Abstract
Combining phylogeographic approaches and hybrid zone inference in a single framework is a robust way to depict respectively the biogeographic history of lineages and the evolutionary processes responsible for speciation. Here, we studied the spatiotemporal patterns of diversification and characterize the hybrid zone between two Atlantic Forest spinetails (Synallaxis ruficapilla and Synallaxis cinerea) using mitochondrial DNA and nuclear (autosomal and Z-linked) genes. We consistently recovered divergence between and within the two species during the late Pliocene and Pleistocene using an isolation with migration model. Also, our results indicate distinct levels of introgression among lineages. Ecological niche models and demographic inferences, used to infer range distributions throughout the late Quaternary, were not consistent with the hypothesis of a large river as a primary barrier responsible for the divergence of the two species. Instead, a scenario of isolation and divergence followed by geographic expansion and admixture as a consequence of Quaternary climatic oscillations was supported. Paleomodels also were not consistent with the idea that the hybrid zone originated in primary differentiation and favor a secondary contact scenario. Model fitting indicated that clines of different loci spanning the hybrid zone are coincident and concordant. The narrow cline for one Z-linked locus could be indicative of some form of post-zygotic selection hindering genetic homogenization between the two species.
Subject terms: Evolution, Population genetics
Introduction
Unveiling biotic evolution in a given region under a spatiotemporal framework helps to understand the origin of biodiversity on Earth. In this context, studies that combine phylogeographic approaches and hybrid zone inference can be crucial to understand the biogeographic processes underlying the origin of lineages and the evolutionary processes responsible for speciation, respectively. Specifically, phylogeographic studies allow one to infer whether landform and/or historical climatic changes have played a major role on the diversification of biotas (Cheviron et al. 2005; Avise 2009; Maldonado‐Coelho et al. 2013; Carnaval et al. 2014). Recent analytical advances in this field also opened the possibility of testing alternative models (e.g., those that include dispersal) to those based on pure vicariance in explaining large scale patterns of diversification across complex landscapes (Smith et al. 2014). In addition, detailed studies in areas of geographical overlap and admixture between genetically divergent populations allow the assessment of the role of selection on new gene combinations that could not be tested in the same genetic background before hybridization. In general, distinct demographic and evolutionary processes can potentially explain narrow clines of genetic and phenotypic traits. Some form of selection is often invoked as an important force hindering the spread of allelic variants across the hybrid zone (Endler 1977; Barton and Hewitt 1985; Barton and Gale 1993). Overall, loci/traits under strong selection and involved in reproductive barriers have limited introgression, resulting in abrupt gradients of allele frequencies and narrow clines. On the other hand, neutral loci or loci under weak selection have more gradual gradients of allele frequencies and broad clines. Thus, hybrid zone inferences are fundamental to understand the evolution of barriers to gene flow. For example, sigmoid/stepped clines and strong levels of linkage disequilibrium (LD) indicate a substantial amount of reproductive isolation (Szymura and Barton 1991; Vines et al. 2016).
In this context, a framework that integrates the possibility to reject at the same time the role of physical geographical barriers (e.g., rivers) versus historical climate changes (e.g., refuges) on population divergence in one hand, and whether hybrid zones originated in primary differentiation as opposed to secondary contact on the other, would be valuable to understand diversification patterns in the buildup of the biological diversity in a given region. More specifically, if genetically divergent sister populations or taxa exhibit signatures of population expansion over time at both sides of a geological barrier, this may imply that they have expanded their ranges historically, which does not allow one to reject a scenario of divergence in refuges (Moritz et al. 2000; Cheviron et al. 2005; Maldonado‐Coelho et al. 2013). Such approach is also relevant when testing alternative scenarios underlying the origin and dynamics of hybrid zones (Morales-Rozo et al. 2017). This has been a contentious issue; one model implies that hybrid zones are simply the result of contact and interbreeding between populations that differentiated genetically in geographic isolation. Alternatively, hybrid zones may have arisen in situ in response to spatial gradients of selection (Mayr 1942; Endler 1977; Harrison 1990; Durrett et al. 2000). In fact, different hybrid zones could have originated under either one of these scenarios and current patterns of genetic and phenotypic variation alone is usually insufficient to differentiate between them (Endler 1977, 1982). However, signatures of population expansion, as inferred from genetic evidence and by paleodistribution modeling, allow to test alternative scenarios. That is, a signature of population expansion could be used in favor of a secondary contact model whereas evidence of constant population size would be consistent with a primary intergradation model (Morales-Rozo et al. 2017). Hence, population expansion in association with other lines of evidence allows one to distinguish, with the same set of analyses, whether (i) the biogeographic processes responsible for the divergence of populations were related either to historical climatic changes or to the formation of a geographic barrier; (ii) the origin of hybrid zones is linked to a primary environmental gradient or if it originated following secondary contact between populations that differentiated in allopatry.
These evolutionary inferences regarding processes leading to population divergence and diversification are extremely valuable in the Neotropics, one of the regions with the highest levels of species richness in the world (Myers et al. 2000), and an understanding of patterns and processes that shape this biological diversity is not a trivial task for biologists. There, the highly diverse and threatened Atlantic Forest (AF) has been the focus of many studies. During the last years, several phylogeographical studies (Cabanne et al. 2008; Carnaval et al. 2009, 2014; Martins et al. 2009; Ribeiro et al. 2010; d’Horta et al. 2011; Maldonado-Coelho 2012; Amaral et al. 2013; Batalha-Filho and Miyaki 2016) have suggested that historical climatic changes (Haffer 1969; Vanzolini and Williams 1970) played a major role on the diversification of the AF biota during the Pleistocene, while other studies indicated other biogeographic processes as drivers of diversification. For example, rivers as barriers (Wallace 1852) were implied as being important in population divergence of lizards (Pellegrino et al. 2005; but see Cabanne et al. (2007) and Maldonado-Coelho (2012)). Additional studies also have suggested neotectonics as a possible diversification driver in some AF organisms (Silva and Straube 1996; Batalha-Filho et al. 2010; Brunes et al. 2010; Thomé et al. 2010, 2014; Amaro et al. 2012). Recently, Leite et al. (2016) presented an alternative model, in which small mammal species have expanded their ranges into the continental shelf during the last glacial maximum (LGM), therefore challenging the traditional scenario of population range fragmentation into distinct continental Pleistocene refugia. These competing scenarios portray a complex evolutionary history of the AF biota (Turchetto-Zolet et al. 2013), and imply that many distinct biogeographic processes have acted continuously since the Tertiary (Rull 2008, 2011).
In discrepancy with this increasing number of phylogeographic studies, an important and neglected issue regarding the evolution of the AF biota is the dynamics of contact and hybrid zones between lineages in some portions of the biome. Some areas of overlap between sister phylogeographic lineages have been identified for some organisms throughout the AF (Martins 2011), with some level of admixture in contact zones in or near river valleys (Maldonado-Coelho 2012). For instance, some authors found evidence for genetic admixture between frog lineages in the Doce and Jequitinhonha river valleys (Rhinella crucifer species complex—Thomé et al. 2012; Phyllomedusa burmeisteri species complex—Brunes et al. 2014; Dendropsophus elegans and Chiasmocleis carvalhoi—Tonini et al. 2013). In birds, contact zones were identified in Paraíba do Sul and Doce River valleys (Xiphorhynchus fuscus—Cabanne et al. 2007; Sclerurus scansor—d’Horta et al. 2011; Pyriglena leucoptera—Maldonado-Coelho 2012; Conopophaga lineata—Dantas et al. 2015) and in the Paraguaçu River Valley (between P. leucoptera and Pyriglena atra—Maldonado-Coelho 2012). Some of these contact zones are thought to have originated during secondary contact between populations that expanded their ranges out of refuges after the onset of warmer periods in the Quaternary. However, there have been no formal tests on the origin of contact zones in the AF. Moreover, some divergence times are older than the Quaternary and challenge the role of more recent (i.e., Pleistocene) historical climatic changes as drivers of population divergence (Thomé et al. 2010, 2012). In turn, this implies that either the same kind of biogeographical process is acting over broad temporal scales or that distinct mechanisms are at play in distinct evolutionary time intervals.
The Rufous-capped Spinetail species complex (Synallaxis ruficapilla) constitutes a good model to shed light on the biogeographic processes underlying the patterns of diversity and speciation in the AF. This suboscine passerine species complex is endemic to the AF (Pacheco and Gonzaga 1995), and comprises S. ruficapilla, the Bahia Spinetail (Synallaxis cinerea) and the Pinto’s Spinetail (Synallaxis infuscata). A taxonomic review based on plumage, morphometric data, and songs suggested that S. cinerea is best regarded as synonymous to S. ruficapilla, being the former only a geographic variant of S. ruficapilla (Stopiglia et al. 2013). However, Batalha-Filho et al. (2013) showed that there is a pronounced mitochondrial divergence (~3%) between these taxa. This study also suggests that the S. ruficapilla complex is not monophyletic. Although S. ruficapilla and S. cinerea form a sister group with high support, S. infuscata seems to be more closely related to Synallaxis moesta (from the Andes and the Tepuis mountain ranges), but this relationship shows weak statistical support. S. ruficapilla occurs from the central part of the AF in Brazil south to Paraguay and northeastern Argentina (Missiones) (Ridgely and Tudor 1994), while S. cinerea ranges from the central part of the AF north to the southern bank of the Paraguaçu River (pers. obs., Silveira 2008). Both species inhabit the understory of evergreen and semi-deciduous forest and are regularly found along forest edges (Ridgely and Tudor 1994; Sick 1997). Furthermore, Batalha-Filho et al. (2013) suggested a possible admixture zone between these species based on the pattern of co-occurrence of divergent mtDNA lineages. Thus, this species complex is a good model to understand the processes underlying diversification in the AF as divergent populations meet and hybridize along a river valley where phylogeographic breaks of other taxa have been described (i.e., in the Jequitinhonha River Valley; Batalha-Filho et al. 2012).
Here, we study this species complex to understand the patterns of diversification in the AF and the dynamics of a hybrid zone between two lineages along a major river system. We used mitochondrial and nuclear (autosomal and sex-linked) genes in a spatiotemporal framework to address the following questions: (i) What is the genetic structure of S. ruficapilla and S. cinerea across the AF? (ii) What is the location and extent of the hybrid zone between these lineages? (iii) Which biogeographic scenario could explain population divergence in this group? (iv) Did the hybrid zone originate by secondary contact or primary divergence? We hope that by combining phylogeographic, hybrid zone inferences, and ecological niche models (ENMs), insights will be gained into the biogeographic processes underlying the origin of the lineages and in the evolution of reproductive barriers that maintain their genetic integrity.
