Abstract
The hook length of the flagellum is controlled to about 55 nm in Salmonella. The flagellar type III protein export apparatus secretes FliK to determine hook length during hook assembly and changes its substrate specificity from the hook protein to the filament protein when the hook length has reached about 55 nm. Salmonella FliK consists of an N-terminal domain (FliKN, residues 1–207), a C-terminal domain (FliKC, residues 268–405) and a flexible linker (FliKL, residues 208–267) connecting these two domains. FliKN is a ruler to measure hook length. FliKC binds to a transmembrane export gate protein FlhB to undergo the export switching. FliKL not only acts as part of the ruler but also contributes to this switching event, but it remains unknown how. Here we report that FliKL is required for efficient interaction of FliKC with FlhB. Deletions in FliKL not only shortened hook length according to the size of deletions but also caused a loose length control. Deletion of residues 206–265 significantly reduced the binding affinity of FliKC for FlhB, thereby producing much longer hooks. We propose that an appropriate length of FliKL is required for efficient interaction of FliKC with FlhB.
Subject terms: Motor protein function, Bacterial secretion
Introduction
The bacterial flagellar hook is a tubular structure composed of the hook protein FlgE and acts as a universal joint to smoothly transmit torque produced by the flagellar motor to the long helical filament that functions as a propeller1. The hook length of the Salmonella flagellum is controlled to about 55 nm2, and the length control is important for the universal joint mechanism3.
The flagellar type III protein export apparatus is located at the flagellar base and transports FlgE subunits from the cytoplasm to the distal end of the growing hook structure for hook assembly4,5. Newly exported FlgE monomers polymerize onto the nascent hook structure with the help of the hook cap (FlgD), which is located at the hook tip6. Interactions between FlgD and FlgE suppress the leakage of FlgE subunits into the culture media so that each FlgE subunit can be efficiently incorporated into the hook structure7,8.
When hook length has reached about 55 nm, the flagellar type III protein export apparatus changes its substrate specificity, thereby terminating hook assembly and initiating filament assembly9,10. FliK, FlhA, FlhB and Flk are directly involved in the export switching process11–14. If certain mutations in FliK, FlhB or FlhA inhibits the export switching of the flagellar type III protein export apparatus, unusually elongated hooks called polyhooks are generated. FlhA and FlhB are transmembrane proteins of the flagellar protein export apparatus15,16, and their C-terminal cytoplasmic domains (FlhAC, FlhBC) project into the cavity within the basal body C ring15,16. FlhAC forms a homo-nonameric ring structure17,18 and provides binding-sites for cytoplasmic export components (FliH, FliI, FliJ)19–21 and flagellar export chaperones (FlgN, FliS, FliT) in with their cognate substrates22–25. Interactions of FlhAC with FliH, FliI and FliJ seems to support the strict order of flagellar assembly26,27. The interaction of FlhBC with FliK induces the export switching of the flagellar type III protein export apparatus28–30. FlhBC binds to FliH, FliI, FliJ and export substrates such as FlgD and FlgE19,31,32. Conformational changes of FlhAC and FlhBC are required for the substrate specificity switching of the flagellar protein export apparatus upon hook completion18,28,33. Flk interferes with premature switching of the protein export apparatus during hook-basal body assembly13,14,34.
The flagellar type III protein export apparatus transports several FliK molecules during hook assembly to determine hook length in a way that the protein export apparatus switches its substrate specificity when the hook length has reached about 55 nm35–38. Salmonella FliK is a 405 amino-acid protein consisting of the N-terminal ruler domain (FliKN, residues 1–207), the C-terminal export switch domain (FliKC, residues 268–405) and a flexible linker (FliKL, residues 208–267) connecting these two domains (Fig. 1a)39–41. FliKN contains a hook-type export signal recognized by the flagellar type III protein export apparatus31,35. Insertions and deletions in FliKN make the hook longer and shorter, respectively, suggesting that FliKN is a molecular ruler42. FliKN binds to FlgD and FlgE43–45, and deletion of residues 129–159 in FliKN reduces the binding affinity of FliK for FlgE, thereby producing longer hooks with the filament attached. This suggests that the interaction of FliKN with FlgE is required for proper hook length measurement44. FliKC consists of a compactly folded core domain consisting of residues 268–352 and an intrinsically disordered C-terminal tail composed of residues 353–405 (FliKCT) (Fig. 1a)40,46. The core domain of FliKC binds to FlhBC to allow the flagellar protein export apparatus to switch its substrate specificity from rod- (FlgB, FlgC, FlgF, FlgG, FlgJ, FliE) and hook-type (FlgD, FlgE, FliK) to filament-type (FlgK, FlgL, FlgM, FliC, FliD)11,12,28,45,47. Genetic analyses of FliKCT have suggested that FliKCT controls the export switching activity of FliKC during hook assembly45,47.
Figure 1.
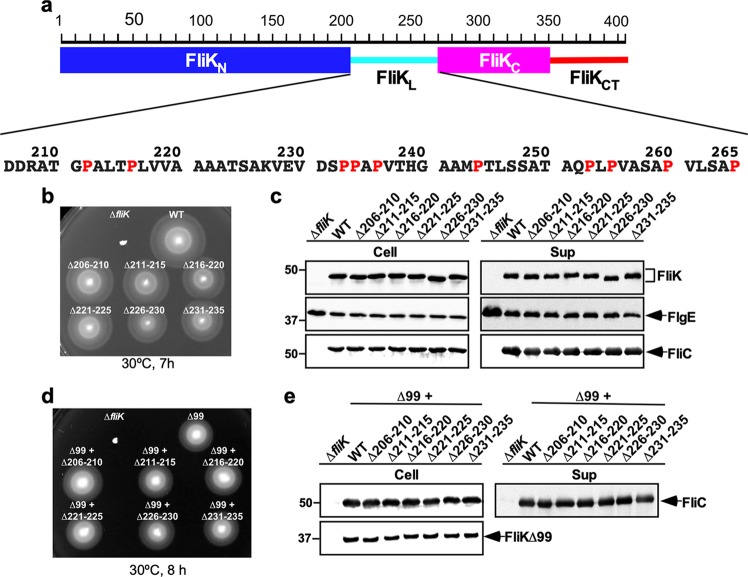
Effect of deletions of five residues within the N-terminal region of FliKL on FliK function. (a) Domain organization of FliK ruler. FliK consists of the N-terminal ruler domain (FliKN), the C-terminal export switch domain (FliKC) and a flexible linker (FliKL) connecting these two domains. FliKC has an intrinsically disordered C-terminal tail (FliKCT). Amino acid sequence of FliKL is shown. Proline residues are highlighted in red. (b) Motility of TH8426 harboring pTrc99AFF4 (∆fliK), pMK002 (WT), pMMK1001 (∆206–210), pMMK1002 (∆211–215), pMMK1003 (∆216–220), pMMK1004 (∆221–225), pMMK1005 (∆226–230) or pMMK1006 (∆231–235) in 0.35% soft agar. Plates were incubated at 30 °C for 7 hours. (c) Secretion assays of FliK, FlgE and FliC. Immunoblotting using polyclonal anti-FliK (1st row), anti-FlgE (2nd row) or anti-FliC (3rd row) antibody, of whole cell proteins (Cell) and culture supernatants (Sup) from the above strains. The regions of interest were cropped from original immunoblots shown in Fig. S6a in the Supplemental information using a software, Photoshop CS6, and then the contrast and brightness were adjusted. The positions of molecular mass markers (kDa) are indicated on the left. (d) Motility of TH8426 harboring pTrc99AFF4 (∆fliK), pNM201 (∆99), pMMK1016 (∆99 + ∆206–210), pMMK1017 (∆99 + ∆211–215), pMMK1018 (∆99 + ∆216–220), pMMK1019 (∆99 + ∆221–225), pMMK1020 (∆99 + ∆226–230) or pMMK1021 (∆99 + ∆231–235) in 0.35% soft agar containing 1 mM IPTG. Plates were incubated at 30 °C for 8 hours. (e) Secretion assay of FliC. Immunoblotting using polyclonal anti-FliC (1st row) or anti-FliK (2nd row) antibody, of whole cell proteins (Cell) and culture supernatants (Sup) from the above strains. The regions of interest were cropped from original immunoblots shown in Fig. S6b using Photoshop CS6, and then the contrast and brightness were adjusted.
