Abstract
We developed an artificial intelligence algorithm (AIA) for smartphones to determine the severity of facial acne using the GEA scale and to identify different types of acne lesion (comedonal, inflammatory) and postinflammatory hyperpigmentation (PIHP) or residual hyperpigmentation. Overall, 5972 images (face, right and left profiles) obtained with smartphones (IOS and/or Android) from 1072 acne patients were collected. Three trained dermatologists assessed the acne severity for each patient. One acne severity grade per patient (grade given by the majority of the three dermatologists from the two sets of three images) was used to train the algorithm. Acne lesion identification was performed from a subgroup of 348 images using a tagging tool; tagged images served to train the algorithm. The algorithm evolved and was adjusted for sensibility, specificity and correlation using new images. The correlation between the GEA grade and the quantification and qualification of acne lesions both by the AIA and the experts for each image were evaluated for all AIA versions. At final version 6, the GEA grading provided by AIA reached 68% and was similar to that provided by the dermatologists. Between version 4 and version 6, AIA improved precision results multiplied by 1.5 for inflammatory lesions, 2.5 for non‐inflammatory lesions and by 2 for PIHP; recall was improved by 2.6, 1.6 and 2.7. The weighted average of precision and recall or F1 score was 84% for inflammatory lesions, 61% for non‐inflammatory lesions and 72% for PIHP.
Keywords: acne, algorithm, artificial intelligence, GEA scale, Global Acne Severity scale, Smartphone, lesion count
1. INTRODUCTION
Acne is a chronic multi‐factorial inflammatory disease of the pilosebaceous follicle that may affect the patient's quality of life. Acne evolves through flare‐ups and affects more than 85% of adolescents. It often continues into adulthood.1 Over the past years, adult acne and acne relapse (such as acne reappearance at regular intervals) have increased in frequency and recent data indicate that, in France, the total number of working days lost due to acne relapses add up to a total of 350 000 days per year.2 Moreover, with a decreasing number of dermatologists over the last decades, getting appointments with dermatologists has become more and more difficult for patients, resulting in irregular patient follow‐up, poor treatment outcome and a positive correlation between the duration of the disease prior to treatment and the development of scars.3, 4, 5 This highlights the need for an appropriate and timely management of acne patients to avoid the development of residual hyperpigmentation and scars or a negative impact on the patient's quality of life. Therefore, the correct grading and identification of acne lesions is the first step in a successful treatment of acne, allowing the most suitable therapeutic approach to be chosen.
To date, health agencies recommend both the inflammatory and non‐inflammatory acne lesion count using acne grading scales such as the Investigator Global Assessment (IGA) for the USA, with up to five ordinal grades (0‐4, ie clear, almost clear, mild, moderate, severe and very severe) or the Global Acne Severity Scale (GEA) for Europe, with a scale ranging from 0 to 5 (no lesions, virtually no lesions, mild, moderate, severe and very severe).6, 7 Computational methods, including the automatic counting of lesions and/or evaluation of the severity grading, similar to that used by doctors in daily clinical practice, are increasing in interest. However, to date, these methods have still not been validated.8, 9, 10
A recent study conducted in the USA assessed the severity of acne using Artificial Intelligence (AI) and the IGA scale.8 AI was able to classify acne lesions according to an IGA ordinal scale with high accuracy, no human intervention and with no need for lesion counting. Moreover, several smartphone applications have been proposed as diagnostic self‐monitoring tools.11
In this article, we present a validated AI algorithm (AIA) which, using smartphones, allows the grading of acne severity based on the GEA scale. It provides a fast determination of the severity of acne and identifies the different lesion types, thus supporting the early therapeutic management of acne patients.
2. METHODS
The AIA was developed in six steps as described in Figure 1.
Figure 1.
