted for HLA serotype and eplet mismatch: Each increase by a unit of one inter‐quartile range exhibited a HR of 1.68 (95% CI 1.17–2.41).
Polymorphic amino acid sequences in immune‐accessible proteins (i.e., transmembrane and secreted) represent plausible epitopes that can be recognized by alloreactive antibodies similar to eplets in HLA (Fig. 3b). As a proof of principle, Reindl‐Schwaighofer et al. designed customized peptide arrays that contained both self and non‐self peptide probes that represented the genetically predicted donor–recipient‐specific amino acid differences. Thereby, a donor‐specific alloimmune response to the mismatched epitopes could be verified. These data, as the initial work done by Mesnard et al. in living donor transplantation suggested, project the concept of HLA epitope mismatch to genome‐wide genetic incompatibility in immune‐accessible transmembrane epitopes.
Loss of gene expression in the recipient
Further evidence for the importance of non‐HLA targets for an alloimmune response comes from a recently published work by Steers et al. 88. The authors tested the association of 44 copy number variants (CNV) that cause complete loss of expression of a specific gene product with the occurrence of biopsy‐proven rejection episodes and defined “genomic collision” as a specific donor–recipient genotype combination in which a recipient who is homozygous for a deletion polymorphism receives a transplant from a donor who has at least one normal allele of this gene (similar to the concept of “LoF mismatch”).
Genomic collision in the LIMS1 locus (based on the single nucleotide variant rs893403 tagging a gene deletion CNV in LIMS1) was independently associated with allograft rejection (HR 1.55; 95% CI, 1.25 to 1.93) in a combined analysis including four independent cohorts. In addition, functional validation of the rs893403 genotype was performed. Reduced LIMS1 mRNA expression levels in recipients who were homozygous for the risk allele were observed. Furthermore, expression of the protein on the cell surface under hypoxic conditions could be verified. The latter makes it a plausible endothelial cell antigen following transplantation. Finally, an antigen‐specific antibody signal against LIMS1 in donor and recipient pairs with acute rejection harboring the “collision genotype” constellation was observed. Together, these findings provide plausible evidence for a directed alloimmune response against the mismatched antigen.
Despite the association with acute rejection, no association between genomic collision at the LIMS1 locus and kidney allograft survival was observed. Interestingly, genome‐wide genetic incompatibility in transmembrane proteins was associated with long‐term graft outcome in the paper by Reindl‐Schwaighofer et al. 10. However, this was not driven by a single variant but an overall mismatch burden. Future studies have to resolve the impact of single highly immunogenic variants (e.g., LoF variants such as LIMS1 that potentially carry multiple non‐self epitopes) and the broad array of polymorphic variants being virtually unique for each donor and recipient pair on kidney transplant outcome.
Defining genome‐wide alloreactive B cell epitopes
The basic concept of all mismatch scores is that the more B cell epitopes are mismatched, the higher the chance for the development of DSA (both HLA and non‐HLA), causing more chronic ABMR and leading to reduced graft survival.
With high‐resolution genetic data available for both the MHC region and the entire genome, identification of molecular mismatches between donor and recipient has become feasible and is represented by the HLA eplet mismatch scoring system and novel genome‐wide non‐HLA mismatch scores (Fig. 2). Both strategies are based on quantification of polymorphic amino acid residues within (i) the HLA molecule or (ii) transmembrane proteins (Fig. 3b). Not every amino acid polymorphism or HLA epitope mismatch has the same impact on the alloimmune response of the recipient. Immunogenicity of these genetically defined epitopes remains unclear, and immunodominant mismatches have to be identified 89. Potential strategies include a focus on T cell help and indirect allorecognition required for DSA formation (e.g., similar to the concept of the PIRCHE‐II score) as well as accessibility of epitopes (e.g., hydrophilicity, extracellular domain of transmembrane peptides). The current approaches based on the number of potential epitope mismatches represent a step in the right direction of matching for proven immunogenic epitopes.