FliKL contains ten proline residues (Fig. 1a)39 and hence is intrinsically disordered41. Hook lengths of the Salmonella fliK(∆238–269) (32 residues deletion) and fliK(∆248–269) (22 residues deletion) mutants are 40.2 ± 6.1 nm [mean ± standard deviation (SD)] and 51.0 ± 8.8 nm, where their average lengths are shorter than that of the wild-type strain (52.7 ± 4.5 nm)42. The length of the hook produced by the fliK(∆161–216) mutant (56 residues deletion) is 48.7 ± 22.3 nm, where the average is also shorter than that of the wild-type strain by 4 nm. However, the SD value is larger than the wild-type one, indicating a much looser length control of the hook structure42. Furthermore, deletions of residues 161–223 and 161–244 cause polyhooks without the filament attached whereas a deletion of residues 208–269 results in the polyhooks with the filament attached (polyhook–filament phenotype)42. These observations raise the possibility that FliKL not only acts as part of the ruler but also contributes to substrate specificity switching of the flagellar protein export apparatus. To clarify this hypothesis, we constructed a series of mutant variants of FliK with in-frame deletions within FliKL. We show that a proper length of FliKL between FliKN and FliKC is required for efficient interaction of FliKC with FlhBC.
Results
Effect of deletions of five amino-acid residues within residues 206–235 on hook length control
It has been shown that the N-terminal portion of FliKL is responsible for proper measurement of hook length42. To clarify the role of residues 206–235 of FliKL in the hook length control, we constructed a series of mutant variants of FliK with sequential 5-amino-acid deletions within a region of residues 206–235, namely FliK(∆206–210), FliK(∆211–215), FliK(∆216–220), (∆221–225), (∆226–230) and FliK(∆231–235) (Table 1). These six fliK deletion variants fully restored motility of the ∆fliK mutant in 0.35% soft agar plates when they were expressed from the pTrc99A-based plasmid (Fig. 1b). Consistently, the levels of FlgE and FliC secreted by these deletion mutants were detected at the wild-type levels (Fig. 1c, 2nd and 3rd rows). These fliK deletions did not affect either protein stability or protein secretion into the culture media (Fig. 1c, 1st row). Therefore, we conclude that these in-frame deletions do not affect FliK function at all.
Table 1.
Strains and plasmids used in this study.
| Strains and Plasmids | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli | ||
| BL21 Star (DE3) | Overexpression of proteins | Novagen |
| Salmonella | ||
| SJW1103 | Wild type for motility and chemotaxis | 61 |
| TH8426 | ∆fliK | 47 |
| MMK1012 | fliK(∆206–235) | This study |
| MMK1013 | fliK(∆206–245) | This study |
| MMK1014 | fliK(∆206–255) | This study |
| MMK1015 | fliK(∆206–265) | This study |
| Plasmids | ||
| pTrc99AFF4 | Modified pTrc expression vector | 62 |
| pKM002 | pTrc99AFF4/FliK | 48 |
| pNM201 | pTrc99AFF4/FliK(∆2–99) | 44 |
| pMMK521 | pETDuet-1/FliK(I304amber) + FlhBC | 45 |
| pMMK1001 | pTrc99AFF4/FliK(∆206–210) | This study |
| pMMK1002 | pTrc99AFF4/FliK(∆211–215) | This study |
| pMMK1003 | pTrc99AFF4/FliK(∆216–220) | This study |
| pMMK1004 | pTrc99AFF4/FliK(∆221–225) | This study |
| pMMK1005 | pTrc99AFF4/FliK(∆226–230) | This study |
| pMMK1006 | pTrc99AFF4/FliK(∆231–235) | This study |
| pMMK1007 | pTrc99AFF4/FliK(∆206–215) | This study |
| pMMK1008 | pTrc99AFF4/FliK(∆216–225) | This study |
| pMMK1009 | pTrc99AFF4/FliK(∆226–235) | This study |
| pMMK1010 | pTrc99AFF4/FliK(∆206–220) | This study |
| pMMK1011 | pTrc99AFF4/FliK(∆221–235) | This study |
| pMMK1012 | pTrc99AFF4/FliK(∆206–235) | This study |
| pMMK1013 | pTrc99AFF4/FliK(∆206–245) | This study |
| pMMK1014 | pTrc99AFF4/FliK(∆206–255) | This study |
| pMMK1015 | pTrc99AFF4/FliK(∆206–265) | This study |
| pMMK1015SP | pTrc99AFF4/FliK(∆206–265SP) | This study |
| pMMK1016 | pTrc99AFF4/FliK(∆2–99 + ∆206–210) | This study |
| pMMK1017 | pTrc99AFF4/FliK(∆2–99 + ∆211–215) | This study |
| pMMK1018 | pTrc99AFF4/FliK(∆2–99 + ∆216–220) | This study |
| pMMK1019 | pTrc99AFF4/FliK(∆2–99 + ∆221–225) | This study |
| pMMK1020 | pTrc99AFF4/FliK(∆2–99 + ∆226–230) | This study |
| pMMK1021 | pTrc99AFF4/FliK(∆2–99 + ∆231–235) | This study |
| pMMK1022 | pTrc99AFF4/FliK(∆2–99 + ∆206–215) | This study |
| pMMK1023 | pTrc99AFF4/FliK(∆2–99 + ∆216–225) | This study |
| pMMK1024 | pTrc99AFF4/FliK(∆2–99 + ∆226–235) | This study |
| pMMK1025 | pTrc99AFF4/FliK(∆2–99 + ∆206–220) | This study |
| pMMK1026 | pTrc99AFF4/FliK(∆2–99 + ∆221–235) | This study |
| pMMK1027 | pTrc99AFF4/FliK(∆2–99 + ∆206–235) | This study |
| pMMK1028 | pTrc99AFF4/FliK(∆2–99 + ∆206–245) | This study |
| pMMK1029 | pTrc99AFF4/FliK(∆2–99 + ∆206–255) | This study |
| pMMK1030 | pTrc99AFF4/FliK(∆2–99 + ∆206–265) | This study |
| pMMK1030SP | pTrc99AFF4/FliK(∆2–99 + ∆206–265SP) | This study |
| pMMK1031 | pETDuet-1/FliK(∆2–99 + I304amber) + FlhBC | This study |
| pMMK1032 | pETDuet-1/FliK(∆2–99 + ∆206–265 + I304amber) + FlhBC | This study |
| pMMK1032SP | pETDuet-1//FliK(∆2–99 + ∆206–265SP + I304amber) + FlhBC | This study |
The length of the most extended polypeptide chain is 0.37 nm per residue. If FliKL adopts a fully extended conformation to act as part of the ruler, we predicted that these 5-amino-acid deletions within residues 206–235 of FliKL would reduce the hook length by 1.9 nm. Therefore, we measured the hook length of these fliK deletion mutants. The average hook length of the fliK(∆206–210), fliK(∆211–215), fliK(∆216–220), fliK(∆221–225), fliK(∆226–230) and fliK(∆231–235) mutants were 48.5 ± 4.5 nm (n = 113), 49.3 ± 5.2 nm (n = 168), 49.0 ± 5.6 nm (n = 130), 48.6 ± 4.2 nm (n = 118), 49.2 ± 4.2 nm (n = 232) and 49.6 ± 5.4 nm (n = 126), respectively, which were shorter than the length of the wild-type hook (53.3 ± 6.5 nm, n = 154) (Fig. S1). Over-expression of FliK slightly shortens the hook length. In contrast, when the expression level of FliK is reduced, the cell produces polyhooks, sometimes with the filament attached27,48. Polyhooks are frequently observed when FlgE is overproduced in wild-type cells27,43. Thus, the balance between the secretion levels of FlgE and FliK seems to be critical for the proper termination of the hook assembly. Since 5-amino-acid deletions within residues 206–235 shorten the hook length by 4 nm, which is shorter than the predicted value, we assume that these deletions may affect not only hook length measurements but also the secretion process of FliK by the type III protein export apparatus and/or the export switching process of the protein export apparatus induced by the interaction of FliKC with FlhBC.
