Abstract
To evaluate the predictive accuracy of the %p2PSA and prostate health index (PHI) in predicting aggressive pathological outcomes in patients with prostate cancer (PCa) undergoing radical prostatectomy (RP), we enrolled 91 patients with organ-confined PCa who were treated with robot-assisted RP. p2PSA levels and the PHI were investigated for their ability to predict pathological results. The %p2PSA and PHI were both significantly higher in patients with ≥pT3 disease, high-risk disease, positive surgical margin, or seminal vesical invasion (SVI). In univariable analysis, p2PSA derivatives were significant predictors of the presence of ≥pT3 disease, high-risk disease, positive surgical margin, and SVI. To predict adverse pathological outcomes at a sensitivity of 90%, p2PSA derivatives had higher specificity than standard PSA derivatives. In multivariable analysis, additional increases in the area under the receiver operating characteristic curve (AUC) were observed with the %p2PSA and PHI for ≥pT3 disease, high-risk disease, and positive surgical margin (8.2% and 2.7%, 6.2% and 4.1%, and 8.6% and 5.4%, respectively). A PHI ≥61.26 enhanced the predictive accuracy of the model for SVI by increasing the AUC from 0.624 to 0.819 (p = 0.009). The preoperative %p2PSA and PHI accurately predict adverse pathological results and are useful for decision-making.
Subject terms: Tumour biomarkers, Tumour biomarkers, Prostate
Introduction
In patients with prostate cancer (PCa), it is vital to avoid overtreatment and procedural complications. Treatment options for localized PCa including active surveillance, radical prostatectomy (RP), and radiation therapy depend on the aggressiveness of the disease1. However, discrepancies often exist between the clinical cancer staging and pathological staging. Approximately half of patients with clinically low-risk PCa at transrectal ultrasonography-guided biopsy of the prostate (TRUSP biopsy) actually have a Gleason score (GS) ≥7 or ≥pathological T3 disease in the final RP pathology2,3.
Multiple preoperative predictive nomograms have been validated for the prediction of pathological outcomes at RP. However, the applicability of these nomograms is limited in the clinical setting due to their difficult accessibility and complexity4. Therefore, magnetic resonance imaging (MRI) has been increasingly used as an alternative diagnostic tool before prostate biopsy or via MRI-targeted biopsy to better identify clinically significant cancer with a GS ≥75. Correct assessment of the local staging by MRI is still being researched. However, the utility of MRI in local cancer staging is limited by its poor sensitivity for detecting extracapsular extension and seminal vesical invasion (SVI)6. Accordingly, we need an accurate and convenient biomarker for preoperatively predicting adverse pathological characteristics and determining who might benefit the most from surgery. Such a biomarker would help physicians in decision-making and to predict prognosis before any intervention.
Total prostate-specific antigen (tPSA) consists of complexed PSA and free PSA (fPSA). Of fPSA forms, p2PSA is one of the isoforms of proPSA, which is considered a promising biomarker that is more cancer-associated7. The prostate health index (PHI), developed by Beckman Coulter, Inc., is a mathematical formula combining three biomarkers as follows: (p2PSA/fPSA) × √tPSA. Numerous studies have shown that the %p2PSA and PHI are the most accurate predictors of PCa at the initial prostate biopsy8–10. Meanwhile, the %p2PSA and PHI are able to identify aggressive PCa with GS ≥7 prior to TRUSP biopsy and predict unfavorable cancer characteristics at the final pathology from RP11,12. However, a lack of a reference range of the %p2PSA and PHI for predicting adverse pathological results limits their clinical utility. In the present study, we aimed to validate the application of the preoperative %p2PSA and PHI to predict adverse pathological outcomes after RP and examine the accuracy of the cut-off values.
Results
The basic characteristics of the study population are listed in (Table 1). Of the 91 patients, the median age was 64 years (IQR, 60–67). The median preoperative tPSA, fPSA, %fPSA, and PSA density were 10.9 ng/ml, 1.2 ng/ml, 12.1%, and 0.4 ng/ml2, respectively. The median preoperative p2PSA, %p2PSA, and PHI were 18.9 pg/ml, 1.9%, and 54.6, respectively. Eventually, 31 (36.3%) and 82 (92.1%) patients were diagnosed with ≥pT3 and pathological GS ≥7 disease, respectively. In addition, 26 patients (28.6%) had an upgraded GS, from GS ≤6 at initial biopsy to a GS sum ≥7 from RP specimens. Overall, 36 patients (39.6%) were classified as having high-risk disease.
