Abstract
The specialties of allergy and clinical immunology have entered the era of precision medicine with the stratification of diseases into distinct disease subsets, specific diagnoses, and targeted treatment options, including biologicals and small molecules. This article reviews recent developments in research and patient care and future trends in the discipline. The section on basic mechanisms of allergic diseases summarizes the current status and defines research needs in structural biology, type 2 inflammation, immune tolerance, neuroimmune mechanisms, role of the microbiome and diet, environmental factors, and respiratory viral infections. In the section on diagnostic challenges, clinical trials, precision medicine and immune monitoring of allergic diseases, asthma, allergic and nonallergic rhinitis, and new approaches to the diagnosis and treatment of drug hypersensitivity reactions are discussed in further detail. In the third section, unmet needs and future research areas for the treatment of allergic diseases are highlighted with topics on food allergy, biologics, small molecules, and novel therapeutic concepts in allergen‐specific immunotherapy for airway disease. Unknowns and future research needs are discussed at the end of each subsection.
Keywords: allergy, exposome, microbiome, neuroimmune, respiratory viral infections
1. INTRODUCTION
The past decades have witnessed extensive progress in unraveling cellular and molecular mechanisms of immune regulation in asthma, allergic diseases, organ transplantation, autoimmune diseases, tumor biology, and chronic infections.1, 2 Consequently, a better understanding of the functions, the reciprocal regulation, and the counterbalance of subsets of immune and inflammatory cells but also structural cells—for example, epithelial and vascular cells, airway smooth muscle cells, neuroendocrine system—that interact via various intercellular messengers will indicate avenues for immune interventions and novel treatment modalities of allergic diseases and immunological disorders. It is generally expected that drug development in the next decades will show a significant shift from chemicals to biologicals. After more than 20 years without any breakthrough drug becoming available for patients, several disciplines including allergology are now experiencing extraordinary times with the recent licensing of several major biological drugs and novel allergen‐specific immunotherapy (AIT) vaccines. Several biological modifiers of the immune response targeting intracellular messengers or their receptors have been developed to date.3, 4, 5, 6, 7, 8 In addition, a number of promising small molecule drugs and vaccines are in the development pipeline.9, 10, 11 This new era is now calling for the development of biomarkers and pheno‐ and endotyping of diseases for customized patient care, which is termed stratified medicine, precision medicine, or personalized medicine.4 Distinguishing phenotypes of a complex disease covers the observable clinically relevant properties of the disease but does not show a direct relationship to disease etiology and pathophysiology. In a complex condition, such as asthma, different pathogenetic mechanisms can induce similar clinical manifestations; however, they may require different treatment approaches.12, 13 These pathophysiological mechanisms underlying disease subgroups are addressed by the term “endotype.”12, 13, 14 Classification of complex diseases based on the concept of endotypes provides advantages for epidemiological, genetic, and drug‐related studies. Accurate endotyping by using reliable biomarkers reflects the natural history of the disease and aims to predict the response to (targeted) treatments.15 Recent studies have focused on better understanding of endotypes and phenotypes of allergic diseases, asthma, allergic and chronic rhinosinusitis ± nasal polyps, chronic obstructive pulmonary disease, and on the development of biomarkers including novel interleukins and microRNAs that regulate their expression to stratify patients.16, 17, 18
2. BASIC MECHANISMS OF ALLERGIC DISEASES—KEY QUESTIONS
2.1. Structural and functional biology of allergens—where are we at?
Cloning of allergen cDNAs and sequencing of purified natural allergens have so far yielded 919 officially accepted allergenic proteins listed in the database of the WHO/IUIS Allergen Nomenclature Sub‐Committee (http://www.allergen.org/; accessed 11/2018). Structures of allergens determined by crystallography or NMR amount to around 100 as summarized in the Structural Database of Allergenic Proteins (http://fermi.utmb.edu/; accessed 11/2018) and by Dall'Antonia et al19 Structural data of allergens allow the study of cross‐reactivities between related allergens,20 or the design of allergens with altered IgE epitopes as vaccine candidates for AIT.21 The location of IgE‐binding epitopes can be determined based on allergen structures and experimental data.22 Various technologies exist for mapping conformational IgE epitopes.23 X‐ray crystallography of an allergen‐antibody complex allows the most precise identification of conformational epitopes. To date, only two structures of cocrystals of IgE and allergen are available, Bos d 524 and Phl p 2,25 both complexed with an IgE Fab. Sequence and structural data have revealed that allergens are members of a limited number of protein families (http://www.meduniwien.ac.at/allfam/). This insight has now become mainstream knowledge and indicates that the biological functions of allergens might be linked to their allergenicity.26
Various explanations for the existence of the allergic immune response have been brought forward including the toxin hypothesis,27, 28 the danger theory,29 and the allergic host defense model.30 Unequivocally, these authors27, 28, 29, 30 argue that it is a common misconception to regard allergens as generally harmless environmental substances. Allergens interact with innate immune receptors (eg, TLR4,31 protease‐activated receptor‐2,32 dectin‐133), disrupt the integrity of membranes (eg, phospholipase A2,34, 35 defensins36, 37), or degrade connective tissues (eg, hyaluronidases38). However, very few studies on why only susceptible individuals raise an allergic immune response have come forward. They indicate that genetic susceptibility is based on altered signal processing39 and mutations of pattern recognition receptors.33
How to expand the understanding of allergens and allergic sensitization?
Provide structures of homologous allergens to study cross‐reactivity
Provide structures of hypoallergenic variants to visualize the effects of allergen design
Provide structures for all major allergen types
Provide structures for allergens complexed with IgE Fabs, IgG Fabs, or single‐chain antibodies
Provide structures of allergens with their ligands
Perform studies on the effect of the biological function of allergens on innate immune cells
Perform studies on signal transduction initiated by allergens in innate immune cells
Define pattern recognition receptors, membrane constituents, or other cellular binding partners of allergens
Define “susceptibility to allergic sensitization” at the molecular and mechanistic level
2.2. Mechanisms of type 2 inflammation and immune tolerance to allergens
2.2.1. Type 2 immune response
Since the discovery of T‐helper (Th) subsets, it was demonstrated in the last three decades that almost all immune cells display functional subsets characterized by distinct signature cytokines and surface receptors. Generally, it is considered that a type 2 immune response is the main player in the pathogenesis of eosinophilic asthma, allergic rhinitis, chronic rhinosinusitis with nasal polyps, eosinophilic esophagitis, and extrinsic atopic dermatitis.40 The type 2 immune response is an immune response to environmental noninfectious proteins and helminths, and involves Th2 cells, type 2 B cells, group 2 innate lymphoid cells, type 2 macrophages, a small fraction of IL‐4–secreting NK cells, IL‐4–secreting NK‐T cells, basophils, eosinophils, and mast cells.41, 42 From a complex network of cytokines, IL‐4, IL‐5, IL‐9, IL‐13, and IL‐31 are mainly secreted from immune system cells and IL‐25, IL‐33, and TSLP from tissue cells, particularly epithelial cells.43, 44 (Figure 1) GATA3 is the key transcription factor for the induction of this response.45 Both the innate and the adaptive immune response contribute to type 2 immune response. Among these cytokines, IL‐4 and IL‐13 play roles in production of allergen‐specific IgE, IL‐5 in eosinophilia,46, 47 IL‐9 and IL‐13 in mucus production, IL‐4 and IL‐13 in tissue migration of Th2 cells and eosinophils, and IL‐4 and IL‐13 in regulation of tight junctions and epithelial barrier integrity.48 Type 1, type 17, type 22, and immune regulatory responses, and nonallergic mechanisms such as environmental factors, psycho‐social stress, activation of metabolic pathways, resident cells in the remodeled phenotype, or epithelial barrier dysfunction further modulate the profile of type 2–driven inflammation. In addition, type 2–driven inflammation is characterized by a high cellular plasticity that enables the cells to adapt to a specific inflammatory milieu. Several subendotypes might exist within the type 2 immune response complex endotype such as the IL‐5‐high, IL‐13‐high, or IgE‐high endotype, and their dominance differs between allergic diseases (Figure 1). Omalizumab targeting IgE, mepolizumab, reslizumab targeting IL‐5, benralizumab targeting the IL‐5 receptor, and dupilumab targeting the IL‐4 and IL‐13 common receptor alpha chain are some of the biologicals currently available to control type 2 inflammation.
Unknowns and future research highlights in type 2 immune response
Which cell is more critical and predominant for general type 2 responses, and in which disease?
Which cytokine is more important for which clinical in vivo situation?
A detailed list of environmental factors that enhance type 2 responses
Mechanisms of viral infections in exacerbation of type 2 diseases
Local immune deficiency caused by a type 2 immune response
Effect of type 2 immune responses to chronicity
Novel biomarkers of type 2 responses for treatment selection, to decide when to stop treatment and to monitor therapy response in type 2 diseases
Role of epithelial barrier leakiness in the development and chronicity of complex type 2 immune response–related diseases
Head‐to‐head comparison of different type 2 immune response‐targeting treatments
Pharmacoeconomics of different type 2 immune response‐targeting treatments in comparison with existing conventional treatments
Disease‐modifying effect of different type 2 immune response‐targeting treatments
Combination treatments with allergen immunotherapy
Figure 1.