To directly test if these 5 amino-acid deletions affect the interaction of FliKC with FlhBC, it is necessary to analyze the protein transport process of FliK and the export switching process of the flagellar type III protein export apparatus separately. To do so, we used a fliK(∆2–99) allele, which is an export deficient variant of FliK, because it retains the ability to catalyze export switching of the flagellar type III protein export apparatus when over-expressed49. We introduced deletions of residues 206–210, 211–215, 216–220, 221–225, 226–230 or 231–235 into the fliK(∆2–99) allele by inverse PCR method and analyzed motility of the ∆fliK mutant cells over-expressing FliK(∆2–99) with these deletions in the presence of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). These additional deletions did not affect motility of the cells over-expressing FliK(∆2–99) (Fig. 1d). Consistently, the inner and extracellular amounts of FliC seen in the fliK(∆2–99 + ∆206–210), fliK(∆2–99 + ∆211–215), fliK(∆2–99 + ∆216–220), fliK(∆2–99 + ∆221–225), fliK(∆2–99 + ∆226–230) and fliK(∆2–99 + ∆231–235) cells were the same as those in the fliK(∆2–99) cells (Fig. 1e, 1st row). Since these deletions did not affect the expression level of FliK(∆2–99) (Fig. 1e, 2nd row), we suggest that they do not affect the interaction of FliK(∆2–99) with FlhBC. Therefore, we propose that these deletions may affect the protein transport process of FliK as well as hook length measurement.
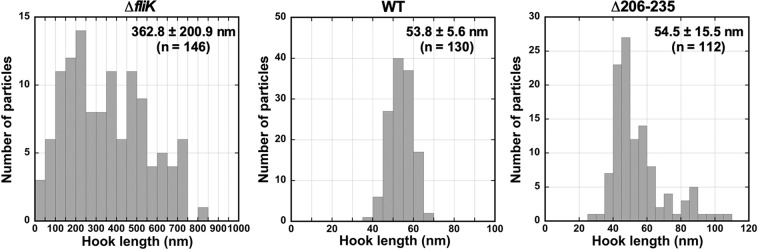
To further understand the ruler function of residues 206–235 in FliKL, we constructed larger deletion variants, FliK(∆206–215), FliK(∆216–225), FliK(∆226–235), FliK(∆206–220), FliK(∆221–235) and FliK(∆206–235) and analyzed their motility in 0.35% soft agar plates (Fig. S2a). Motility of the fliK(∆206–215), fliK(∆226–235), fliK(∆206–220), fliK(∆221–235) and fliK(∆206–235) cells was almost the same as wild-type motility whereas that of the fliK(∆216–225) mutant was slightly less than the wild-type level (Fig. S2a). The cellular and secretion levels of these deletion variants were essentially the same as the wild-type levels (Fig. S2b). Because these deletion mutants were expressed from the pTrc99AFF4 vector, it is also possible that their over-expression results in motility comparable to the wild-type level. To verify this possibility, the wild-type fliK allele on the chromosomal DNA was replaced by the fliK(∆206–235) allele. Motility of the fliK(∆206–235) mutant was slightly less than that of wild-type cells (Fig. S2c). To test whether the deletion of residues 206–235 affect hook length, we isolated hook-basal bodies from the fliK(∆206–235) mutant and measured the hook length (Figs. 2 and S3). A major peak of the hook length distribution was shifted to a shorter value than that of the wild-type (Fig. 2). While the majority of wild-type hook length was distributed within a range from 50 nm to 60 nm, the hook length distribution of the fliK(∆206–235) mutant showed a major peak population between 41 nm and 50 nm. However, longer hooks were also observed albeit shorter than those of polyhooks produced by the ∆fliK mutant [362.8 ± 200.9 nm (N = 146)]. As a result, the average hook length of the fliK(∆206–235) mutant was 54.5 ± 15.5 nm (n = 112), compared to 53.8 ± 5.6 nm (n = 130) for the wild-type. The SD value of the fliK(∆206–235) mutant was larger than the wild-type one, indicating that the deletion of residues 206–235 in FliKL cause a looser hook length control than the wild-type. Therefore, we suggest that a deletion of residues 206–235 not only shortens the hook length according to the size of deletion but also affects the interaction of FliKC with FlhBC during hook assembly.
Figure 2.
Effect of deletion of residues 206–235 of FliKL on hook length control. Histograms of hook length distribution of TH8426 (∆fliK), SJW1103 (WT) and MMK1012 (∆206–235).
Effect of much larger deletions within FliKL on hook length control and substrate specificity switching
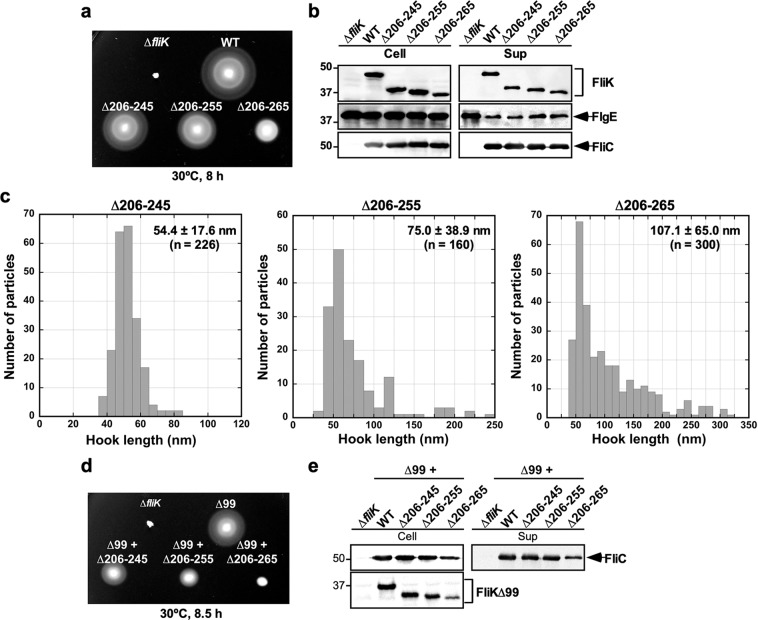
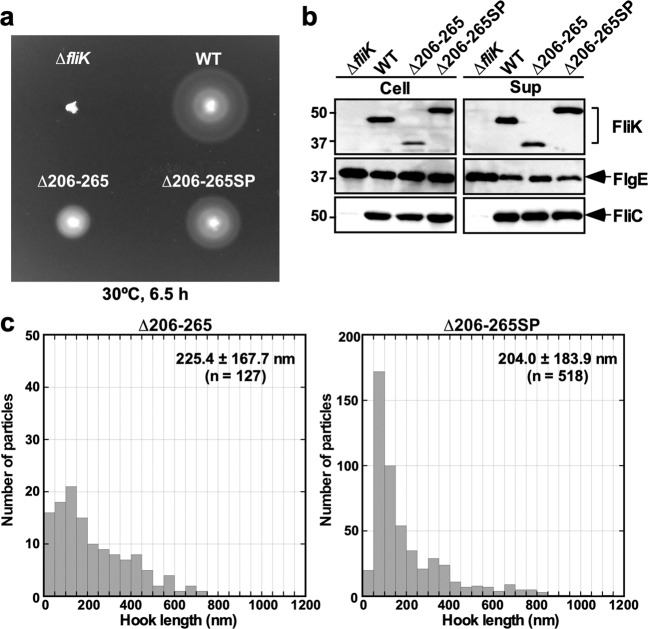
To clarify the role of FliKL in the export switching process of the flagellar type III protein export apparatus, the wild-type fliK allele on the chromosome was replaced by the fliK(∆206–245), fliK(∆206–255) or fliK(∆206–265) allele. Motility of these mutants were worse than wild-type motility (Fig. 3a) although the cellular and secreted amounts of FlgE and FliC were detected almost at the wild-type levels (Fig. 3b, 2nd and 3rd rows). Because neither cellular nor extracellular FliK level was affected by these larger deletions within FliKL (Fig. 3b, 1st row), this suggests that these deletions reduce FliK function.
Figure 3.
