Abstract
To elucidate R-gene evolution, we compared the genomic compositions and structures of chromosome regions carrying R-gene clusters among cultivated and wild rice species. Map-based sequencing and gene annotation of orthologous genomic regions (1.2 to 1.9 Mb) close to the terminal end of the long arm of rice chromosome 11 revealed R-gene clusters within six cultivated and ancestral wild rice accessions. NBS-LRR R-genes were much more abundant in Asian cultivated rice (O. sativa L.) than in its ancestors, indicating that homologs of functional genes involved in the same pathway likely increase in number because of tandem duplication of chromosomal segments and were selected during cultivation. Phylogenetic analysis using amino acid sequences indicated that homologs of paired Pikm1–Pikm2 (NBS-LRR) genes conferring rice-blast resistance were likely conserved among all cultivated and wild rice species we examined, and the homolog of Xa3/Xa26 (LRR-RLK) conferring bacterial blight resistance was lacking only in Kasalath.
Subject terms: Genome evolution, Comparative genomics, Molecular evolution, Genetic variation, Evolutionary genetics
Introduction
Resistance genes (R-genes) confer disease resistance on plants, often forming clusters and showing frequent changes in copy number among genomes1–3. As the sequences of cluster members are highly homologous, it is believable that the individual genes have evolved possibly through duplication events4,5. Considerably, clustering of similar R-genes with the highly conserved sequences may lead to the creation of new resistance specificities via unequal crossing-over, gene conversion, or both which can be an important genetic resource4,6. Models including the birth-and-death model and the balancing model are often used to explain the evolutionary process of R-genes. Based on a prediction that defeated R-genes get lost rapidly from the host population due to a metabolic cost associated with the gene maintenance, the birth-and-death model proposes that new disease-resistance genes are born by gene duplication4,7. In plants, several R-genes such as wheat Pm3, Arabidopsis thaliana RPP13, flax L and capsicum eIF4E, for example, seem to have evolved in this way since they are likely involved in co-evolutionary relationships with pathogens8–11. The balancing model suggests that genetic variation in disease resistance is maintained, even though there exists a fitness cost associated with the preservation of temporarily non-functional R-genes12. R-genes of RPM1, RPS2 and RPS5 in Arabidopsis and Lr21 in wheat have been studied, revealing that both functional and non-functional alleles show the coexistence over a long period of evolutionary time in wild populations12–15. Factors that explain the differences about the evolutionary patterns of R-genes in these two distinct models are still not well known.
Cultivated rice varieties are either Asian or African in origin. The Asian cultivated rice Oryza sativa L. consists of two main subspecies, indica and japonica, each of which has a wild rice as its ancestor. The wild rice that was the source of O. sativa ssp. japonica is O. rufipogon (a perennial wild rice); the one that was the source of O. sativa ssp. indica is O. nivara (an annual wild rice, also called annual-growth-type O. rufipogon); and the one that was the source of the African cultivated rice O. glaberrima is O. barthii16–19. It remains controversial whether or not japonica and indica were domesticated independently20,21.
Gene annotation from the whole genomic sequence of the japonica rice cultivar Nipponbare has revealed the existence of more than 500 R-gene models along its 12 chromosomes, encoding nucleotide binding site (NBS) – leucine-rich repeat (LRR) domains22,23. In particular, rice chromosome 11 is highly enriched in R-genes; up to 201 loci encode the domains of not only NBS-LRR but also LRR – receptor-like kinase (LRR-RLK) or wall-associated serine/threonine protein kinase (WAK), mostly in clusters24.
Breeding for a wide range of disease-resistance qualities is one of the major goals of rice improvement. To achieve this goal, various alleles of R-genes have been discovered and put to practical use. The rice gene Xa3/Xa26, identified on the long arm of chromosome 11 (Chr11L) in indica rice, encodes an LRR-RLK conferring resistance to Xanthomonas oryzae, which causes bacterial blight disease25,26. Another major blast-resistance locus located on Chr11L is Pik, derived from the japonica rice Tsuyuake. The blast resistance provided by one of the Pik alleles, Pikm, is conferred not by a single gene, but rather by a combination of two adjacent NBS-LRR genes (Pikm1-TS, Pikm2-TS)27. Although a few R-genes have been found to confer disease resistance, the functions of most of the clustered R-genes are still unknown.
To clarify how R-genes arise and evolve, it is desirable to compare gene duplication among plant accessions exposed and not exposed to disease for a long time. A comprehensive comparison of the sequences and structures of R-gene cluster regions between domesticated rice species and their wild ancestors would be an optimal way to give insights into the molecular evolution of R-genes of plants grown in different areas for a long time in cultivated fields or in wild environments. Current whole-genome resequencing with advanced next-generation sequencing (NGS) technology has promoted large-scale and genome-wide comparative genomics among different rice cultivars and wild rice accessions18. However, simply mapping the NGS short-read sequences onto the reference genome created from the japonica rice Nipponbare23 will likely not be effective for uncovering the structural complexity of R-gene cluster regions because of the rapid and dynamic changes in genomic composition and structure, caused particularly by chromosomal rearrangement events such as duplication and insertion or deletion of DNA segments during the evolution of rice species and even cultivated accessions. Therefore, to uncover the dynamics and evolution of R-gene cluster formation we need to obtain all of the relevant genomic information by completely sequencing these kinds of special regions independently in both cultivated and wild rice species.