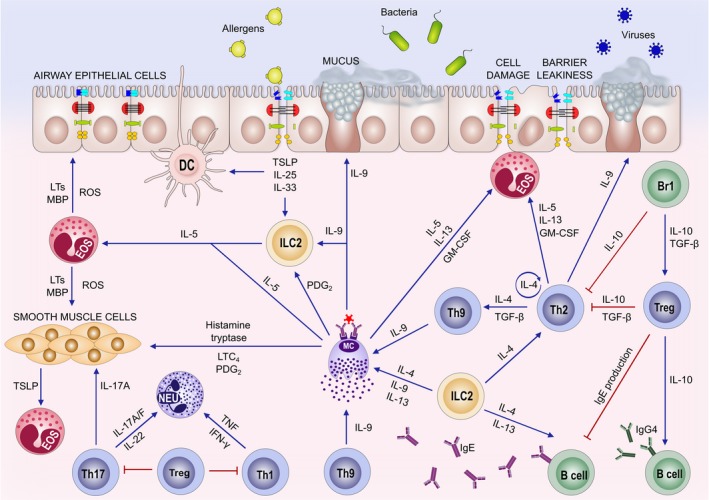
Molecular mechanisms in allergic inflammation. Epithelial leakiness and activation and their proinflammatory cytokine and chemokine (TNF‐α, IL‐13, TSLP, IL‐25, IL‐33) production induce inflammation and contribute to the Th2 response. Highly activated epithelial cells undergo apoptosis and shedding takes place. Chemokines are essential players for the recruitment of inflammatory cells followed by survival and reactivation of migrating inflammatory cells and their interaction with resident tissue cells and other inflammatory cells. Innate lymphoid cells (ILC2) play a role in T‐ and B‐cell activation and recruitment and are early providers of type 2 cytokines and T‐cell recruitment‐related chemokines. The Th2 type of an immune microenvironment is characterized by IL‐4, IL‐5, IL‐9, IL‐13, IL‐25, IL‐33 production by Th2 cells, ILCs, mast cells, and tissue cells. Eosinophilia is induced by IL‐5, IL‐25, and IL‐33. Local and systemic IgE production takes place in allergic patients with the involvement of IL‐4 and IL‐13. Other effector T‐cell subsets, such as Th9, Th17, and Th22 cells, also play partial roles in inflammation, mucus production, and tissue healing. Smooth muscle, myofibroblast activation, and bronchial hyperreactivity are related to IL‐4, IL‐9, IL‐13, IL‐25, and IL‐33. Several chemokines, and arachidonic acid pathway molecules and other small molecules play roles in the inflammatory cell recruitment and further augmentation of the inflammatory cascades. Treg and Breg cells play a role on control of inflammation and extensive cellular activation by using IL‐10 and TGF‐β as well as many other suppressive mechanisms
2.2.2. T‐regulatory and B‐regulatory cells
Immune regulation is an important function of the immune system to tolerate self‐tissues and non–self‐environmental allergens. T‐regulatory (Treg) cell subsets have distinct phenotypes and include constitutive and inducible subsets of CD4+CD25+ Forkhead box P3 (FOXP3)+ Treg cells and type 1 Treg cells (Tr1). As a second major player in immune regulation, IL‐10–producing B‐regulatory (Breg) cells have also been demonstrated to suppress allergen‐specific responses and promote IgG4 isotype antibodies.49 Allergen tolerance in high‐dose–exposed individuals such as beekeepers and cat owners, the AIT response, and protective effects of farm exposure make up one of the most representative areas where Treg and Breg cells display their major roles.49, 50, 51 IL‐10, IL‐35, and TGF‐beta are the major suppressor cytokines with immune regulatory functions within multiple complex mechanisms.42, 50, 52 Different subsets have been defined in different disease conditions, and research should further identify their regulation and in vivo relevance.53 T‐ and B‐regulatory cells suppress many functions of type 2 inflammation including type 2 innate lymphoid cells.54 Extensive research is ongoing in this area. To date, there are no biologicals that induce T‐ and B‐regulatory responses in patients; however, various modes of AIT represent major stimulators of these cells in an allergen‐specific manner in vivo.
Unknowns and future research highlights in Treg and Breg response
Life span of allergen immunotherapy and natural exposure‐induced Treg and Breg cells in vivo
Effect of Treg and Breg cells on tissue cells
Functional comparison of different subsets of Treg and Breg cells
Molecular mechanisms of Treg and Breg cell generation in vivo
Adjuvants that promote Treg and Breg cells in vivo
Relationship of resident tissue cells and their interaction with Treg and Breg cells in allergen immunotherapy‐induced immune tolerance
Early biomarkers and predictors for the generation of Treg and Breg responses
Mechanisms of long‐term maintenance of allergen tolerance and the link to Treg and Breg responses
Mechanisms of inducing high‐affinity IgG4 and low‐affinity IgE antibodies
2.3. Neuroimmune mechanisms in allergic inflammation
It is becoming increasingly clear that immune cells do not act alone and that cross talk and reciprocal regulation between neural and immune systems are essential in the pathophysiology of allergic diseases including allergic asthma, atopic dermatitis, and food allergies.55, 56 Immune and neuronal cell types are found in large numbers at skin and mucosal barrier surfaces and are in close contact with each other forming a neuronal‐immune cell network.57, 58, 59, 60 Both immune and neural cells detect and respond to environmental threats and harmful stimuli including allergens. Innate and adaptive immune responses mediate proinflammatory responses by secretion of cytokines (eg, IL‐4, IL‐5, IL‐9. IL‐13, IL‐25, IL‐31, IL‐33, TSLP), chemokines (eg, histamine), and other lipid mediators (eg, leukotrienes) on encountering allergens. Within these cytokines, a biological targeting IL‐31 was shown to treat itch in atopic dermatitis. In addition to mediating allergic responses via immune responses, these proinflammatory mediators also directly activate sensory neurons that regulate itch, cough, sneezing, bronchoconstriction, and alterations in gastrointestinal motility.59
On stimulation, sensory and autonomic neurons release neuropeptides and neurotransmitters such as substance P, neurokinin A, neuromedin U (NMU), calcitonin gene–related peptide, vasoactive intestinal peptide, acetylcholine, and norepinephrine that signal immune cells.55 In the airways, calcitonin gene–related peptide is released by sensory nerves, which has been shown to inhibit dendritic cell maturation and allergen‐specific T‐cell responses.61 In the gut, ILC2 cells were shown to express Nmur1, a receptor for the neuropeptide NMU. ILC2s live in close proximity to NMU‐producing nerve cells and become proinflammatory when exposed to NMU. NMU signaling can significantly amplify allergic inflammation when high levels of IL‐25, IL‐33, and TSLP are present.62 In a mouse model of allergic asthma, norepinephrine was found to stimulate IgE production on binding β2‐adrenergic receptors and activating B cells.63, 64 A positive feedback loop between neurotransmitters and neuropeptides and immune cells exists. However, our understanding of these interactions and the signals that mediate their responses to the ever‐changing physiological and pathological conditions is still very limited.
Future prospects for research on neuroimmune regulation of allergic diseases
The mechanisms underlying allergen‐induced release of proinflammatory mediators and neural activation (reflexes)
Colocalization and direct and local communications between neuronal and immune cells and their role in mediating allergic response and tolerance
Identification of neuropeptides and neurotrophins that directly act on immune cells via receptors
Development of pharmacological compounds targeting neuropeptides and neurotrophins that mediate allergic response
Feedback loop between neuronal and immune cells in mediating immune homeostasis
2.4. Exposome, environmental factors and allergy
The rising trend in allergies is associated with changes in lifestyle and the control of infections which, taken together, seem to result in an “under‐challenged” immune system.65, 66 On the other hand, lifestyle changes and indoor and outdoor environmental pollutants67, 68, 69 are suspected to keep our immune system in a constant state of low‐grade inflammation. Apart from direct effects of outdoor pollutants on humans, pollen‐producing plants are themselves subject to modification by anthropogenic pollutants.70, 71 Presumably, beneficial factors include growing up in a rural environment, traditional lifestyle, and a nutrition rich in dietary fibers and of a high diversity. It has become evident that environmental factors induce epigenetic changes which are associated with allergic diseases (summarized in Ref.72).
The exposome includes the entire environmental exposures that a person experiences, from conception throughout the whole life (Figure 2).73 A clear missing knowledge is the lack of thorough epidemiological studies encompassing a holistic approach with exposome and reactome (response patterns) over a life span. This wide gap further opens especially because validated methods for exposome assessment, especially the personal one, are lacking. Furthermore, a clear bias emerges since biogenic and anthropogenic pollutants are measured outdoors while people spend a considerable part of their lives indoors. The research focus in environmental health and allergy should therefore be to study the impact of indoor and outdoor pollutants focusing on the role of combined exposures to air pollution, microclimate, green spaces, and allergens. Innovative approaches to characterize environmental exposures including satellite data and stationary and personal monitoring should be developed.74 This further requires the development of new informatics tools and data analytics to analyze the large and complex generated datasets. The aim should be to understand the impact of environment through the entire life span on the complete disease spectrum to unravel the interaction of the environment with the barrier organs including their microbiomes, the immune system, and the whole body. Climate change exhibits direct and indirect effects on human health–related aspects. Climate variability modifies the abundance and occurrence of plants and fungi, noticeably those with high allergological importance. The effect of climate change on human health is both a threat and foremost a research focus especially in the field of allergy. A thorough understanding of the molecular mechanisms of the interactions of environment with the human body but also environment‐environment interactions will enable us to develop prevention strategies for allergies.
Hot spots in environmental health research
Moving from associations to causalities and molecular mechanisms
Understanding environment‐gene interactions and especially the role of epigenetic changes
Develop innovative methods for exposome assessment, especially the personal one
Development of devices for personal monitoring of real‐time pollen and fungal spore abundance spatiotemporal information
Understanding additive and summative effects of environmental factors on health and disease
Define personal thresholds for environmental triggers for allergic symptoms
Figure 2.