Effect of deletion of residues 206–245, 206–255 or 206–265 on FliK function. (a) Motility of TH8426 (∆fliK), SJW1103 (WT), MMK1013 (∆206–245), MMK1014 (∆206–255) and MMK1015 (∆206–265) in 0.35% soft agar. Plates were incubated at 30 °C for 8 hours. (b) Secretion assays of FliK, FlgE and FliC. Immunoblotting using polyclonal anti-FliK (1st row), anti-FlgE (2nd row) or anti-FliC (3rd row) antibody, of whole cell proteins (Cell) and culture supernatants (Sup) from the above strains. The regions of interest were cropped from original immunoblots shown in Fig. S7a in the Supplemental information using Photoshop CS6, and then the contrast and brightness were adjusted. The positions of molecular mass markers (kDa) are indicated on the left. (c) Histograms of hook length distribution of MMK1013 (∆206–245), MMK1014 (∆206–255) and MMK1015 (∆206–265). (d) Motility of TH8426 harboring pTrc99AFF4 (∆fliK), pNM201 (∆99), pMMK1028 (∆99 + ∆206–245), pMMK1029 (∆99 + ∆206–255) or pMMK1030 (∆99 + ∆206–265) in 0.35% soft agar containing 1 mM IPTG. Plates were incubated at 30 °C for 8 hours. (e) Secretion assay of FliC. Immunoblotting using polyclonal anti-FliC (1st row) or anti-FliK (2nd row) antibody, of whole cell proteins (Cell) and culture supernatants (Sup) from the above strains. The regions of interest were cropped from original immunoblots shown in Fig. S7b in the Supplemental information using Photoshop CS6, and then the contrast and brightness were adjusted. The positions of molecular mass markers (kDa) are indicated on the left.
To test whether these larger deletions affect the hook length control, we measured the hook length of the fliK(∆206–245), fliK(∆206–255) and fliK(∆206–265) mutants (Figs. 3c and S3). The hook length distribution of the fliK(∆206–245) mutant shows a major peak population between 45 nm and 55 nm. Longer hooks and polyhooks were seen for the fliK(∆206–255) and fliK(∆206–265) mutants although shorter hooks were also observed (Fig. S3). As a result, the average hook lengths of the fliK(∆206–245), fliK(∆206–255) and fliK(∆206–265) mutants were 54.4 ± 17.6 nm (n = 226), 75.0 ± 38.9 nm (n = 160) and 107.1 ± 65.0 nm (n = 300), respectively, indicating that the hook length control becomes worse in the fliK(∆206–255) and fliK(∆206–265) mutants.
To investigate whether these larger deletions directly affect the export switching function of FliKC, we introduces deletions residues 206–245, 206–255 or 206–265 into the fliK(∆2–99) allele. Motility of the cells over-expressing FliK(∆2–99) with these three deletions was reduced significantly (Fig. 3d), and especially the deletion of residues 206–265 reduced the cellular and extracellular levels of FliC by about 3-fold, thereby reducing motility considerably (Fig. 3e). This suggests that these three deletions reduce the export switching function of FliKC.
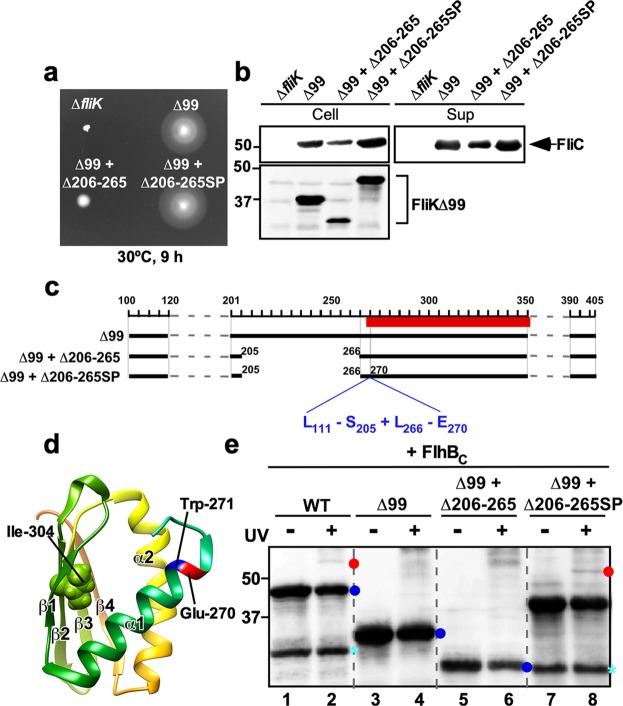
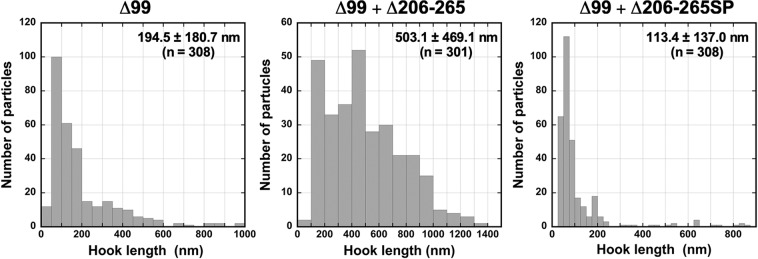
To clarify why the deletion of residues 206–265 considerably reduces the export switching activity of FliKC, we isolated fourteen pseudorevertants from the cells over-expressing FliK(∆2–99) with an in-frame deletion of residues 206–265. Motility of these pseudorevertants was much better than that of its parent cells and was slightly better than that of the cells over-expressing FliK(∆2–99) (Fig. 4a). Consistently, the cellular and extracellular levels of FliC seen in these pseudorevertants were much higher than those in the fliK(∆2–99 + ∆206–265) cells and slightly larger than those in the fliK(∆2–99) cells (Fig. 4b). These results indicate that the suppressor mutations enhance the probability of export switching to occur. DNA sequence analysis revealed that all suppressor mutations are insertion mutation of a DNA fragment encoding 100 amino-acid residues corresponding to residues 111–270 without residues 206–265 due to gene duplication (Fig. 4c). As a result, this intragenic FliK(∆2–99 + ∆206–265) suppressor mutation variant, namely FliK(∆2–99 + ∆206–265SP), showed much slower mobility on SDS-PAGE than FliK(∆2–99) and FliK(∆2–99 + ∆206–265) (Fig. 4b). The inserted sequence of the suppressor mutant was located between Glu-270 and Trp-271 residues in helix α1 of the core domain of FliKC (Fig. 4d). The fliK(∆2–99 + ∆206–265) mutant cells produced polyhooks sometimes with filaments attached (Fig. S4). In contrast, most of the pseudorevertant cells had a few flagella (Fig. S4). The pseudorevertant cells produced normal hooks or much shorter polyhooks compared to the fliK(∆2–99) and fliK(∆2–99 + ∆206–265) cells (Fig. S4). Careful measurements of the hook length indicated that the average hook length of the pseudorevertant were 113.4 ± 136.7 nm (n = 308), compared to 194.5 ± 180.4 nm (n = 308) for the fliK(∆2–99) cells and 503.1 ± 281.3 nm (n = 301) for the fliK(∆2–99 + ∆206–265) cells (Figs. 5 and S5). These results led to a conclusion that FliKL is required for efficient substrate specificity switching of the flagellar type III protein export apparatus.
Figure 4.
Isolation of pseudorevertants from the fliK(∆99 + ∆206–265) mutant. (a) Motility of TH8426 transformed with pTrc99A (∆fliK), pNM201 (∆99), pMMK1030 (∆99 + ∆206–265) or pMMK1030SP (∆99 + ∆206–265SP) in 0.35% soft agar containing 1 mM IPTG. Plates were incubated at 30 °C for 9 hours. (b) Secretion assay of FliC. Immunoblotting using polyclonal anti-FliC (1st row) or anti-FliK (2nd row) antibody, of whole cell proteins (Cell) and culture supernatants (Sup) prepared from the above strains. The regions of interest were cropped from original immunoblots shown in Fig. S8a in the Supplemental information using Photoshop CS6, and then the contrast and brightness were adjusted. The positions of molecular mass markers (kDa) are indicated on the left. (c) Location of the intragenic suppressor insertion mutation on FliK(∆99 + ∆206–265) and schematic bar diagram representations of FliK∆99 products caused by a deletion of residues 206–265 and the intragenic suppressor insertion of residues 111–270 lacking residues 206–265 (L111– S205 + L266–E 270) between Glu-270 and Trp-271 residues. Red bar indicates a core domain of FliKC. (d) NMR structure of a core domain of FliKC (PDB ID: 2RRL). The Cα backbone is color-coded from green to orange, going through the rainbow colors from the N- to the C-terminus. A highly conserved Ile-304 residue is involved in the interaction with FlhB. The inserted suppressor sequence is located between Glu-270 and Trp-271 residues. (e) Photo-crosslinking between FliK(∆2–99) and FlhBC. E. coli BL21(DE3) cells co-expressing FliK(I304pBPA), FliK(∆99 + I304pBPA), FliK(∆99 + ∆206–265 + I304pBPA) or FliK(∆99 + ∆206–265SP + I304pBPA) with FhBC were UV-irradiated for 5 min (+) or not irradiated (−), and then analyzed by immunoblotting with polyclonal anti-FliK antibody. The regions of interest were cropped from original immunoblots shown in Fig. S8b in the Supplemental information using Photoshop CS6, and then the contrast and brightness were adjusted. The positions of molecular mass markers (kDa) are indicated on the left. The positions of free FliK and FliK-FlhBC photo-crosslinked products are shown by blue and red balls, respectively. C-terminal truncated variants of FliK are shown by cyan asterisk.