Here, by map-based sequencing, we performed a comprehensive comparison among different cultivars and their ancestral wild accessions of the genomic composition and structure of a genomic region that is known to contain R-gene clusters and is located close to the terminal end of rice Chr11L. In the japonica rice cultivar Nipponbare, this chromosomal region, flanked by the two DNA markers R0251 and E50301, carries about 1.35 Mb of genomic sequences and at least 38 genes annotated to hold NBS-LRR or LRR-RLK domains24,28. Known as the largest R-gene cluster detected in the rice genome, this region includes important loci of the disease resistance genes Pikm1 and Pikm2 as well as Xa3/Xa2626,27,29.
Results
Overall comparison of chromosomal composition and structure
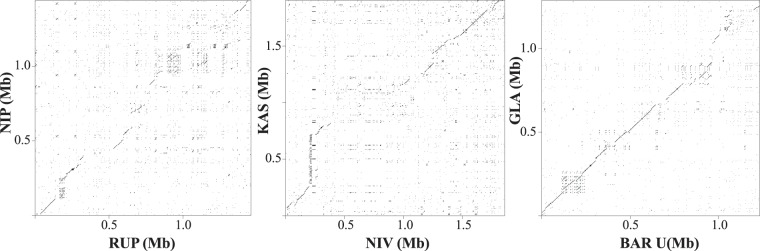
By screening BAC libraries using nine DNA markers as well as BAC-end sequences, we constructed BAC-based chromosomal physical maps that completely covered the R-gene cluster region on each accession, with the sole exception of O. rufipogon W1943 (RUF), which had one gap remaining (Supplementary Fig. S1). Complete sequencing of the minimum-tiling-path BAC clones from each accession finally yielded non-redundant DNA sequences ranging from 1.23 to 1.91 Mb in length (Table 1, Supplementary Table S1). Sequence comparison by BLAST analysis proved the sequence conservation and chromosomal synteny of the above genomic region among cultivated and wild species, although substantial structural variations (as represented by large-scale insertions and deletions) were present, causing differences in the region’s physical length (Fig. 1, Supplementary Table S2). The Asian cultivated rice O. sativa ssp. indica Kasalath (KAS) and its wild ancestor O. nivara W0106 (NIV) had the most abundant DNA sequences on the orthologous region flanked by the two conserved DNA markers R0251 and E50301, amounting to 1.74 Mb and 1.69 Mb, respectively, of DNA (Fig. 2, Table 1). The Asian cultivated rice O. sativa ssp. japonica Nipponbare (NIP) and its wild ancestor RUF had 1.35 and 1.32 Mb, respectively—less than those of KAS and NIV. The African cultivated rice O. glaberrima IRGC104038 (GLA) and its wild ancestor O. barthii W1588 (BAR) had the smallest sequences, both with 1.17 Mb—smaller than in the Asian cultivated and wild accessions. Very clearly, the two Asian rice cultivars and their wild ancestors—particularly the annual-growth-type wild rice NIV—had a higher (34.3% to 50.1%) repeat content than the African cultivated rice GLA and its wild ancestor BAR (30.9% and 30.7%, respectively). The high content of repetitive sequences detected in the Asian rice species was probably due to the frequent amplification of retrotransposable elements in them (Supplementary Table S3). Annotation of transcripts including non-coding genes and pseudogenes within the above genomic sequences resulted in a total of 62 to 97 gene models (Table 1, Supplementary Table S4). At least 40.3% of the total predicted genes belonged to the R-gene family, because they encoded domains highly similar to those of the NBS-LRR and LRR-RLK gene sequences in public databases. KAS had many more R-gene loci than any of the other cultivated or wild rice accessions had, including 53 NBS-LRR genes and 4 LRR-RLK genes, which accounted for 58.8% of its total number of genes. The total numbers of R-genes were increased in both NIP and KAS but were slightly decreased in GLA relative to the numbers in their respective wild ancestors.
Table 1.
Summary of map-based sequences and gene annotation determined from the R-gene cluster region on rice chromosome 11L.
| Name | Oryza sativa Nipponbare | Oryza rufipogon W1943 | Oryza sativa Kasalath | Oryza nivara W0106 | Oryza glaberrima IRGC104038 | Oryza barthii W1588 |
|---|---|---|---|---|---|---|
| Abbreviation | NIP | RUF | KAS | NIV | GLA | BAR |
| Region | Asia | Asia | Asia | Asia | Africa | Africa |
| Cultivated or wild | cultivated | wild | cultivated | wild | cultivated | wild |
| Total length of non-redundant DNA sequences (bp)a | 1,354,172 | 1,320,785b | 1,740,657 | 1,690,167 | 1,172,253 | 1,165,008 |
| GC content (%) | 43.0 | 43.8 | 43.2 | 44.4 | 42.6 | 42.8 |
| Repeat content (%) | 34.3 | 39.4 | 38.2 | 50.1 | 30.9 | 30.7 |
| Total number of annotated genes | 84 | 79 | 97 | 62 | 82 | 95 |
| Number of NBS-LRR R-genesc | 26 (8) | 20 (5) | 53 (17) | 18 (7) | 29 (9) | 33 (14) |
| Number of LRR-RLK R-genesc | 12 (2) | 13 (10) | 4 (2) | 7 (3) | 10 (7) | 11 (5) |
aCalculated from the orthologous chromosomal region flanked by the two conserved DNA markers R0251 and E50301 (see Fig. 2) in all cultivated and wild rice accessions. bIncluding 200 “Ns” used to represent the single physical gap remaining unclosed in O. rufipogon. cArabic numbers in blankets indicate pseudogenes.