Material and methods
Sampling and molecular procedures
Tissue samples (muscle and blood) of 87 specimens of S. ruficapilla and 22 specimens of S. cinerea were sampled at 34 localities across their range in the AF (Fig. 1 and Table S1). Most samples (with two exceptions) are associated with vouchered specimens (Table S1). Molecular laboratory procedures of DNA extraction, amplification, and sequencing followed Batalha-Filho et al. (2012). We obtained sequences of two mitochondrial genes: cytochrome b (cytb) with primers L-14841 and H-16065 (Lougheed et al. 2000); and NADH dehydrogenase subunit 2 (ND2) with primers Lmet (Hackett 1996) and H6313 (Johnson and Sorenson 1998). We also sequenced two autosomal nuclear introns: β-fibrinogen intron 5 (FIB5) with primers FIB5 and FIB6 (Marini and Hackett 2002) and myoglobin intron 2 (myo2) using primers Myo2 and Myo3F (Slade et al. 1993; Heslewood et al. 1998, respectively). Some sequences (17 of cytb, 16 of ND2, 17 of FIB5, and 16 of myo2) for these genes were generated in a previous study (Batalha-Filho et al. 2013). To investigate the hybrid zone we generated sequences of three Z-chromosome-linked genes from a subset of 48 samples from the contact zone between S. ruficapilla and S. cinerea as indicated by the mtDNA variation (Fig. 2): BRM15 with primers BRM-15F and BRM-15R (Borge et al. 2005), CHDZ18 with primers CHDZ-18F and CHDZ-18R (Borge et al. 2005) and PLAA with primers PLAA-F and PLAA-R (Backström et al. 2010). PCR conditions for all genes followed Batalha-Filho et al. (2013), but the annealing temperatures of the Z-linked loci were different: BRM15—60 °C, CHDZ18—56 °C, and PLAA—57 °C. Primers sequences are given in Table S2.
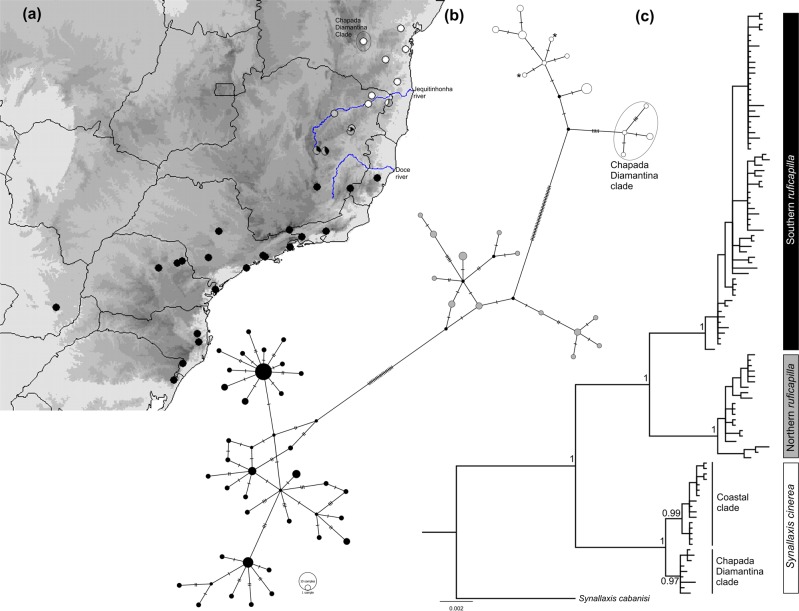
Fig. 1.
a Localities sampled for Synallaxis ruficapilla and S. cinerea. The darker the area in the map, the higher the altitude. The colors of the localities indicate the mtDNA lineages recovered in the haplotype network and the phylogeny: black—southern S. ruficapilla; gray—northern S. ruficapilla; white—S. cinerea. b Haplotype network based on concatenated cytb (1001 bp) and ND2 (1041 bp) from 98 individuals with both sequences available. Each circle corresponds to one haplotype, and its size is proportional to frequency; each mark on lines connecting haplotypes refers to a mutational step. The smallest black circles depict median vectors. Colors correspond to mitochondrial phylogroups. Haplotype with asterisks depicts S. ruficapilla individuals that fall in the S. cinerea clade. c Bayesian phylogeny based on concatenated cytb and ND2 genes. Node supports are posterior probabilities of Bayesian inference
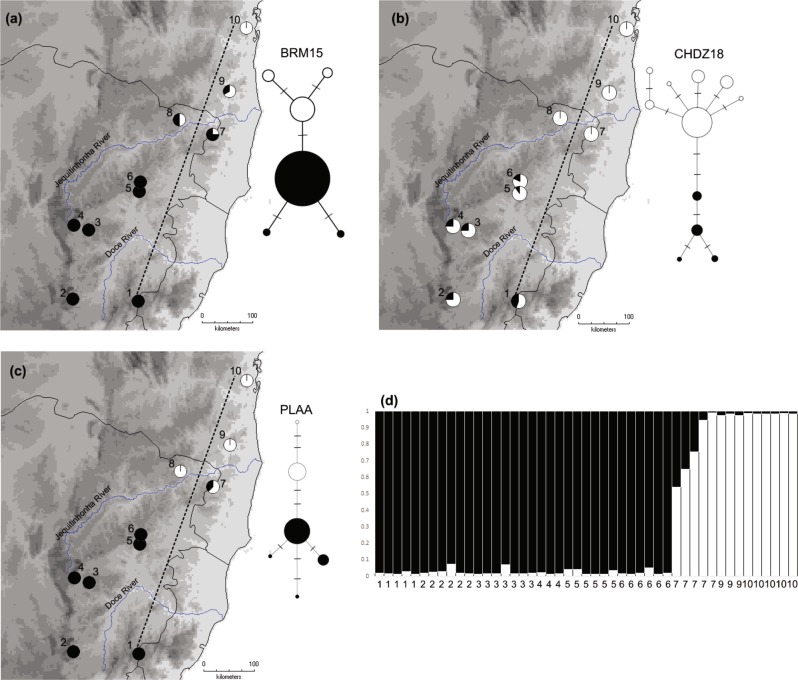
Fig. 2.
Haplotype networks (a–c) and Structure results (d) based on Z-linked loci for S. ruficapilla and S. cinerea populations spanning the hybrid zone. Maps are showing distribution of segregating alleles of BRM15 (a, 356 bp), CHDZ18 (b, 286 bp), and PLAA (c, 587 bp) genes. Dashed lines in the maps depict the 2D hybrid zone transect (Table S2). d Plot of membership coefficients of the best clustering (k = 2) from Structure. Black bars represent S. ruficapilla and white bars indicate S. cinerea
As Synallaxis does not exhibit plumage sexual dimorphism, we performed molecular sexing of the 48 individuals from the hybrid zone to identify the number of gene copies of sex-linked (Z-linked) loci. We used M5 (Bantock et al. 2008) and P8 (Griffiths et al. 1998) primers. PCR (10 µL) contained template DNA (50 ng), 1× of Taq buffer (GE Healthcare), 2 mM of MgCl2, dNTPs (0.32 µM), 0.5 µM of each primer and 0.5 U of Taq polymerase (GE Healthcare). PCR conditions were an initial denaturation step at 94 °C for 8 min; followed by 40 cycles at 94 °C for 1 min, 54 °C for 30 s, and 72 °C for 30 s plus a final extension step at 72 °C for 2 min. PCR products were checked for size and quality of the band in 3% agarose gels.
Sequence edition, alignment, phasing, and recombination
Electropherograms were inspected and assembled in contigs using CodonCode Aligner v. 3.7 (CodonCode Inc.). Heterozygous sites of nuclear loci were coded according to IUPAC code when double peaks were present in both strands of the same individual’s electropherograms. Nuclear sequences that contained heterozygous indels were analyzed using the algorithm Process Heterozygous Indels in CodonCode Aligner v. 3.7. Sequences were aligned using the CLUSTAL W method (Higgins et al. 1994) in MEGA5 (Tamura et al. 2011). All alignments were inspected and corrected visually. The gametic phase of heterozygote individuals was resolved using the algorithm PHASE (Stephens et al. 2001) with default settings in DnaSP 5 (Librado and Rozas 2009) and 0.6 as the minimum probability. Individuals with lower probabilities were removed from further analyses. We used the PHI test in SPLITSTREE4 (Bruen et al. 2006; Huson and Bryant 2006) to check for recombination in nuclear loci. This test was used due to its power to distinguish recombination events from homoplasy (Bruen et al. 2006).
Population structure tests
We generated median-joining networks (Bandelt et al. 1999) using PopART 1.7 (Leigh and Bryant 2015) for each locus to infer the relationships between haplotypes and their geographic distribution. For further analyses, we concatenated mtDNA genes (ND2 and cytb). Z-linked genes were not included in coalescent demographic analyses (EBSP and IMa2) because they may be under selection and these analyses assume neutral variation.
We reconstructed the gene genealogies of populations and taxa based on the mtDNA genes concatenated using a Bayesian inference with MrBayes 3.2.3 (Ronquist et al. 2012) at the CIPRES Science Gateway (Miller et al. 2010). The best fit model for each gene (GTR+I for cytb and HKY+G for ND2) was selected using MrModeltest 2.2 (Nylander 2004) based on the Akaike Information Criterion (AIC) in conjunction with PAUP* (Swofford 1998). To root the tree we used Synallaxis cabanisi (GenBank accession numbers: cytb, KC437438; ND2, KC437514), following Batalha-Filho et al. (2013). We performed two independent Bayesian runs with 10 million of generations and four chains of Markov chain Monte Carlo (MCMC) each. The first one million generations were discarded as burn-in, after which trees were sampled every 500 generations. Chain convergence (effective sample size—ESS values > 200) was checked using the likelihood plots for each run with Tracer 1.6 (http://beast.bio.ed.ac.uk/Tracer). The potential scale reduction factor was also used to check chain convergence and burn-in; values close to one indicate good convergence between runs (Gelman and Rubin 1992). Post burn-in trees were summarized in a 50% majority-rule Bayesian consensus tree. Tree was visualized in FigTree 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
To assess the level of population genetic structure among localities we performed an analysis of molecular variance (AMOVA, Excoffier et al. 1992) with three hierarchical levels for each locus using ARLEQUIN 3.5.1.2 (Excoffier and Lischer 2010). We also estimated the fixation indices ΦCT (structure among groups) and ΦST (structure among localities within groups). For the third level of this analysis, we considered two groups comprising each species, and four groups comprising mitochondrial phylogroups, as indicated in Fig. 1. Statistical significance was obtained with 1000 permutations. For this analysis, we used only localities with at least two samples.
Historical demography
We estimated nucleotide diversity per site (π) and number of haplotypes (h) for each locus using DnaSP5. To test for historical demographic size changes, we applied the neutrality test Fu’s Fs (Fu 1997) and the R2 test (Ramos-Onsins and Rozas 2002) in DnaSP5. Significance levels were obtained based on 10,000 coalescent simulations.
We also inferred the population size dynamics through time for all loci combined by using the Extended Bayesian Skyline Plot method (EBSP; Heled and Drummond 2008) implemented in BEAST 1.6.2 (Drummond and Rambaut 2007). This method reduces the stochastic effect from the coalescent process by combining different genes trees in a non-parametric approach of coalescence to estimate fluctuation of the effective population size through time (Ho and Shapiro 2011). Two independent EBSP runs were obtained with 150–250 million of steps of MCMC for each mitochondrial phylogroup using the following parameters: an initial UPGMA tree, a linear model, parameters sampled every 10,000 steps and a burn-in of 20%. The hLRT test implemented in MEGA5 did not reject the molecular clock hypothesis (p < 0.05) for all loci in both clades, except for mtDNA in the southern clade and in myo2 for the northern clade of the S. ruficapilla. Thus, we applied an uncorrelated lognormal relaxed clock prior in partitions that the hLRT rejected the clock and a strict clock prior for the remaining loci. The best-fit model for each gene was selected using MrModeltest 2.2 based on the AIC in conjunction with PAUP*. We used 1.05% (s.d. 0.05%) per lineage per million years as the mutation rate for mtDNA (Weir and Schluter 2008) under a normal prior distribution. Based on the probabilities obtained, BEAST estimated the mutation rate for nuclear introns individually under a default lognormal prior distribution. To check the convergence of parameters between runs and the analysis performance (ESS values > 200) we used TRACER 1.6. After removing burn-in we combined independent runs for each phylogroup by using LogCombiner.