Figure 5.
Effects of deletion of residues 206–265 and its intragenic suppressor insertion mutation on length distribution of the hooks produced by the fliK(∆2–99) mutant. Histograms of hook length distribution of TH8426 harboring pNM201 (∆99), pMMK1030 (∆99 + ∆206–265) or pMMK1030SP (∆99 + ∆206–265SP).
Interaction between FliK(∆2–99) and FlhBC
Targeted photo-crosslinking experiments have shown that Val-302 and Ile-304 of FliKC are in relatively close proximity of FlhBC, allowing FliK to form a photo-crosslinked product with FlhBC45. To investigate whether the deletion of residues 206–265 of FliKL and its suppressor insertion mutation reduces and increases the binding affinity of FliKC for FlhBC, respectively, we introduced an amber mutation at position 304 of FliK(∆2–99), FliK(∆2–99 + ∆206–265) and FliK(∆2–99 + ∆206–265SP) to incorporate p-benzoyl-phenylalanine (pBPA), which is a photo-reactive phenylalanine and carried out photo-crosslinking experiments. We used FliK(I304amber) as a positive control. FliK(I304pBPA) produced a ca. 53 kDa photo-crosslinked product with FlhBC after UV irradiation (Fig. 4e, lane 2), in agreement with a previous report45. However, FliK(∆2–99 + I304pBPA) did not form any photo-crosslinked products with FlhBC (Fig. 4e, lane 4), indicating that the binding affinity of FliK(∆2–99) for FlhBC is lower than that of wild-type FliK. FliK(∆2–99 + ∆206–265SP + I304pBPA) reproducibly formed a 51 kDa photo-crosslinked product along with FlhBC (Fig. 4e, lane 8) whereas FliK(∆2–99 + ∆206–265 + I304pBPA) did not (lane 6). This suggests that the inserted sequence of the pseudorevertant increases the binding affinity of FliK(∆2–99) for FlhBC so that the export switching of the type III protein export apparatus occurs more efficiently.
Effect of the 100 residues suppressor insertion on the length of hook produced by the fliK(∆206–265) mutant
To investigate whether the insertion mutation of the fliK(∆2–99 + ∆206–265) suppressor mutant also improves the export switching function of the fliK(∆206–265) mutant, we introduced the extra 100 amino-acid insertion sequence of the fliK(∆2–99 + ∆206–265SP) mutant into the fliK(∆206–265) allele by the overlap PCR method to generate the fliK(∆206–265SP) allele. Motility of the ∆fliK mutant harboring pMMK1015SP [FliK(∆206–265SP)] was better than that of the ∆fliK mutant harboring pMMK1015 [FliK(∆206–265)] although it was worse than that of the ∆fliK mutant transformed with pKM002 (wild-type FliK) (Fig. 6a). Neither cellular nor extracellular FliK level was affected by the inserted sequence (Fig. 6b, 1st row). The amounts of FlgE secreted by the fliK(∆206–265SP) mutant were slightly less than that by the fliK(∆206–265) mutant and almost the same as the wild-type level (Fig. 6b, 2nd and 3rd rows). There was no difference in the cellular and extracellular FliC levels between the fliK(∆206–265) and fliK(∆206–265SP) mutants. These results indicates that the inserted sequence of the fliK(∆2–99 + ∆206–265) suppressor mutant is also capable of improving the export switching function of FliK(∆206–265).
Figure 6.
Effects of the inserted sequence of the intragenic fliK(∆2–99 + ∆206–265) suppressor mutant on length distribution of the hooks produced by the fliK(∆206–265) mutant. (a) Motility of TH8426 harboring pTrc99A (∆fliK), pKM002 (WT), pMMK1015 (∆206–265) or pMMK1015SP (∆206–265SP) in 0.35% soft agar. Plates were incubated at 30 °C for 6.5 hours. (b) Secretion assays of FlgE and FliC. Immunoblotting using polyclonal anti-FliK (1st row), anti-FlgE (2nd row) or anti-FliC (3rd row) antibody, of whole cell proteins (Cell) and culture supernatants (Sup) prepared from the above strains. The regions of interest were cropped from original immunoblots shown in Fig. S9 using Photoshop CS6, and then the contrast and brightness were adjusted. The positions of molecular mass markers (kDa) are indicated on the left. (c) Histograms of hook length distribution of TH8426 harboring pMMK1015 or pMMK1015SP.
The amino acid sequence of FliK(∆206–265SP) is longer by 40 amino-acids than that of wild-type FliK, from which the average hook length is predicted to be about 70 nm, thereby reducing motility. To verify this hypothesis, we isolated hook-basal bodies from the ∆fliK mutant carrying pMMK1015 [FliK(∆206–265)] or pMMK1015SP [FliK(∆206–265SP)] and measured their hook length (Figs. 6c and S5). The average hook length of the ∆fliK mutant carrying pMMK1015 was 225.4 ± 167.7 nm (n = 127), which were longer than that of the MMK1015 strain (107.1 ± 65.0 nm). Since FliK(∆206–265) was expressed from the pTrc99AFF4 vector, we assume that such a length difference may be a consequence of the multicopy effect of FliK(∆206–265SP). The hook length distribution of the ∆fliK mutant carrying pMMK1015SP showed a major peak population between 61 nm and 100 nm, but much longer hooks and polyhooks were observed as well. As a result, the average hook length of the ∆fliK mutant carrying pMMK1015SP was 204.0 ± 183.9 nm (n = 518), which was shorter than that of the ∆fliK mutant carrying pMMK1015. This suggests that the suppressor insertion mutation increases the probability of the interaction between FliKC with a deletion of residue 206–265 and FlhBC, thereby increasing the export switching probability of the flagellar type III protein export apparatus.
Discussion
The bacterial injectisome directly transports virulence effector proteins into the cytosol of host cells for bacterial infection. The injectisome consists of basal body rings and a tubular structure called the needle and looks similar to the flagellar hook-basal body50. The injectisome uses a secreted molecular ruler, SctP (originally referred to as YscP and InvJ in the Yeshinia and Salmonella injectisomes, respectively) to determine the needle length in a way similar to FliK51,52. The core domain of FliKC is conserved among FliK/SctP family53 and possesses a fold similar to the C-terminal domain of SctP of the injectisome of Pseudomonas aeruginosa54. It has been shown that residues 301–350 of FliKC are directly involved in substrate specificity switching of the flagellar type III protein export apparatus47. Recent photo-crosslinking experiments have demonstrated that the conserved core domain of FliKC directly binds to FlhBC45. Similar protein-protein interactions have been observed in the injectisome55. These suggest that length control and substrate specificity switching mechanisms are conserved in both flagellar and injectisome systems. However, it remains unknown how the length measurement process by the secreted ruler is linked to the substrate specificity switching process of the type III protein export apparatus.