Table 1.
Basic characteristics.
| Variables, median | n = 91 patients |
|---|---|
| Age, years (IQR) | 64 (60, 67) |
| Abnormal DRE, n (%) | 35 (38.5%) |
| Prior to radical prostatectomy | |
| Prostate volume, ml (IQR) | 30.5 (23.7, 41.3) |
| Prostate weight, g (IQR) | 35.2 (27.6, 46.4) |
| tPSA, ng/ml (IQR) | 10.9 (7.5, 16.6) |
| fPSA, ng/ml (IQR) | 1.2 (0.7, 2.1) |
| %fPSA (IQR) | 12.1 (8.0, 16.1) |
| p2PSA, pg/ml (IQR) | 18.9 (11.9, 31.5) |
| %p2PSA (IQR) | 1.9 (1.2, 2.3) |
| PHI (IQR) | 54.6 (36.2, 79.2) |
| PSAD, ng/ml2 (IQR) | 0.4 (0.2, 0.6) |
| Clinical cancer stage, n (%) | |
| ≤T1c | 53 (58.2%) |
| T2a-2c | 38 (41.8%) |
| GS at TRUSP biopsy, n (%) | |
| ≤6 | 30 (33.0%) |
| 7 | 40 (44.0%) |
| ≥8 | 21 (23.1%) |
| Percentage of positive cores, % (IQR) | 20 (10, 40) |
| Maximum % of positive cores, (IQR) | 40 (16, 80) |
| Pathological results of radical prostatectomy | |
| Pathological stage, n (%) | |
| pT0 | 2 (2.2%) |
| pT2 | 58 (63.7%) |
| pT3 | 30 (35.2%) |
| pT4 | 1 (1.1%) |
| Pathological GS, n (%) | |
| ≤6 | 7 (7.9%) |
| 7 | 66 (74.2%) |
| ≥8 | 16 (18.0%) |
| Positive lymph node, n (%) | 10 (11.9%) |
| Positive surgical margin, n (%) | 43 (47.3%) |
| Upgrade of GS compared with the biopsy, n (%) | 26 (28.6%) |
| High-risk disease, n (%) | 36 (39.6%) |
IQR: interquartile range; DRE: digital rectal examination; PSA: prostate specific antigen; tPSA = total PSA; fPSA = free PSA; %fPSA = percentage of free to total PSA; p2PSA: [−2]pro PSA; %p2PSA = (p2PSA/fPSA × 1000) × 100; PHI: Prostate Health Index; PSAD: PSA density; GS: Gleason score; TRUSP: transrectal ultrasonography of the prostate.
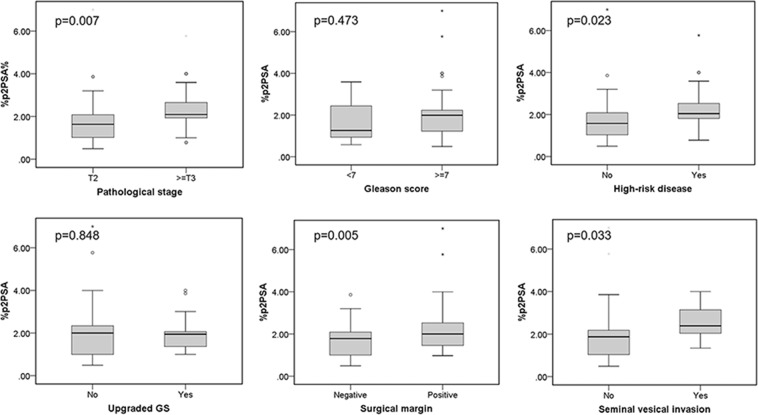
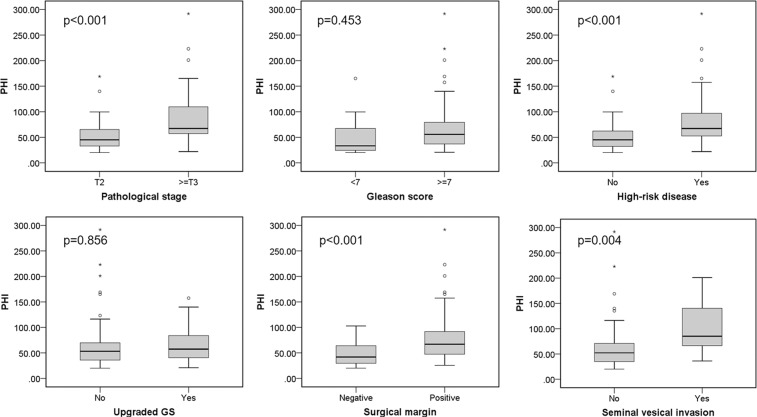
As shown in (Figs. 1 and 2), the %p2PSA and PHI were significantly higher in patients with ≥pT3 disease, high-risk disease, positive surgical margin, and presence of SVI (%p2PSA: p = 0.007, 0.023, 0.005, and 0.033, respectively; PHI: p < 0.001, <0.001, <0.001, and 0.004, respectively). In the univariable analysis, the p2PSA, %p2PSA, and PHI were accurate parameters for predicting ≥pT3 disease, high-risk disease, and a positive surgical margin (Table 2). The PHI was the only significant predictor of SVI (odds ratio [OR]: 1.01, 95% confidence interval [CI] 1.00–1.03, p = 0.014). Meanwhile, tPSA was also a predictor of ≥pT3 disease, high-risk disease, and positive surgical margin. On the other hand, GS ≥7 at TRUSP biopsy was a predictor of ≥pT3 disease and high-risk disease.