Isolation and migration
We obtained estimates of divergence time and gene flow among lineages of S. ruficapilla complex based on a data set including all loci under the multilocus Bayesian coalescent method of isolation with migration (Nielsen and Wakeley 2001; Hey and Nielsen 2004) that is implemented in the software IMa2 version 27/08/2012 (Hey 2010). IMa2 implements the isolation with migration method for more than two populations in the same analysis. Then, we implemented this run for the three well supported lineages of S. ruficapilla and S. cinerea (coastal clade) (Fig. 1). The Chapada Diamantina clade of S. cinerea was not included in this analysis, as only mtDNA was obtained for this group. In the analysis with three populations, the software estimates 13 demographic parameters (Fig. S1): effective population sizes of each population (θ1, θ2, and θ3) and of their ancestral populations (θ4 and θ5), asymmetrical migration rates among current populations (m1→2, m2→1, m1→3, m3→1, m2→3, and m3→2) and divergence time among populations (t0 and t1). We used the HKY model for all loci and inheritance scalars of 0.25 and 1.0 for the mtDNA and nuclear loci, respectively. Migration rates were estimated only for modern populations, assuming zero migration for ancestral populations. We used the topology recovered by MrBayes (Fig. 1): ((southern ruficapilla, northern ruficapilla), coastal cinerea). We allowed the program to estimate independent values of θ and m for each population. We performed many preliminary runs to adjust the maximum value of the prior distribution for each parameter and the number and heating scheme of MCMCs. The analyses were then performed with 80 MCMCs and geometric heating (ha = 0.999 and hb = 0.3). The maximum values of priors for the demographic parameters were θ = 25, m = 10, t = 8. We implemented 4 runs with different random seeds with 5 million generations of MCMC each, and having the first million discarded as burn-in. We assumed a generation time of 1 year as adopted in Cabanne et al. (2008). The parameters were scaled in demographic units using the same mutation rate of mitochondrial genes implemented in EBSP analyses. A mutation rate of 0.135% per site per lineage per million years for nuclear introns was applied (Ellegren 2007). We assumed errors in the mutation rates as priors in the calculation of scalar mutation of 0.96–1.13% and of 0.12–0.15% for mtDNA and nDNA, respectively (Ellegren 2007; Weir and Schluter 2008). To check the convergence of parameters in each run, we made sure that all ESS values were higher than 50.
Hybrid zone analyses
To evaluate levels of introgression between S. ruficapilla and S. cinerea in a subset of 48 samples for Z-linked genes (Fig. 2), we generated median-joining networks using PopART 1.7 for each locus, Bayesian clustering with admixture model and cline model fitting. We used the software Structure 2.3.4 (Pritchard et al. 2000) to test the number of clusters (k) ranging from one to five and repeating each run 5 times. Each run consisted of 5 million generations of MCMC, discarding the first 500,000 generations as burn-in. We used the admixture models to evaluate the level of introgression between species through sampled localities. The most likely estimate of K was obtained using Evanno’s method (Evanno et al. 2005) implemented in Structure Harvester (Earl and vonHoldt 2012). Runs for the most likely K were combined using Clumpp 1.1.2 (Jakobsson and Rosenberg 2007).
For cline fitting we considered only the two lineages of S. ruficapilla and S. cinerea in geographic contact, that is, the southern and northern clades of the former and the coastal clade of the latter. The northern clade of S. ruficapilla was not recovered based on Z-linked loci (see Results). Thus, northern and southern clades of S. ruficapilla were grouped together in cline fit analyses based on Z-linked loci. Additionally, these clades were also combined in a cline analysis for mtDNA, as they share many segregating sites in mitochondrial genes. Cline fit inferences were based in SNP frequencies along the transect. For each locus we identified a segregating SNP in samples from parental populations with frequency higher than 0.8 at the southern tail and lower than 0.2 at the northern tail. The only exception was the CHDZ18 locus that did not present high frequency of any segregating SNP (maximum 0.44) at the southern tail of the transect. The segregating SNPs selected for each locus are as follows: mtDNA—site 95 in cytb, site 723 in ND2; site 110 in CHDZ18; site 319 in BRM15; site 180 in PLAA.
A similar approach to previous analyses (Macholán et al. 2007) was used to reduce the dimensionality of the hybrid zone transect. A smoothing procedure was performed in SigmaPlot 13.0 using the 3D Contour Plot, in which each site was defined by a combination of its geographical coordinates and average allele frequency (over all loci). The zone centre was defined as the 0.5 isocline and a linear transect was generated along a long axis of accumulating distances from the southernmost sampling site (locality 1; Fig. 2) to the intersection of shortest perpendicular straight-lines drawn from the long axis to each site.
Tests on the deviation from the Hardy–Weinberg equilibrium were performed for all Z-linked loci in GENEPOP (http://genepop.curtin.edu.au/; Rousset 2008). Population coefficient of ancestry (q) estimated in Structure for all loci with k = 2 and population SNP frequencies for each of the Z-linked and the mtDNA loci were fit to equilibrium geographic clines (Szymura and Barton 1986; Gay et al. 2008) using the package hzar (Derryberry et al. 2014) in R 3.4.4 (R Core Team). The MCMC algorithm was employed in a set of 15 models available in hzar (Derryberry et al. 2014), which differ in estimated cline shape parameters. All models estimated cline centre (c, in km from sampling locality 1, Fig. 2) and cline width (w, 1/maximum slope). In addition, models varied in (i) the fit of exponential decay tail parameters δ and τ (none, mirrored tails, both tails separately, right tail only and left tail only), where δ and τ represent the distance from the cline centre to the tail and the tail slope, respectively and (ii) in the scaling of maximum (pmax) and minimum (pmin) allele frequencies (fixed or free to vary); i.e., some models had no scaling (pmin = 0, pmax = 1), fixed scaling (pmin and pmax using observed minimum and maximum frequencies) or allowed pmin and pmax to be estimated. These models were compared to a null model with no clinal transition. In all models, we constrained the MCMC to sample the geographic distance encompassed by the length of our transect. Three independent chains for 1.0 × 106 generations were run for each model. The fit of these 15 models was then compared using AIC corrected for small sample size (AICc). Adequate mixing and convergence in cline parameter estimation were evaluated by visualizing MCMC sampling trajectories. Coincidence and concordance among loci clines were evaluated using two log-likelihood confidence intervals from the best cline model for each locus. Non-overlapping confidence intervals were taken as evidence of differences in cline position and width.
In addition, these clines were contrasted to a model of neutral diffusion (Endler 1977; Barton and Gale 1993) that estimates the width of a cline under no selection or barriers to gene flow: w = 2.51σ√t. Here, the width of a cline (w) is a function of the number of generations since secondary contact (t) and parent-offspring dispersal distance (σ). A neutral cline wider than the clines estimated for a given trait indicates that selection is preventing genetic homogenization of populations. The mean natal dispersal distance of 0.931 km estimated for another suboscine bird (Woltmann et al. 2012) was employed. The time since secondary contact was based on our paleomodeling showing a northward expansion of the southern species during the LGM (ca. 0.021 mya). Generation time was assumed to be 1 year as above.
Inferring the paleodistribution
To infer the distribution of AF spinetails through the late Quaternary we generated ENMs using the package BIOMOD (Thuiller et al. 2009) in R 3.4.4. We implemented the Ensemble forecasting approach (Araújo and New 2007) that combines different modeling algorithms while considering for variation among their results and statistical support for each model. We implemented all 10 algorithms available in BIOMOD: Generalized Linear Model, Generalized Additive Model, Generalized Boosting Model, Classification Tree Analysis, Artificial Neural Network, Surface Range Envelop—BIOCLIM, Flexible Discriminant Analysis, Multiple Adaptive Regression Splines, Random Forest and Maximum Entropy. The occurrence points were the same as used for genetic sampling (Table S1), as for these records we could confirm whether localities are from S. ruficapilla or S. cinerea based on the genetic data. We generated ENMs using three different strategies to take into account possible niche divergence between species: combined records of S. ruficapilla and S. cinerea (34 localities); only records of S. ruficapilla (27 localities); and only records of S. cinerea (7 localities). Localities with admixture between species were also included and assigned to either species based on the species with the highest frequency of mtDNA lineages. We gathered environmental variables from WorldClim online database v1.4 (www.worldclim.org/bioclim). Based on a factor analysis on a correlation matrix to minimize collinearity problems when constructing models (Terribile et al. 2012) we selected seven out of 19 Bioclim variables available: mean diurnal range, temperature seasonality, maximum temperature of warmest month, precipitation of wettest month, precipitation of driest month, precipitation seasonality, and precipitation of warmest quarter. Bioclim variables were used in 2.5 arc-min resolution (approximately 4.5 km at the Equator) and cropped to South America extent (between latitudes 14°N and 57°S, and longitudes 30°W and 85°W). We used layers for (i) current conditions, (ii) Medium Holocene (MH, ~0.006 mya), (iii) LGM (~0.022 mya), and (iv) Last Interglacial (LIG; ~0.12–0.14 mya). As MH and LGM periods have more than one Atmospheric Oceanic Global Circulation Model (AOGCM) available, we restricted our AOGCM dataset to those present in both these periods: CCSM4, MIROC-ESM, and MPI-ESM-P, developed through Coupled Model Intercomparison Project Phase 5 (CMIP5) (http://cmip-pcmdi.llnl.gov/cmip5/). We calculated the mean value for each cell in the spatial variable of different AOGCMs for a period using the raster package (Hijmans and van Etten 2014) in R 3.4.4. We chose this approach in order to yield a single paleodistribution model for each period (instead of a different hypothesis for each AOGCM). Model calibration was based on 20 replicates for each algorithm: the first ten replicates randomly split occurrence points into training data (75%) and testing data (25%) for model evaluation (partitioned models) and the remaining ten replicates were performed using the total data set (full models). Evaluation was based on the TSS (True Skill Statistic; Allouche et al. 2006), and full models presenting TSS > 0.9 were then used to generate the final ensembled outputs. Paleoclimate scenarios were generated by projecting layers of the past on the current distribution model. Resulting grid layers were visualized in DIVA-GIS 7.5 (https://www.diva-gis.org/).
Results
Sequence features
We obtained 1001 bp for cytb (N = 99), 1041 bp for ND2 (N = 93), 525 bp for FIB5 (N = 97), and 601 bp for myo2 (N = 89). In addition, we obtained sequences for all 48 samples from the contact zone between S. ruficapilla and S. cinerea of three Z-linked genes: 356 bp of BRM15, 286 bp of CHDZ18, and 587 bp of PLAA. In the mtDNA genes, 66 and 50 variable sites were respectively observed for cytb and ND2, and no indels were detected in these genes. We did not detect the presence of stop codons, indicating their mitochondrial origin. We observed 29 and 28 polymorphic sites in the autosomal introns FIB5 and myo2, respectively. We did not observe indels in myo2, but in FIB5 we found a 7 bp indel in five individuals. For the Z-linked genes we observed five, ten, and eight variable sites for BRM15, CHDZ18, and PLAA, respectively. We observed a 1 and a 5 bp long indel in two samples in the BRM15 gene. Indels were not considered in further analyses.
PHASE resolved alleles with high probability for all nuclear loci (p > 0.6 for autosomal loci; p > 0.9 for Z-linked loci). PHI test did not find significant evidence of recombination in any nuclear loci (FIB5, p = 0.939; myo2, p = 0.221; BRM15, p = 1.0; CHDZ18, p = 0.122; PLAA, p = 1.0).