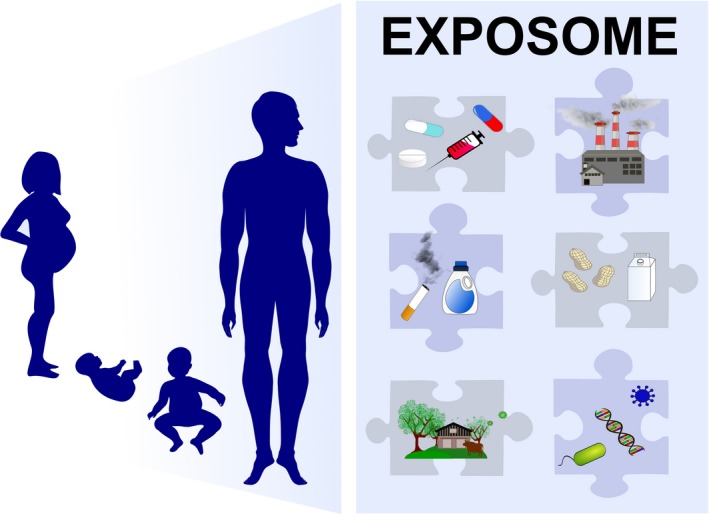
The exposome: The exposome includes the entire environmental exposures of an individual from conception throughout the whole life. Early‐life events such as mode of delivery, breastfeeding, mother's diet, lifestyle and health status, antibiotics, and other drug usage in pregnancy and early childhood, early‐life environment (ie, siblings, pets at home, proximity to farm animals and green areas, usage of primary farm products) can significantly influence the epigenetic regulation of immune system and tissue cells
2.5. Role of the microbiome and diet in immune responses
Enormous varieties of microbes colonize the skin and mucosal body surfaces. These microbes are organized within complex community structures, whose composition is dependent on the specific body site examined. It is increasingly appreciated that the microbiome interacts intimately with mucosal immune processes and disrupted communication between the microbiome and the host due to altered microbiome composition and/or metabolism is thought to negatively influence immune homeostatic networks and may play a role in immune hypersensitivity to environmental exposures, such as allergens.75
A number of studies have consistently demonstrated that an altered gut, lung, nasal, or skin microbiome is associated with, and sometimes precedes, allergic sensitization and inflammation.76, 77 In particular, early‐life events such as mode of delivery, breastfeeding, mother's diet and health status, antibiotics, and other drug usage in pregnancy and early childhood, early‐life environment (ie, siblings, pets at home, proximity to farm animals and green areas, usage of primary farm products) can significantly influence the timing of bacterial colonization and establishment.78, 79, 80 However, one of the most potent modulators of microbiota composition is diet, as consumed foods provide the fuel for microbial metabolic activities.81 For example, microbiota‐accessible carbohydrates (MAC) are complex carbohydrates found in dietary fibers that contribute to microbial diversity and generation of metabolites, such as short‐chain fatty acids (SCFAs).82 SCFAs promote regulatory immune responses, and high SCFA levels early in life are associated with protection from atopic sensitization.83 In contrast, a high‐fat diet is associated with negative effects on microbiota composition and metabolism.
Despite increasing awareness of the importance of microbiome‐diet interactions in health and disease, the molecular basis for these multidirectional functional interactions is only beginning to be described. Although diet‐microbiome interventions are exciting and attractive approaches, many unknown factors still limit the successful translation of these potentially game‐changing interventions into the clinic.
Unknowns in the microbiome area for future research
Contribution of the virome (viral communities) and mycobiome (fungal communities) to immune tolerance networks
Mechanistic pathways linking diet diversity, microbial metabolism, and allergy prevention
Role of the diet in modulating microbial communities outside of the gut
The importance of baseline microbial populations or historical dietary patterns in determining the response to microbiota or diet‐based interventions
The influence of microbiota on the clinical response to allergen‐specific immunotherapy and its mechanisms
Definition of a healthy microbiota and ways to achieve it
Possibility of intervention in the microbiome of a diseased individual
2.6. Respiratory viral infections and allergy
Respiratory viruses are the most common causes of respiratory diseases, which can be linked with the potentiation of acute and chronic respiratory mucosal inflammation. This usually occurs through mechanisms including upregulation of cell adhesion molecules, pathogen sensing receptors, and Toll‐like receptors, which are common immunopathogenic factors mediating or involved in virus‐ and allergen‐induced mucosal inflammation.84, 85, 86, 87, 88, 89, 90, 91 In addition, respiratory viruses were suggested to impact cilia and tight junction integrity in airway epithelial cells through the modulation of ZO‐1, claudin‐1, and occludin in the airway epithelial barrier,92, 93, 94 which may be linked to pathophysiology of airway diseases. The nasal epithelium is the primary portal of entry for respiratory viruses and immediate target for viral replication in the airways.87, 95 It is also an active component of initial host responses against viral infection. Such nasal epithelial‐specific transcriptomic alterations may significantly influence the downstream immune responses and homeostasis that define the pathology of respiratory infection and complications.87, 95, 96, 97 This is evident in the case of most respiratory viral infections, which, while self‐limiting, could trigger chronic type 2 inflammatory responses via excessive release of chemokines and cytokines into the airways. The resulting recruitment of the immune cells (ie, neutrophils, eosinophils, mast cells, and T cells) may then ultimately predispose the airway to remodeling.98, 99, 100, 101 In addition, a recent study showed that H3N2 infection of the nasal epithelium was associated with significant increase in interferons (IFN‐α, IFN‐γ, IL‐29), proinflammatory cytokines (TNF‐α, BDNF, IL‐3), and viral‐associated chemokines (IP‐10, MCP‐3, I‐TAC, MIG), detectable as early as 24 hours postinfection.102 This translates into rapid monocyte, NK‐cell, and innate T‐cell (MAIT and γδ T cells) activation, evident with CD38+ and/or CD69+ upregulation.102 Therefore, an understanding of the predominant type and underlying mechanisms of mucosal inflammation triggered by common viral infections will allow identification of targets for better management of chronic airway inflammatory diseases.
Unknowns and future prospects for research in viral infections and allergic diseases
The predominant type and underlying mechanisms of mucosal inflammation (eg, type 2 or non‐type 2) triggered by infection of different types of respiratory viruses
Mechanistic role of viral infections in chronicity
Mechanisms of viral infections in exacerbations
Mechanisms of viral infections in breaking of allergen tolerance
Novel mechanisms to prevent or avoid viral infections
Novel vaccines for various viruses
Novel anti‐viral treatments based on newly identified mechanisms
3. DIAGNOSTIC CHALLENGES AND REGULATORY CONSIDERATIONS
3.1. Clinical trials for the treatment of allergic diseases
In 2008, the Committee for Medicinal Products for Human Use (CMPH) of the European Medicine Agency (EMA) has implemented the “Guideline on the Clinical Development of Products for Specific Immunotherapy for the Treatment of Allergic Diseases (CHMP/EWP/18504/2006)” (Available from: http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2009/09/WC500003605.pdf and ref.103) and by this has set methodological standards for clinical trial designs for AIT regarding phase I‐III performances and outcomes. This guidance has been followed in “Therapy Allergen Ordinance (TAO)” which has been initiated for future registration and marketing authorization in Germany of a group of allergen extracts of specific species (details in Ref.104). An increasing number of AIT products fulfill the regulatory demands and have been authorized in different countries on the basis of proven efficacy in the clinical documentation.105, 106 However, the regulatory guidance leaves some space for interpretation regarding certain specificities of clinical trial design in both early and late phases of clinical development programs for AIT, and harmonization of methodological principles in the design of these trials would be preferable for all parties involved.107 Hence, the Immunotherapy Interest Group (IT‐IG) of the European Academy of Allergy and Clinical Immunology (EAACI) has elaborated different task‐force projects regarding improvement of methodological study design in AIT (https://www.eaaci.org/organisation/eaaci-interest-groups/ig-on-immunotherapy/activities/2880-task-forces-of-the-immunotherapy-interest-group.html; accessed on 07 Dec 2018). In an EAACI Position Paper, this group has aimed to summarize and standardize clinical endpoint measures and has elaborated a combined symptom and medication score (CSMS) as standard primary endpoint for future (pivotal) trials in AIT.108 Another example is an EAACI Position Paper, which overviews current concepts in tolerance‐inducing mechanisms aimed to highlight potential biomarkers which may be of predictive value for determining responders to AIT in clinical trials.15 However, there is an urgent need for further harmonization and clinical validation of methodological determinants in AIT clinical study design, which can only be achieved by international collaboration of clinical experts, methodologists, and regulatory authorities.109, 110
Examples of unknowns and future prospects for harmonization of AIT trial design and interpretation of trial results (modified to references107, 109, 110)
Evaluation and validation of possible biomarkers of predictive value for efficacy15
Further validation of clinical meaningful primary and secondary endpoints108
Clinically justified definitions of relevant treatment effect sizes
Potential of allergen exposure chambers for AIT product development107
Minimal level of evidence needed for the clinical documentation of efficacy in the pediatric population110
Better understanding of placebo effects in SCIT and SLIT107
Defining clinical endpoints and establishing the effectiveness, disease‐modifying properties, and the duration of both in asthmatic patients
3.2. Precision medicine and immune monitoring of allergic diseases
Precision medicine (providing the right treatment to the right patient and the right dose at the right time) requires an accurate diagnosis and monitoring of the treatment response. While precision medicine has been practiced in allergology for over a century since the advent of grass pollen‐specific immunotherapy,111 it currently infers (often synonymous with “personalized medicine”) use of the new “omics” technologies to identify genes or biomarkers for diagnosis or monitoring of treatment efficacy (Figure 3).13, 112 The “omics” revolution is based on platform technologies in genomics (by far the most robust), metabolomics, proteomics, epigenomics, transcriptomics, lipidomics, and microbiomics to generate vast global datasets, and advanced bioinformatics to interrogate and interpret the datasets using machine learning and artificial intelligence (Figure 4).113 Such analysis of population‐based datasets can reveal novel insights to underpin therapeutic selection from an expanded range of precise biologicals.114 Examples are emerging from patients with inborn errors of immunity (IEI) in whom the genetically defined defect can be specifically targeted with therapeutics.115, 116 The functional utility of data from the omics platforms will be further enhanced by the public release of omics datasets including Genotype‐Tissue Expression (GTEx)117 and Encyclopaedia of DNA Elements (ENCODE).118
Figure 3.