Overall, high‐resolution genetic data have broadened our understanding of alloimmunity that now incorporates genome‐wide genetic mismatch. These novel concepts will help to identify immunological high‐risk constellations and may provide an evidence‐based strategy for individualized immunosuppression.
Funding
The authors have declared no funding.
Conflict of interest
The authors have declared no conflicts of interest.
Acknowledgements
This work was supported by the Vienna Science and Technology Fund WWTF (grant# WWTF LS16‐019 & # LS18‐031) and the ÖNB (grant# 17289).
References
- 1. The MHC sequencing consortium . Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing consortium. Nature 1999; 401: 921. [DOI] [PubMed] [Google Scholar]
- 2. Haas M. An updated Banff schema for diagnosis of antibody‐mediated rejection in renal allografts. Curr Opin Organ Transplant 2014; 19: 315. [DOI] [PubMed] [Google Scholar]
- 3. Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015; 43: D423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kramer CSM, Israeli M, Mulder A, et al The long and winding road towards epitope matching in clinical transplantation. Transpl Int 2019; 32: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roopenian D, Choi EY, Brown A. The immunogenomics of minor histocompatibility antigens. Immunol Rev 2002; 190: 86. [DOI] [PubMed] [Google Scholar]
- 6. Dierselhuis MP, Spierings E, Drabbels J, et al Minor H antigen matches and mismatches are equally distributed among recipients with or without complications after HLA identical sibling renal transplantation. Tissue Antigens 2013; 82: 312. [DOI] [PubMed] [Google Scholar]
- 7. Bertinetto FE, Dall'Omo AM, Mazzola GA, et al Role of non‐HLA genetic polymorphisms in graft‐versus‐host disease after haematopoietic stem cell transplantation. Int J Immunogenet 2006; 33: 375. [DOI] [PubMed] [Google Scholar]
- 8. 1000 Genomes Project Consortium , Abecasis GR, Auton A, et al An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wiebe C, Pochinco D, Blydt‐Hansen TD, et al Class II HLA epitope matching‐A strategy to minimize de novo donor‐specific antibody development and improve outcomes. Am J Transplant 2013; 13: 3114. [DOI] [PubMed] [Google Scholar]
- 10. Reindl‐Schwaighofer R, Heinzel A, Kainz A, et al Contribution of non‐HLA incompatibility between donor and recipient to kidney allograft survival: genome‐wide analysis in a prospective cohort. Lancet 2019; 393: 910. [DOI] [PubMed] [Google Scholar]
- 11. Tambur AR, Claas FH. HLA epitopes as viewed by antibodies: what is it all about? Am J Transplant 2015; 15: 1148. [DOI] [PubMed] [Google Scholar]
- 12. Lim WH, Wong G, Heidt S, Claas FHJ. Novel aspects of epitope matching and practical application in kidney transplantation. Kidney Int 2018; 93: 314. [DOI] [PubMed] [Google Scholar]
- 13. Dragun D, Catar R, Philippe A. Non‐HLA antibodies against endothelial targets bridging allo‐ and autoimmunity. Kidney Int 2016; 90: 280. [DOI] [PubMed] [Google Scholar]
- 14. Zhang Q, Reed EF. The importance of non‐HLA antibodies in transplantation. Nat Rev Nephrol 2016; 12: 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matsuda Y, Sarwal MM. Unraveling the role of allo‐antibodies and transplant injury. Front Immunol 2016; 7: 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sypek M, Kausman J, Holt S, Hughes P. HLA epitope matching in kidney transplantation: an overview for the general nephrologist. Am J Kidney Dis 2018; 71: 720. [DOI] [PubMed] [Google Scholar]
- 17. Reindl‐Schwaighofer R, Heinzel A, Signorini L, Thaunat O, Oberbauer R. Mechanisms underlying human genetic diversity: consequence for antigraft antibody responses. Transpl Int 2018; 31: 239. [DOI] [PubMed] [Google Scholar]
- 18. Montgomery RA, Tatapudi VS, Leffell MS, Zachary AA. HLA in transplantation. Nat Rev Nephrol 2018; 14: 558. [DOI] [PubMed] [Google Scholar]