Figure 1.
%p2PSA relative to pathological cancer stage, Gleason score, high-risk disease, upgraded Gleason score, surgical margin status, seminal vesical invasion. GS: Gleason score; p2PSA: [−2]pro PSA; p2PSA% = percentage of p2PSA to fPSA ratio; * = Extreme outliers; ° = mild outliers.
Figure 2.
PHI relative to pathological cancer stage, Gleason score, high-risk disease, upgraded Gleason score, surgical margin status, seminal vesical invasion. GS: Gleason score; PHI: Prostate Health Index; * = Extreme outliers; ° = mild outliers.
Table 2.
Univariable analyses to predict pathological outcomes in patients who underwent radical prostatectomy.
| Cancer stage ≥ pT3 | Gleason score ≥ 7 | High-risk disease | ||||
|---|---|---|---|---|---|---|
| OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | |
| Age | 1.01 (0.93, 1.11) | 0.754 | 1.18 (1.02, 1.38) | 0.031 | 1.01 (0.92, 1.10) | 0.866 |
| Prostate volume | 0.99 (0.97, 1.01) | 0.428 | 0.98 (0.96, 1.01) | 0.170 | 0.99 (0.96, 1.01) | 0.274 |
| Biopsy GS ≥7 | 5.91 (1.59, 22.06) | 0.008 | 2.24 (0.59, 8.44) | 0.233 | 5.17 (1.75, 15.27) | 0.003 |
| tPSA | 1.07 (1.01, 1.14) | 0.028 | 1.02 (0.93, 1.13) | 0.614 | 1.07 (1.01, 1.14) | 0.030 |
| fPSA | 1.42 (0.92, 2.18) | 0.109 | 0.86 (0.48, 1.54) | 0.606 | 1.41 (0.92, 2.16) | 0.117 |
| %fPSA | 0.95 (0.87, 1.03) | 0.199 | 0.91 (0.81, 1.01) | 0.078 | 0.94 (0.87, 1.02) | 0.157 |
| p2PSA | 1.03 (1.01, 1.06) | 0.005 | 1.01 (0.98, 1.03) | 0.702 | 1.03 (1.01, 1.06) | 0.007 |
| %p2PSA | 1.79 (1.08, 2.95) | 0.023 | 1.38 (0.61, 3.10) | 0.438 | 1.62 (1.01, 2.61) | 0.047 |
| PHI | 1.02 (1.01, 1.04) | 0.002 | 1.01 (0.99, 1.03) | 0.452 | 1.02 (1.01, 1.04) | 0.003 |
| Upgraded GS sum | Positive surgical margin | Seminal vesical invasion | ||||
| OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | |
| Age | 1.06 (0.96, 1.18) | 0.213 | 1.00 (0.92, 1.09) | 0.964 | 0.99 (0.87, 1.13) | 0.874 |
| Prostate volume | 1.01 (0.99, 1.03) | 0.291 | 0.98 (0.96, 1.01) | 0.182 | 1.00 (0.96, 1.03) | 0.839 |
| Biopsy GS ≥7 | 0.00 (0.00, 0.03) | <0.001 | 1.55 (0.64, 3.76) | 0.333 | 2.42 (0.49, 12.00) | 0.278 |
| tPSA | 1.01 (0.96, 1.07) | 0.654 | 1.11 (1.03, 1.18) | 0.004 | 1.07 (1.00, 1.15) | 0.055 |
| fPSA | 1.23 (0.80, 1.89) | 0.337 | 1.34 (0.87, 2.05) | 0.183 | 1.36 (0.80, 2.32) | 0.250 |
| %fPSA | 1.00 (0.92, 1.09) | 0.987 | 0.94 (0.86, 1.01) | 0.098 | 0.92 (0.81, 1.06) | 0.247 |
| p2PSA | 1.01 (0.99, 1.02) | 0.421 | 1.04 (1.01, 1.07) | 0.006 | 1.02 (1.00, 1.03) | 0.085 |
| %p2PSA | 0.96 (0.62, 1.50) | 0.861 | 2.06 (1.18, 3.60) | 0.011 | 1.55 (0.95, 2.55) | 0.080 |
| PHI | 1.00 (0.99, 1.01) | 0.858 | 1.03 (1.01, 1.05) | 0.001 | 1.01 (1.00, 1.03) | 0.014 |
CI: confidence interval; GS: Gleason score; OR: odds ratio; PHI: Prostate Health Index; PSA: prostate specific antigen; tPSA: total PSA; fPSA: free PSA; %fPSA = percentage of free to total PSA; p2PSA: [−2]pro PSA; %p2PSA = (p2PSA/fPSA × 1000) × 100.