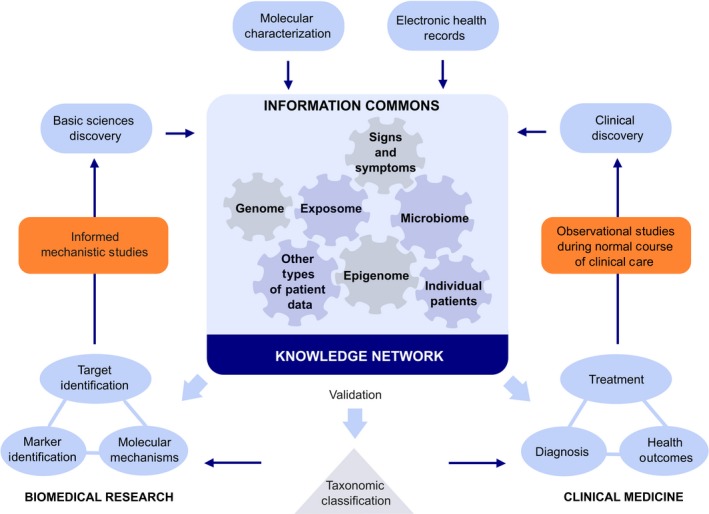
Precision medicine: Precision medicine requires the network of knowledge from both biomedical and clinical research. It includes all of the omics areas and exposome from molecular characterization and biomarker development to electronic health records, and clinical discoveries in diagnosis and treatment. The introduction of a new taxonomy is needed to ensure that all the stakeholders speak the same language
Figure 4.
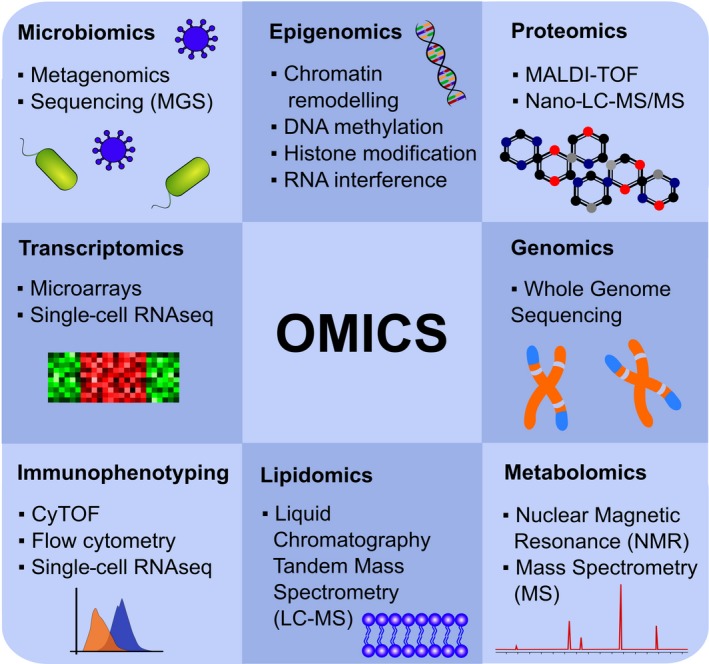
Omics: The omics revolution was one of the major driving forces of recent developments that enabled investigation of almost everything at the molecular level of proteins, lipids, and small molecules including innumerable DNA and RNA sequencings with a hypothesis‐free approach
Technological advances in immune monitoring capability are augmented by highly standardized or chimeric recombinant allergens and peptides (B‐ and T‐cell epitope‐based). Exponential advances in microarrays, time‐of‐flight mass cytometry (CyTOF), basophil activation tests, next‐generation gene sequencing, and RNA‐seq are generating huge enabling datasets.15, 115, 116, 119, 120, 121, 122, 123 The risk that small populations of highly pathogenic cells might be masked by abundant signatures of more frequent or more active cells is combated by the emergence of enhanced single B‐ and T‐cell immunophenotyping using flow cytometry–based assays. This allows longitudinal immunoprofiling of relevant cell subsets in individuals before and during AIT. Better dissection of mechanisms underlying allergic diseases informs better tailoring of therapies.115, 116, 119, 120, 121, 122, 124, 125
Unmet needs in precision medicine
Consensus on endotypes and clinically applicable biomarkers for distinct allergic disorders
Accurate targeted activity; even, widespread or specific as needed
Increased availability of oral formulations: solid or liquid
More favorable dosing intervals
Minimization of adverse side effects (especially anaphylactic/allergic reactions, antibody induction, complement activation)
Economic feasibility enhancement
Determination of long‐term effects
“Large dataset” protection and interpretation, including ethical considerations
Development of a precision medicine‐specific nomenclature
Development of treatment and diagnostic algorithms
Development of precision medicine–focused guidelines
3.3. Allergic rhinitis and nonallergic rhinitis
Chronic rhinitis (CR) is one of the most common diseases globally, with a considerable financial burden.126, 127 At present, CR is simplistically subclassified as allergic rhinitis (AR) and nonallergic rhinitis (NAR).128 Although phenotyping of rhinitis has important consequences in the treatment of the entity,129 the presently employed phenotypes cannot meet the needs of precision medicine; suggesting an urgent need for the CR phenotypes to be updated with the progress of diagnostic methods. In this regard, Meng and colleagues have recently investigated the phenotypes of CR based on a cluster analysis of 12 clinical variables.130 In this study, AR was subclassified as allergic rhinitis with or without asthma, while NAR was subclassified as nonallergic rhinitis with eosinophilia syndrome (NARES) without asthma, NARES with asthma, local allergic rhinitis (LAR), and idiopathic rhinitis. The finding of the LAR cluster was interesting because none of these patients had a history of asthma, but demonstrated high levels of local eosinophils and local production of specific IgE (local IgE), of which the latter has been used in studies of LAR over the last few years.131, 132, 133 Indeed, another study by Meng and colleagues has suggested that local IgE is a reliable noninvasive alternative to serum IgE for the diagnosis of AR,134 and there is emerging evidence that local IgE could also be used instead of nasal allergen provocation test (NAPT) for the diagnosis of LAR. Besides local IgE, nasal cytology has also been shown to be useful in the diagnosis of CR A recent study by She and colleagues assessed nasal cytology in chronic sinusitis patients with rhinitis, using the liquid‐based ThinPrep Cytology Test (TCT) and demonstrated that this technique has higher sensitivity, specificity, and positive predictive value for inflammation in the inferior turbinates than for inflammation in the maxillary sinus.135 Thus, the TCT might also be used in the studies on CR, especially NARES.
In conclusion, there is increasing evidence that local IgE and nasal cytology are useful clinical diagnostic markers in CR and might represent the way forward especially for studies differentiating the endotypes of AR and NAR in the future.
Future research questions and prospects for chronic rhinosinusitis
Could local IgE measurement be used for the diagnosis of local allergic rhinitis?
Could nasal cytology be used in the study of chronic rhinitis, especially for the differential diagnosis of chronic rhinitis?
Can we provide the diagnostic standard of local IgE determination for allergic rhinitis and local allergic rhinitis?
Can we provide the diagnostic standard of nasal eosinophil count for nonallergic rhinitis with eosinophilia syndrome?