Figure 1.
Comparative analysis of genomic sequences among R-gene cluster regions between cultivated and wild rice species. Sequence alignment between the Asian japonica rice variety Nipponbare (NIP) and its ancestor wild rice species O. rufipogon (RUP), the Asian indica rice variety Kasalath (KAS) and its ancestor wild rice species O. nivara (NIV) as well as the Africa cultivated rice O. glaberrima (GLA) and its ancestor wild rice species O. barthii (BAR) is shown respectively from the left to right. The position of matched sequences detected by BLASTZ is dot-plotted.
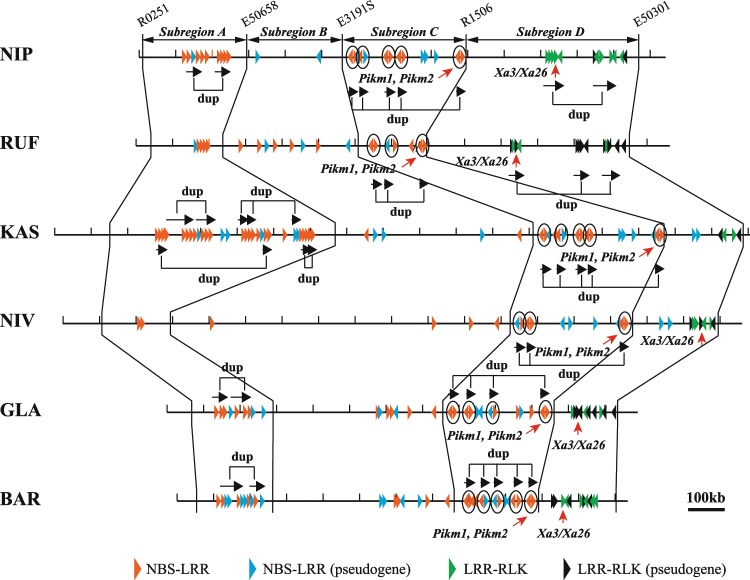
Figure 2.
Detection of location and distribution pattern of R-genes. On the basis of the positions of DNA markers, the whole R-gene cluster region examined is divided into four subregions, A to D. Arrowheads indicate intact or pseudo NBS-LRR and LRR-RLK genes. Subregion C contains the Pik/Pikm locus responsible for rice blast resistance and subregion D contains the Xa3/Xa26 locus responsible for bacterial blight disease resistance indicated by red arrows. The black arrows grouped by “dup” indicates the unit of segmental duplication. The duplicated gene pairs in opposite directions were circled.
Chromosomal organization of R-genes
To describe and compare the constitutions and patterns of organization of all R-gene loci, for convenience we divided the whole genomic region into four subregions, A to D, on the basis of the conserved marker positions (Fig. 2, Supplementary Table S4). Within subregion A (between R0251 and E50658) we found 10 R-genes encoding the NBS-LRR domain of NIP (282 kb), but only five in its wild ancestor RUF (198 kb). We found up to 30 NBS-LRR family genes in KAS (618 kb), but only two in its wild ancestor NIV (183 kb). Thus, there were many more NBS-LRR genes in the Asian cultivated rices, particularly KAS, than in their ancestors. We found 9 NBS-LRR-family genes in the African GLA (219 kb) and 11 in its wild ancestor BAR (219 kb), indicating comparatively high conservation of R-gene-family numbers between the two.
Within subregion B (between E50658 and E3191S) we found only two NBS-LRR-family genes in NIP (250 kb), but eight within the same subregion of RUF (366 kb) (Fig. 2). KAS (542 kb) had five and NIV (926 kb) had four, whereas GLA (462 kb) had seven and BAR (494 kb) had eight, reflecting the lack of substantial change in copy number of R-genes between each cultivated rice and its wild ancestor. The big change in physical length between KAS and NIV was due to the marked difference in the amount of repetitive sequences abundant in long-terminal-repeat retrotransposons (Table 1, Supplementary Table S3).
Subregion C (between E3191S and R1506) corresponded to the region in rice that contains the Pik/Pikm locus, which confers blast resistance. We found 14 NBS-LRR-family genes in NIP (340 kb) and seven in RUP (181 kb), and 16 in KAS (348 kb) and 10 in NIV (335 kb), indicating that there was a marked increment in R-gene copy numbers in the Asian cultivated rices relative to their ancestors, as in subregion A. Pikm-specific resistance is conferred by the cooperation of the Pimk1-TS and Pikm2-TS genes, which are adjacent to each other and are transcribed in opposite directions27. Very interestingly, many adjacent NBS-LRR gene pairs with opposite transcription directions were found within this region, including five pairs in NIP, three in RUF, five in KAS, three in NIV, four in GLA, and five in BAR (Fig. 2). On the other hand, there was almost no change in NBS-LRR gene numbers between the two African rice accessions, with 13 in GLA (309 kb) and 14 (221 kb) in BAR. No LRR-RLK genes were detected within subregion A, B or C.
Subregion D (between R1506 and E50301) carries the bacterial blight resistance gene locus Xa3/Xa2626,29. Only R-genes encoding the LRR-RLK domain were observed within subregion D of four of the accessions, at 12 in NIP (459 kb), 13 in RUF (554 kb), 10 in GLA (164 kb) and 11 in BAR (208 kb). In contrast, two NBS-LRR genes, together with four and seven LRR-RLK family genes, were detected respectively within the KAS (209 kb) and NIV (228 kb). There was thus a reduction in R-gene numbers in the indica rice relative to its ancestor.