- 19. Richie RE, Johnson HK, Tallent MB, Turner B, Vaughn W, Niblack G. The role of HLA tissue matching in cadaveric kidney transplantation. Ann Surg 1979; 189: 581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ekberg H, Tedesco‐Silva H, Demirbas A, et al Reduced exposure to calcineurin inhibitors in renal transplantation. N Engl J Med 2007; 357: 2562. [DOI] [PubMed] [Google Scholar]
- 21. Shi X, Lv J, Han W, et al What is the impact of human leukocyte antigen mismatching on graft survival and mortality in renal transplantation? A meta‐analysis of 23 cohort studies involving 486,608 recipients. BMC Nephrol 2018; 19: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ali JM, Bolton EM, Bradley JA, Pettigrew GJ. Allorecognition pathways in transplant rejection and tolerance. Transplantation 2013; 96: 681. [DOI] [PubMed] [Google Scholar]
- 23. DeWolf S, Grinshpun B, Savage T, et al Quantifying size and diversity of the human T cell alloresponse. JCI Insight 2018; 3: pii: 121256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DeWolf S, Sykes M. Alloimmune T cells in transplantation. J Clin Invest 2017; 127: 2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lamb KE, Lodhi S, Meier‐Kriesche HU. Long‐term renal allograft survival in the United States: a critical reappraisal. Am J Transplant 2011; 11: 450. [DOI] [PubMed] [Google Scholar]
- 26. Wekerle T, Segev D, Lechler R, Oberbauer R. Strategies for long‐term preservation of kidney graft function. Lancet 2017; 389: 2152. [DOI] [PubMed] [Google Scholar]
- 27. Sellares J, de Freitas DG, Mengel M, et al Understanding the causes of kidney transplant failure: the dominant role of antibody‐mediated rejection and nonadherence. Am J Transplant 2012; 12: 388. [DOI] [PubMed] [Google Scholar]
- 28. Terasaki PI. Humoral theory of transplantation. Am J Transplant 2003; 3: 665. [DOI] [PubMed] [Google Scholar]
- 29. Colvin RB. Antibody‐mediated renal allograft rejection: diagnosis and pathogenesis. J Am Soc Nephrol 2007; 18: 1046. [DOI] [PubMed] [Google Scholar]
- 30. Valenzuela NM, Reed EF. Antibodies in transplantation: the effects of HLA and non‐HLA antibody binding and mechanisms of injury. Meth Mol Biol 2013; 1034: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Halloran PF, Famulski KS, Reeve J. Molecular assessment of disease states in kidney transplant biopsy samples. Nat Rev Nephrol 2016; 12: 534. [DOI] [PubMed] [Google Scholar]
- 32. Lefaucheur C, Loupy A. Antibody‐mediated rejection of solid‐organ allografts. N Engl J Med 2018; 379: 2580. [DOI] [PubMed] [Google Scholar]
- 33. Heidt S, Haasnoot GW, Witvliet MD, et al Allocation to highly sensitized patients based on acceptable mismatches results in low rejection rates comparable to non‐sensitized patients. Am J Transplant 2019; 19: 2926‐2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. El‐Awar NR, Akaza T, Terasaki PI, Nguyen A. Human leukocyte antigen class I epitopes: update to 103 total epitopes, including the C locus. Transplantation 2007; 84: 532. [DOI] [PubMed] [Google Scholar]
- 35. Duquesnoy RJ. A structurally based approach to determine HLA compatibility at the humoral immune level. Hum Immunol 2006; 67: 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Duquesnoy RJ, Marrari M. Correlations between Terasaki's HLA class I epitopes and HLAMatchmaker‐defined eplets on HLA‐A, ‐B and ‐C antigens. Tissue Antigens 2009; 74: 117. [DOI] [PubMed] [Google Scholar]
- 37. Rodey GE, Fuller TC. Public epitopes and the antigenic structure of the HLA molecules. Crit Rev Immunol 1987; 7: 229. [PubMed] [Google Scholar]
- 38. Duquesnoy RJ. The eplet load concept in clinical transplantation. Pediatr Transplant 2016; 20: 884. [DOI] [PubMed] [Google Scholar]