Population structure
The Bayesian phylogenetic tree based on mtDNA data evidenced four clades with high support (Fig. 1c): (1) a southern clade of S. ruficapilla from the south to the central AF; (2) a northern clade of S. ruficapilla from the central region of the AF that is geographically restricted at the interfluvium of Doce and Jequitinhonha rivers; (3) a coastal clade of S. cinerea from the Jequitinhonha River Valley in central AF to the northern range of this species; and (4) an inland S. cinerea clade from the forests flanking the Chapada Diamantina Mountain Range (hereafter Chapada Diamantina clade). Haplotypes of the northern clade of S. ruficapilla were found at many localities where there are also haplotypes of both the southern clade of S. ruficapilla and the coastal clade of S. cinerea. The mitochondrial haplotype network (Fig. 1b) recovered the same population structure observed in the Bayesian tree. Haplotype networks based on autosomal loci did not detect a population structure as identified by the mtDNA (Fig. S1).
Our study recovered a contact zone among the coastal clade of S. cinerea and the two clades of S. ruficapilla at the interfluvium of the Doce and Jequitinhonha rivers (Figs. 1 and 2). Specifically, the mtDNA haplotypes of S. ruficapilla and S. cinerea co-occur in four localities along this contact zone (localities 3, 4, 5, and 7; Figs. 1 and 2). Furthermore, in one locality (5 in Figs. 1 and 2) all the three mtDNA lineages co-occur, while in the other three localities we either observed in sympatry only haplotypes of the northern and southern S. ruficapilla or northern S. ruficapilla and costal S. cinerea clades.
The AMOVA results based on mtDNA significantly supported two and four groups (k = 2, k = 4) in the third hierarchical level as the best population clustering for the S. ruficapilla complex (Table 1). Nevertheless, clustering considering four groups retained more genetic variation in the third level, which is in accordance with the phylogenetic and haplotype network results (Fig. 1). AMOVA for autosomal introns did not recover any signal of population structure, as most of the variation was observed within localities (Table 1).
Table 1.
Analysis of molecular variance (AMOVA) of S. ruficapilla and S. cinerea lineages
| Locus | Source of variation | DF | Percentage of variation | Fixation index |
|---|---|---|---|---|
|
mtDNA k = 2 |
Among clades | 1 | 69.25 | фCT = 0.6925** |
| Among localities within clades | 20 | 16.07 | фST = 0.8531** | |
| Within localities | 75 | 16.68 | ||
| Total | 96 | |||
|
mtDNA k = 4 |
Among clades | 3 | 75.44 | фCT = 0.7544** |
| Among localities within clades | 18 | 4.15 | фST = 0.7958** | |
| Within localities | 75 | 20.41 | ||
| Total | 96 | |||
| FIB5 | Among clades | 2 | 16.62 | фCT = 0.1662* |
| Among localities within clades | 20 | 23.51 | фST = 0.4012** | |
| Within localities | 155 | 59.87 | ||
| Total | 177 | |||
| Myo2 | Among clades | 2 | 5.49 | фCT = 0.0549** |
| Among localities within clades | 20 | −11.97 | фST = −0.0648ns | |
| Within localities | 131 | 106.48 | ||
| Total | 153 |
DF degrees of freedom
nsnon-significant; *p < 0.01; **p < 0.000001
Historical demography
Summary statistics for all markers are shown in Table 2. The R2 test indicates population expansion based on mtDNA and FIB5 for the southern clade of S. ruficapilla (Table 2). The Fs test indicated population expansion for all S. ruficapilla clades based on mtDNA and FIB5. However, no signature of population expansion was recovered for the myo2 locus. Inference based on all loci combined in EBSP indicated population growth for all clades during the late Pleistocene (Fig. 3): the southern and northern clades of S. ruficapilla respectively at ca. 0.05 and 0.10 mya, and the coastal clade of S. cinerea at ca. 0.02 mya. However, confidence intervals of all estimates overlapped, which indicate that population growth is best regarded between 0.1 and 0.02 mya in all clades. EBSP for the Chapada Diamantina clade of S. cinerea was not generated as only mtDNA data was available for this group.
Table 2.
Summary statistics for clades of S. ruficapilla and S. cinerea
| Clade | Locus | N | h | π/s.d. | R 2 | F s |
|---|---|---|---|---|---|---|
| Southern | mtDNA | 57 | 34 | 0.00252/0.00018 | 0.0355** | −26.3685** |
| S. ruficapilla | FIB5 | 108 | 17 | 0.00236/0.00023 | 0.0382* | −11.2400** |
| myo2 | 98 | 16 | 0.00687/0.00020 | 0.1351ns | −0.9707ns | |
| Northern | mtDNA | 20 | 15 | 0.00397/0.00119 | 0.1262ns | −3.8942* |
| S. ruficapilla | FIB5 | 42 | 6 | 0.00118/0.00023 | 0.0706ns | −2.7487* |
| myo2 | 32 | 12 | 0.00914/0.00043 | 0.1926ns | −0.3702ns | |
| Coastal | mtDNA | 16 | 8 | 0.00123/0.00011 | 0.1289ns | −2.3834ns |
| S. cinerea | FIB5 | 28 | 8 | 0.00159/0.00036 | 0.0746* | −5.0094* |
| myo2 | 24 | 11 | 0.00887/0.00054 | 0.1983ns | −0.7640ns | |
| CD S. cinerea | mtDNA | 5 | 4 | 0.00115/0.00037 | 0.2408ns | 0.8482ns |
| Total | mtDNA | 98 | 57 | 0.01332/0.00105 | – | – |
| FIB5 | 178 | 22 | 0.00200/0.00018 | – | – | |
| myo2 | 154 | 25 | 0.00781/0.00018 | – | – | |
| BRM15 | 84 | 6 | 0.00142/0.00025 | – | – | |
| CHDZ18 | 81 | 11 | 0.00504/0.00065 | – | – | |
| PLAA | 84 | 6 | 0.00194/0.00019 | – | – |
N sample size, h number of haplotypes, π nucleotide diversity per site, s.d. standard deviation, CD Chapada Diamantina clade
**p < 0.0001; *p < 0.05; nsnon-significant
Fig. 3.
Historical demographic estimates from Extended Bayesian Skyline Plot (EBSP) for lineages of S. ruficapilla and S. cinerea: a southern clade of S. ruficapilla; b same plot but zoomed on the last 0.20 mya; c northern clade of S. ruficapilla; d coastal clade of S. cinerea. The line in the middle shows the median estimate of the EBSP, and the upper and lower lines show the upper and lower 95% highest posterior density limits, respectively. The Y-axis is in ln scale. The X-axis represents time in millions of years
Isolation with migration
Our estimates of divergence time between clades of the S. ruficapilla complex obtained by IMa2 (Fig. S2 and Table 3) indicated that diversification took place during the late Pliocene to the Pleistocene. The oldest divergence, between S. cinerea and the ancestor of S. ruficapilla clades, occurred at ca. 0.902 mya (95% highest posterior density [HPD]: 0.541–3.461 mya). The split between the southern and northern clades of S. ruficapilla occurred at ca. 0.721 mya (95% HPD: 0.077–1.885 mya). Migration rate estimates were higher between the southern and the northern clades of S. ruficapilla. Migration rates were asymmetrical between the southern clade of S. ruficapilla and the coastal S. cinerea clade, being higher from S. cinerea into S. ruficapilla (Fig. S3 and Table 4). Our estimates of effective population size (Ne) evidenced that the southern clade had a higher Ne than the other clades. However, Ne likelihood curves of S. cinerea and S. ruficapilla overlapped (Fig. S2 and Table 3).
Table 3.
Estimations of effective population size (number of individuals) and divergence time (in years) by IMa2
| θ1 | θ2 | θ3 | θ4 | θ5 | t0 | t1 | |
|---|---|---|---|---|---|---|---|
| Highest point | 1,275,539 | 455,840 | 293,523 | 279,996 | 523,472 | 721,985 | 902,048 |
| Lower HPD of 95% | 885,979 | 244,828 | 131,206 | 0a | 0a | 77,912 | 521,145a |
| Upper HPD of 95% | 1,819,300 | 875,158 | 623,567 | 2,427,988a | 2,395,525a | 1,885,471 | 3,461,026a |
θ: Effective population size in number of individuals for southern S. ruficapilla (θ1), northern S. ruficapilla (θ2), coastal S. cinerea (θ3), ancestor of S. ruficapilla clades (θ4), and ancestor of S. ruficapilla and coastal S. cinerea (θ5). t: Divergence time in years between northern and southern clades of S. ruficapilla (t0), and between S. ruficapilla and coastal S. cinerea (t1)
95% of HPD: confidence intervals of 95% of posterior distribution after burn-in
aHPD values had low precision and were influenced by priors because the likelihood distribution was flat at the end and did not touch 0
Table 4.
Estimations of migration rates (2Nm: number of migrants per generation) estimated by IMa2. Parameters are according to Fig. 5
| m1→2 | m2→1 | m1→3 | m3→1 | m2→3 | m3→2 | |
|---|---|---|---|---|---|---|
| Highest point | 0.9978 | 2.575 | 0.1408 | 0.2945 | 0.04222 | 1.104 |
| Lower HPD of 95% | 0 | 0.5703 | 0 | 0 | 0 | 0 |
| Upper HPD of 95% | 5.739 | 6.792 | 3.420 | 2.813 | 5.362 | 7.196 |
Arrows depict direction of migration (i.e., m1→2 means the rate which population 2 received migrants from population 1): m1→2 is migration rate of southern to northern S. ruficapilla clades and m2→1 is vice versa; m2→3 is migration rate of northern S. ruficapilla to coastal S. cinerea and m3→2 is vice versa; m1→3 is migration rate of southern S. ruficapilla to coastal S. cinerea and m3→1 is vice versa
95% of HPD: confidence intervals of 95% of posterior distribution after burn-in
Hybrid zone
Haplotype networks and the Structure results based on the Z-linked genes recovered two lineages that corresponded to S. ruficapilla and S. cinerea (Figs. 2 and S4). The mtDNA subdivision of S. ruficapilla in southern and northern clades was not recovered by the Z-linked genes. These results confirmed the presence of a hybrid zone between the two species, which is also located near the Jequitinhonha River (Fig. 2 and Table S3). In addition, we found heterozygous males in localities 7–9 that exhibit alleles from both species in BRM15 and PLAA loci (three in PLAA from locality 7; four in BRM15 from localities 7–9). Structure results indicated a higher level of admixture in locality 7, with an abrupt decrease in introgression levels away from this population (Fig. 2d).
AICc indicated Model 1 (with fixed scaling for pmin and pmax and no tail parameters estimated) as the best-fit model for all loci. Clines for mtDNA and Z-linked loci BRM15 and PLAA were coincident (i.e., same centre) with overlapping log-likelihood confidence intervals (Fig. 4 and Table S4): mtDNA c = 462.92 (397.74–551.90) km; BRM15 c = 529.25 (475.65–593.93) km; PLAA c = 457.54 (413.10–485.57) km. These results were corroborated by the estimated cline centre for the coefficient of ancestry qc = 435.85 (370.93–486.06) km. Corroborating the Structure results, cline centre for these loci were around locality 7 (Figs. 2 and 4). Clines of these three loci were also concordant (i.e., overlapping widths). However, the Z-linked gene PLAA showed the narrowest cline: mtDNA w = 197.66 (95.04–415.21) km; BRM15 w = 179.37 (96.42–329.94) km; PLAA w = 28.32 (0.61–196.05) km. Cline width for the coefficient of ancestry q also presented a similar pattern and overlapped with all loci: w = 78.29 (2.26–279.34) km. Confidence intervals for cline centre of the CHDZ18 gene did not overlap with those from the other loci (Fig. S5), with a mean cline centre displaced southwards: c = 218.12 (21.43–345.82) km. This locus had a smoother allelic transition and a broader cline relative to the other loci: w = 403.20 (62.94–779.92) km. The expected cline width estimated under a neutral diffusion model was 339 km wide.