Phylogenetic tree of R-genes
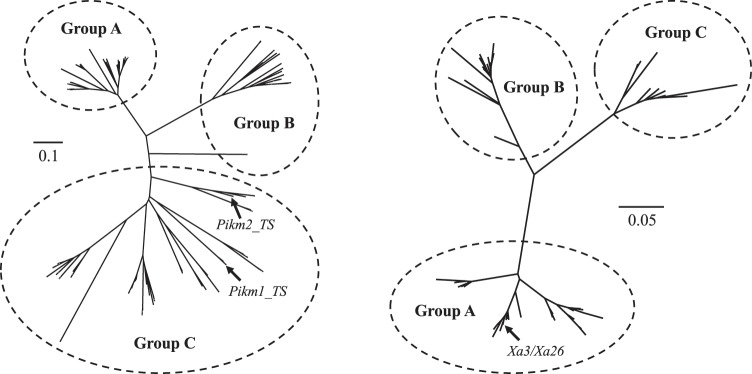
Phylogenetic analysis of all R-genes by using amino acid sequences from the NBS domain identified three major groups of NBS-LRR genes (Fig. 3 also see Supplementary Fig. S2 for details). Group A consisted of 67 NBS-LRR family genes, all derived from subregion A. Group B had 34 genes, all from subregion B. Group C had 78 genes, most from subregion C, and four (two in KAS and two in NIV) from subregion D. These data suggest that gene duplication was restricted to within each subregion. Clearly, the pair of Pikm1-TS and Pikm-TS2, both necessary for disease resistance, fell into Group C; their orthologs are conserved within the genomes of all of the cultivated and wild rice accessions: respectively RG025 and RG026 in NIP, RG050 and RG051 in KAS, RG019 and RG020 in RUP, RG015 and RG016 in NIV, RG028 and RG029 in GLA, and RG032 and RG033 in BAR. The phylogenetic tree revealed low sequence identity between the pairs of adjacent R-genes, clearly indicating that they are not paralogs generated directly by gene duplication but that they were instead duplicated in pairs.
Figure 3.
Phylogenetic analysis of R-genes. Phylogenetic trees are created by the neighbour-joining method from amino acid sequences derived from the domain region of NBS-LRR (left) and LRR-LRK (right) genes in all species analyzed. Arrows indicate locations of R-genes with known functions.
Phylogenetic analysis of R-genes by using amino acid sequences from the kinase domains also yielded three major groups of LRR-RLK family genes, all within subregion D of the accessions (Fig. 3; also see Supplementary Fig. S3 for details). Xa3/Xa26 was a member of Group A; its ortholog was likely conserved in NIP (RLK004), RUP (RLK003), NIV (RLK004), GLA (RLK003) and BAR (RLK003), but it seems to have been lost in KAS.
The sequence identities (phylogenetic tree) and chromosomal organization shown by the R-genes provide strong evidence for the evolutionary event of segmental duplication in all rice accessions we investigated. For example, at least five segmental duplication events seem to have happened so far within subregion A of KAS, unlike in its wild ancestor NIV (Fig. 2). Only one duplication of R-genes, however, likely occurred in the African cultivated GLA within the same subregion; it obviously had its origin in the wild ancestor BAR, indicating that the event took place before the divergence of the two closely related species. Within subregion C, carrying the paired genes Pikm1 and Pikm2 and their homologs, on the other hand, NIP and KAS both seem to have experienced gene duplications four times, two of which could be traced back to their wild rice ancestors, RUF and NIV (Fig. 2). Some of the duplications revealed species specificity: for example, the segmental duplications carrying LRR-RLK genes within subregion D were likely restricted to NIP and its ancestor RUP, because no strong evidence indicating segmental duplication with multiple R-genes was found in the same subregion of KAS, NIV, GLA or BAR.
Discussion
Domestication and subsequent repetitive cultivation by humans have increased the number of R-genes
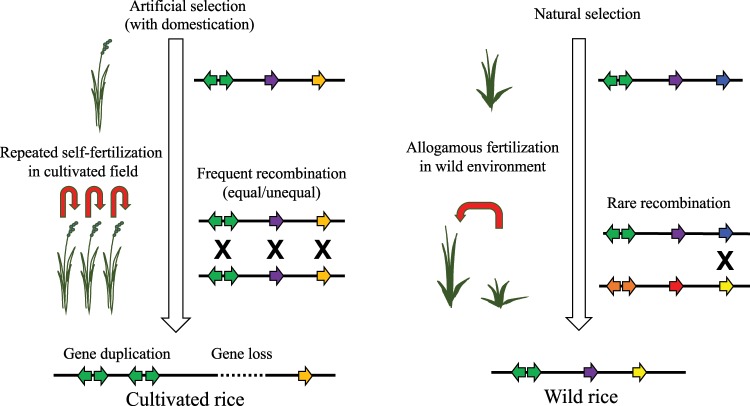
The numbers of NBS-LRR family genes were substantially greater in the Asian cultivated rices than in their wild ancestors (Table 1). This difference is probably due to the effects of domestication and of repeated cultivation on genotype (cultivar vs. wild: homozygous or heterozygous), fertilisation process (self-fertilising vs. allogamous), plant population density (dense vs. sparse), and generation time (annual single or multiple harvests vs. perennial or annual). In cultivated rice, it is desirable for alleles to be homozygous and for plants to be self-fertilising so as to maintain the phenotype. The copy number of tandem R-genes increases or decreases with chromosomal recombination, but the rate of these changes will increase if genetically homozygous, self-fertilising individuals are raised repeatedly in densely planted fields. In addition, the life cycle of cultivated rice is annual even with multiple cropping, whereas that of wild rice is either annual or perennial. Cultivated rice has been artificially selected for adaptation to each cultivation area. We consider that these factors have promoted the duplication of R-genes in cultivated rice: the ready potential of R-gene clusters to multiply has been promoted markedly by domestication and repeated cultivation (Fig. 4).