- 39. Duquesnoy RJ. HLAMatchmaker: a molecularly based algorithm for histocompatibility determination. I. Description of the algorithm. Hum Immunol 2002; 63: 339. [DOI] [PubMed] [Google Scholar]
- 40. Duquesnoy RJ, Mulder A, Askar M, Fernandez‐Vina M, Claas FH. HLAMatchmaker‐based analysis of human monoclonal antibody reactivity demonstrates the importance of an additional contact site for specific recognition of triplet‐defined epitopes. Hum Immunol 2005; 66: 749. [DOI] [PubMed] [Google Scholar]
- 41. Lachmann N, Niemann M, Reinke P, et al Donor‐recipient matching based on predicted indirectly recognizable HLA epitopes independently predicts the incidence of de novo donor‐specific HLA antibodies following renal transplantation. Am J Transplant 2017; 17: 3076–3086. [DOI] [PubMed] [Google Scholar]
- 42. Wiebe C, Kosmoliaptsis V, Pochinco D, et al HLA‐DR/DQ molecular mismatch: a prognostic biomarker for primary alloimmunity. Am J Transplant 2019; 19: 1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Claas FH, Witvliet MD, Duquesnoy RJ, Persijn GG, Doxiadis II. The acceptable mismatch program as a fast tool for highly sensitized patients awaiting a cadaveric kidney transplantation: short waiting time and excellent graft outcome. Transplantation 2004; 78: 190. [DOI] [PubMed] [Google Scholar]
- 44. Kosmoliaptsis V, Sharples LD, Chaudhry AN, Halsall DJ, Bradley JA, Taylor CJ. Predicting HLA class II alloantigen immunogenicity from the number and physiochemical properties of amino acid polymorphisms. Transplantation 2011; 91: 183. [DOI] [PubMed] [Google Scholar]
- 45. Wiebe C, Kosmoliaptsis V, Pochinco D, Taylor CJ, Nickerson P. A Comparison of HLA molecular mismatch methods to determine HLA immunogenicity. Transplantation 2018; 102: 1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Otten HG, Calis JJ, Kesmir C, van Zuilen AD, Spierings E. Predicted indirectly recognizable HLA epitopes presented by HLA‐DR correlate with the de novo development of donor‐specific HLA IgG antibodies after kidney transplantation. Hum Immunol 2013; 74: 290. [DOI] [PubMed] [Google Scholar]
- 47. Geneugelijk K, Niemann M, Drylewicz J, et al PIRCHE‐II is related to graft failure after kidney transplantation. Front Immunol 2018; 9: 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stegall MD, Cornell LD, Park WD, Smith BH, Cosio FG. Renal allograft histology at 10 years after transplantation in the tacrolimus era: evidence of pervasive chronic injury. Am J Transplant 2018; 18: 180. [DOI] [PubMed] [Google Scholar]
- 49. Einecke G, Reeve J, Halloran PF. Hyalinosis lesions in renal transplant biopsies: time‐dependent complexity of interpretation. Am J Transplant 2017; 17: 1346. [DOI] [PubMed] [Google Scholar]
- 50. Wiebe C, Nevins TE, Robiner WN, Thomas W, Matas AJ, Nickerson PW. The synergistic effect of class II HLA epitope‐mismatch and nonadherence on acute rejection and graft survival. Am J Transplant. 2015; 15: 2197. [DOI] [PubMed] [Google Scholar]
- 51. Hricik DE, Formica RN, Nickerson P, et al Adverse outcomes of tacrolimus withdrawal in immune‐quiescent kidney transplant recipients. J Am Soc Nephrol 2015; 26: 3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Senev A, Coemans M, Lerut E, et al Histological picture of antibody‐mediated rejection without donor‐specific anti‐HLA antibodies: clinical presentation and implications for outcome. Am J Transplant 2019; 19: 763. [DOI] [PubMed] [Google Scholar]
- 53. Luque S, Lucia M, Melilli E, et al Value of monitoring circulating donor‐reactive memory B cells to characterize antibody‐mediated rejection after kidney transplantation. Am J Transplant 2019; 19: 368. [DOI] [PubMed] [Google Scholar]
- 54. Grafft CA, Cornell LD, Gloor JM, et al Antibody‐mediated rejection following transplantation from an HLA‐identical sibling. Nephrol Dial Transplant 2010; 25: 307. [DOI] [PubMed] [Google Scholar]