Fig. 4.

Maximum likelihood clines for the Z-linked loci and mtDNA across the hybrid zone of Atlantic Forest spinetails (Synallaxis ruficapilla and S. cinerea). Clines depict frequency of a given S. ruficapilla allele as a function of an accumulating distance from the southernmost site (locality 1, Fig. 1). Larger values of the coefficient of ancestry represent a higher proportion of genetic ancestry from S. ruficapilla. Distinct colors correspond to mtDNA (green), BRM15 (red), PLAA (purple), CHDZ18 (gray), and coefficient of ancestry (black). The maximum likelihood 95% credibility intervals of these clines are presented in Fig. S5
Paleodistribution of Atlantic Forest spinetails
Our ENMs exhibited good accuracy, as evidenced by true skill statistic values (TSS ≥ 0.9). ENMs for combined species produced similar results of models only using S. ruficapilla records, as models only with S. cinerea records generated maps with suitable areas in the southern AF, where this species does not occur, in all periods (Fig. 5). While current models for S. ruficapilla and combined species were similar to known modern distribution of AF spinetails, the current model of S. cinerea overestimates its distribution (Fig. 5). Despite differences in ENMs for combined and individual species, all models were consistent with a scenario in which S. ruficapilla and S. cinerea were isolated during the LIG and resumed secondary contact during the LGM (Fig. 5).
Fig. 5.
Ecological niche models (ENMs) of Atlantic Forest spinetails (records for both species and those from each species separately) for current time, mid Holocene (MH), last glacial maximum (LGM), and last interglacial (LIG). Warmer colors indicate a higher probability of species occurrence, as depicted in the legend. Dashed lines represent Doce (further south) and Jequitinhonha (further north) Rivers
Discussion
Diversification of Atlantic Forest spinetails
Our analyses based on mtDNA data revealed the presence of four lineages within the S. ruficapilla species complex (0.5–2.8% divergence; Fig. 1 and Table 1), with divergence times taking place during the late Pliocene to Pleistocene (Fig. S2 and Table 3). Z-linked genes corroborated the genetic differentiation between the two species, with a hybrid zone found along the Jequitinhonha River Valley. Despite morphological and vocal overlap between these species (Stopiglia et al. 2013), our analyses consistently support a clear genetic divergence between the two taxa described in an earlier analysis (Batalha-Filho et al. 2013). The split between the southern and northern clades in S. ruficapilla was not recovered based on autosomal (Fig. S1 and Table 1) and Z-linked genes (Fig. 2), probably due to recent population divergence. Also, the genetic divergence between the two species observed in mtDNA (Fig. 1) and Z-linked (Fig. 2) genes was not recovered in autosomal loci (Fig. S1). In theory, we expect weak or no divergence in autosomal genes due to incomplete lineage sorting for recent divergences (Palumbi et al. 2001; Zink and Barrowclough 2008). Additionally, differences in effective population sizes between mtDNA, sex-linked and autosomal loci could have led to a faster rate of fixation of mtDNA and Z-linked alleles due to drift and differential introgression in these loci between the two species could have contributed to genetic homogenization in autosomal loci (i.e., Haldane's rule; Haldane 1922; Borge et al. 2005; Macholán et al. 2007; Carling and Brumfield 2008, 2009).
The geographic distribution of lineages partially concurs with previous reports of range distributions and phenotypic diagnoses for S. ruficapilla and S. cinerea (Pacheco and Gonzaga 1995; Ribon et al. 2002; Vasconcelos and Silva 2004). However, the cryptic mtDNA lineage of S. ruficapilla from central AF (i.e., the northern clade, Fig. 1) does not coincide with any previously described phenotypic variation. This clade seems to be restricted to forests along the Espinhaço Mountain Range in the middle of the Jequitinhonha River Basin. On the other hand, while the Chapada Diamantina clade of S. cinerea exhibits low (0.5%) mitochondrial divergence from the coastal clade (Fig. 1), it seems to present distinct vocal variation (S.S. Santos et al. in prep). Additional analyses with a dense vocal sampling are needed to investigate this issue.
Our results were not consistent with the role of Jequitinhonha River as a barrier responsible for the split between S. ruficapilla and S. cinerea (Vasconcelos and Silva 2004), since we observed (i) mtDNA haplotypes and Z-linked alleles of S. cinerea in both banks of this river, (ii) Z-linked alleles of S. ruficapilla in both banks of the river, and (iii) paleodistribution models indicate range fragmentation and isolation due to historical climatic change events. While Amazonian rivers play an important role in avian population divergence (Ribas et al. 2012; Fernandes et al. 2012; Maldonado‐Coelho et al. 2013), AF rivers seem to be more permeable barriers to gene flow (Cabanne et al. 2007; Maldonado-Coelho 2012). In a scenario of a river as a primary barrier one would expect absence of population expansion in populations flanking the river (Cheviron et al. 2005) and a temporal congruence between population divergence and the origin of the current river course (Moritz et al. 2000). Thus, evidence from the geographic distribution of genetic variation and paleomodeling (Figs. 1, 3, and 5) were not consistent with rivers as drivers of differentiation of S. ruficapilla complex, but were consistent with climatic oscillations as drivers of population divergence in this group. However, an alternative and yet untested explanation is that the hybrid zone between the two spinetails, as well as other contact zones in the AF, rest on population density troughs (Barton 1979; Barton and Turelli 2011). Hence, contact zones coinciding with rivers valleys in AF could simply reflect a low population density region in which hybrid zones become “trapped”. Future demographic studies should address this issue.
The presence of a hybrid zone in the Doce-Jequitinhonha interfluve, as indicated by evolutionary relationships (Bayesian tree and haplotype network, Fig. 1), three independent population genetic analyses (IMa2, Structure and cline fitting, Figs. 2, 4, S3, and S5; Tables 4 and S3), the signature of late Pleistocene demographic expansion (Fig. 3) and ENMs (Fig. 5), seems to be consistent with a scenario of isolation and divergence during the late Pleistocene (last 0.1 mya). Indeed, our results of population size changes (between 0.1 and 0.02 mya) and ENMs were consistent with a scenario of fragmentation during the LIG and expansion in the LGM, which is in accordance with a recent hypothesis that postulates population growth during the latter period (Leite et al. 2016). According to this scenario, AF small mammal species expanded their range into the emerged continental shelf during the LGM, instead of the traditional view of population fragmentation during this period (Haffer 1969). Population growth during the LGM has also been documented for other AF passerines (Batalha-Filho et al. 2012; Cabanne et al. 2013).
The role of Quaternary climatic changes in shaping diversification in S. ruficapilla and S. cinerea might also be advocated to explain the evolutionary history of this group. Indeed, the refugia hypothesis has not been rejected by many studies of AF organisms (e.g., Cabanne et al. 2007, 2008; Carnaval et al. 2009; Martins et al. 2009; Ribeiro et al. 2010; d’Horta et al. 2011; Maldonado-Coelho 2012; Batalha-Filho and Miyaki 2016). Our data revealed Plio-Pleistocene splits and recent (last 0.1 mya) demographic expansion in all lineages (Fig. 3), which is in accordance with expectations of the refugia hypothesis (Moritz et al. 2000). However, climatic oscillations extend back into the Tertiary (Haffer 1997; Jansson and Dynesius 2002). Thus, we suggest that isolation and divergence due to forest fragmentation during the last ca. 2 mya is the most likely scenario explaining the evolution of S. ruficapilla and S. cinerea in the AF.
Hybrid zone dynamics
Clustering analysis based on Z-linked genes recovered two groups that correspond to the two spinetail species (Fig. 2). Also, this and cline fitting results indicate that the centre of the hybrid zone lies either near or at locality 7 (Fig. 2). Finally, these analyses suggest that introgression is somewhat geographically limited, despite hybrids being common at this locality.
There is an inverse relationship between cline width and the amount of selection on a given loci (Endler 1977; Barton 1983; Barton and Gale 1993), and cline width is a valuable information to infer which traits are involved in barriers to gene flow. The PLAA, BRM15, and CHDZ18 genes have been shown to be under selection in hybrid zones of other passerine birds (Borge et al. 2005; Backström et al. 2010; Elgvin et al. 2011), and particularly the narrower cline of the PLAA gene (Fig. 4 and Table S4) concurs with the idea of sex chromosomes being involved in reproductive isolation in vertebrates (e.g., Macholán et al. 2007; Qvarnström and Bailey 2009; Irwin 2018). Indeed, as the PLAA gene had the narrowest cline, it is possible that this locus is under stronger selection. A previous study suggested that this gene is likely under diversifying selection and often has higher divergence rates than other Z-linked loci (Backström et al. 2010); thus, this gene might not function well in a hybrid background. Thus, even though the log-likelihood confidence intervals of loci overlap, the narrower cline of the PLAA gene may suggest differential selection acting on different regions of the Z chromosome in the spinetail hybrid zone. In fact, distinct introgression patterns in different sex-linked genes seem to be a common feature in hybrid zones (Carling and Brumfield 2009; Macholán et al. 2011). On the other hand, CHDZ18 had a broader cline than the other two Z-linked genes and it is possible that the fact that it is not fixed at the end of the transect is due to either retention of ancient polymorphism or introgressive hybridization due to weaker selection acting on this gene.
Our results of cline slope and position suggest that the hybrid zone is likely maintained by a balance between selection and dispersal (Barton and Hewitt 1985; Gay et al. 2008). Although both endogenous and exogenous selection will result in a balance between selection and dispersal and consequently clines of similar shape (Barton and Gale 1993), we suggest that some form of endogenous selection underlies the width and position of this hybrid zone (i.e., the tension zone model), as there is no evidence of any apparent environmental gradient (pers. observ.). In this regard, post-zygotic selection likely maintains the width of the PLAA cline. Alternatively, the cline width observed for this locus could be the result of a recent admixture event (Endler 1977). However, the PLAA cline is more than 100 km narrower than a cline estimated under a neutral diffusion process (Table S4), thereby providing further evidence that some form of selection is hindering genetic homogenization.
Our reduced sample size prevents an estimation of LD among loci. However, as clines of distinct loci were both coincident and concordant, it is possible that these loci are statistically associated. In our analyses, the simple sigmoid cline model (Model 1; only centre and slope fitted) was favored over stepped cline models (i.e., with centre, slope, and exponential tails fitted) for all loci. This may suggest two things. First, LD may be responsible to have brought these clines together (except the CHDZ18 locus) into geographic coincidence and concordance. When alleles at different loci come into LD, clines tend to be spatially clustered and selection tends to act on them as a unit, augmenting selection against hybrids and reducing cline width (Bazykin 1969; Barton 1983; Szymura and Barton 1991; Nürnberger et al. 1995; Kruuk et al. 1999). Second, selection pressures on each individual locus may also have played a significant role, as loci influenced by this process are expected to have sigmoid rather than stepped clines (Barton 1983; Vines et al. 2016). Overall, these results suggest that (i) both the indirect effect of loci in LD and the effect of direct selection on individual loci may influence the fate of the spinetail hybrid zone and that (ii) the Jequitinhonha River is a permeable barrier to dispersal, as strong geographic barriers are expected to produce stepped clines (Barton and Hewitt 1989; Kawakami and Butlin 2012).