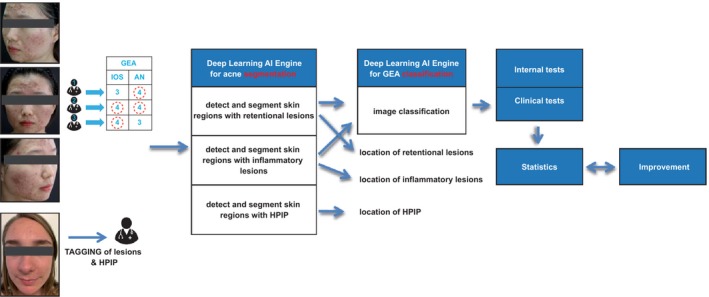
Development and validation process of artificial intelligence algorithm. Internal data testing was performed using Krippendorff's alpha and Cohen's kappa. Clinical tests were performed using an interclass correlation coefficient and the Cicchetti interpretation table. The F1 score (0‐1) equalled the weighted average of precision and recall. PIHP, postinflammatory hyperpigmentation
2.1. Step 1—Data collection
A total of 5972 images from the face, and from right and left profiles of 1072 acne patients from different racial groups having signed an informed consent, were collected from France, South Africa, China and India. All patients had to have different acne severity and images were taken using, if possible, at the same time, two smartphones equipped with IOS or Android systems, as described in Table 1.
Table 1.
Overall patient demographics and acne severity (1072 acne patients—5972 images)
| Male (%) | Female (%) | Age ± SD (y) | IOS (n) | AN (n) | GEA 0 (n) | GEA 1 (n) | GEA 2 (n) | GEA 3 (n) | GEA 4 (n) | GEA 5 (n) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Caucasian | 32 | 67 | 24.1 ± 9.0 | 1430 | 1537 | 42 | 248 | 160 | 106 | 13 | 2 |
| African | 36 | 64 | 22.2 ± 8.2 | 882 | 882 | 7 | 125 | 129 | 32 | 1 | |
| Asian | 30 | 70 | 28.0 ± 11.7 | 429 | 429 | 6 | 67 | 47 | 22 | 1 | |
| Latin | 38 | 62 | 21.7 ± 7.1 | 39 | 39 | 2 | 7 | 4 | |||
| Indien | 31 | 69 | 21.4 ± 3.7 | 153 | 153 | 15 | 33 | 3 | |||
| Total | 35 | 65 | 23.9 ± 9.2 | 2933 | 3039 | 55 | 440 | 338 | 182 | 52 | 5 |
Abbreviations: AN, Android; GEA, Group of experts in acne; IOS, Apple system; SD, standard deviation.
2.2. Step 2—Grading of data and statistical analysis
Three trained dermatologists with expertise in acne graded each patient's acne severity using the European GEA scale and the three image views as described inFigure 1.6 For each patient, the final GEA grade chosen was that confirmed by at least 2 out of the 3 dermatologists (Table 1).
In addition, inflammatory and non‐inflammatory lesions and PIHP were tagged by one of the three dermatologists on 348 images from 117 acne patients of the three main racial groups, and from GEA 0 to 4 for their identification and recognition (Figure 1 and Table 2).
Table 2.
Patient demographics and acne severity used for tagging by a dermatologist (117 acne patients—348 images)
| Male (%) | Female (%) | Age ± SD (y) | IOS (n) | AN (n) | GEA 0 (n) | GEA 1 (n) | GEA 2 (n) | GEA 3 (n) | GEA 4 (n) | GEA 5 (n) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Caucasian (n = 46) | 48 | 52 | 19.7 ± 7.2 | 90 | 45 | 0 | 9 | 16 | 18 | 3 | 0 |
| African (n = 36) | 22 | 78 | 19.9 ± 6.0 | 48 | 60 | 0 | 7 | 21 | 8 | 0 | 0 |
| Asian (n = 35) | 26 | 74 | 24.1 ± 12.5 | 54 | 51 | 3 | 10 | 11 | 10 | 1 | 0 |
| Total (n = 117) | 33 | 67 | 21.6 ± 9.3 | 192 | 156 | 3 | 26 | 48 | 36 | 4 | 0 |
Abbreviations: GEA, Group of experts in acne; SD, standard deviation.