- 55. Kalil J, Guilherme L, Neumann J, et al Humoral rejection in two HLA identical living related donor kidney transplants. Transplant Proc 1989; 21: 711. [PubMed] [Google Scholar]
- 56. Terasaki PI. Deduction of the fraction of immunologic and non‐immunologic failure in cadaver donor transplants. Clin Transplants 2003: 449. [PubMed] [Google Scholar]
- 57. Opelz G, Collaborative Transplant Study . Non‐HLA transplantation immunity revealed by lymphocytotoxic antibodies. Lancet 2005; 365: 1570. [DOI] [PubMed] [Google Scholar]
- 58. Zou Y, Stastny P, Susal C, Dohler B, Opelz G. Antibodies against MICA antigens and kidney‐transplant rejection. N Engl J Med 2007; 357: 1293. [DOI] [PubMed] [Google Scholar]
- 59. Tan JC, Wadia PP, Coram M, et al H‐Y antibody development associates with acute rejection in female patients with male kidney transplants. Transplantation 2008; 86: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jackson AM, Delville M, Lamarthee B, Anglicheau D. Sensitization to endothelial cell antigens: unraveling the cause or effect paradox. Hum Immunol 2019; 80:614. [DOI] [PubMed] [Google Scholar]
- 61. Paul LC, Claas FH, van Es LA, Kalff MW, de Graeff J. Accelerated rejection of a renal allograft associated with pretransplantation antibodies directed against donor antigens on endothelium and monocytes. N Engl J Med 1979; 300: 1258. [DOI] [PubMed] [Google Scholar]
- 62. Moraes JR, Stastny P. A new antigen system expressed in human endothelial cells. J Clin Invest 1977; 60: 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sun Q, Liu Z, Chen J, et al Circulating anti‐endothelial cell antibodies are associated with poor outcome in renal allograft recipients with acute rejection. Clin J Am Soc Nephrol 2008; 3: 1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Breimer ME, Rydberg L, Jackson AM, et al Multicenter evaluation of a novel endothelial cell crossmatch test in kidney transplantation. Transplantation 2009; 87: 549. [DOI] [PubMed] [Google Scholar]
- 65. Dragun D, Muller DN, Brasen JH, et al Angiotensin II type 1‐receptor activating antibodies in renal‐allograft rejection. N Engl J Med 2005; 352: 558. [DOI] [PubMed] [Google Scholar]
- 66. Philogene MC, Johnson T, Vaught AJ, Zakaria S, Fedarko N. Antibodies against angiotensin II type 1 and endothelin A receptors: relevance and pathogenicity. Hum Immunol 2019; 80: 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lefaucheur C, Viglietti D, Bouatou Y, et al Non‐HLA agonistic anti‐angiotensin II type 1 receptor antibodies induce a distinctive phenotype of antibody‐mediated rejection in kidney transplant recipients. Kidney Int 2019; 96: 189. [DOI] [PubMed] [Google Scholar]
- 68. Ceolotto G, Papparella I, Bortoluzzi A, et al Interplay between miR‐155, AT1R A1166C polymorphism, and AT1R expression in young untreated hypertensives. Am J Hypertens 2011; 24: 241. [DOI] [PubMed] [Google Scholar]
- 69. Kosior‐Jarecka E, Wrobel‐Dudzinska D, Lukasik U, et al Plasma endothelin‐1 and single nucleotide polymorphisms of endothelin‐1 and endothelin type A receptor genes as risk factors for normal tension glaucoma. Mol Vis 2016; 22: 1256. [PMC free article] [PubMed] [Google Scholar]
- 70. Zinn AR, Alagappan RK, Brown LG, Wool I, Page DC. Structure and function of ribosomal protein S4 genes on the human and mouse sex chromosomes. Mol Cell Biol 1994; 14: 2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gratwohl A, Dohler B, Stern M, Opelz G. H‐Y as a minor histocompatibility antigen in kidney transplantation: a retrospective cohort study. Lancet 2008; 372: 49. [DOI] [PubMed] [Google Scholar]