3.4. New approaches to the diagnosis and treatment of drug hypersensitivity reactions
Drug hypersensitivity reactions (DHRs) are defined as adverse effects of pharmaceutical formulations that clinically resemble allergy. Drug allergies are defined as DHRs for which a definite immunological mechanism, IgE‐ or T cell–mediated, is demonstrated.136 DHRs constitute an important health problem, affecting more than 7% of the population, 137 for whom drugs, such as beta‐lactam antibiotics and nonsteroidal anti‐inflammatory drugs, are essential for treatment of common diseases.138, 139 Based on the clinical characteristics of DHRs, different phenotypes have been identified,137 although the lack in understanding the underlying mechanisms of many DHRs has hampered the definition of endotypes and identification of biomarkers.140, 141
The classification of DHRs based on the time elapsed between drug administration and development of symptoms is still a matter of debate, because it is difficult to establish a cutoff point to distinguish between immediate and nonimmediate DHRs.13, 136, 141 These data are relevant for defining phenotypes and establishing an accurate diagnosis and specific treatment. An important recent advance has been the inclusion of “Drug hypersensitivity” as a subsection in the International Classification of Diseases (ICD)‐11.140
The diagnosis of DHR is mainly based on skin tests and drug provocation tests, methods that are not free of risk, still lack standardization, and differ depending on the drug, mechanisms, and even the health system.142, 143, 144 There is an urgent need for developing new in vitro diagnostic tests or improving those already existing,138 such as basophil activation test,145, 146 to improve the diagnostic workup. The complexity of DHR diagnosis and its lack of optimal specificity lead to an over‐diagnosis. This is an important problem, as patients “labeled” as allergic receive alternative treatments that are usually less effective and more toxic, so “de‐labeling” constitutes a public health measure.147 Primary care physicians are often the first point of contact for patients with DHRs; thus, they have a key role in diagnosis and need specific training.148
Although the specific treatment of DHRs is avoiding the drug involved and those chemically related, desensitization is nowadays a frequent option.149, 150, 151 In that sense, rapid drug desensitization is a cost‐effective technique that activates inhibitory mechanisms and permits patients to receive the first‐choice medications to which they are allergic.149, 150
Research needs for DHRs
The prevalence and incidence of DHRs
Underlying mechanisms of DHRs
The most adequate classification of DHRs
Definition of endotypes and biomarker identification
-
The most accurate diagnostic approach:
i Skin test standardization, sensitivity, specificity, and predictive values for most drugs
ii A standardized protocol for performing drug provocation test
iii The role of in vitro tests for diagnosis
The mechanism of inhibition in rapid drug desensitization procedure
4. UNMET NEEDS AND FUTURE RESEARCH AREAS IN TREATMENT OF ALLERGIC DISEASES
4.1. How to treat food allergy in the future: new developments and concepts
We are observing a pandemic increase in food allergy and approaching an era of efficient treatments. In peanut allergy oral immunotherapy (OIT), phase III studies on AR101152 in peanut allergic patients and phase II(b) and III studies on epicutaneous immunotherapy (EPIT) for milk and peanut153, 154 have been conducted. The FDA application for AR101 is submitted, while peanut EPIT submission has been retracted to provide additional technical information. Different ways of application differ with regard to efficacy of desensitization; however, all current applications are linked to an avoidance regimen and it is unclear how long the individual treatment needs to be applied. Consistent data from conventional high‐dose milk, peanut, and egg OIT report good efficacy with regard to desensitization.155, 156 Therefore, in addition to these highly standardized products, OIT using conventional food sources may become a more frequent treatment offered by clinicians in the community as a result of excessive demand in the absence of guidelines and recommendations. As a first step, the European Academy of Allergy and Clinical Immunology (EAACI) stated to consider OIT for these three foods in settings with the appropriate infrastructure and experience.155
The major issues in treating food allergy by immunotherapy are safety, the low rate of tolerance induction,155 a high rate of side effects and dropouts,153 a lack of understanding of the optimal dose and time of treatment, and the existence of only few, suboptimal biomarkers that predict treatment response and how to perform multifood OIT.13, 157, 158 These limitations are addressed in numerous treatment approaches: (a) peptide immunotherapy targeting the T‐cell compartment and lacking IgE cross‐linking159, 160, 161; (b) hypoallergenic variants of allergens or extracts by chemical or thermal modification162, 163 or mutations which combine reduced desensitization with a minimally altered T‐cell epitope diversity164; (c) the usage of immunomodulatory substances and/or particles165; (d) the addition of prebiotics and/or probiotics76, 166; (e) the application of biologics either alone167, 168 or as adjuvants of OIT169, 170; (g) very low dose OIT171, 172; and (h) sublingual OIT.
Recent methodological developments on cloning and antibody generation from single‐cell sorting of allergen‐specific B cells will allow novel insights on the nature of peanut‐specific B‐cell responses and may give rise to novel high‐affinity blocking antibody treatments.173
Unknowns in the treatment of food allergy
Which markers predict treatment response?
Which markers can be used to monitor tolerance development?
What is the optimal dose and time of treatment?
Is there a role for biologics to improve safety and efficacy of immunotherapeutic approaches?
What is the best route to apply immunotherapy?
How can we implement oral immunotherapy safely in a community setting?
How to modify allergen formulations for tolerance induction?
4.2. Treatment of allergic diseases with biologics
Molecular mechanisms of type 2 inflammation in allergic disease are discussed above. Treatment of allergic disease with biologicals particularly targets type 2 inflammation. For several years, omalizumab was the only broadly applied biological in allergic diseases in childhood and adult asthma174 and chronic urticaria.8, 175 Recently, phase III trials demonstrated efficacy by blocking the IL‐4/IL‐13 pathway in glucocorticoid‐dependent severe asthma, moderate‐to‐severe uncontrolled asthma,176, 177 CRSwNP,178 and atopic dermatitis179 (dupilumab), by blocking IL‐5 in severe eosinophilic asthma (mepolizumab,180 reslizumab181) and CRSwNP and severe uncontrolled asthma by blocking the IL‐5 receptor (benralizumab182, 183).184 FDA and EMA approved mepolizumab, reslizumab, and benralizumab for adult uncontrolled asthma and dupilumab for atopic dermatitis in adolescents and adults, and these biologics are integrated in current guidelines and position papers.12, 175, 185, 186 New indications for these biologicals can be expected in the near future.184, 187
Novel data arising from a long pipeline of cytokine and chemokine receptor targeting drugs will lead to additional treatment options and change the landscape of therapeutics in other atopic diseases including food allergy, chronic rhinosinusitis with nasal polyps,12, 188 and systemic mastocytosis.189 Biologics may also increase efficacy and safety of AIT.
Phase II trials of biologics targeting type‐2 pathways beyond IL‐4, IL‐5, and IL‐13 are encouraging. Tezepelumab blocking the TSLP receptor showed efficacy in uncontrolled asthma independent of eosinophil counts.190 Nemolizumab blocks the IL‐31R‐alpha and reduces pruritus and to a certain extent also dermatitis severity.191 It is a good example for biologicals with the potential to be combined with a second to achieve better disease control. Another important group of emerging biologics will address mucosal inflammation44, 192 and upstream events which are key for innate lymphoid cells such as anti–IL‐25 and anti–IL‐33.
Costs are an important factor when prescribing biologics. Currently, direct treatment expenses only partially contribute to the overall disease‐associated financial burden.193, 194 Thus, costs related to comorbidities195 and the impact of biologics on these factors will be key. The development of biomarkers, prediction models,5, 196 the design of trials comparing different biologics and the implementation of strategies to investigate the safety, function, and efficacy in children, the elderly, and pregnant women represent additional crucial challenges that need to be answered in the near future.
Gaps in the treatment of allergic disease with biologics
How to predict treatment response?
Will new biologics help to promote tolerance induction?
How to define precision medicine approaches to treat severe and complex atopic phenotypes?
Long‐term side effects of biologics?
Safety and efficacy of biologics in childhood, in pregnancy, and in elderly?
Novel biomarkers and sets of biomarkers will be needed.
Treatment algorithms and guidelines for biologics usage are needed.
4.3. Small molecules for the treatment of allergic asthma
Several targeted therapeutic options for asthma and related conditions have been licensed in the past two decades. Apart from parenteral monoclonal antibodies directed against key inflammatory targets, small molecules comprise another class of systemic medication interfering with inflammatory pathways underlying these disorders.197 Leukotriene modifiers (LM), and specifically cysteinyl leukotriene (CysLT) receptor 1 antagonists (LTRA), are the first small molecule agents widely applied for targeted treatment of asthma and comorbid AR both in adults and in children.198 Being launched in an evolving era and lacking adequate biomarkers, initial positioning of anti‐leukotrienes in asthma treatment has been mainly based on their efficacy in clinical models and not on adequate patient stratification which may have delayed proper positioning of this targeted therapy.199
More recently, another class of lipid mediator antagonists entered clinical development: antagonists of the prostaglandin D2 (PGD2) receptor DP2 also known as chemoattractant receptor‐homologous molecule expressed on Th2 cells (CRTH2).200 DP2/CRTH2 receptors are present on several inflammatory cells including mast cells, T‐helper 2 cells, type 2 ILCs, and eosinophils, and hence, PGD2 plays an important role in linking both the innate and adaptive immune system through type 2 responses.201 Although two compounds showed (modest) efficacy in allergen challenge,202, 203 many CRTH2 antagonists failed in later clinical phases, possibly due to inadequate (non‐type 2) patient populations. With the emerging evidence of an upregulated PGD2 pathway and its association with type 2 inflammation in uncontrolled severe eosinophilic asthma,204 more recently, several CRTH2 antagonists have been tested in type 2 conditions, including allergic and/or refractory eosinophilic asthma, showing improvements in several clinical outcomes.205, 206, 207, 208, 209 In a post hoc analysis, CRTH2 antagonist OC000459 (Timapiprant) appeared most effective in younger (≤40 years) patients with uncontrolled atopic eosinophilic asthma (blood eosinophils ≥ 250 cells/μL).209 Currently, several CRTH2 antagonist programs are in phase III studies which should help to consolidate phenotypes and biomarkers responding to these targeted drugs. Additionally, while the same immune/inflammatory cells express both CysLT1 and DP2/CRTH2 receptors, further research is warranted on potential synergistic effects of LTRA and CRTH2 antagonists in T2 inflammatory conditions.
Research needs for treatment with novel small molecules
Sensitive and reliable point‐of‐care biomarkers to identify potential responders and to monitor (long‐term) effects of anti‐lipid mediator small molecules (LM, LTRA, CRTH2 antagonists).
Combining LTRA and CRTH2 antagonists may be beneficial in patients with type 2 inflammatory conditions and, hence, warrants further clinical investigation.