2.3. Step 3—Training Of The AIA
A total of 4958 images (from the 5972 pool) corresponding to 903 acne participants were used to develop the algorithm (Table 3). Each set of three images associated with one GEA grade and the images with tagged lesions and PIHP were used to train the algorithm. As the difficulty to obtain images from patients with severe acne resulted in a low number of Grade 4 and 5, participants with an acne grade of ‘4’ or ‘5’ were pooled and graded ‘4+’.
Table 3.
Patient demographics and acne severity to develop the algorithm (903 acne patients—4958 images)
| Male (%) | Female (%) | Age ± SD (y) | IOS (n) | AN (n) | GEA 0 (n) | GEA 1 (n) | GEA 2 (n) | GEA 3 (n) | GEA 4 (n) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Caucasian | 33 | 67 | 24.5 ± 9.4 | 1223 | 1329 | 42 | 223 | 138 | 83 | 15 |
| African | 33 | 67 | 22.9 ± 8.6 | 723 | 723 | 7 | 115 | 100 | 18 | 1 |
| Asian | 30 | 70 | 28.0 ± 11.7 | 429 | 429 | 6 | 67 | 47 | 22 | 1 |
| Latin | 33 | 67 | 28.3 ± 7.8 | 9 | 9 | 3 | ||||
| Indian | 21 | 79 | 23.1 ± 5.8 | 42 | 42 | 15 | ||||
| Total | 33 | 67 | 24.6 ± 9.7 | 2426 | 2532 | 55 | 405 | 285 | 123 | 35 |
Abbreviations: AN, Android; GEA, Group of experts in acne; IOS, Apple system; SD, standard deviation.
From these images, image classification and segmentation techniques based on deep learning provided by Perfect Mobile Corp (New Taipei City, Taiwan) were applied to develop the algorithm. Image classification techniques were used for classifying the input image into five different groups (GEA 0, 1, 2, 3, 4+) in order to define the final GEA score. In addition, image detection/segmentation techniques were used to localize the different lesion types and PIHP on the target areas and results were submitted to a dermatologist for correction and improvement.
2.4. Step 4—Testing of the AIA versions
The algorithm was tested on a set of 1014 new images from 169 different acne patients than in step 2.3 (Table 4).
Table 4.
Patient demographics and acne severity for internal testing of the algorithm (169 acne patients—1014 images)
| Male (%) | Female (%) | Age ± SD (y) | IOS (n) | AN (n) | GEA 0 (n) | GEA 1 (n) | GEA 2 (n) | GEA 3 (n) | GEA 4 (n) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Caucasian | 29 | 71 | 22.0 ± 5.9 | 210 | 210 | 26 | 21 | 23 | ||
| African | 47 | 53 | 18.7 ± 5.3 | 159 | 159 | 11 | 29 | 13 | ||
| Asian | ||||||||||
| Latin | 40 | 60 | 19.7 ± 5.8 | 30 | 30 | 2 | 7 | 1 | ||
| Indian | 33 | 67 | 21.0 ± 2.4 | 108 | 108 | 15 | 21 | |||
| Total | 43 | 57 | 20.6 ± 5.3 | 507 | 507 | 0 | 37 | 52 | 58 | 22 |
Abbreviations: AN, Android; GEA, Group of experts in acne; IOS, Apple system; SD, standard deviation.
2.5. Step 5—Clinical testing of AIA in acne patients
Given good internal test results, the 4th version of the algorithm was tested through a clinical evaluation performed in 53 acne patients from the department of Dermatology Nantes France with a severity grade ranging from ‘1’ to ‘3’ on the GEA scale 6 (Table 5). Face‐to‐face evaluations and from photographs of GEA grades and lesion counts by the dermatologists and by the AIA were compared.
Table 5.
Patient demographics and acne severity in the clinical test (53 acne patients—159 images)
| Male (%) | Female (%) | Age ± SD (y) | Min | Max | IOS Photograph 1 (n) | IOS Photograph 2 (n) | GEA0 (n) | GEA1 (n) | GEA2 (n) | GEA3 (n) | GEA4 (n) | GEA5 (n) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 28% (n = 15) | 72% (n = 38) | 21.8 ± 7.3 | 13 | 47 | 159 | 159 | 0 | 31 | 13 | 9 | 0 | 0 |
Abbreviations: GEA, Group of experts in acne; IOS, Apple system; SD, standard deviation.