- 72. Baranwal AK, Mehra NK. Major histocompatibility complex class I chain‐related A (MICA) molecules: relevance in solid organ transplantation. Front Immunol 2017; 8: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lemy A, Andrien M, Lionet A, et al Posttransplant major histocompatibility complex class I chain‐related gene A antibodies and long‐term graft outcomes in a multicenter cohort of 779 kidney transplant recipients. Transplantation 2012; 93: 1258. [DOI] [PubMed] [Google Scholar]
- 74. Porcheray F, DeVito J, Yeap BY, et al Chronic humoral rejection of human kidney allografts associates with broad autoantibody responses. Transplantation 2010; 89: 1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Thaunat O, Graff‐Dubois S, Fabien N, et al A stepwise breakdown of B‐cell tolerance occurs within renal allografts during chronic rejection. Kidney Int 2012; 81: 207. [DOI] [PubMed] [Google Scholar]
- 76. See SB, Aubert O, Loupy A, et al Post‐transplant natural antibodies associate with kidney allograft injury and reduced long‐term survival. J Am Soc Nephrol 2018; 29: 1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zorn E, See SB. Is there a role for natural antibodies in rejection following transplantation? Transplantation 2019; 103: 1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li L, Sigdel T, Vitalone M, Lee SH, Sarwal M. Differential immunogenicity and clinical relevance of kidney compartment specific antigens after renal transplantation. J Proteome Res 2010; 9: 6715. [DOI] [PubMed] [Google Scholar]
- 79. Sigdel TK, Li L, Tran TQ, et al Non‐HLA antibodies to immunogenic epitopes predict the evolution of chronic renal allograft injury. J Am Soc Nephrol 2012; 23: 750. [DOI] [PubMed] [Google Scholar]
- 80. Jackson AM, Sigdel TK, Delville M, et al Endothelial cell antibodies associated with novel targets and increased rejection. J Am Soc Nephrol 2015; 26: 1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Delville M, Lamarthee B, Pagie S, et al Early acute microvascular kidney transplant rejection in the absence of anti‐HLA antibodies is associated with preformed IgG antibodies against diverse glomerular endothelial cell antigens. J Am Soc Nephrol 2019; 30: 692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. International HapMap 3 Consortium , Altshuler DM, Gibbs RA, et al Integrating common and rare genetic variation in diverse human populations. Nature 2010; 467: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shen H, Li J, Zhang J, et al Comprehensive characterization of human genome variation by high coverage whole‐genome sequencing of forty four Caucasians. PLoS ONE 2013; 8: e59494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. MacArthur DG, Balasubramanian S, Frankish A, et al A systematic survey of loss‐of‐function variants in human protein‐coding genes. Science 2012; 335: 823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Martin PJ, Levine DM, Storer BE, et al Genome‐wide minor histocompatibility matching as related to the risk of graft‐versus‐host disease. Blood 2017; 129: 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mesnard L, Muthukumar T, Burbach M, et al Exome sequencing and prediction of long‐term kidney allograft function. PLoS Comput Biol 2016; 12: e1005088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Pineda S, Sigdel TK, Chen J, Jackson AM, Sirota M, Sarwal MM. Novel non‐histocompatibility antigen mismatched variants improve the ability to predict antibody‐mediated rejection risk in kidney transplant. Front Immunol. 2017; 8: 1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Steers NJ, Li Y, Drace Z, et al Genomic mismatch at LIMS1 locus and kidney allograft rejection. N Engl J Med 2019; 380: 1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kramer C, Heidt S, Claas FHJ. Towards the identification of the relative immunogenicity of individual HLA antibody epitopes. Hum Immunol 2019; 80: 218. [DOI] [PubMed] [Google Scholar]