Figure 4.
Differences in the evolution of R-genes between cultivated and wild rice species. Cultivated rice loses its heterozygosity during domestication as self-fertile individuals are cultivated repeatedly. As a result, duplication of R-genes tends to occur, and sometimes loss of R-genes happens in parallel. The characteristics of the birth-and-death model are therefore notable in cultivated rice. In contrast, wild rice maintains non-functional alleles. Allele introgression from other individuals occurs, causing the accumulation of mutations, thus potentially leading to the creation of functional alleles under natural conditions. The characteristics of the balancing model are therefore notable in wild rice.
The region including the NBS-LRR genes showed comparatively high sequence identity and similar R-gene numbers in the African cultivated rice GLA and its wild ancestor BAR, indicating an extremely high level of conservation of chromosomal synteny (Fig. 1, Table 1). In Africa, O. glaberrima was domesticated from O. barthii about 3000 years ago, much later than domestication of O. sativa approximately 9000 years ago30. In future, it will be important to perform a comprehensive analysis and comparison of the population structures of the African and Asian rice Accessions to determine whether the above differences in evolutionary and domestication history have had a marked influence on the changes in R-gene copy numbers in rice genomes.
Features of the birth-and-death model predominate in R-genes of Asian cultivated rice
The major mechanism of R-gene birth was tandem duplication, which we found frequently in Asian cultivated rice (Fig. 2). Respectively, 26.3% and 33.3% of all R-genes in NIP and KAS were pseudogenes (Table 1). This phenomenon therefore represents a birth-and-death model that occurred in a relatively short period of time after rice was domesticated. To have duplicated genes is costly, even if the genes are intact. However, because in the cultivated environment there is little or no competition for survival against neighbouring plants, this disadvantage may not be an issue.
Features of the balancing model predominate in R-genes of wild rice
Variations in functional alleles were conserved intact. Within subregion C, the paired resistance genes Pikm1 and Pikm2 were conserved intact across all six accessions, and pseudogenes were observed from their paired homologs (Fig. 2). This finding suggests that the paralogs remain intact, and non-functional alleles are retained over relatively long periods of time. Such a situation is consistent with the balancing model. This model proposes that variation in disease resistance is maintained, even though there is a fitness cost associated with the maintenance of R-genes in the absence of their matching pathogens. Examples of the acquisition of resistance after the start of cultivation are known: at least five alleles—Pik, Pikm, Pikh, Pikp and Piks—have been identified on chromosome 11 as blast resistance genes31–33. Pik and Pikm appear to have emerged after the domestication of rice, whereas Pikp emerged before34. Many intact but non-functional alleles—especially in wild rice—have the potential to change through mutation to functional alleles conferring disease resistance.
Large deletion carries a risk of irreversible change and the loss of a specific gene family. Deletion of a region and changes to a pseudogene are both irreversible changes that can lead to the loss of an R-gene. In KAS, the gene corresponding to Xa3/Xa26 was lost, probably owing to a deletion within subregion D (Fig. 2). Irreversible loss of an allele of a disease-resistance gene is contrary to the balancing model. KAS is a drought tolerant upland landrace traditionally grown in the short summer in Bangladesh under rainfed conditions. We consider, therefore, that the loss of R-genes may not be critical in some cultivated rice plants that are grown under environmental conditions that lack the pressures that might alter their disease resistance to a given pathogen.
Differences between amplifiable and difficult-to-amplify R-genes
Duplication of the NBS-LRR gene was restricted within each subregion (Fig. 2). Some subregions showed greater sequence identity between orthologs than between paralogs and maintained chromosomal synteny. Other subregions showed copy number variation and disruption of chromosomal synteny. Why do contiguous R-genes have such different evolutionary patterns?
Within subregion A, the substantial amplification of R-gene copies in Asian cultivated rice—particularly in KAS—compared with their wild ancestors could be easily regarded as being due to multiple periods of segmental duplication. A similar case was present within subregion C: each duplication event was clearly associated with two paired genes showing opposite transcription directions, either orthologous or paralogous to the paired genes Pik1 and Pik2 of known function27. The RPP2 locus in Arabidopsis is another example in which two NBS-LRR family genes, RPP2A and RPP2B, have developed to work with disease resistance by pairing, thus cooperating to provide target recognition or signalling functions lacked by either partner protein35. Our results thus demonstrate that neighbouring genes that provide disease resistance cooperatively were often duplicated together. Unlike in subregions A and C, no marked changes in R-gene copies have occurred within subregions B and D between the species of cultivated and wild rice. The number of R-genes present within subregion B in NIP has even dropped to a quarter of that in its wild ancestor RUP.