At a sensitivity of 90% to predict the presence of ≥pT3 cancer, high-risk disease, positive surgical margin, or SVI, the %p2PSA or PHI had a higher specificity in comparison with tPSA (Table 3). In this setting, the most appropriate cut-off values for %p2PSA were ≥1.21, ≥1.12, and ≥1.17 for the presence of ≥pT3 cancer, high-risk disease, and positive surgical margin, respectively. In contrast, the most appropriate cut-off values for PHI were determined to be ≥33.92, ≥33.92, ≥33.92, and ≥61.26 for the presence of ≥pT3 cancer, high-risk disease, positive surgical margin, and SVI, respectively.
Table 3.
The cut-off values of p2PSA derivatives and the PHI in predicting ≥pT3 cancer, high-risk disease, a positive surgical margin, and seminal vesical invasion at a sensitivity of 90%.
| Outcome | PSA derivatives | Cut-off | Specificity |
|---|---|---|---|
| Cancer stage ≥pT3 | tPSA | ≥5.95 | 16.7% |
| p2PSA | ≥10.51 | 15.0% | |
| %p2PSA | ≥1.21 | 30.0% | |
| PHI | ≥33.92 | 26.7% | |
| High-risk disease | tPSA | ≥5.95 | 18.2% |
| p2PSA | ≥10.51 | 16.4% | |
| %p2PSA | ≥1.12 | 29.1% | |
| PHI | ≥33.92 | 29.1% | |
| Positive surgical margin | tPSA | ≥5.34 | 16.7% |
| p2PSA | ≥10.46 | 14.6% | |
| %p2PSA | ≥1.17 | 33.3% | |
| PHI | ≥33.92 | 31.3% | |
| Seminal vesical invasion | tPSA | ≥7.22 | 25.0% |
| PHI | ≥61.26 | 66.3% |
PSA: prostate specific antigen; tPSA: total PSA; p2PSA: [−2]pro PSA; %p2PSA = (p2PSA/fPSA × 1000) × 100; PHI: Prostate Health Index.
In the multivariable analysis, base models for each predictive end point were selected according to the univariable analytical results. Thus, age, prostate volume, tPSA, and biopsy GS ≥7 were selected as the base model for a cancer stage ≥pT3 and high-risk disease, whereas age, prostate volume, and tPSA acted as the base model for a positive surgical margin and SVI (Supplementary Table). We separately tested the predictive accuracy of the p2PSA, %p2PSA, or PHI by adding them individually to the base model. For predicting ≥pT3 cancer, a %p2PSA ≥1.21 had a statistically significant OR of 5.41 (95% CI 1.33–22.04, p = 0.019) and increased the AUC from 0.687 to 0.768 (p = 0.073). To predict the presence of high-risk disease, a %p2PSA ≥1.12 and PHI ≥33.92 had significant ORs of 6.94 (95% CI 1.66–29.06, p = 0.008) and 4.52 (95% CI 1.08–19.00, p = 0.039), respectively. The addition of the %p2PSA and PHI to the base model increased the AUC for predicting high-risk disease by 0.062 (p = 0.195) and 0.041 (p = 0.104), respectively. For predicting a positive surgical margin, a %p2PSA ≥1.17 and PHI ≥33.92 had significant ORs of 4.04 (95% CI 1.24–13.15, p = 0.020) and 3.91 (95% CI 1.12–13.63, p = 0.032), respectively. The addition of the %p2PSA and PHI increased the AUCs by 0.086 (p = 0.097) and 0.054 (p = 0.156), respectively. A PHI ≥61.26 significantly predicted SVI with an OR of 20.85 (95% CI 2.26–191.91, p = 0.007). Inclusion of the PHI in the base model significantly increased the AUC from 0.624 to 0.819 (p = 0.009).