2.6. Step 6—Improvement and validation of the algorithm
Any mistake in the GEA evaluation and location of the different lesion types and PIHP was indicated to the developer in order to improve the AIA, and the AIA was to be tested again.
2.7. Statistics
Inter‐dermatologist reproducibility was checked using Krippendorff's alpha and Cohen's kappa statistical tests.12 Intra‐class correlation coefficient or ICC and a descriptive statistic test were used to measure the reproducibility of numerical measurements made by the different examiners. Similar tests were used to compare lesion counts by dermatologists and the algorithm. Furthermore, for their location, precision, defined as the correct number and type of lesions identified by the AIA, and recall, defined as the number and type of lesions correctly identified by the dermatologists and by the AIA, were evaluated. Tags made by dermatologists were compared with algorithm tags by identifying true (TP)‐ and false (FP)‐positive tags and true (TN)‐ and false (FN)‐negative tags and precision (TP/ (TP + FP)) and recall (TP/(TP + FN)). The weighted average of precision and recall or F1 score corresponding to the ratio between 2 (precision x recall)/(precision + recall) was calculated for all the images. Coefficients were interpreted using the Cicchetti interpretation table (ICC ≥ 0.75 = Excellent; [0.60‐0.75] = Good; [0.40‐0.60] = Fair and <0.40 = poor).13
3. RESULTS
Full face and right and left profile images of 1072 acne patients from different racial groups (10 to 58 years old) were collected and graded for their severity by three dermatologists (Table 1). The weighted kappa for the evaluation performed by the three dermatologists using the GEA grade for the three main racial groups of different phototype exceeded 0.6, corresponding to a substantial correlation. When comparing the GEA grade obtained for the same patient, a substantial agreement for African Black (for IOS [0.69 (95%: −0.64 to 0.75; P < .0001)] and for Android [0.65 (95%: −0.6 to 0.71; P < .0001)]), a moderate agreement for Caucasian White (for IOS [0.59 (95%: −0.54 to 0.64; P < .0001)] and Android [0.52 (95%: −0.47 to 0.57; P < .0001)]) and a moderate/fair agreement for Asian participants (moderate for IOS [0.52 (95%: −0.43 to 0.6; P < .0001)] and fair for Android [0.38 (95%: −0.3 to 0.45; P < .0001)]) was obtained. In addition, inflammatory and non‐inflammatory lesions and PIHP were tagged on 348 photographs from 117 patients by a dermatologist (Table 2).
Results indicate that the highest accuracy among the three dermatologists was obtained for Black Africans using ‘IOS images’ (‘Substantial’—Alpha = 0.69, P < .0001) and the lowest agreement was obtained for Chinese, using ‘Android images’ (‘Fair’—Alpha = 0.38, P < .0001). Thus, agreement depends on the type of device using IOS or Android, even if they were equipped with cameras of similar quality, as the difference in colour rendition influenced on the acne severity assessment (more red for IOS than Android). Moreover, results obtained for Asians have to be considered with caution, due to the low number of participants. A total of 4958 images corresponding to 903 acne participants with the GEA grade given by the majority of the 3 dermatologists from the two sets of three images (Table 3) associated with a total of 2939 tagged inflammatory lesions, 7603 tagged non‐inflammatory lesions and 5702 tagged PIHP were used to develop the algorithm. During the development, tags performed by the algorithm were submitted to the same dermatologist for correction; 1014 new images from 169 acne patients (Table 4) were used for testing using the GEA scale and counting of inflammatory lesions, non‐inflammatory lesions and PIHP.