Geographic origin of the hybrid zone
The primary divergence and secondary contact scenarios were proposed as alternatives to account for the origin of hybrid zones long ago (Mayr 1942), but relatively few studies have contrasted them in a spatial–temporal framework. Most of the empirical evidence that hybrid zones arose through secondary contact comes from studies in the temperate zone (Barton and Hewitt 1985; Swenson 2008), but recent analyses also provide evidence supporting this model for the Neotropical region (Arias et al. 2012; Morales-Rozo et al. 2017). In our study, signatures of population expansion were found in both ENM (Fig. 5) and historical demographic analyses (Fig. 3 and Table 2) and suggest that the spinetail hybrid zone arose as secondary contact between populations that differentiated in allopatry in distinct refugia. Although the credibility interval associated with the divergence time is wide (Pleistocene to late Pliocene, Table 3), and that previous secondary contact periods not modeled here are likely, our paleodistribution models sets a minimum and recent age for the origin of the spinetail hybrid zone (i.e., during the LGM). This scenario is also consistent with evidence based on fossil pollen (e.g., Behling 1995, 1997, 2002; Behling and Negrelle 2001), phylogeographic studies (e.g., Cabanne et al. 2008; Maldonado-Coelho 2012; Batalha-Filho and Miyaki 2016), and other paleodistribution analyses (e.g., Carnaval et al. 2014; Leite et al. 2016), which indicate climatically induced fragmentation and range shifts of the AF itself and its associated organisms in the Quaternary.
Evolutionary forces maintaining the hybrid zone
Although it is not possible to distinguish between intrinsic and extrinsic mechanisms maintaining the narrow cline width at this stage, our results provide some insights that could be tested with further studies. Alternative hypotheses that account for narrow clines include the role of pre- or post-zygotic selection (i.e., due to sexual or ecological divergence), or a combination of both. For example, ENMs suggest that when the ecological niches of S. ruficapilla and S. cinerea are inferred separately, they remain geographically restricted relative to the model when they are inferred together. This may suggest a different range of environmental conditions for either species, which in turn could indicate that ecological divergence is a source of selection causing lower hybrid fitness. However, it is also possible that this difference in ENMs is a consequence of some limitation in sampling sites. Future analyses will allow to contrast these possibilities.
Final remarks
Here, we provide a spatiotemporal diversification scenario of S. ruficapilla and S. cinerea spinetails in the AF, as well as a portrait of evolutionary processes shaping the hybrid zone between them. Divergence between species was recovered in mtDNA and Z-linked loci and suggests that isolation and differentiation in Quaternary refugia was a main cause of diversification. The narrow cline of one Z-linked locus relative to other loci suggests that some form of selection may shape this hybrid zone. Detailed studies on vocal, ecological, and genomic variation across this hybrid zone are warranted to understand further the speciation process of these taxa.
Supplementary information
Acknowledgements
The authors thank Marcos Raposo (MNRJ), Fabrício R. Santos (UFMG), Jon Fjeldså (ZMUC), Carla Fontana (MCP), and Luis F. Silveira (MZUSP) for providing some of the tissues used in this study. The authors thank Fábio S. R. Amaral, Claydson P. Assis, Luciano M. Lima, Ana C. Fazza, Tiago S. Ribeiro, and Sidnei S. Santos for their immense help in fieldwork. The authors are grateful to Rilquer Mascarenhas for help with ecological niche model analyses. The authors thank the Reserva Ecológica Michelin (Kevin Flesher) for logistic support during sampling in its area. This study was funded by Fundação de Amparo à Pesquisa do Estado de São Paulo—FAPESP (2008/05705-4; BIOTA 2013/50297-0), NSF (DEB 1343578), NASA, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 443249/2014-8, 303713/2015-1), PROPCI/UFBA (PRODOC-2013/5813), and Fundação de Amparo à Pesquisa do Estado da Bahia (FAPESB RED0045/2014, JCB0026/2016). H.B.-F. also acknowledges the CNPq Research Productivity Fellowships (307037/2018-5). Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis and Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) provided permits to collect the samples. This work was developed in the Research Center on Biodiversity and Computing (BioComp) of the Universidade de São Paulo (USP), supported by the USP Provost’s Office for Research. The authors thank two anonymous reviewers for their comments that improved this work.
Data archiving
The sequences generated in this study were deposited in GenBank under the following accession numbers: cytb, MK920451–MK920538; ND2, MK920539–MK920623; FIB5, MK920697–MK920776; myo2, MK920624–MK920696; BRM15, MK920873–MK920920; CHDZ18, MK920825–MK920872; PLAA, MK920777–MK920824.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41437-019-0234-y) contains supplementary material, which is available to authorized users.
References
- Allouche O, Tsoar A, Kadmon R. Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS) J Appl Ecol. 2006;43:1223–1232. doi: 10.1111/j.1365-2664.2006.01214.x. [DOI] [Google Scholar]
- Amaral FR, Albers PK, Edwards SV, Miyaki CY. Multilocus tests of Pleistocene refugia and ancient divergence in a pair of Atlantic Forest antbirds (Myrmeciza) Mol Ecol. 2013;22:3996–4013. doi: 10.1111/mec.12361. [DOI] [PubMed] [Google Scholar]
- Amaro RC, Rodrigues MT, Yonenaga-Yassuda Y, Carnaval AC. Demographic processes in the montane Atlantic rainforest: molecular and cytogenetic evidence from the endemic frog Proceratophrys boiei. Mol Phylogenet Evol. 2012;62:880–888. doi: 10.1016/j.ympev.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Araújo MB, New M. Ensemble forecasting of species distributions. Trends Ecol Evol. 2007;22:42–47. doi: 10.1016/j.tree.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Arias CF, Rosales C, Salazar C, Castaño J, Bermingham E, Linares M, et al. Sharp genetic discontinuity across a unimodal Heliconius hybrid zone. Mol Ecol. 2012;21:5778–5794. doi: 10.1111/j.1365-294X.2012.05746.x. [DOI] [PubMed] [Google Scholar]
- Avise JC. Phylogeography: retrospect and prospect. J Biogeogr. 2009;36:3–15. doi: 10.1111/j.1365-2699.2008.02032.x. [DOI] [Google Scholar]
- Backström N, Lindell J, Zhang Y, Palkopoulou E, Qvarnström A, Saetre G-P, et al. A high-density scan of the Z chromosome in Ficedula flycatchers reveals candidate loci for diversifying selection. Evolution. 2010;64:3461–3475. doi: 10.1111/j.1558-5646.2010.01082.x. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Bantock TM, Prys-Jones RTP, Lee PLM. New and improved molecular sexing for museum bird specimens. Mol Ecol Notes. 2008;8:519–528. doi: 10.1111/j.1471-8286.2007.01999.x. [DOI] [PubMed] [Google Scholar]
- Barton NH. The dynamics of hybrid zones. Heredity. 1979;43:341–359. doi: 10.1038/hdy.1979.87. [DOI] [Google Scholar]
- Barton NH. Multilocus clines. Evolution. 1983;37:454–471. doi: 10.1111/j.1558-5646.1983.tb05563.x. [DOI] [PubMed] [Google Scholar]
- Barton NH, Hewitt GM. Analysis of hybrid zones. Annu Rev Ecol Evol Syst. 1985;16:113–148. doi: 10.1146/annurev.es.16.110185.000553. [DOI] [Google Scholar]
- Barton NH, Hewitt GM. Adaptation, speciation and hybrid zones. Nature. 1989;341:497–503. doi: 10.1038/341497a0. [DOI] [PubMed] [Google Scholar]
- Barton NH, Gale KS. Genetic analysis of hybrid zones. In: Harrison R, editor. Hybrid zones and the evolutionary process. New York, NY: Oxford University Press; 1993. pp. 13–45. [Google Scholar]
- Barton NH, Turelli M. Spatial waves of advance with bistable dynamics: cytoplasmic and genetic analogues of Allee effects. Am Nat. 2011;178:E48–E75. doi: 10.1086/661246. [DOI] [PubMed] [Google Scholar]
- Batalha-Filho H, Cabanne GS, Miyaki CY. Phylogeography of an Atlantic forest passerine reveals demographic stability through the last glacial maximum. Mol Phylogenet Evol. 2012;65:892–902. doi: 10.1016/j.ympev.2012.08.010. [DOI] [PubMed] [Google Scholar]
- Batalha-Filho H, Irestedt M, Fjeldså J, Ericson PGP, Silveira LF, Miyaki CY. Molecular systematics and evolution of the Synallaxis ruficapilla complex (Aves: Furnariidae) in the Atlantic Forest. Mol Phylogenet Evol. 2013;67:86–94. doi: 10.1016/j.ympev.2013.01.007. [DOI] [PubMed] [Google Scholar]
- Batalha-Filho H, Miyaki CY. Late Pleistocene divergence and postglacial expansion in the Brazilian Atlantic Forest: multilocus phylogeography of Rhopias gularis (Aves: Passeriformes) J Zool Syst Evol Res. 2016;54:137–147. doi: 10.1111/jzs.12118. [DOI] [Google Scholar]
- Batalha-Filho H, Waldschmidt AM, Campos LAO, Tavares MG, Fernandes-Salomão TM. Phylogeography and historical demography of the Neotropical stingless bee Melipona quadrifasciata (Hymenoptera, Apidae): incongruence between morphology and mitochondrial DNA. Apidologie. 2010;41:534–547. doi: 10.1051/apido/2010001. [DOI] [Google Scholar]
- Bazykin AD. Hypothetical mechanism of speciation. Evolution. 1969;23:685–687. doi: 10.1111/j.1558-5646.1969.tb03550.x. [DOI] [PubMed] [Google Scholar]
- Behling H. Investigations into the Late Pleistocene and Holocene history of vegetation and climate in Santa Catarina (S Brazil) Veg Hist Archaeobot. 1995;4:127–152. doi: 10.1007/BF00203932. [DOI] [Google Scholar]
- Behling H. Late Quaternary vegetation, climate and fire history of the Araucaria forest and campos region from Serra Campos Gerais, Paraná State (South Brazil) Rev Palaeobot Palynol. 1997;97:109–121. doi: 10.1016/S0034-6667(96)00065-6. [DOI] [Google Scholar]
- Behling H. South and southeast Brazilian grasslands during Late Quaternary Times: a synthesis. Palaeogeogr Palaeoclim Palaeoecol. 2002;177:19–27. doi: 10.1016/S0031-0182(01)00349-2. [DOI] [Google Scholar]
- Behling H, Negrelle RRB. Tropical rain forest and climate dynamics of the Atlantic lowland, southern Brazil, during the late Quaternary. Quat Res. 2001;56:383–389. doi: 10.1006/qres.2001.2264. [DOI] [Google Scholar]