Since both DP2/CRTH2 receptors and CysLT1 receptors are present on both immune/inflammatory and structural cells, apart from anti‐inflammatory activity, blocking these receptors may potentially have disease‐modifying effects (“anti‐remodeling”).
4.4. Novel therapeutic concepts in AIT for airway disease
Allergen‐specific immunotherapy not only reduces symptoms in patients with AR,106 LAR,1 and asthma,106, 210 but there is also evidence that AIT can reduce the development of asthma and new sensitizations,210, 211, 212 thus being the only available disease‐modifying treatment. Altogether, albeit not all of the highest quality, there is evidence that AIT can halt the allergic march in patients with AR.213 Moreover, there is some evidence that AIT is cost‐effective in AR with or without asthma.214
Allergen‐specific immunotherapy is usually given as subcutaneous injections215, 216 or sublingually,217, 218, 219 but novel treatment forms such as peptide immunotherapy,11, 220 intralymphatic immunotherapy11, 220 and use of recombinant allergens, and immune‐modulating adjuvants and nanoparticles21, 220, 221 are under development.
Despite this positive profile, AIT is only used for highly selected patient groups in most countries in Europe.109 The reasons for this limited penetration are multifold16, 103, 222 but the long duration of the treatment, the potential side effects especially in groups that could most benefit from AIT, and the inability to predict development of allergic disease and response to AIT treatment are among the most important ones. Recently, EAACI has been very active in providing guidelines for immunotherapy109, 223 to help physicians and patients in their decisions. However, for further expansion of AIT, we must influence the balance between allergenicity and immunogenicity, which can improve both duration of treatment and create a better side effect profile. Furthermore, we need greater understanding of the molecular mechanisms underlying the development of respiratory allergic disease and of AIT at the level of the individual patient, facilitating better patient stratification for AIT to further improve optimal personalized treatment.16, 107
Research needs for novel AIT vaccines
Short, effective, and safe treatment
Prediction of individual success before or early during the treatment
Better understanding of the underlying molecular and immunological mechanisms
Optimal AIT clinical trials that reduce bias and heterogeneity
Collaboration between physicians, patient organizations, companies, and regulators
5. CONCLUSIONS
Our specialty has been evolving at full speed with the introduction of several novel concepts such as knowledge of structures and biological functions of allergens to better understand what makes them allergenic, molecular mechanisms of the type 2 immune response and immune tolerance. Due to the omics revolution and harnessing artificial intelligence to handle huge global datasets to facilitate accurate diagnoses and precise and personalized monitoring of disease, novel treatments are highly expected to further evolve. Like many other disciplines, we are experiencing the early days of the development of new biologicals that have entered the clinic. Small molecules and combinations may offer a rational alternative for the treatment of specific subtypes of asthma and related diseases. Future studies and head‐to‐head comparisons with the more expensive biologics should provide the answer. AIT is the only disease‐modifying treatment option for allergic patients. Despite its overall favorable profile, the use of AIT in many countries is still limited. For further dissemination of AIT, we must influence the balance between allergenicity and immunogenicity, improve the vaccines with the hope for long‐term cure in many patients, and develop novel prevention modalities for early intervention, which can overall improve the efficacy of treatment and create a better side effect profile. As shown in the text boxes in each of the sections, still many questions are waiting to be answered.
CONFLICTS OF INTEREST
Author ZD reports personal fees from Aquilon, ALK, AstraZeneca, Boehringer Ingelheim, Gilead Hal Allergy, MSD, and Sanofi‐Genzyme, during the conduct of the study. Apart from academic affiliations, ZD works at a phase I/II unit performing clinical studies for different biotech and pharma companies. Author TE reports other from DBV, grants from the Innovation fund Denmark, outside the submitted work; TE is the Co‐I or scientific lead in three investigator‐initiated oral immunotherapy trials supported by the Allergy and Anaphylaxis Program Sickkids. Author KN reports personal fees from Regeneron, grants from NIAID, FARE, and EAT, outside the submitted work; other from Novartis, Sanofi, Astellas, Nestle, BeforeBrands, Alladapt, ForTra, Genentech, AImmune Therapeutics, and DBV Technologies, outside the submitted work. Author REO’H reports other potential financial activities from Aravax Pty Ltd and Paranta Bio Pty Ltd, outside the submitted work. Author OP reports personal fees from Novartis Pharma, MEDA Pharma, Mobile Chamber Experts (a GA2LEN Partner), Pohl‐Boskamp, Indoor Biotechnologies, and Astellas Pharma Global, outside the submitted work; grants and personal fees from ALK‐Abelló, Allergopharma, Stallergenes Greer, HAL Allergy Holding B.V./HAL Allergie GmbH, Bencard Allergie GmbH/Allergy Therapeutics, Lofarma, ASIT Biotech Tools S.A., Laboratorios LETI/LETI Pharma, and Anergis S.A., outside the submitted work; grants from Biomay, Nuvo, Circassia, and Glaxo Smith Kline, outside the submitted work. Author CAA reports grants from Allergopharma, Idorsia, Scibase, Swiss National Science Foundation, Christine Kühne‐Center for Allergy Research and Education, European Commission’s Horison's 2020 Framework Programme, Cure, advisory board of Sanofi‐Aventis/Regeneron, grants from Novartis Research Institutes, and Astra Zeneca, outside the submitted work. Authors HB, WF, LO’M, MJT, CTH, DYW, and LZ report no conflicts of interest in relation to this work.
Breiteneder H, Diamant Z, Eiwegger T, et al. Future research trends in understanding the mechanisms underlying allergic diseases for improved patient care. Allergy. 2019;74:2293–2311. 10.1111/all.13851
REFERENCES
- 1. Rondon C, Blanca‐Lopez N, Campo P, et al. Specific immunotherapy in local allergic rhinitis: a randomized, double‐blind placebo‐controlled trial with Phleum pratense subcutaneous allergen immunotherapy. Allergy. 2018;73(4):905‐915. [DOI] [PubMed] [Google Scholar]
- 2. Eckl‐Dorna J, Fröschl R, Lupinek C, et al. Intranasal administration of allergen increases specific IgE whereas intranasal omalizumab does not increase serum IgE levels‐A pilot study. Allergy. 2018;73(5):1003‐1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boligan KF, von Gunten S. Innate lymphoid cells in asthma: cannabinoids on the balance. Allergy. 2017;72(6):839‐841. [DOI] [PubMed] [Google Scholar]
- 4. Terl M, Sedlák V, Cap P, et al. Asthma management: a new phenotype‐based approach using presence of eosinophilia and allergy. Allergy. 2017;72(9):1279‐1287. [DOI] [PubMed] [Google Scholar]
- 5. Casale TB, Chipps BE, Rosén K, et al. Response to omalizumab using patient enrichment criteria from trials of novel biologics in asthma. Allergy. 2018;73(2):490‐497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaplan AP, Gimenez‐Arnau AM, Saini SS. Mechanisms of action that contribute to efficacy of omalizumab in chronic spontaneous urticaria. Allergy. 2017;72(4):519‐533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paganin F, Mangiapan G, Proust A, et al. Lung function parameters in omalizumab responder patients: an interesting tool? Allergy. 2017;72(12):1953‐1961. [DOI] [PubMed] [Google Scholar]
- 8. Staubach P, Metz M, Chapman‐Rothe N, et al. Omalizumab rapidly improves angioedema‐related quality of life in adult patients with chronic spontaneous urticaria: X‐ACT study data. Allergy. 2018;73(3):576‐584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Okubo K, Hashiguchi K, Takeda T, et al. A randomized controlled phase II clinical trial comparing ONO‐4053, a novel DP1 antagonist, with a leukotriene receptor antagonist pranlukast in patients with seasonal allergic rhinitis. Allergy. 2017;72(10):1565‐1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zellweger F, Gasser P, Brigger D, Buschor P, Vogel M, Eggel A. A novel bispecific DARPin targeting FcgammaRIIB and FcepsilonRI‐bound IgE inhibits allergic responses. Allergy 2017;72(8):1174‐1183. [DOI] [PubMed] [Google Scholar]
- 11. Mosges R, Kasche EM, Raskopf E, et al. A randomized, double‐blind, placebo‐controlled, dose‐finding trial with Lolium perenne peptide immunotherapy. Allergy. 2018;73(4):896‐904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hellings PW, Fokkens WJ, Bachert C, et al. Positioning the principles of precision medicine in care pathways for allergic rhinitis and chronic rhinosinusitis ‐ A EUFOREA‐ARIA‐EPOS‐AIRWAYS ICP statement. Allergy. 2017;72(9):1297‐1305. [DOI] [PubMed] [Google Scholar]