The clinical evaluation conducted in the 53 acne patients (Table 5) by three dermatologists showed a correspondence between the 4th version of the AIA and the different assessments made by the dermatologists using the GEA scale reaching 48%; conversely, it was 59% between dermatologists using images and 67% during face‐to‐face assessments. The intra‐class correlation coefficient (ICC) for the face‐to‐face counting by the three dermatologists varied from 0.68 (Good) to 0.8 (excellent) for inflammatory lesions, from 0.47 (fair) to 0.89 (excellent) for non‐inflammatory lesions and from 0.16 (poor) to 0.55 (fair) for PIHP. In comparison, for the algorithm, the ICC obtained for images counting were lower and varied from 0.51 (fair) to 0.74 (good) for inflammatory lesions, from 0.2 (poor) to 0.51 (fair) for non‐inflammatory lesions and from 0.1 (poor) to 0.57 (fair) for PIHP. During this validation step, we also evaluated the reproducibility of the algorithm through two sets of three images of the same participants and correspondence rate of 74% was noted.
After improvement of the AIA, when testing final version 6, the GEA grading provided by the algorithm reached 68% and was similar to that provided by the dermatologists. Furthermore, the algorithm provided improved precision results multiplied by 1.5 for inflammatory lesions, 2.5 for non‐inflammatory lesions and 2 for PIHP; recall was improved by 2.6, 1.6 and 2.7, respectively, as shown in Table 6. The F1 score calculated at the end of the process on the global image basis for the sixth version considered accurate was 84% for inflammatory lesions, 61% for non‐inflammatory lesions and 72% for PIHP.
Table 6.
Precision and recall provided by the different version of the algorithm
| Precision | Recall | |||||
|---|---|---|---|---|---|---|
| AIA V4 vs dermatologist (%) | AIA V5 vs dermatologist (%) | AIA V6 vs dermatologist (%) | AIA V4 vs dermatologist (%) | AIA V5 vs dermatologist (%) | AIA V6 vs dermatologist (%) | |
| Comedonal lesions | 19 | 24 | 48 | 14 | 32 | 37 |
| Inflammatory lesions | 47 | 59 | 72 | 43 | 64 | 73 |
| PIHP | 25 | 32 | 49 | 22 | 41 | 60 |
Abbreviations: AIA, Artificial intelligence algorithm; PIHP, postinflammatory hyperpigmentation.
4. DISCUSSION
Even though acne is a very common disease, only little time or no time at all is spent on medical and dermatology education.14 For this reason, it is important for clinicians to regularly update their practice to reflect current diagnostic and treatment standards.
Moreover, clinical assessment of acne severity does not always correlate with the patient's perception of its severity.15, 16, 17, 18, 19, 20 Because the correct grading and identification of acne lesions is the first step in a successful treatment of acne, this AIA may help dermatologists and other medical practitioners to choose the most suitable treatment options according to their patients’ acne.15
We have developed an AIA able to assess acne severity and to identify and to count acne lesions from three images (face and right and left profiles) of acne patients using smartphones with IOS and Android systems, which both are used worldwide, thus also allowing global use of our AIA.
The present AIA was able to classify acne patients according to the severity of their disease based on the GEA scale, a scale that is widely used in Europe to diagnose acne severity and which has been validated through direct clinical and image grading.6
Moreover, we have studied the suitability of the GEA scale for images of Black African and Asian (Chinese) in which the problem of PIHP is more important than in the Caucasian population.
Research studies use a multitude of scales to grade acne, and there is still no standard evaluation tool.21 The present AIA may be an interesting and easy‐to‐use tool for standardized clinical investigations in acne. Moreover, this AIA, when using geo‐localization, may also allow to assess the impact of external factors that influence the occurrence, duration and severity of acne such as pollution and climatic condition.22
Non‐adherence to acne treatment is a common problem worldwide.3, 4 Acne requires continued and regular treatment, with topical and oral therapies, but the use of both is currently associated with a high level of non‐adherence.23 Adherence may be improved by early management, including an increased frequency of visits and an improved more frequent evaluation of improvement and by simplifying treatment regimens. Effective cosmetic management alongside pharmaceutical treatment was shown to increase adherence.15, 24 Other work has already been done to develop an automatic classification system of acne patients, focusing on the automatization of acne lesion counts or for the severity measurement using the IGA scale, widely used in the USA.8, 9, 10 However, to our knowledge, this is the first time that an AIA for smartphones has been developed that assesses acne severity based on the GEA scale and that allows the identification and counting of acne lesions with reliability comparable to that of a trained dermatologist. Moreover, the present algorithm may be suitable for the three main racial groups, suggesting a high generalizability.