Discussion
In this cohort, we confirmed the accuracy of the preoperative %p2PSA and PHI in predicting adverse pathological results for patients with clinically organ-confined PCa undergoing RP. The %p2PSA and PHI had excellent ability to predict the four major pathological outcomes considered: ≥pT3 cancer, high-risk disease, positive surgical margin, and SVI. An additional predictive benefit was provided by the addition of the %p2PSA and PHI to the base model in the multivariable analysis, with particular benefit of the PHI in predicting SVI. There has been no consensus on the most appropriate cut-off value for the %p2PSA and PHI in cancer detection due to the use of different study designs11. However, prior studies investigating the predictive value of the %p2PSA and PHI in patients treated with RP did not provide a reference range for clinical utility8,12–14. The major strength of the current study is that we offered cut-off values of p2PSA derivatives and tested their predictive accurracy in a multivariable model (Table 3 and Supplementary Table). As a result, it will be easier for physicians to have a clear threshold of p2PSA derivatives at clinical application.
A meta-analysis showed that %p2PSA and PHI could detect more aggressive PCa with GS ≥7 at the initial prostate biopsy (AUCs of 0.54 and 0.67 for %p2PSA and PHI, respectively)11. In patients indicated for the first prostate biopsy or repeated biopsy, NCCN guidelines suggest that a PHI >35 indicates a higher probability of high-grade PCa. Such an association with cancer aggressiveness has been extended to patients with PCa undergoing active surveillance. Tosoian et al.15 revealed that both baseline and longitudinal %p2PSA and PHI provided outstanding predictive value in biopsy reclassification and upgraded the GS in men under active surveillance. To further extend these results, examination of the relationship between p2PSA derivatives and the final pathology is warranted.
Although our findings failed to confirm the prediction of pathological GS ≥7 PCa, the ability of p2PSA derivatives to predict aggressive PCa was still confirmed. The potential causes of the failure to predict a pathological GS ≥7 include the small sample size of our cohort and the high proportion of patients (82 of 92; 89.1%) with pT3 disease or a GS ≥7. However, our cohort could not represent a comprehensive patient group with organ-confined PCa due to treatment indications. Secondly, the results may suggest that p2PSA derivatives better correlate with the extent of cancer invasion than the cancer grade. Heidegger et al.16 revealed that the highest p2PSA was seen in patients with a GS ≥8 at RP and the lowest in those with a GS ≤6. A significant difference was seen in p2PSA values between a GS ≥8 and GS ≤7 (p < 0.01). Guazzoni et al.17 confirmed that the %p2PSA and PHI were accurate biomarkers of pT3 disease, a pathological GS ≥7, an upgraded GS, and tumor volume < 0.5 ml in men undergoing RP. Another multicenter study by Fossati et al.12 also supported their accurate prediction of pT3 disease and/or a pathological GS ≥7. However, the %p2PSA and PHI seemed to provide slight benefit to the traditional predictive models. The increase in the AUC with the PHI for predictive accuracy was actually low for pT3 cancer (2.0–2.5%) and a pathological GS ≥7 (3–6%).
Previous reports have indicated that the reference ranges of biomarkers should be adjusted for different ethnic groups. Rhodes et al.18 showed that p2PSA derivatives were slightly higher in black men than in white men. Lower PHI values with a higher AUC for cancer detection at the initial TRUSP biopsy were observed in an Asian series compared with European studies10,19. At a cut-off range of the PHI between 35 and 55, Asian men had a lower detection rate of PCa and high-grade PCa than European men. Lower cut-off values were thus applied to Asian men for detecting PCa at the initial TRUSP biopsy19. Chiu et al. showed that the %p2PSA and PHI in Asian men had a higher AUC increase over the base model in predicting pT3 or a pathological GS ≥7 than in the Western studies (7.9% and 7.2% vs. 1.2% and 2.3%, respectively)12,13. The net clinical benefit of PHI in predicting pT3 or a pathological GS ≥7 was demonstrated in decision curve analysis when the threshold probability ranged between 20% and 45%13. Our findings seem to be consistent with these results, with a PHI at a lower cut-off value of 33.92 predicting the presence of ≥pT3 PCa, high-risk disease, and a positive surgical margin.