- 13. Muraro A, Lemanske RF, Castells M, et al. Precision medicine in allergic disease‐food allergy, drug allergy, and anaphylaxis‐PRACTALL document of the European Academy of Allergy and Clinical Immunology and the American Academy of Allergy, Asthma and Immunology. Allergy. 2017;72(7):1006‐1021. [DOI] [PubMed] [Google Scholar]
- 14. Agache I, Strasser DS, Klenk A, et al. Serum IL‐5 and IL‐13 consistently serve as the best predictors for the blood eosinophilia phenotype in adult asthmatics. Allergy. 2016;71(8):1192‐1202. [DOI] [PubMed] [Google Scholar]
- 15. Shamji MH, Kappen JH, Akdis M, et al. Biomarkers for monitoring clinical efficacy of allergen immunotherapy for allergic rhinoconjunctivitis and allergic asthma: an EAACI Position Paper. Allergy. 2017;72(8):1156‐1173. [DOI] [PubMed] [Google Scholar]
- 16. Bonertz A, Roberts GC, Hoefnagel M, et al. Challenges in the implementation of EAACI guidelines on allergen immunotherapy: a global perspective on the regulation of allergen products. Allergy. 2018;73(1):64‐76. [DOI] [PubMed] [Google Scholar]
- 17. Brooks CR, van Dalen CJ, Hermans IF, Gibson PG, Simpson JL, Douwes J. Sputum basophils are increased in eosinophilic asthma compared with non‐eosinophilic asthma phenotypes. Allergy. 2017;72(10):1583‐1586. [DOI] [PubMed] [Google Scholar]
- 18. Caillaud D, Chanez P, Escamilla R, et al. Asthma‐COPD overlap syndrome (ACOS) vs 'pure' COPD: a distinct phenotype? Allergy. 2017;72(1):137‐145. [DOI] [PubMed] [Google Scholar]
- 19. Dall'antonia F, Pavkov‐Keller T, Zangger K, Keller W. Structure of allergens and structure based epitope predictions. Methods. 2014;66(1):3‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meno KH, Kastrup JS, Kuo IC, Chua KY, Gajhede M. The structure of the mite allergen Blo t 1 explains the limited antibody cross‐reactivity to Der p 1. Allergy. 2017;72(4):665‐670. [DOI] [PubMed] [Google Scholar]
- 21. Pfaar O, Lou H, Zhang Y, Klimek L, Zhang L. Recent developments and highlights in allergen immunotherapy. Allergy. 2018;73(12):2274‐2289. [DOI] [PubMed] [Google Scholar]
- 22. Devanaboyina SC, Cornelius C, Lupinek C, et al. High‐resolution crystal structure and IgE recognition of the major grass pollen allergen Phl p 3. Allergy. 2014;69(12):1617‐1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Breiteneder H. Mapping of conformational IgE epitopes of food allergens. Allergy. 2018;73(11):2107‐2109. [DOI] [PubMed] [Google Scholar]
- 24. Niemi M, Jylha S, Laukkanen ML, et al. Molecular interactions between a recombinant IgE antibody and the beta‐lactoglobulin allergen. Structure. 2007;15(11):1413‐1421. [DOI] [PubMed] [Google Scholar]
- 25. Padavattan S, Flicker S, Schirmer T, et al. High‐affinity IgE recognition of a conformational epitope of the major respiratory allergen Phl p 2 as revealed by X‐ray crystallography. J Immunol. 2009;182(4):2141‐2151. [DOI] [PubMed] [Google Scholar]
- 26. Matricardi PM, Kleine‐Tebbe J, Hoffmann HJ, et al. EAACI molecular allergology user's guide. Pediatr Allergy Immunol. 2016;27(suppl 23):1‐250. [DOI] [PubMed] [Google Scholar]
- 27. Profet M. The function of allergy: immunological defense against toxins. Q Rev Biol. 1991;66(1):23‐62. [DOI] [PubMed] [Google Scholar]
- 28. Mukai K, Tsai M, Starkl P, Marichal T, Galli SJ. IgE and mast cells in host defense against parasites and venoms. Semin Immunopathol. 2016;38(5):581‐603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matzinger P. The evolution of the danger theory. Interview by Lauren Constable, Commissioning Editor. Expert Rev Clin Immunol. 2012;8(4):311‐317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Palm NW, Rosenstein RK, Medzhitov R. Allergic host defences. Nature. 2012;484(7395):465‐472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Trompette A, Divanovic S, Visintin A, et al. Allergenicity resulting from functional mimicry of a Toll‐like receptor complex protein. Nature. 2009;457(7229):585‐588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Florsheim E, Yu S, Bragatto I, et al. Integrated innate mechanisms involved in airway allergic inflammation to the serine protease subtilisin. J Immunol. 2015;194(10):4621‐4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gour N, Lajoie S, Smole U, et al. Dysregulated invertebrate tropomyosin‐dectin‐1 interaction confers susceptibility to allergic diseases. Sci Immunol. 2018;3(20):eaam9841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marichal T, Starkl P, Reber L, et al. A beneficial role for immunoglobulin E in host defense against honeybee venom. Immunity. 2013;39(5):963‐975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Palm NW, Rosenstein RK, Yu S, Schenten DD, Florsheim E, Medzhitov R. Bee venom phospholipase A2 induces a primary type 2 response that is dependent on the receptor ST2 and confers protective immunity. Immunity. 2013;39(5):976‐985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pablos I, Eichhorn S, Machado Y, et al. Distinct epitope structures of defensin‐like proteins linked to proline‐rich regions give rise to differences in their allergenic activity. Allergy. 2018;73(2):431‐441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Järvå M, Lay FT, Phan TK, et al. X‐ray structure of a carpet‐like antimicrobial defensin‐phospholipid membrane disruption complex. Nat Commun. 2018;9(1):1962 10.1038/s41467-018-04434-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stern R. Hyaluronidases in cancer biology. Semin Cancer Biol. 2008;18(4):275‐280. [DOI] [PubMed] [Google Scholar]
- 39. Smole U, Radauer C, Lengger N, et al. The major birch pollen allergen Bet v 1 induces different responses in dendritic cells of birch pollen allergic and healthy individuals. PLoS One. 2015;10(1):e0117904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Iinuma T, Okamoto Y, Morimoto Y, et al. Pathogenicity of memory Th2 cells is linked to stage of allergic rhinitis. Allergy. 2018;73(2):479‐489. [DOI] [PubMed] [Google Scholar]
- 41. Gori S, Vermeulen M, Remes‐Lenicov F, et al. Acetylcholine polarizes dendritic cells toward a Th2‐promoting profile. Allergy. 2017;72(2):221‐231. [DOI] [PubMed] [Google Scholar]
- 42. Kortekaas Krohn I, Shikhagaie MM, Golebski K, et al. Emerging roles of innate lymphoid cells in inflammatory diseases: clinical implications. Allergy. 2018;73(4):837‐850. [DOI] [PubMed] [Google Scholar]
- 43. Flayer CH, Haczku A. The Th2 gene cluster unraveled: role of RHS6. Allergy. 2017;72(5):679‐681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hong H‐Y, Chen F‐H, Sun Y‐Q, et al. Local IL‐25 contributes to Th2‐biased inflammatory profiles in nasal polyps. Allergy. 2018;73(2):459‐469. [DOI] [PubMed] [Google Scholar]
- 45. Hwang SS, Jang SW, Lee KO, Kim HS, Lee GR. RHS6 coordinately regulates the Th2 cytokine genes by recruiting GATA3, SATB1, and IRF4. Allergy. 2017;72(5):772‐782. [DOI] [PubMed] [Google Scholar]
- 46. Ravanetti L, Dijkhuis A, Sabogal Pineros YS, et al. An early innate response underlies severe influenza‐induced exacerbations of asthma in a novel steroid‐insensitive and anti‐IL‐5‐responsive mouse model. Allergy. 2017;72(5):737‐753. [DOI] [PubMed] [Google Scholar]
- 47. Prakash Babu S, Chen Y‐y k, Bonne‐Annee S, et al. Dysregulation of interleukin 5 expression in familial eosinophilia. Allergy. 2017;72(9):1338‐1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Weller K, Church MK, Hawro T, et al. Updosing of bilastine is effective in moderate to severe chronic spontaneous urticaria: a real life study. Allergy. 2018;73(10):2073‐2075. [DOI] [PubMed] [Google Scholar]
- 49. Boonpiyathad T, Meyer N, Moniuszko M, et al. High‐dose bee venom exposure induces similar tolerogenic B‐cell responses in allergic patients and healthy beekeepers. Allergy. 2017;72(3):407‐415. [DOI] [PubMed] [Google Scholar]
- 50. Akdis CA, Akdis M. Advances in allergen immunotherapy: aiming for complete tolerance to allergens. Sci Transl Med. 2015;7(280):280ps6‐280ps6. [DOI] [PubMed] [Google Scholar]
- 51. Schröder PC, Illi S, Casaca VI, et al. A switch in regulatory T cells through farm exposure during immune maturation in childhood. Allergy. 2017;72(4):604‐615. [DOI] [PubMed] [Google Scholar]
- 52. Ferstl R, Frei R, Barcik W, et al. Histamine receptor 2 modifies iNKT cell activity within the inflamed lung. Allergy. 2017;72(12):1925‐1935. [DOI] [PubMed] [Google Scholar]
- 53. Wirz OF, Głobińska A, Ochsner U, et al. Comparison of regulatory B cells in asthma and allergic rhinitis. Allergy. 2019;74(4):815‐818. [DOI] [PubMed] [Google Scholar]
- 54. Aron JL, Akbari O. Regulatory T cells and type 2 innate lymphoid cell‐dependent asthma. Allergy. 2017;72(8):1148‐1155. [DOI] [PubMed] [Google Scholar]
- 55. Voisin T, Bouvier A, Chiu IM. Neuro‐immune interactions in allergic diseases: novel targets for therapeutics. Int Immunol. 2017;29(6):247‐261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Veres TZ, Rochlitzer S, Braun A. The role of neuro‐immune cross‐talk in the regulation of inflammation and remodelling in asthma. Pharmacol Ther. 2009;122(2):203‐214. [DOI] [PubMed] [Google Scholar]