The main limitation of our work is the fact that the algorithm has been built on a defined aim and variables. GEA evaluation and lesion tagging were performed by trained dermatologists on low‐quality smartphone images, using visible light exposure without following any precise protocol. Therefore, automatization of these evaluations with any smartphone working for three racial groups may reduce the variability. This is critical because of the higher reproducibility of results generated by the algorithm compared with those provided by the dermatologists.
Despite these limitations, we believe that the present artificial intelligence algorithm is a promising tool for an easy and reproducible evaluation of acne severity and lesion identification, allowing in the near future to adapt acne treatment to each patient as best as possible.
CONFLICT OF INTERESTS
S. Seité and Dominique Moyal are employees of La Roche‐Posay, France. Brigitte Dréno received consultancy honoraria from La Roche‐Posay, Galderma, Pierre Fabre. Amir Khammari received consultancy honoraria from La Roche‐Posay. Michael Benzaquen received consultancy honoraria from La Roche‐Posay.
AUTHOR CONTRIBUTIONS
SS, DM and BD have participated in the conception and design of the studies, AK and MB in the acquisition and analysis of data, and all authors in the preparation and critical review of the manuscript. All read and approved the final submitted version.
ACKNOWLEDGEMENTS
The authors acknowledge the editing assistance of Karl Patrick Göritz, SMWS, France, and the great technical support of G. Le Dantec and K. Abidi.
Seité S, Khammari A, Benzaquen M, Moyal D, Dréno B. Development and accuracy of an artificial intelligence algorithm for acne grading from smartphone photographs. Exp Dermatol. 2019;28:1252–1257. 10.1111/exd.14022
Funding information
This work was supported by La Roche‐Posay Dermatological Laboratories, France.
REFERENCES
- 1. Dréno B, Ann. Dermatol. Venereol. 2010, 137(Suppl 2), S49. [DOI] [PubMed] [Google Scholar]
- 2. Dreno B, Bordet C, Seite S, Taieb C, J. Eur. Acad. Dermatol. Venereol. 2019, 33, 937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pawin H, Beylot C, Chivot M, Faure M, Poli F, Revuz J, Dréno B, Dermatology 2009, 218, 26. [DOI] [PubMed] [Google Scholar]
- 4. Dréno B, Thiboutot D, Gollnick H, Finlay AY, Layton A, Leyden JJ, Leutenegger E, Perez M, Global Alliance to Improve Outcomes in Acne , Int. J. Dermatol. 2010, 49, 448. [DOI] [PubMed] [Google Scholar]
- 5. Layton AM, Seukeran D, Cunliffe WJ, Dermatology 1997, 195(Suppl 1), 15; discussion 38‐40. [DOI] [PubMed] [Google Scholar]
- 6. Dréno B, Poli F, Pawin H, Beylot C, Faure M, Chivot M, Auffret N, Moyse D, Ballanger F, Revuz J, J. Eur. Acad. Dermatol. Venereol. 2011, 25, 43. [DOI] [PubMed] [Google Scholar]
- 7. FDA . Acne vulgaris: establishing effectiveness of drugs intended for treatment guidance for industry May 2018 2018, https://www.fda.gov/downloads/Drugs/.../Guidances/UCM071292.pdf (accessed February 2019).