Several novel biomarkers have been simultaneously compared with the PHI for predicting pathological results from RP, such as prostate cancer antigen 3 (PCA3) or TMPRSS2:ERG fusions20,21. Both the PHI and PCA3 significantly increased the predictive accuracy of the base model for extracapsular extension, whereas the PHI added only incremental value for predicting a pathological GS ≥7 and SVI20. Tallon et al.21 suggested that the PHI was a more reliable biomarker than the other two markers to predict a pathological GS ≥7. Both the PHI and the TMPRSS2:ERG test could significantly predict extracapsular extension. A notable 14% increase in the AUC was seen when these three biomarkers were combined with the base model.
The performance of the PHI and MRI in predicting significant PCa after RP were compared by Porpiglia et al.22. A 4% increase in the AUC over the base model was added by the PHI (AUC = 0.75, p < 0.01) and a 7% increase in the AUC over the base model was seen with MRI (AUC = 0.78, p < 0.01). Nevertheless, the optimal sequences for combining serum markers and imaging studies in the clinical setting should be further investigated. Further cost-effectiveness analysis should also be taken into consideration, given that, compared with the PHI, MRI is less suitable in many ways; for instance, it is more expensive, has a greater demand for professional radiological interpretation, and has more limited equipment availability23.
Several limitations are found in our study. Although the small sample size of our cohort may limit the statistical significance, we demonstrated the outstanding predictive accuracy of the %p2PSA and PHI. Second, the patient group included in this study could not be used as a surrogate of patients with organ-confined PCa. A selection bias may exist because surgical indications are affected by factors such as patients’ decisions, age, or comorbidities. Finally, the pathological specimens were reviewed by different urogenital pathologists instead of via centralized evaluation.
In conclusion, the %p2PSA and PHI accurately predict aggressive pathological features in RP specimens, including the presence of ≥pT3 cancer, high-risk disease, positive surgical margin, and SVI. The cut-off values of p2PSA derivatives improve their application in clinical practice. In particular, a PHI ≥61.26 significantly increases the predictive accuracy of the model for identifying the presence of SVI.
Methods
Between February 2017 and June 2018, 91 men with biopsy-proven clinically organ-confined PCa who underwent robot-assisted RP were prospectively enrolled from the National Taiwan University Hospital, a tertiary medical institution. Clinical data were obtained, including age, digital rectal examination, prostate volume, prostate weight, PSA derivatives, GS from TRUSP biopsy, number of positive biopsy cores, and percentage of positive biopsy cores. Cancer staging was completed with bone scintigraphy and MRI. Exclusion criteria included any possible factors that might alter PSA values: (1) active urinary tract infection; (2) use of 5-alpha reductase inhibitors such as finasteride or dutasteride; (3) preoperative androgen-deprivation therapy; and (4) transurethral resection of the prostate prior to the RP.
The novel biomarkers p2PSA, %p2PSA [(p2PSA/fPSA × 1000) × 100], and PHI were compared with the widely accepted standard tests: tPSA, fPSA, and %fPSA. Blood samples were drawn prior to the RP after informed consent was obtained from patients. Within 3 hours of blood collection, the serum samples were processed by centrifugation at 1500 × g for 15 minutes and stored at −20 °C until analysis, as reported by Semjonow et al.24. The blood samples were analyzed with a Beckman Coulter Access 2 immunoassay analyzer (Beckman Coulter Taiwan Inc.) with Beckman Coulter Access Hybritech reagent and calibrators. Specimens from TRUSP biopsy and RP were evaluated by experienced genitourinary pathologists who were blinded to the serum results.
The primary objective of our study focused on investigating the accuracy of the p2PSA, %p2PSA, and PHI in predicting adverse pathological features from RP, specifically: (1) extracapsular disease (pT3), (2) pathological GS ≥7 cancer, (3) high-risk disease (defined as pT3 and/or GS ≥8 based on the risk stratification of the NCCN guidelines in 2018), (4) upgrading of the GS sum from ≤6 at biopsy to ≥7 at RP specimen analysis, (5) positive surgical margin, and (6) SVI.