- Borge T, Webster MT, Andersson G, Saetre G-P. Contrasting patterns of polymorphism and divergence on the Z chromosome and autosomes in two Ficedula flycatcher species. Genetics. 2005;171:1861–1873. doi: 10.1534/genetics.105.045120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunes TO, Alexandrino J, Baêta D, Zina J, Haddad CFB, Sequeira F. Species limits, phylogeographic and hybridization patterns in Neotropical leaf frogs (Phyllomedusinae) Zool Scr. 2014;43:586–604. doi: 10.1111/zsc.12079. [DOI] [Google Scholar]
- Brunes TO, Sequeira F, Haddad CF, Alexandrino J. Gene and species trees of a Neotropical group of treefrogs: genetic diversification in the Brazilian Atlantic Forest and the origin of a polyploid species. Mol Phylogenet Evol. 2010;57:1120–1133. doi: 10.1016/j.ympev.2010.08.026. [DOI] [PubMed] [Google Scholar]
- Cabanne GS, d’Horta FM, Sari EHR, Santos FR, Miyaki CY. Nuclear and mitochondrial phylogeography of the Atlantic forest endemic Xiphorhynchus fuscus (Aves: Dendrocolaptidae): biogeography and systematic implications. Mol Phylogenet Evol. 2008;49:760–773. doi: 10.1016/j.ympev.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Cabanne GS, Santos FR, Miyaki CY. Phylogeography of Xiphorhynchus fuscus (Passeriformes, Dendrocolaptidae): vicariance and recent demographic expansion in southern Atlantic forest. Biol J Linn Soc. 2007;91:73–84. doi: 10.1111/j.1095-8312.2007.00775.x. [DOI] [Google Scholar]
- Cabanne GS, Sari EHR, Meyer D, Santos FR, Miyaki CY. Matrilineal evidence for demographic expansion, low diversity and lack of phylogeographic structure in the Atlantic forest endemic Greenish Schiffornis Schiffornis virescens (Aves: Tityridae) J Ornithol. 2013;154:371–384. doi: 10.1007/s10336-012-0901-8. [DOI] [Google Scholar]
- Carling MD, Brumfield RT. Haldane’s rule in an avian system: using cline theory and divergence population genetics to test for differential introgression of mitochondrial, autossomal, and sex-linked loci across the Passerina bunting hybrid zone. Evolution. 2008;62:2600–2615. doi: 10.1111/j.1558-5646.2008.00477.x. [DOI] [PubMed] [Google Scholar]
- Carling MD, Brumfield RT. Speciation in Passerina buntings: introgression patterns of sex-linked loci identify a candidate gene region for reproductive isolation. Mol Ecol. 2009;18:834–847. doi: 10.1111/j.1365-294X.2008.04038.x. [DOI] [PubMed] [Google Scholar]
- Carnaval AC, Hickerson MJ, Haddad CFB, Rodrigues MT, Moritz C. Stability predicts genetic diversity in the Brazilian Atlantic forest hotspot. Science. 2009;323:785–789. doi: 10.1126/science.1166955. [DOI] [PubMed] [Google Scholar]
- Carnaval AC, Waltari E, Rodrigues MT, Rosauer D, VanDerWal J, Damasceno R, et al. Prediction of phylogeographic endemism in an environmentally complex biome. Proc R Soc B. 2014;281:20141461. doi: 10.1098/rspb.2014.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheviron ZA, Hackett SJ, Capparella AP. Complex evolutionary history of a Neotropical lowland forest bird (Lepidothrix coronata) and its implication for historical hypotheses of the origin of Neotropical avian diversity. Mol Phylogent Evol. 2005;36:338–357. doi: 10.1016/j.ympev.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Dantas GPM, Sari EHR, Cabanne GS, Pessoa RO, Marini MA, Miyaki CY, et al. Population genetic structure of the Atlantic Forest endemic Conopophaga lineata (Passeriformes: Conopophagidae) reveals a contact zone in the Atlantic Forest. J Ornithol. 2015;156:85–99. doi: 10.1007/s10336-014-1106-0. [DOI] [Google Scholar]
- Derryberry EP, Derryberry GE, Maley JM, Brumfield RT. hzar: hybrid zone analysis using an R software package. Mol Ecol Res. 2014;14:652–663. doi: 10.1111/1755-0998.12209. [DOI] [PubMed] [Google Scholar]
- d’Horta F, Cabanne GS, Meyer D, Miyaki CY. The genetic effects of Late Quaternary climatic changes over a tropical latitudinal gradient: diversification of an Atlantic Forest passerine. Mol Ecol. 2011;20:1932–1935. doi: 10.1111/j.1365-294X.2011.05063.x. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett R, Buttel L, Harrison R. Spatial models for hybrid zones. Heredity. 2000;84:9–19. doi: 10.1046/j.1365-2540.2000.00566.x. [DOI] [PubMed] [Google Scholar]
- Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- Elgvin TO, Hermansen JS, Fijarczyk A, Bonnet T, Borge T, Sæther SA, et al. Hybrid speciation in sparrows II: a role for sex chromosomes? Mol Ecol. 2011;20:3823–3837. doi: 10.1111/j.1365-294X.2011.05182.x. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Molecular evolutionary genomics of birds. Cytogenet Genome Res. 2007;117:120–130. doi: 10.1159/000103172. [DOI] [PubMed] [Google Scholar]
- Endler JA. Geographic variation, speciation, and clines. Monogr Popul Biol. 1977;10:201–246. [PubMed] [Google Scholar]
- Endler JA. Problems in distinguishing historical from ecological factors in biogeography. Am Zool. 1982;22:441–452. doi: 10.1093/icb/22.2.441. [DOI] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analyses of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes AM, Wink M, Aleixo A. Phylogeography of the chestnut-tailed antbird (Myrmeciza hemimelaena) clarifies the role of rivers in Amazonian biogeography. J Biogeogr. 2012;39:1524–1535. doi: 10.1111/j.1365-2699.2012.02712.x. [DOI] [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay L, Crochet P-A, Bell DA, Lenormand T. Comparing clines on molecular and phenotypic traits in hybrid zones: a window on tension zone models. Evolution. 2008;62:2789–2806. doi: 10.1111/j.1558-5646.2008.00491.x. [DOI] [PubMed] [Google Scholar]
- Gelman A, Rubin D. Inference from iterative simulation using multiple sequences. Stat Sci. 1992;7:457–511. doi: 10.1214/ss/1177011136. [DOI] [Google Scholar]
- Griffiths R, Double M, Orr KCY, Dawson RJG. A DNA test to sex most birds. Mol Ecol. 1998;7:1071–1075. doi: 10.1046/j.1365-294x.1998.00389.x. [DOI] [PubMed] [Google Scholar]
- Hackett SJ. Molecular phylogenetics and biogeography of tanagers in the genus Ramphocelus (Aves) Mol Phylogenet Evol. 1996;5:368–382. doi: 10.1006/mpev.1996.0032. [DOI] [PubMed] [Google Scholar]
- Haffer J. Speciation in Amazonian forest birds. Science. 1969;165:131–137. doi: 10.1126/science.165.3889.131. [DOI] [PubMed] [Google Scholar]
- Haffer J. Alternative models of vertebrate speciation in Amazonia: an overview. Biodivers Conserv. 1997;6:451–476. doi: 10.1023/A:1018320925954. [DOI] [Google Scholar]
- Haldane JBS. Sex ratio and unisexual sterility in animal hybrids. J Genet. 1922;12:101–109. doi: 10.1007/BF02983075. [DOI] [Google Scholar]
- Harrison RG. Hybrid zones: windows on evolutionary process. In: Futuyma D, Antonovics J, editors. Surveys in evolutionary biology. New York, NY: Oxford University Press; 1990. pp. 69–128. [Google Scholar]
- Heled J, Drummond AJ. Bayesian inference of population size history from multiple loci. BMC Evol Biol. 2008;8:289. doi: 10.1186/1471-2148-8-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslewood MM, Elphinstone MS, Tidemann SC, Baverstock PR. Myoglobin intron variation in the Gouldian finch Erythrura gouldiae assessed by temperature gradient gel electrophoresis. Electrophoresis. 1998;19:142–151. doi: 10.1002/elps.1150190203. [DOI] [PubMed] [Google Scholar]
- Hey J. Isolation with migration models for more than two populations. Mol Biol Evol. 2010;27:905–920. doi: 10.1093/molbev/msp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167:747–60. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans RJ, van Etten J (2014) raster: Geographic data analysis and modeling. R package version, 2(8)
- Ho SY, Shapiro B. Skyline-plot methods for estimating demographic history from nucleotide sequences. Mol Ecol Resour. 2011;11:423–434. doi: 10.1111/j.1755-0998.2011.02988.x. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Irwin DE. Sex chromosomes and speciation in birds and other ZW systems. Mol Ecol. 2018;27:3831–3851. doi: 10.1111/mec.14537. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Jansson R, Dynesius M. The fate of clades in a world of recurrent climatic change: Milankovitch oscillations and evolution. Annu Rev Ecol Syst. 2002;33:741–77. doi: 10.1146/annurev.ecolsys.33.010802.150520. [DOI] [Google Scholar]
- Johnson KP, Sorenson MD. Comparing molecular evolution in two mitochondrial coding genes (cytochrome b and ND2) in the dabbling ducks (tribe: Anatini) Mol Phylogenet Evol. 1998;10:82–94. doi: 10.1006/mpev.1997.0481. [DOI] [PubMed] [Google Scholar]
- Kawakami T, Butlin, RK (2012) Hybrid zones. In: eLS. John Wiley & Sons Ltd., Chichester. 10.1002/9780470015902.a0001752.pub2
- Kruuk LE, Baird SJE, Gale KS, Barton NH. A comparison of multilocus clines maintained by environmental adaptation or by selection against hybrids. Genetics. 1999;153:1959–1971. doi: 10.1093/genetics/153.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JW, Bryant D. PopART: full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6:1110–1116. doi: 10.1111/2041-210X.12410. [DOI] [Google Scholar]
- Leite YL, Costa LP, Loss AC, Rocha RG, Batalha-Filho H, Bastos AC, et al. Neotropical forest expansion during the last glacial period challenges refuge hypothesis. Proc Natl Acad Sci USA. 2016;113:1008–1013. doi: 10.1073/pnas.1513062113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSPv5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lougheed SC, Freeland JR, Handford P, Boag PT. A molecular phylogeny of warbling-finches (Poospiza): paraphyly in a Neotropical emberizid genus. Mol Phylogenet Evol. 2000;17:367–378. doi: 10.1006/mpev.2000.0843. [DOI] [PubMed] [Google Scholar]
- Macholán M, Baird SJE, Dufková P, Munclinger P, Bímová BV, Piálek J. Assessing multilocus introgression patterns: a case study on the mouse X chromosome in central Europe. Evolution. 2011;65:1428–1446. doi: 10.1111/j.1558-5646.2011.01228.x. [DOI] [PubMed] [Google Scholar]
- Macholán M, Munclinger P, Sugerkova M, Dufková P, Bímová B, Bozikova E, et al. Genetic analysis of autosomal and x-linked markers across a mouse hybrid zone. Evolution. 2007;61:746–771. doi: 10.1111/j.1558-5646.2007.00065.x. [DOI] [PubMed] [Google Scholar]
- Maldonado-Coelho M. Climatic oscillations shape the phylogeographical structure of Atlantic Forest fire-eye antbirds (Aves: Thamnophilidae) Biol J Linn Soc. 2012;105:900–924. doi: 10.1111/j.1095-8312.2011.01823.x. [DOI] [Google Scholar]
- Maldonado‐Coelho M, Blake JG, Silveira LF, Batalha‐Filho H, Ricklefs RE. Rivers, refuges and population divergence of fire‐eye antbirds (Pyriglena) in the Amazon Basin. J Evol Biol. 2013;26:1090–1107. doi: 10.1111/jeb.12123. [DOI] [PubMed] [Google Scholar]