- 8. Melina A, Dinh NN, Tafuri B, Schipani G, Nistico S, Cosentino C, Amato F, Thiboutot D, Cherubini A, J. Drugs Dermatol. 2018, 17(9), 1006. [PubMed] [Google Scholar]
- 9. Malik AS, Humayun J, Kamel N, Yap FB, Skin Res. Technol. 2014, 20, 322. [DOI] [PubMed] [Google Scholar]
- 10. Min S, Kong HJ, Yoon C, Kim HC, Suh DH, Skin Res. Technol. 2013, 19, e423. [DOI] [PubMed] [Google Scholar]
- 11. Scott IA, Scuffham P, Gupta D, Harch TM, Borchi J, Richards B, Aust. Health. Rev. 2018. 10.1071/AH18064. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 12. McHugh ML, Biochem. Med. (Zagreb) 2012, 22, 276. [PMC free article] [PubMed] [Google Scholar]
- 13. Russell CM, Williamson DF, Bartko JJ, Bradley EL, Am. J. Hum. Biol. 1994, 6, 311. [DOI] [PubMed] [Google Scholar]
- 14. Jamil A, Muthupalaniappen L, Md Nor N, Siraj HH, Salam A, Malays. J. Med. Sci. 2016, 23, 78. [PMC free article] [PubMed] [Google Scholar]
- 15. Thiboutot DM, Dreno B, Abanmi A, Alexis AF, Araviiskaia E, Barona Cabal MI, Bettoli V, Casintahan F, Chow S, da Costa A, El Ouazzani T, Goh CL, Gollnick H, Gomez M, Hayashi N, Herane MI, Honeyman J, Kang S, Kemeny L, Kubba R, Lambert J, Layton AM, Leyden JJ, López‐Estebaranz JL, Noppakun N, Ochsendorf F, Oprica C, Orozco B, Perez M, Piquero‐Martin J, See JA, Suh DH, Tan J, Lozada VT, Troielli P, Xiang LF, J. Am. Acad. Dermatol. 2018;78(2s1):, S1.e1. [DOI] [PubMed] [Google Scholar]
- 16. Agnew T, Furber G, Leach M, Segal L, J. Clin. Aesthet. Dermatol. 2016, 9, 40. [PMC free article] [PubMed] [Google Scholar]
- 17. Stein Gold L, Tan J, Kircik L, J. Drugs Dermatol. 2016, 15, 79. [PubMed] [Google Scholar]
- 18. Nast A, Dreno B, Bettoli V, Bukvic Mokos Z, Degitz K, Dressler C, Finlay AY, Haedersdal M, Lambert J, Layton A, Lomholt HB, López‐Estebaranz JL, Ochsendorf F, Oprica C, Rosumeck S, Simonart T, Werner RN, Gollnick H, J. Eur. Acad. Dermatol. Venereol. 2016, 30(8), 1261. [DOI] [PubMed] [Google Scholar]
- 19. Zaenglein AL, Pathy AL, Schlosser BJ, Alikhan A, Baldwin HE, Berson DS, Bowe WP, Graber EM, Harper JC, Kang S, Keri JE, Leyden JJ, Reynolds RV, Silverberg NB, Stein Gold LF, Tollefson MM, Weiss JS, Dolan NC, Sagan AA, Stern M, Boyer KM, Bhushan R, J. Am. Acad. Dermatol. 2016, 74, 945. [DOI] [PubMed] [Google Scholar]
- 20. Seth D, Cheldize K, Brown D, Freeman EF, Curr Dermatol Rep. 2017, 6, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tan J, Wolfe B, Weiss J, Stein‐Gold L, Bikowski J, Del Rosso J, Webster GF, Lucky A, Thiboutot D, Wilkin J, Leyden J, Chren MM J. Am. Acad. Dermatol. 2012, 67(2), 187. [DOI] [PubMed] [Google Scholar]
- 22. Dreno B, Bettoli V, Araviiskaia E, Sanchez Viera M, Bouloc A, J. Eur. Acad. Dermatol. Venereol. 2018, 32, 812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tan X, Al‐Dabagh A, Davis SA, Lin HC, Balkrishnan R, Chang J, Feldman SR, Am. J. Clin. Dermatol. 2013, 14, 243. [DOI] [PubMed] [Google Scholar]
- 24. Tanghetti EA, Skin Therapy Lett. 2011, 16, 4. [PubMed] [Google Scholar]