It has been reported that FliKL forms part of the ruler to determine hook length, but a deletion of residues 208–269 results in polyhooks with the filament attached42, having led to a hypothesis that residues 208–235 may contribute to efficient substrate specificity switching of the flagellar type III protein export apparatus. To verify this hypothesis, we introduced systematic deletions into FliKL and found that a deletion of residues 206–235 not only shortened hook length according to the size of deletion but also caused a loose hook length control (Fig. 2). The hook length control became much worse in the fliK(∆206–255) and fliK(∆206–265) mutants (Fig. 3c). The deletion of residues 206–265 considerably reduced the export switching function of FliKC (Fig 3d,e). An insertion of 100 amino-acids between Glu-270 and Trp-271 residues in the core domain of FliKC considerably improved the switching function of FliK(∆2-99 + ∆206-265), thereby shortening the hook length considerably and increasing the probability of filament formation significantly (Figs. 4 and 5). Consistently, this inserted sequence allowed FliK(∆2-99 + ∆206-265 + I304pBPA) to form a photo-crosslinked product with FlhBC in a way similar to FliK(I304pBPA) (Fig. 4e). Therefore, we suggest that the inserted sequence of the suppressor mutant significantly increases the binding affinity of the core domain of FliKC for FlhBC. Although FliK(∆2-99) is not secreted via the flagellar type III protein export apparatus during hook assembly, it retains the ability to catalyze the substrate specificity switching of the flagellar type III protein export apparatus to a considerable degree49. Therefore, we suggest that FliKL is also required for efficient interaction between the core domain of FliKC and FlhBC. When the length of FliKL was shortened by deletions, the export switching activity of FliKC was reduced depending on the size of deletions (Fig. 3). Furthermore, when the linker length became longer by 40 amino-acids compared to the wild-type length, the switching function of FliKC became worse (Fig. 6). Therefore, we propose that a proper length of FliKL between FliKN and FliKC may be important for FliKC to bind to FlhBC to switch the substrate specificity of the flagellar type III protein export apparatus. Assuming that FliKN suppresses the switching activity of FliKC when FliKN gets close to FliKC via deletion of residues in FliKL, it is also possible that FliKL may push FliKN away from FliKC to allow these two domains to fully exert their own functions.
The core domain of FliKC consists of four β-strands, β1, β2, β3 and β4 and two α-helices, α1 and α2. Three parallel β1, β3 and β4 strands and one anti-parallel β2 strand form a hydrophobic core with the α1 and α2 helices (Fig. 4d)46. Highly conserved Val-302 and Ile-304 residues in the β2 strand, which are critical for the switching function of FliK47, form hydrophobic interaction networks in FliKC46. Photo-crosslinking experiments have shown that Val-302 and Ile-304 are in very close proximity to FlhBC, suggesting that these two residues are exposed on the molecular surface of FliKC upon binding to FlhBC45. Since FliK(∆2-99 + ∆206-265SP + I304pBPA) formed a photo-crosslinked product with FlhBC whereas neither FliK(∆2-99 + I304pBPA) nor FliK(∆2-99 + ∆206-265 + I304pBPA) did (Fig. 4e), this suggests that the inserted sequence of the suppressor mutant induces a conformational change of the N-terminal portion of the core domain of FliKC to allow Ile-304 to bind to FlhBC. Therefore, we propose that FliKL may be required for efficient conformational rearrangements of FliKC to interact with FlhBC. However, it is also possible that deletion of residues 206–265 makes FliKN very close to FliKC to suppress the interaction of FliKC with FlhBC and that the inserted sequence of the suppressor mutant may push FliKN away from FliKC, allowing FliK to bind to FlhBC.
FliKC associates with and dissociates from FliKN in solution41. When FliKL adopts a fully extended conformation, the N-terminal portion of the core domain of FliKC becomes disordered41. Because the length of the most extended polypeptide is 0.37 nm per residue, the stretch of FliK sequence inside the channel of the hook-basal body must be longer than 250 residues to measure the hook length of about 55 nm together with the rod length of 35 nm. Therefore, we propose that FliKL may adopt a fully extended conformation when the hook length reaches about 55 nm, allowing Val-302 and Ile-304 in the hydrophobic core domain of FliKC to be in very close proximity to FlhBC to catalyze substrate specificity switching of the flagellar type III protein export apparatus.
Methods
Bacterial strains, plasmids and media
Bacterial strains and plasmids used in this study are listed in Table 1. To construct the Salmonella fliK(∆206–235), fliK(∆206–245), fliK(∆206–255) and fliK(∆206–265) mutant strains, the fliK gene on the chromosome was replaced by the fliK(∆206–235), fliK(∆206–245), fliK(∆206–255) and fliK(∆206–265) alleles, using the λ Red homologous recombination system56. L-broth contained 10 g of Bacto-Tryptone, 5 g of yeast extract and 5 g of NaCl per liter. Soft agar plates contained 10 g of Bacto Tryptone, 5 g of NaCl and 0.35% Bacto-Agar per liter. Ampicillin was added at a final concentration of 100 μg/ml if necessary.
DNA manipulations
DNA manipulations were carried out as described57. A series of mutant variants of FliK with deletions within FliKL were generated by inverse PCR using pKM00248 or pNM20144 as a template. The fliK(∆206–265SP) allele were generated by overlap PCR method. All of the fliK deletions were confirmed by DNA sequencing. DNA sequencing reactions were carried out using BigDye v3.1 (Applied Biosystems) and then the reaction mixtures were analyzed by a 3130 Genetic Analyzer (Applied Biosystems).
Motility assays in soft agar
Fresh colonies were inoculated onto 0.35% soft tryptone agar plates and incubated at 30 °C. At least seven independent measurements were carried out.
Secretion assays
Secretion assays were performed as described previously58. Salmonella cells were grown in 5 ml L-broth containing 100 μg/ml ampicillin at 30 °C with shaking until the cell density had reached an OD600 of ca. 1.2–1.6. 1.5 ml of each culture was transferred into a 1.5 ml Eppendorf tube. After centrifugation (15,000 g, 5 min, 4 °C), cell pellets and culture supernatants were collected, separately. The cells were resuspended in OD600 × 250 μl of SDS-loading buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.001% bromophenol blue) containing 1 μl of 2-mercaptoethanol and heated at 95 °C for 3 min. Trichloroacetic acid was added to each culture supernatant at a final concentration of 10%. After leaving on ice for 1 h, proteins in the culture supernatants were precipitated by centrifugation at 20,000 g for 20 min. Pellets were suspended in OD600 x 25 μl of a Tris-SDS loading buffer (one volume of 1 M Tris, nine volumes of 1 × SDS loading buffer) containing 1 μl of 2-mercaptoethanol and heated at 95 °C for 3 min. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), immunoblotting with polyclonal anti-FlgE, anti-FliC or anti-FliK antibody was carried out as described previously16. Detection was done with an ECL prime western blotting detection reagent (GE Healthcare). Chemiluminescence signals were captured by a Luminoimage analyzer LAS-3000 (GE Healthcare). The regions of interest were cropped from original immunoblots shown in the Supplemental information using a software, Photoshop CS6, and then the contrast and brightness were adjusted. At least three independent experiments were performed.
Electron microscopy
Osmotically shocked Salmonella cells were prepared described previously45. After centrifugation (18,500 g, 30 min), the cell pellets were resuspended in 200 μl of H2O. Samples were applied to carbon-coated copper grids and were negatively stained with 1.0% (W/V) phosphotungstic acid, pH 7.0. Micrographs were recorded at a magnification of × 5,000 with a JEM-1200EXII transmission electron microscope (JEOL, Tokyo, Japan) operating at 80 kV.
Hook-basal bodies and polyhook-basal bodies were isolated as described before27. Salmonella cells were grown in 5 l L-broth containing ampicillin at 30 °C with shaking until the cell density had reached an OD600 of ca. 1.0. The cells were harvested by centrifugation (10,000 g, 10 min, 4 °C) and suspended in 20 ml of ice-cold 0.1 M Tris-HCl pH 8.0, 0.5 M sucrose, followed by addition of EDTA and lysozyme at the final concentrations of 10 mM and 0.1 mg/ml, respectively. The cell suspensions were stirred for 30 min at 4 °C and then were solubilized on ice for 1 hour by adding Triton X-100 and MgSO4 at final concentrations of 1% and 10 mM, respectively. The cell lysates were adjusted to pH 10.5 with 5 M NaOH and then centrifuged (10,000 g, 20 min, 4 °C) to remove cell debris. After ultracentrifugation (45,000 g, 60 min, 4 °C), pellets were resuspended in 10 mM Tris-HCl, pH 8.0, 5 mM EDTA, 1% Triton X-100, and the solution was loaded a 20–50% (w/w) sucrose density gradient in 10 mM Tris-HCl, pH 8.0, 5 mM EDTA, 1% Triton X-100. After ultracentrifugation (49,100 g, 13 h, 4 °C), intact flagella were collected and ultracentrifuged (60,000 g, 60 min, 4 °C). Pellets were suspended in 50 mM glycine, pH 2.5, 0.1% Triton X100, and were incubated at room temperature for 30 min to depolymerize flagellar filaments. After ultracentrifugation (60,000 g, 60 min, 4 °C), pellets were resuspended in 50 μl of 10 mM Tris-HCl, pH 8.0, 5 mM EDTA, 0.1% Triton X100. Samples were negatively stained with 2%(w/v) uranyl acetate. Samples were applied to carbon-coated copper grids and were negatively stained with 2%(w/v) uranyl acetate. Electron micrographs were recorded with a JEM-1011 transmission electron microscope (JEOL, Tokyo, Japan) operated at 100 kV and equipped with a F415 CCD camera (TVIPS, Gauting, Germany). Hook length was measured by ImageJ version 1.48 (National Institutes of Health).