In conclusion, the R-gene-cluster region located close to the terminal end of the long arm of rice chromosome 11 harbours genomic sequences on a megabase scale; this region carries subregions that clearly show differences in R-gene-amplification ability. We believe that subregions A and C hold chromosomal sites with high frequency of unequal recombination. These were probably induced by retrotransposon activity that promoted segmental duplications carrying R-genes that were selectively retained by humans during the long period of domestication and regional adaptation of rice.
The functions of most of R-genes are unknown
Exploration and use of R-genes are among the most important goals of crop breeding. Although there are many genes in the R-gene-cluster regions predicted to carry NBS-LRR or LRR-RLK domains, the real functions of most of them are unknown. An important future task will be to elucidate whether these R-gene paralogs have no function or whether simply their target fungi have not yet been identified. Genes that already endow resistance to specific pathogens are conserved, but highly mutated genes represent a potential reserve against future pathogens. This concept is consistent with the observation that clustered RGC2 genes in lettuce can exhibit heterogeneous rates of evolution36. It is not known why only specific genes among an abundance of paralogs are responsible for disease resistance. Pik and Pikm are the same gene27. Xa3 and Xa26 are also the same gene29. In the case of Cf-9 in tomato, sequence exchange occurs more frequently between orthologs than between paralogs37; this is also true of the Pi2/9 locus in rice38 and Arabidopsis4. This process would be expected to conserve the sequence identity and function of alleles, and thereby to promote the rapid development of new pathogen-recognition specificities under selection. This might be particularly important for cultivars when they are moved to new planting areas to face different environmental conditions.
It is very important to consider the chromosomal rearrangement events of single- or multiple-gene duplication, indels and sequence mutations that could lead to changes in gene number and to pseudogenisation among different genomes. Comprehensive comparison on a large scale in an effort to understand in detail the contents and evolution of R-gene clusters in rice is not possible simply by mapping short reads onto one reference genome. Currently, whole genome reference sequences have been assembled from several rice varieties through map-based sequencing and/or whole-genome sequencing with the NGS technology39,40. Comparing the sequence data located within the orthologous region of R-gene cluster between the two Chinese elite indica rice varieties, Minghui 63 and Shuhui498, and the six cultivated and wild rice species reported here by BLAST analysis confirmed the presence of dramatic genome divergence between or even within a single Oryza species, providing furthermore the genomic variations for future understanding of the evolutionary mechanisms on R-genes (Supplementary Figs. S4 and S5). The genomic sequences obtained at the present study thereafter can be used as a platform for further comparative studies of R-gene clusters within cultivated and wild rice populations and will certainly help to elucidate those nucleotide substitutions that are critical to disease resistance. To our knowledge, this study is the first to systematically obtain referenced genomic sequences from the large R-gene cluster in both cultivated and wild rice species. These findings should have a substantial impact on our understanding of the evolution and function of disease-resistance genes in plants.
Methods
Construction of BAC-based physical map
Besides the japonica rice cultivar Nipponbare (NIP; acc. no. WRC001, Japan), the whole genomic sequence of which is publicly available, we used the indica rice cultivar Kasalath (KAS; acc. no. WRC002, Bangladesh), two Asian wild rice accessions, O. rufipogon (RUF; acc. no. W1943, China; ancestor of japonica) and O. nivara (NIV; accession no. W0106, India; ancestor of indica), one accession of African cultivated rice O. glaberrima (GLA; accession no. IRGC104038, Senegal), and one of African wild rice O. barthii (BAR; accession no. W1588, Cameroon). The close genetic relationships of the cultivated rices with their wild ancestors have been confirmed both at the gene level41–43 and at a population structure level, as analysed by using genome-wide retrotransposon markers (in preparation). BAC libraries of each cultivar or accession were constructed as described before41,44. Positive BAC clones respectively covering the targeted R-gene cluster region on Chr11L of KAS, GLA, RUF, NIV and BAR orthologous to NIP Chr11L were screened by using a number of genetic or EST markers, or both, with a polymerase-chain reaction (PCR) method similar to that reported previously45,46. End sequences of positive BAC clones were also used for chromosomal walking with the same PCR method to select new BAC clones covering any of the physical gaps remaining within the above genomic region.
BAC sequencing and gene annotation
Minimum-tiling-path BAC clones were chosen from the BAC physical maps and sequenced by using a shotgun sequencing approach described before23. Genes were predicted by using an annotation system we developed previously23,47. Genes predicted to encode a hypothetical protein or transposable element were ignored. Pseudogenes were defined when coding sequences identical to functional genes were truncated because of the presence of disruptive mutations such as frame shifts or premature stop codons. Total amounts of repetitive sequences were calculated in RepeatMasker v. 1.332 software (http://www.repeatmasker.org) with the Oryza Repeat Database (http://rice.plantbiology.msu.edu/annotation_oryza.shtml) as a reference.
Sequences and structural comparison
Genomic sequences on the R-gene cluster region were compared between each cultivated and wild rice pair by BLAST algorithm48. BLAST alignments were dot-plotted in Dottup software (http://www.bioinformatics.nl/cgi-bin/emboss/help/dottup) to explore the conservation of chromosomal composition and structure between cultivated and wild rice accessions. To analyse the development and evolution of R-genes, we performed a phylogenetic analysis using amino acid sequences spanning the region from the P-loop to the GLPL motif of NBS-LRR genes and the whole kinase domain of LRR-RLK genes, including those present within pseudogenes. Sequences were aligned in MAFFT software49 to generate neighbour-joining trees in MEGA7 software50.