- Marini MA, Hackett SJ. A multifaceted approach to the characterization of an intergeneric hybrid manakin (Pipridae) from Brazil. Auk. 2002;119:1114–1120. doi: 10.1093/auk/119.4.1114. [DOI] [Google Scholar]
- Martins FM. The Brazilian Atlantic forest historical biogeography and the Carnaval–Moritz model of Pleistocene refugia: what do the phylogeographical studies tell us? Biol J Linn Soc. 2011;104:499–509. doi: 10.1111/j.1095-8312.2011.01745.x. [DOI] [Google Scholar]
- Martins FM, Templeton AR, Pavan ACO, Kohlbach BC, Morgante JS. Phylogeography of the common vampire bat (Desmodus rotundus): marked population structure, Neotropical Pleistocene vicariance and incongruence between nuclear and mtDNA markers. BMC Evol Biol. 2009;9:294. doi: 10.1186/1471-2148-9-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. Systematics and the origin of species. New York, NY: Columbia University Press; 1942. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, 14 November 2010, p 1–8
- Morales-Rozo A, Tenorio EA, Carling MD, Cadena CD. Origin and cross-century dynamics of an avian hybrid zone. BMC Evol Biol. 2017;17:257. doi: 10.1186/s12862-017-1096-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz C, Patton JL, Schneider CJ, Smith TB. Diversification of rainforest faunas: an integrated molecular approach. Annu Rev Ecol Syst. 2000;31:533–563. doi: 10.1146/annurev.ecolsys.31.1.533. [DOI] [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Wakeley J. Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics. 2001;158:885–896. doi: 10.1093/genetics/158.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger B, Barton NH, MacCallum CJ, Gilchrist J, Appleby M. Natural selection on quantitative traits in a Bombina hybrid zone. Evolution. 1995;49:1224–1238. doi: 10.1111/j.1558-5646.1995.tb04449.x. [DOI] [PubMed] [Google Scholar]
- Nylander JAA (2004) MrModeltest 2.2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University
- Pacheco JF, Gonzaga LP. A new species of Synallaxis of the ruficapila/infuscata complex form eastern Brazil (Passeriformes: Funariidae) Ararajuba. 1995;3:3–11. [Google Scholar]
- Palumbi SR, Cipriano F, Hare MP. Predicting nuclear gene coalescence from mitochondrial data: the three-times rule. Evolution. 2001;55:859–868. doi: 10.1554/0014-3820(2001)055[0859:PNGCFM]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Pellegrino KCM, Rodrigues MT, Waite AN, Morando M, Yassuda YY, Sites JW., Jr Phylogeography and species limits in the Gymnodactylus darwinii complex (Gekkonidae, Squamata): genetic structure coincides with river systems in the Brazilian Atlantic Forest. Biol J Linn Soc. 2005;85:13–26. doi: 10.1111/j.1095-8312.2005.00472.x. [DOI] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qvarnström A, Bailey RI. Speciation through evolution of sex-linked genes. Heredity. 2009;102:4–15. doi: 10.1038/hdy.2008.93. [DOI] [PubMed] [Google Scholar]
- Ramos-Onsins S, Rozas J. Statistical properties of new neutrality test against population growth. BMC Evol Biol. 2002;19:2092–2100. doi: 10.1093/oxfordjournals.molbev.a004034. [DOI] [PubMed] [Google Scholar]
- Ribas CC, Aleixo A, Nogueira ACR, Miyaki CY, Cracraft J. A palaeobiogeographic model for biotic diversification within Amazonia over the past three million years. Proc R Soc B. 2012;279:681–689. doi: 10.1098/rspb.2011.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro RA, Lemos-Filho JP, Ramos ACS, Lovato MB. Phylogeography of the endangered rosewood Dalbergia nigra (Fabaceae): insights into the evolutionary history and conservation of the Brazilian Atlantic Forest. Heredity. 2010;106:46–57. doi: 10.1038/hdy.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribon R, Whitney BM, Pacheco JF. Discovery of Bahia spinetail Synallaxis cinerea in north-east Minas Gerais, Brasil, with additional records of some rare and threatened montane Atlantic forest birds. Cotinga. 2002;17:46–50. [Google Scholar]
- Ridgely R, Tudor G. The birds of South America, volume II: the suboscine passerines. Austin: University of Texas Press; 1994. [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Hohna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. Genepop'007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Res. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Rull V. Speciation timing and Neotropical biodiversity: the Tertiary–Quaternary debate in the light of molecular phylogenetic evidence. Mol Ecol. 2008;17:2722–2729. doi: 10.1111/j.1365-294X.2008.03789.x. [DOI] [PubMed] [Google Scholar]
- Rull V. Neotropical biodiversity: timing and potential drivers. Trends Ecol Evol. 2011;26:508–513. doi: 10.1016/j.tree.2011.05.011. [DOI] [PubMed] [Google Scholar]
- Sick H. Ornitologia Brasileira. Rio de Janeiro: Nova Fronteira; 1997. [Google Scholar]
- Silva JMC, Straube FC. Systematics and biogeography of Scaled Woodcreepers (Aves: Dendrocolaptidae) Study Neotrop Fauna Environ. 1996;31:3–10. doi: 10.1076/snfe.31.1.3.13321. [DOI] [Google Scholar]
- Silveira LF. Synallaxis cinerea Wied, 1831. In: Machado ABM, Drummond GM, Paglia AP, editors. Livro Vermelho da Fauna Brasileira Ameaçada de Extinção, vol 1. Belo Horizonte: Fundação Biodiversitas; 2008. pp. 578–579. [Google Scholar]
- Slade RW, Moritz C, Heideman A, Hale PT. Rapid assessment of single-copy nuclear DNA variation in diverse species. Mol Ecol. 1993;2:359–373. doi: 10.1111/j.1365-294X.1993.tb00029.x. [DOI] [PubMed] [Google Scholar]
- Smith BT, McCormack JE, Cuervo AM, Hickerson MJ, Aleixo A, Cadena CD, et al. The drivers of tropical speciation. Nature. 2014;515:406–409. doi: 10.1038/nature13687. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith N, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopiglia R, Raposo MA, Teixeira DM. Taxonomy and geographic variation of the Synallaxis ruficapilla Vieillot, 1819 species-complex (Aves: Passeriformes: Furnariidae) J Ornithol. 2013;154:191–207. doi: 10.1007/s10336-012-0886-3. [DOI] [Google Scholar]
- Swenson NG. The past and future influence of geographic information systems on hybrid zone, phylogeographic and speciation research. J Evol Biol. 2008;21:421–434. doi: 10.1111/j.1420-9101.2007.01487.x. [DOI] [PubMed] [Google Scholar]
- Swofford DL. Phylogenetic analysis using parsimony (* and other methods), v. 4. Sunderland: Sinauer; 1998. [Google Scholar]
- Szymura JM, Barton NH. Genetic analysis of a hybrid zone between the fire-bellied toads, Bombina bombina and Bombina variegata, near Cracow in southern Poland. Evolution. 1986;40:1141–1159. doi: 10.1111/j.1558-5646.1986.tb05740.x. [DOI] [PubMed] [Google Scholar]
- Szymura JM, Barton NH. The genetic structure of the hybrid zone between the fire-bellied toads Bombina bombina and B. variegata: comparisons between transects and between loci. Evolution. 1991;45:237–261. doi: 10.1111/j.1558-5646.1991.tb04400.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terribile LC, Lima-Ribeiro MS, Araújo MB, Bizão N, Collevatti RG, Dobrovolski R, et al. Areas of climate stability of species ranges in the Brazilian Cerrado: disentangling uncertainties through time. Nat Conserv. 2012;10:152–159. doi: 10.4322/natcon.2012.025. [DOI] [Google Scholar]
- Thomé MTC, Zamudio KR, Giovanelli JGR, Haddad CFB, Baldissera FA, Jr, Alexandrino JMB. Phylogeography of endemic toads and post-Pliocene persistence of the Brazilian Atlantic Forest. Mol Phylogenet Evol. 2010;55:1018–1031. doi: 10.1016/j.ympev.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Thomé MTC, Zamudio KR, Haddad CFB, Alexandrino J. Delimiting genetic units in Neotropical toads under incomplete lineage sorting and hybridization. BMC Evol Biol. 2012;12:242. doi: 10.1186/1471-2148-12-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomé MTC, Zamudio KR, Haddad CFB, Alexandrino J. Barriers, rather than refugia, underlie the origin of diversity in toads endemic to the Brazilian Atlantic Forest. Mol Ecol. 2014;23:6152–6164. doi: 10.1111/mec.12986. [DOI] [PubMed] [Google Scholar]
- Thuiller W, Lafourcade B, Engler R, Araújo MB. BIOMOD—a platform for ensemble forecasting of species distributions. Ecography. 2009;32:369–373. doi: 10.1111/j.1600-0587.2008.05742.x. [DOI] [Google Scholar]
- Tonini JFR, Costa LP, Carnaval AC. Phylogeographic structure is strong in the Atlantic Forest; predictive power of correlative paleodistribution models, not Always. J Zool Syst Evol Res. 2013;51:114–121. doi: 10.1111/jzs.12014. [DOI] [Google Scholar]
- Turchetto-Zolet AC, Pinheiro F, Salgueiro F, Palma-Silva C. Phylogeographical patterns shed light on evolutionary process in South America. Mol Ecol. 2013;22:1193–1213. doi: 10.1111/mec.12164. [DOI] [PubMed] [Google Scholar]
- Vanzolini PE, Williams EE. South American anoles: the geographic differentiation and evolution of the Anolis chrysolepis species group (Sauria: iguanidae) Arq Zool. 1970;19:1–298. doi: 10.11606/issn.2176-7793.v19i1-2p1-176. [DOI] [Google Scholar]
- Vasconcelos MF, Silva JF. Limite norte da distribuição do pichororé Synallaxis ruficapilla (Aves, Furnariidae) Bol Mus Biol Mello Leitão. 2004;17:65–68. [Google Scholar]
- Vines TH, Dalziel AC, Albert AYK, Veen T, Schulte PM, Schluter D. Cline coupling and uncoupling in a stickleback hybrid zone. Evolution. 2016;70:1023–1038. doi: 10.1111/evo.12917. [DOI] [PubMed] [Google Scholar]
- Wallace AR. On the monkeys of the Amazon. Proc Zool Soc Lond. 1852;20:107–110. [Google Scholar]
- Weir JT, Schluter D. Calibrating the avian molecular clock. Mol Ecol. 2008;17:2321–2328. doi: 10.1111/j.1365-294X.2008.03742.x. [DOI] [PubMed] [Google Scholar]
- Woltmann S, Sherry TW, Kreiser BR. A genetic approach to estimate natal dispersal distances and self-recruitment in resident rainforest birds. J Avian Biol. 2012;43:33–42. doi: 10.1111/j.1600-048X.2011.05572.x. [DOI] [Google Scholar]
- Zink RM, Barrowclough GF. Mitochondrial DNA under siege in avian phylogeography. Mol Ecol. 2008;17:2107–2121. doi: 10.1111/j.1365-294X.2008.03737.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences generated in this study were deposited in GenBank under the following accession numbers: cytb, MK920451–MK920538; ND2, MK920539–MK920623; FIB5, MK920697–MK920776; myo2, MK920624–MK920696; BRM15, MK920873–MK920920; CHDZ18, MK920825–MK920872; PLAA, MK920777–MK920824.