Statistical analyses were conducted with SPSS version 22.0 (IBM Corp, Inc., Chicago, IL, USA). The median and interquartile range (IQR) are presented for non-nominal variables. First, univariable analysis was used to test the ability of the parameters to predict pathological outcomes. Then, the cut-off values of each significant PSA or p2PSA derivative were selected at 90% sensitivity based on the area under the receiver operating characteristic curve (AUC). Finally, we examined the cut-off values of p2PSA derivatives separately in multivariable logistic regression models for their ability to predict aggressive pathological results. The AUCs of different predictive models were compared separately with the basic model. A two-sided p < 0.05 was considered statistically significant.
The present cohort was supported by the Institutional Review Board and Research Ethics Committee of National Taiwan University Hospital (Approval Code: 201612091RIPD). All methods were performed in accordance with the relevant guidelines and regulations. The informed consent was obtained from all participants and/or their legal guardians. The biomarker reagents were sponsored by Beckman Coulter Taiwan Inc., which had no participation in the study design or statistical analysis and was blinded to the results.
Supplementary information
Acknowledgements
The Beckman Coulter Access 2 immunoassay analyzer, access Hybritech reagent, and calibrators for all assays were provided by Beckman Coulter Taiwan Inc., which was blinded to the study design, data collection, statistical results, decision to publish and preparation of the manuscript. The authors declare no anticipated employment or personal financial interests.
Author contributions
All the authors contributed to the paper. Chao-Yuan Huang, Yung-Ting Cheng and Chih-Hung Chiang conceived the research idea and designed the study protocol. The data was collected by Yung-Ting Cheng, Chao-Yuan Huang, Chung-Hsin Chen, Shih-Ting Chiu, Jian-Hua Hong, Yeong-Shiau Pu, Shih-Ping Liu, Yu-Chuan Lu, Yi-Kai Chang, Hong-Chiang Chang, Kuo-How Huang, Yuan-Ju Lee, Po-Ming Chow, I-Ni Chiang, and Shih-Chun Hung. Yung-Ting Cheng, Chao-Yuan Huang, Jian-Hua Hong and Shih-Ting Chiu carried out the data analysis. Manuscript was written by Yung-Ting Cheng and critically reviewed by Chao-Yuan Huang and Chih-Hung Chiang. All the authors provided critical comments and approved the submission of the manuscript.This manuscript has been copyedited by native English speakers with a related biomedical background in BioMed Proofreading® LLC. We thank the Department of Medical Research at the National Taiwan University Hospital for helpful discussions during manuscript preparation. We thank the research assistants: I-Ting Teng, Pei-Yu Hsu for blood specimen collection and analysis. We thank Yun-Chieh Liang for the contributions of statistical analysis.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-57618-2.
References
- 1.Hamdy FC, et al. 10-Year Outcomes after Monitoring, Surgery, or Radiotherapy for Localized Prostate Cancer. N. Engl. J. Med. 2016;375:1415–1424. doi: 10.1056/NEJMoa1606220. [DOI] [PubMed] [Google Scholar]
- 2.Dinh KT, et al. Incidence and Predictors of Upgrading and Up Staging among 10,000 Contemporary Patients with Low Risk Prostate Cancer. J. urology. 2015;194:343–349. doi: 10.1016/j.juro.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Chun FK, et al. Development and internal validation of a nomogram predicting the probability of prostate cancer Gleason sum upgrading between biopsy and radical prostatectomy pathology. Eur. urology. 2006;49:820–826. doi: 10.1016/j.eururo.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Lughezzani G, et al. Predictive and prognostic models in radical prostatectomy candidates: a critical analysis of the literature. Eur. urology. 2010;58:687–700. doi: 10.1016/j.eururo.2010.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasivisvanathan V, et al. MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N. Engl. J. Med. 2018;378:1767–1777. doi: 10.1056/NEJMoa1801993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Rooij M, et al. Accuracy of Magnetic Resonance Imaging for Local Staging of Prostate Cancer: A Diagnostic Meta-analysis. Eur. urology. 2016;70:233–245. doi: 10.1016/j.eururo.2015.07.029. [DOI] [PubMed] [Google Scholar]
- 7.Mikolajczyk SD, Rittenhouse HG. Pro PSA: a more cancer specific form of prostate specific antigen for the early detection of prostate cancer. Keio J. medicine. 2003;52:86–91. doi: 10.2302/kjm.52.86. [DOI] [PubMed] [Google Scholar]