Supplementary information
Acknowledgements
We thank Dr. Rod A. Wing at the University of Arizona for supplying the BAC library of O. nivara, and Mrs Hiroko Fujisawa and Mr Wataru Karasawa at the Institute of Crop Science for technical support. This work was supported by grants from the Ministry of Agriculture, Forestry and Fisheries of Japan (Green Technology Project GD-2007, Genomics for Agricultural Innovation QTL5003).
Author contributions
J.W. and T.M. designed this work. S.K. and H.K. contributed to the BAC clone screening and sequencing. J.W., H.M. and Y.M. performed the data analysis. J.W., H.M. and T.S. wrote the manuscript.
Data availability
Genomic sequences of each BAC clone are available at NCBI (see Supplementary Table S1 for the accession number). Pseudomolecule sequences created from the above BAC sequences for each cultivated rice and wild rice species, and amino acid sequences of each predicted R-gene are also included within the supplementary Data on line.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-57729-w.
References
- 1.Collins N, et al. Molecular characterization of the maize Rp1-D rust resistance haplotype and its mutants. Plant Cell. 1999;11:1365–1376. doi: 10.1105/tpc.11.7.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parniske M, et al. Homologues of the Cf-9 disease resistance gene (Hcr9s) are present at multiple loci on the short arm of tomato chromosome 1. Mol. Plant-Microbe Interact. 1999;12:93–102. doi: 10.1094/MPMI.1999.12.2.93. [DOI] [PubMed] [Google Scholar]
- 3.Meyers BC, Kozik A, Griego A, Kuang HH, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:1683–1683. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michelmore RW, Meyers BC. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- 5.Leister D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends in Genet. 2004;20:116–122. doi: 10.1016/j.tig.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Hulbert SH, Webb CA, Smith SM, Sun Q. Resistance gene complexes: Evolution and utilization. Annu. Rev. Phytopathol. 2001;39:285–312. doi: 10.1146/annurev.phyto.39.1.285. [DOI] [PubMed] [Google Scholar]
- 7.Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes RW. Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell. 2004;16:309–318. doi: 10.1105/tpc.016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rose LE, et al. The maintenance of extreme amino acid diversity at the disease resistance gene, RPP13, in Arabidopsis thaliana. Genetics. 2004;166:1517–1527. doi: 10.1534/genetics.166.3.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dodds PN, et al. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yahiaoui N, Brunner S, Keller B. Rapid generation of new powdery mildew resistance genes after wheat domestication. Plant J. 2006;47:85–98. doi: 10.1111/j.1365-313X.2006.02772.x. [DOI] [PubMed] [Google Scholar]
- 11.Charron C, et al. Natural variation and functional analyses provide evidence for co-evolution between plant eIF4E and potyviral VPg. Plant J. 2008;54:56–68. doi: 10.1111/j.1365-313X.2008.03407.x. [DOI] [PubMed] [Google Scholar]
- 12.Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature. 1999;400:667–671. doi: 10.1038/23260. [DOI] [PubMed] [Google Scholar]
- 13.Tian D, Araki H, Stahl E, Bergelson J, Kreitman M. Signature of balancing selection in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2002;99:11525–11530. doi: 10.1073/pnas.172203599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mauricio R, et al. Natural selection for polymorphism in the disease resistance gene Rps2 of Arabidopsis thaliana. Genetics. 2003;163:735–746. doi: 10.1093/genetics/163.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang L, et al. Evolution of new disease specificity at a simple resistance locus in a crop-weed complex: Reconstitution of the Lr21 gene in wheat. Genetics. 2009;182:595–602. doi: 10.1534/genetics.108.099614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge S, Sang T, Lu BR, Hong D-Y. Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proc. Nalt. Acad. Sci. USA. 1999;96:14400–14405. doi: 10.1073/pnas.96.25.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Londo JP, Chiang Y-C, Hung K-H, Chiang T-Y, Schaal BA. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc. Natl. Acad. Sci. USA. 2006;103:9578–9583. doi: 10.1073/pnas.0603152103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang X, et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490:497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stein JC, et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 2018;50:285–296. doi: 10.1038/s41588-018-0040-0. [DOI] [PubMed] [Google Scholar]
- 20.Yang CC, et al. Independent domestication of Asian rice followed by gene flow from japonica to indica. Mol. Biol. Evol. 2012;29:1471–1479. doi: 10.1093/molbev/msr315. [DOI] [PubMed] [Google Scholar]
- 21.Choi JY, et al. The Rice paradox: Multiple origins but single domestication in Asian rice. Mol. Bio. Evol. 2017;34:969–979. doi: 10.1093/molbev/msx049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou T, et al. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol. Genet. Genomics. 2004;271:402–415. doi: 10.1007/s00438-004-0990-z. [DOI] [PubMed] [Google Scholar]