Photo-crosslinking
E. coli BL21(DE3) cells harboring pEVOL59 and a pETDuet-based plasmid encoding both FliK with an amber mutation and FlhBC were exponentially grown at 30 °C in L-broth containing 1 mM pBPA. Then, 100 μM IPTG and 0.02% arabinose were added and the incubation was continued until the culture density had reached an OD600 of ca. 1.4–1.5. Photo-crosslinking was carried out as described previously60. The cell pellets were harvested by centrifugation, suspended in SDS-loading buffer, and heated at 95 °C for 3 min. After SDS-PAGE, immunoblotting with polyclonal anti-FliK antibody was carried out. Detection was done with an ECL prime western blotting detection reagent. Chemiluminescence signals were captured by a Luminoimage analyzer LAS-3000. The regions of interest were cropped from original immunoblots shown in the Supplemental information using a software, Photoshop CS6, and then the contrast and brightness were adjusted. At least three independent experiments were performed.
Supplementary information
Acknowledgements
We acknowledge Prof. P.G. Schultz for his kind gift of pEVOL. This work was supported in part by JSPS KAKENHI Grant Numbers JP18K14638 (to M.K.), JP25000013 (to K.N.), and JP26293097 and JP19H03182 (to T.M.) and MEXT KAKENHI Grant Number JP15H01640 (to T.M.). This work has also been partially supported by JEOL YOKOGUSHI Research Alliance Laboratories of Osaka University to K.N.
Author contributions
M.K., N.K., S.I.A. and T.M. conceived and designed research; M.K., S.T., Y.I., S.I.A. and T.M. preformed research; M.K., S.T., Y.I., S.I.A. and T.M. analysed the data; and M.K., K.N., S.I.A. and T.M. wrote the paper based on discussion with other authors.
Data availability
All data generated or analyzed during this study are included in this published article and its Supplementary Information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-57782-5.
References
- 1.Nakamura S, Minamino T. Flagella-driven motility of bacteria. Biomolecules. 2019;9:279. doi: 10.3390/biom9070279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirano T, Yamaguchi S, Oosawa K, Aizawa S-I. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J. Bacteriol. 1994;176:5439–5449. doi: 10.1128/JB.176.17.5439-5449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spöring I, et al. Hook length of the bacterial flagellum is optimized for maximal stability of the flagellar bundle. PLOS Biol. 2018;16:e2006989. doi: 10.1371/journal.pbio.2006989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minamino T. Protein export through the bacterial flagellar type III export pathway. Biochim. Biophys. Acta. 2014;1843:1642–1648. doi: 10.1016/j.bbamcr.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Ohnishi K, Ohto Y, Aizawa S, Macnab RM, Iino T. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J. Bacteriol. 1994;176:2272–2281. doi: 10.1128/JB.176.8.2272-2281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moriya N, Minamino T, Imada K, Namba K. Genetic analysis of the bacterial hook-capping protein FlgD responsible for hook assembly. Microbiology. 2011;157:1354–1362. doi: 10.1099/mic.0.047100-0. [DOI] [PubMed] [Google Scholar]
- 7.Sakai T, Inoue Y, Terahara N, Namba K, Minamino T. A triangular loop of domain D1 of FlgE is essential for hook assembly but not for the mechanical function. Biochem. Biophys. Res. Commun. 2018;495:1789–1794. doi: 10.1016/j.bbrc.2017.12.037. [DOI] [PubMed] [Google Scholar]
- 8.Ferris HU, Minamino T. Flipping the switch: bringing order to flagellar assembly. Trends Microbiol. 2006;14:519–526. doi: 10.1016/j.tim.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Minamino T, Imada K, Namba K. Mechanisms of type III protein export for bacterial flagellar assembly. Mol. BioSyst. 2008;4:1105–1115. doi: 10.1039/b808065h. [DOI] [PubMed] [Google Scholar]
- 10.Minamino T. Hierarchical protein export mechanism of the bacterial flagellar type III protein export apparatus. FEMS Microbiol. lett. 2018;365:fny117. doi: 10.1093/femsle/fny117. [DOI] [PubMed] [Google Scholar]
- 11.Kutsukake K, Minamino T, Yokoseki T. Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium. J. Bacteriol. 1994;176:7625–7629. doi: 10.1128/JB.176.24.7625-7629.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams AW, et al. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J. Bacteriol. 1996;178:2960–2970. doi: 10.1128/JB.178.10.2960-2970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kutsukake K. Hook-length control of the export-switching machinery involves a double-locked gate in Salmonella typhimurium flagellar morphogenesis. J. Bacteriol. 1997;179:1268–1273. doi: 10.1128/JB.179.4.1268-1273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirano T, Mizuno S, Aizawa S-I, Hughes KT. Mutations in Flk, FlgG, FlhA, and FlhE that affect the flagellar type III secretion specificity switch in Salmonella enterica. J. Bacteriol. 2009;181:3938–3949. doi: 10.1128/JB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minamino T, Iino T, Kutsukake K. Molecular characterization of the Salmonella typhimurium flhB operon and its protein products. J. Bacteriol. 1994;176:7630–7637. doi: 10.1128/JB.176.24.7630-7637.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minamino T, Macnab RM. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 1999;181:1388–1394. doi: 10.1128/JB.181.5.1388-1394.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abrusci P, et al. Architecture of the major component of the type III secretion system export apparatus. Nat. Struct. Mol. Biol. 2013;20:99–104. doi: 10.1038/nsmb.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terahara N, et al. Insight into structural remodeling of the FlhA ring responsible for bacterial flagellar type III protein export. Sci. Adv. 2018;4:eaao7054. doi: 10.1126/sciadv.aao7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minamino T, Macnab RM. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol. Microbiol. 2000;35:1052–1064. doi: 10.1046/j.1365-2958.2000.01771.x. [DOI] [PubMed] [Google Scholar]
- 20.Minamino T, et al. Role of the C-terminal cytoplasmic domain of FlhA in bacterial flagellar type III protein export. J. Bacteriol. 2010;192:1929–1936. doi: 10.1128/JB.01328-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minamino T, Morimoto YV, Hara N, Namba K. An energy transduction mechanism used in bacterial flagellar type III protein export. Nat. Commun. 2011;2:475. doi: 10.1038/ncomms1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bange G, et al. FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc. Natl. Acad. Sci. USA. 2010;107:11295–11300. doi: 10.1073/pnas.1001383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minamino T, et al. Interaction of a bacterial flagellar chaperone FlgN with FlhA is required for efficient export of its cognate substrates. Mol. Microbiol. 2012;83:775–788. doi: 10.1111/j.1365-2958.2011.07964.x. [DOI] [PubMed] [Google Scholar]
- 24.Kinoshita M, Hara N, Imada K, Namba K, Minamino T. Interactions of bacterial chaperone-substrate complexes with FlhA contribute to co-ordinating assembly of the flagellar filament. Mol. Microbiol. 2013;90:1249–1261. doi: 10.1111/mmi.12430. [DOI] [PubMed] [Google Scholar]
- 25.Xing, Q. et al. Structure of chaperone-substrate complexes docked onto the export gate in a type III secretion system. Nat. Commun. 9, 1773. [DOI] [PMC free article] [PubMed]
- 26.Minamino T, et al. FliH and FliI ensure efficient energy coupling of flagellar type III protein export in Salmonella. MicrobiologyOpen. 2016;5:424–435. doi: 10.1002/mbo3.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inoue Y, Morimoto YV, Namba K, Minamino T. Novel insights into the mechanism of well-ordered assembly of bacterial flagellar proteins in Salmonella. Sci. Rep. 2018;8:1787. doi: 10.1038/s41598-018-20209-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minamino T, Macnab RM. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J. Bacteriol. 2000;182:4906–4919. doi: 10.1128/JB.182.17.4906-4914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fraser GM, et al. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol. Microbiol. 2003;48:1043–1057. doi: 10.1046/j.1365-2958.2003.03487.x. [DOI] [PubMed] [Google Scholar]