- 8.Lazzeri M, et al. Serum isoform [−2]proPSA derivatives significantly improve prediction of prostate cancer at initial biopsy in a total PSA range of 2-10 ng/ml: a multicentric European study. Eur. urology. 2013;63:986–994. doi: 10.1016/j.eururo.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Lughezzani G, et al. Multicenter European external validation of a prostate health index-based nomogram for predicting prostate cancer at extended biopsy. Eur. urology. 2014;66:906–912. doi: 10.1016/j.eururo.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, Y. T. et al. The application of p2PSA% and prostate health index in prostate cancer detection: A prospective cohort in a Tertiary Medical Center. Journal of the Formosan Medical Association = Taiwan yi zhi (2018). [DOI] [PubMed]
- 11.Wang W, et al. Diagnostic ability of %p2PSA and prostate health index for aggressive prostate cancer: a meta-analysis. Sci. reports. 2014;4:5012. doi: 10.1038/srep05012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fossati N, et al. Preoperative Prostate-specific Antigen Isoform p2PSA and Its Derivatives, %p2PSA and Prostate Health Index, Predict Pathologic Outcomes in Patients Undergoing Radical Prostatectomy for Prostate Cancer: Results from a Multicentric European Prospective Study. Eur. urology. 2015;68:132–138. doi: 10.1016/j.eururo.2014.07.034. [DOI] [PubMed] [Google Scholar]
- 13.Chiu PK, et al. Prostate Health Index and %p2PSA Predict Aggressive Prostate Cancer Pathology in Chinese Patients Undergoing Radical Prostatectomy. Ann. surgical oncology. 2016;23:2707–2714. doi: 10.1245/s10434-016-5183-6. [DOI] [PubMed] [Google Scholar]
- 14.Mearini L, et al. Use of the Prostate Health Index for the Detection of Aggressive Prostate Cancer at Radical Prostatectomy. Urologia internationalis. 2015;95:390–399. doi: 10.1159/000379758. [DOI] [PubMed] [Google Scholar]
- 15.Tosoian JJ, et al. Association of [−2]proPSA with biopsy reclassification during active surveillance for prostate cancer. J. urology. 2012;188:1131–1136. doi: 10.1016/j.juro.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heidegger I, et al. [−2]proPSA is an early marker for prostate cancer aggressiveness. Prostate cancer prostatic diseases. 2014;17:70–74. doi: 10.1038/pcan.2013.50. [DOI] [PubMed] [Google Scholar]
- 17.Guazzoni G, et al. Preoperative prostate-specific antigen isoform p2PSA and its derivatives, %p2PSA and prostate health index, predict pathologic outcomes in patients undergoing radical prostatectomy for prostate cancer. Eur. urology. 2012;61:455–466. doi: 10.1016/j.eururo.2011.10.038. [DOI] [PubMed] [Google Scholar]
- 18.Rhodes T, et al. Distribution and associations of [−2]proenzyme-prostate specific antigen in community dwelling black and white men. J. urology. 2012;187:92–96. doi: 10.1016/j.juro.2011.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiu, P. K. et al. A Multicentre Evaluation of the Role of the Prostate Health Index (PHI) in Regions with Differing Prevalence of Prostate Cancer: Adjustment of PHI Reference Ranges is Needed for European and Asian Settings. European urology (2018). [DOI] [PubMed]
- 20.Cantiello F, et al. Prognostic accuracy of Prostate Health Index and urinary Prostate Cancer Antigen 3 in predicting pathologic features after radical prostatectomy. Urologic oncology. 2015;33:163.e115–123. doi: 10.1016/j.urolonc.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Tallon L, et al. Comparative evaluation of urinary PCA3 and TMPRSS2: ERG scores and serum PHI in predicting prostate cancer aggressiveness. Int. J. Mol. sciences. 2014;15:13299–13316. doi: 10.3390/ijms150813299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porpiglia F, et al. In-parallel comparative evaluation between multiparametric magnetic resonance imaging, prostate cancer antigen 3 and the prostate health index in predicting pathologically confirmed significant prostate cancer in men eligible for active surveillance. BJU international. 2016;118:527–534. doi: 10.1111/bju.13318. [DOI] [PubMed] [Google Scholar]
- 23.Sathianathen NJ, et al. Incorporating Biomarkers into the Primary Prostate Biopsy Setting: A Cost-Effectiveness Analysis. J. urology. 2018;200:1215–1220. doi: 10.1016/j.juro.2018.06.016. [DOI] [PubMed] [Google Scholar]
- 24.Semjonow A, et al. Pre-analytical in-vitro stability of [−2]proPSA in blood and serum. Clin. biochemistry. 2010;43:926–928. doi: 10.1016/j.clinbiochem.2010.04.062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.