- 23.International Rice Genome sequencing Project The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 24.Rice Chromosomes 11 and 12 Sequencing Consortia The sequence of rice chromosomes 11 and 12, rich in disease resistance genes and recent gene duplications. BMC Biol. 2005;3:20. doi: 10.1186/1741-7007-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z, Sun X, Wang S, Zhang Q. Genetic and physical mapping of a new gene for bacterial blight resistance in rice. Theor. Appl. Genet. 2003;106:1467–1472. doi: 10.1007/s00122-003-1205-4. [DOI] [PubMed] [Google Scholar]
- 26.Sun Y, et al. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 2004;37:517–527. doi: 10.1046/j.1365-313X.2003.01976.x. [DOI] [PubMed] [Google Scholar]
- 27.Ashikawa I, et al. Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics. 2008;180:2267–2276. doi: 10.1534/genetics.108.095034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choisne, N. et al. The sequence of rice chromosomes 11 and 12, rich in disease resistance genes and recent gene duplications. BMC Biology3 (2005). [DOI] [PMC free article] [PubMed]
- 29.Xiang Y, Cao YL, Xu CG, Li XH, Wang SP. Xa3, conferring resistance for rice bacterial blight and encoding a receptor kinase-like protein, is the same as Xa26. Theor Appl. Genet. 2006;113:1347–1355. doi: 10.1007/s00122-006-0388-x. [DOI] [PubMed] [Google Scholar]
- 30.Vaughan DA, Lu B-R, Tomooka N. Was Asian rice (Oryza sativa) domesticated more than Once? Rice. 2008;1:16–24. doi: 10.1007/s12284-008-9000-0. [DOI] [Google Scholar]
- 31.Inukai T, et al. Allelism of blast resistance genes in near-isogenic lines of rice. Phytopathology. 1994;84:1278–1283. doi: 10.1094/Phyto-84-1278. [DOI] [Google Scholar]
- 32.Hayashi K, Yoshida H, Ashikawa I. Development of PCR-based allele-specific and InDel marker sets for nine rice blast resistance genes. Theor. Appl. Genet. 2006;113:251–260. doi: 10.1007/s00122-006-0290-6. [DOI] [PubMed] [Google Scholar]
- 33.Li LY, et al. Wang L, Jing JX, Li ZQ, Lin F, Huang LF, Pan QH. The Pik(m) gene, conferring stable resistance to isolates of Magnaporthe oryzae, was finely mapped in a crossover-cold region on rice chromosome 11. Mol. Breeding. 2007;20:179–188. doi: 10.1007/s11032-007-9118-6. [DOI] [Google Scholar]
- 34.Zhai C, et al. The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytologist. 2011;189:321–334. doi: 10.1111/j.1469-8137.2010.03462.x. [DOI] [PubMed] [Google Scholar]
- 35.Sinapidou E, et al. Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J. 2004;38:898–909. doi: 10.1111/j.1365-313X.2004.02099.x. [DOI] [PubMed] [Google Scholar]
- 36.Kuang H, Woo SS, Meyers BC, Nevo E, Michelmore RW. Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell. 2004;16:2870–2894. doi: 10.1105/tpc.104.025502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parniske M, Jones JDG. Recombination between diverged clusters of the tomato Cf-9 plant disease resistance gene family. Proc. Natl. Acad. Sci. USA. 1999;96:5850–5855. doi: 10.1073/pnas.96.10.5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou B, Dolan M, Sakai H, Wang GL. The genomic dynamics and evolutionary mechanism of the Pi2/9 locus in rice. Mol. Plant-Insect Interact. 2007;20:63–71. doi: 10.1094/MPMI-20-0063. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, et al. Extensive sequences divergence between the reference genomes of two elite indica rice varieties Zhenshan 97 and Minhui 63. Proc. Natl. Acad. Sci. USA. 2016;113(E35):E5163–E5171. doi: 10.1073/pnas.1611012113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du H, et al. Sequencing and de novo assembly of a near complete indica rice genome. Nat. Commun. 2017;8:15324. doi: 10.1038/ncomms15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J, Mizuno H, Sasaki T, Matsumoto T. Comparative analysis of rice genome sequence to understand the molecular basis of genome evolution. Rice. 2008;1:119–126. doi: 10.1007/s12284-008-9021-8. [DOI] [Google Scholar]
- 42.Yamane H, et al. Molecular and evolutionary analysis of the Hd6 photoperiod sensitivity gene within Genus Oryza. Rice. 2009;2:56–66. doi: 10.1007/s12284-008-9019-2. [DOI] [Google Scholar]
- 43.Matsumoto, et al. The Nipponbare genome and the next- generation of rice genomics research in Japan. Rice. 2016;9:33. doi: 10.1186/s12284-016-0107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katagiri S, et al. End sequencing and chromosomal in silico mapping of BAC clones derived from an indica rice cultivar, Kasalath. Breeding Sci. 2004;54:273–279. doi: 10.1270/jsbbs.54.273. [DOI] [Google Scholar]
- 45.Wu J, et al. A comprehensive rice transcript map containing 6591 expressed sequence tag stes. Plant Cell. 2002;14:525–535. doi: 10.1105/tpc.010274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanamori H, et al. A BAC physical map of aus rice cultivar ‘Kasalath’, and the map-based genomic sequence of ‘Kasalath’ chromosome 1. Plant J. 2013;76:699–708. doi: 10.1111/tpj.12317. [DOI] [PubMed] [Google Scholar]
- 47.Rice Annotation Project The annotation project database (RAP-DB): 2008 updated. Nucleic Acids Res. 2007;36:1028–1033. doi: 10.1093/nar/gkm978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altschul SF, et al. Gapped BLAST and PSIBLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 50.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Bio. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic sequences of each BAC clone are available at NCBI (see Supplementary Table S1 for the accession number). Pseudomolecule sequences created from the above BAC sequences for each cultivated rice and wild rice species, and amino acid sequences of each predicted R-gene are also included within the supplementary Data on line.