- 30.Ferris HU, et al. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J. Biol. Chem. 2005;280:41236–41242. doi: 10.1074/jbc.M509438200. [DOI] [PubMed] [Google Scholar]
- 31.Evans LD, Poulter S, Terentjev EM, Hughes C, Fraser GM. A chain mechanism for flagellum growth. Nature. 2013;504:287–290. doi: 10.1038/nature12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMurry JL, et al. Weak interactions between Salmonella enterica FlhB and other flagellar export apparatus proteins govern type III secretion dynamics. PLOS One. 2015;10:e0134884. doi: 10.1371/journal.pone.0134884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inoue Y, et al. Structural insight into the substrate specificity switch mechanism of the type III protein export apparatus. Structure. 2019;27:965–976. doi: 10.1016/j.str.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 34.Aldridge P, Karlinsey JE, Becker E, Chevance FF, Hughes KT. Flk prevents premature secretion of the anti sigma factor FlgM into the preiplasm. Mol. Microbiol. 2006;60:630–642. doi: 10.1111/j.1365-2958.2006.05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minamino T, González-Pedrajo B, Yamaguchi K, Aizawa S-I, Macnab RM. FliK, the protein responsible for flagellar hook length control in Salmonella, is exported during hook assembly. Mol. Microbiol. 1999;34:295–304. doi: 10.1046/j.1365-2958.1999.01597.x. [DOI] [PubMed] [Google Scholar]
- 36.Erhardt M, et al. The role of the FliK molecular ruler in hook-length control in Salmonella enterica. Mol. Microbiol. 2010;75:1272–1284. doi: 10.1111/j.1365-2958.2010.07050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erhardt M, Singer HM, Wee DH, Keener JP, Hughes KT. An infrequent molecular ruler controls flagellar hook length in Salmonella enterica. EMBO J. 2011;30:2948–2961. doi: 10.1038/emboj.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uchida K, Aizawa S-I. The flagellar soluble protein FliK determines the minimal length of the hook in Salmonella enterica serovar Typhimurium. J. Bacteriol. 2014;196:1753–1758. doi: 10.1128/JB.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawagishi I, Homma M, Williams AW, Macnab RM. Characterization of the flagellar hook length control protein of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 1996;178:2954–2959. doi: 10.1128/JB.178.10.2954-2959.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minamino T, et al. Domain organization and function of Salmonella FliK, a flagellar hook-length control protein. J. Mol. Biol. 2004;341:491–502. doi: 10.1016/j.jmb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 41.Kodera N, Uchida K, Ando T, Aizawa S-I. Two-ball structure of the flagellar hook-length control protein FliK as revealed by high-speed atomic force microscopy. J. Mol. Biol. 2015;427:406–414. doi: 10.1016/j.jmb.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Shibata S, et al. FliK regulates flagellar hook length as an internal ruler. Mol. Microbiol. 2007;64:1404–1415. doi: 10.1111/j.1365-2958.2007.05750.x. [DOI] [PubMed] [Google Scholar]
- 43.Moriya N, Minamino T, Hughes KT, Macnab RM, Namba K. The type III flagellar export specificity switch is dependent on FliK ruler and a molecular clock. J. Mol. Biol. 2006;359:466–477. doi: 10.1016/j.jmb.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 44.Minamino T, Moriya N, Hirano T, Hughes KT, Namba K. Interaction of FliK with the bacterial flagellar hook is required for efficient export specificity switching. Mol. Microbiol. 2009;74:239–251. doi: 10.1111/j.1365-2958.2009.06871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kinoshita M, Aizawa S-I, Inoue Y, Namba K, Minamino T. The role of intrinsically disordered C-terminal region of FliK in substrate specificity switching of the bacterial flagellar type III export apparatus. Mol. Microbiol. 2017;105:572–588. doi: 10.1111/mmi.13718. [DOI] [PubMed] [Google Scholar]
- 46.Mizuno S, Amida H, Kobayashi N, Aizawa S-I, Tate S. The NMR structure of FliK, the trigger for the switch of substrate specificity in the flagellar type III secretion apparatus. J. Mol. Biol. 2011;409:558–573. doi: 10.1016/j.jmb.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 47.Minamino T, Ferris HU, Morioya N, Kihara M, Namba K. Two parts of the T3S4 domain of the hook-length control protein FliK are essential for the substrate specificity switching of the flagellar type III export apparatus. J. Mol. Biol. 2006;362:1148–1158. doi: 10.1016/j.jmb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 48.Muramoto K, Makishima S, Aizawa S-I, Macnab RM. Effect of cellular level of FliK on flagellar hook and filament assembly in Salmonella typhimurium. J. Mol. Biol. 1998;277:871–882. doi: 10.1006/jmbi.1998.1659. [DOI] [PubMed] [Google Scholar]
- 49.Hirano T, Shibata S, Ohnishi K, Tani T, Aizawa S. N-terminal signal region of FliK is dispensable for length control of the flagellar hook. Mol. Microbiol. 2005;56:346–360. doi: 10.1111/j.1365-2958.2005.04615.x. [DOI] [PubMed] [Google Scholar]
- 50.Kubori T, et al. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 51.Journet L, Agrain C, Broz P, Cornelis GR. The needle length of bacterial injectisomes is determined by a molecular ruler. Science. 2003;302:1757–1760. doi: 10.1126/science.1091422. [DOI] [PubMed] [Google Scholar]
- 52.Wee DH, Hughes KT. Molecular ruler determines needle length for the Salmonella Spi-1 injectisome. Proc. Natl. Acad. Sci. USA. 2015;112:4098–103. doi: 10.1073/pnas.1423492112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agrain C, et al. Characterization of a type III secretion substrate specificity switch (T3S4) domain in YscP from Yersinia enterocolitica. Mol. Microbiol. 2005;56:54–67. doi: 10.1111/j.1365-2958.2005.04534.x. [DOI] [PubMed] [Google Scholar]
- 54.Bergeron JR, et al. The structure of a type 3 secretion system (T3SS) ruler protein suggests a molecular mechanism for needle length sensing. J. Biol. Chem. 2016;291:1676–1691. doi: 10.1074/jbc.M115.684423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ho O, et al. Characterization of the ruler protein interaction interface on the substrate specificity switch protein in the Yersinia type III secretion system. J. Biol. Chem. 2017;292:3299–3311. doi: 10.1074/jbc.M116.770255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl, Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saijo-Hamano Y, Minamino T, Macnab RM, Namba K. Structural and functional analysis of the C-terminal cytoplasmic domain of FlhA, an integral membrane component of the type III flagellar protein export apparatus in Salmonella. J. Mol. Biol. 2004;343:457–466. doi: 10.1016/j.jmb.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 58.Minamino T, Kinoshita M, Namba K. Fuel of the bacterial flagellar type III protein export apparatus. Methods Mol. Biol. 2017;1593:3–16. doi: 10.1007/978-1-4939-6927-2_1. [DOI] [PubMed] [Google Scholar]
- 59.Young TS, Ahmad I, Yin JA, Schultz PG. An enhanced system for unnatural amino acid mutagenesis in E. coli. J. Mol. Biol. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 60.Hara N, Morimoto YV, Kawamoto A, Namba K, Minamino T. Interaction of the extreme N-terminal region of FliH with FlhA is required for efficient bacterial flagellar protein export. J. Bacteriol. 2012;194:5353–5360. doi: 10.1128/JB.01028-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamaguchi S, Fujita H, Sugata K, Taira T, Iino T. Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J. Gen. Microbiol. 1984;130:255–265. doi: 10.1099/00221287-130-2-255. [DOI] [PubMed] [Google Scholar]
- 62.Ohnishi K, Fan F, Schoenhals GJ, Kihara M, Macnab RM. The FliO, FliP, FliQ, and FliR proteins of Salmonella typhimurium: putative components for flagellar assembly. J. Bacteriol. 1997;179:6092–6099. doi: 10.1128/JB.179.19.6092-6099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Supplementary Information files.