- 57. Chavan SS, Tracey KJ. Essential neuroscience in immunology. J Immunol. 2017;198(9):3389‐3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chesne J, Cardoso V, Veiga‐Fernandes H. Neuro‐immune regulation of mucosal physiology. Mucosal Immunol. 2019;12(1):10‐20. [DOI] [PubMed] [Google Scholar]
- 59. Nassenstein C, Krasteva‐Christ G, Renz H. New aspects of neuroinflammation and neuroimmune crosstalk in the airways. J Allergy Clin Immunol. 2018;142(5):1415‐1422. [DOI] [PubMed] [Google Scholar]
- 60. Nassenstein C, Kutschker J, Tumes D, Braun A. Neuro‐immune interaction in allergic asthma: role of neurotrophins. Biochem Soc Trans. 2006;34(Pt 4):591‐593. [DOI] [PubMed] [Google Scholar]
- 61. Rochlitzer S, Veres TZ, Kuhne K, et al. The neuropeptide calcitonin gene‐related peptide affects allergic airway inflammation by modulating dendritic cell function. Clin Exp Allergy. 2011;41(11):1609‐1621. [DOI] [PubMed] [Google Scholar]
- 62. Wallrapp A, Riesenfeld SJ, Burkett PR, et al. The neuropeptide NMU amplifies ILC2‐driven allergic lung inflammation. Nature. 2017;549(7672):351‐356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lorton D, Bellinger DL. Molecular mechanisms underlying beta‐adrenergic receptor‐mediated cross‐talk between sympathetic neurons and immune cells. Int J Mol Sci. 2015;16(3):5635‐5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pongratz G, McAlees JW, Conrad DH, Erbe RS, Haas KM, Sanders VM. The level of IgE produced by a B cell is regulated by norepinephrine in a p38 MAPK‐ and CD23‐dependent manner. J Immunol. 2006;177(5):2926‐2938. [DOI] [PubMed] [Google Scholar]
- 65. Gilles S, Akdis C, Lauener R, et al. The role of environmental factors in allergy: a critical reappraisal. Exp Dermatol. 2018;27(11):1193‐1200. [DOI] [PubMed] [Google Scholar]
- 66. Bieber T, Akdis C, Lauener R, et al. Global Allergy Forum and 3rd Davos Declaration 2015: atopic dermatitis/Eczema: challenges and opportunities toward precision medicine. Allergy. 2016;71(5):588‐592. [DOI] [PubMed] [Google Scholar]
- 67. Wang XD, Zheng M, Lou HF, et al. An increased prevalence of self‐reported allergic rhinitis in major Chinese cities from 2005 to 2011. Allergy. 2016;71(8):1170‐1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Morgenstern V, Zutavern A, Cyrys J, et al. Atopic diseases, allergic sensitization, and exposure to traffic‐related air pollution in children. Am J Respir Crit Care Med. 2008;177(12):1331‐1337. [DOI] [PubMed] [Google Scholar]
- 69. Fuertes E, Standl M, Cyrys J, et al. A longitudinal analysis of associations between traffic‐related air pollution with asthma, allergies and sensitization in the GINIplus and LISAplus birth cohorts. PeerJ. 2013;1 10.7717/peerj.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhao F, Durner J, Winkler JB, et al. Pollen of common ragweed (Ambrosia artemisiifolia L.): illumina‐based de novo sequencing and differential transcript expression upon elevated NO2/O3. Environ Pollut. 2017;224:503‐514. [DOI] [PubMed] [Google Scholar]
- 71. Beck I, Jochner S, Gilles S, et al. High environmental ozone levels lead to enhanced allergenicity of birch pollen. PLoS One. 2013;8(11):e80147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Traidl‐Hoffmann C. [Allergy ‐ an environmental disease]. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2017;60(6):584‐591. [DOI] [PubMed] [Google Scholar]
- 73. Agache I, Miller R, Gern JE, et al. Emerging concepts and challenges in implementing the exposome paradigm in allergic diseases and asthma. Allergy. 2019;74(3):449‐463. [DOI] [PubMed] [Google Scholar]
- 74. Damialis A, Häring F, Gökkaya M, et al. Human exposure to airborne pollen and relationships with symptoms and immune responses: indoors versus outdoors, circadian patterns and meteorological effects in alpine and urban environments. Sci Total Environ. 2018;653:190‐199. [DOI] [PubMed] [Google Scholar]
- 75. Jatzlauk G, Bartel S, Heine H, Schloter M, Krauss‐Etschmann S. Influences of environmental bacteria and their metabolites on allergies, asthma, and host microbiota. Allergy. 2017;72(12):1859‐1867. [DOI] [PubMed] [Google Scholar]
- 76. Lunjani N, Satitsuksanoa P, Lukasik Z, Sokolowska M, Eiwegger T, O'Mahony L. Recent developments and highlights in mechanisms of allergic diseases: microbiome. Allergy. 2018;73(12):2314‐2327. [DOI] [PubMed] [Google Scholar]
- 77. Sokolowska M, Frei R, Lunjani N, Akdis CA, O'Mahony L. Microbiome and asthma. Asthma Res Pract. 2018;4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Birzele LT, Depner M, Ege MJ, et al. Environmental and mucosal microbiota and their role in childhood asthma. Allergy. 2017;72(1):109‐119. [DOI] [PubMed] [Google Scholar]
- 79. Stein MM, Hrusch CL, Gozdz J, et al. Innate immunity and asthma risk in Amish and Hutterite farm children. N Engl J Med. 2016;375(5):411‐421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bokulich NA, Chung J, Battaglia T, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8(343):343ra82‐343ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gentile CL, Weir TL. The gut microbiota at the intersection of diet and human health. Science. 2018;362(6416):776‐780. [DOI] [PubMed] [Google Scholar]
- 82. Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet‐induced extinctions in the gut microbiota compound over generations. Nature. 2016;529(7585):212‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Roduit C, Frei R, Ferstl R, et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy. 2019;74(4):799‐809. [DOI] [PubMed] [Google Scholar]
- 84. Bianco A, Whiteman SC, Sethi SK, Allen JT, Knight RA, Spiteri MA. Expression of intercellular adhesion molecule‐1 (ICAM‐1) in nasal epithelial cells of atopic subjects: a mechanism for increased rhinovirus infection? Clin Exp Immunol. 2000;121(2):339‐345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Braciale TJ, Sun J, Kim TS. Regulating the adaptive immune response to respiratory virus infection. Nat Rev Immunol. 2012;12(4):295‐305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kolesnikova L, Heck S, Matrosovich T, Klenk HD, Becker S, Matrosovich M. Influenza virus budding from the tips of cellular microvilli in differentiated human airway epithelial cells. J Gen Virol. 2013;94(Pt 5):971‐976. [DOI] [PubMed] [Google Scholar]
- 87. Wang DY, Li Y, Yan Y, Li C, Shi L. Upper airway stem cells: understanding the nose and role for future cell therapy. Curr Allergy Asthma Rep. 2015;15(1):490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yan Y, Tan KS, Li C, et al. Human nasal epithelial cells derived from multiple subjects exhibit differential responses to H3N2 influenza virus infection in vitro. J Allergy Clin Immunol. 2016;138(1):276‐281. [DOI] [PubMed] [Google Scholar]
- 89. Tan KS, Yan Y, Ong HH, Chow V, Shi L, Wang DY. Impact of respiratory virus infections in exacerbation of acute and chronic rhinosinusitis. Curr Allergy Asthma Rep. 2017;17(4):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ravanetti L, Dijkhuis A, Dekker T, et al. IL‐33 drives influenza‐induced asthma exacerbations by halting innate and adaptive antiviral immunity. J Allergy Clin Immunol. 2019;143(4):1355‐1370. [DOI] [PubMed] [Google Scholar]
- 91. Kim MJ, Shim DH, Cha HR, et al. Chitinase 3‐like 1 protein plays a critical role in respiratory syncytial virus‐induced airway inflammation. Allergy. 2019;74(4):685‐697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kast JI, McFarlane AJ, Głobińska A, et al. Respiratory syncytial virus infection influences tight junction integrity. Clin Exp Immunol. 2017;190(3):351‐359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yeo NK, Jang YJ. Rhinovirus infection‐induced alteration of tight junction and adherens junction components in human nasal epithelial cells. Laryngoscope. 2010;120(2):346‐352. [DOI] [PubMed] [Google Scholar]
- 94. Tian T, Zi X, Peng Y, et al. H3N2 influenza virus infection enhances oncostatin M expression in human nasal epithelium. Exp Cell Res. 2018;371(2):322‐329. [DOI] [PubMed] [Google Scholar]
- 95. Tan KS, Ong HH, Yan Y, et al. In vitro model of fully differentiated human nasal epithelial cells infected with rhinovirus reveals epithelium‐initiated immune responses. J Infect Dis. 2018;217(6):906‐915. [DOI] [PubMed] [Google Scholar]
- 96. Tan KS, Yan Y, Koh WLH, et al. Comparative transcriptomic and metagenomic analyses of influenza virus‐infected nasal epithelial cells from multiple individuals reveal specific nasal‐initiated signatures. Front Microbiol. 2018;9:2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Aab A, Wirz O, van de Veen W, et al. Human rhinoviruses enter and induce proliferation of B lymphocytes. Allergy. 2017;72(2):232‐243. [DOI] [PubMed] [Google Scholar]
- 98. Jurak LM, Xi Y, Landgraf M, Carroll ML, Murray L, Upham JW. Interleukin 33 selectively augments rhinovirus‐induced type 2 immune responses in asthmatic but not healthy people. Front Immunol. 2018;9:1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Fedele G, Schiavoni I, Nenna R, et al. Analysis of the immune response in infants hospitalized with viral bronchiolitis shows different Th1/Th2 profiles associated with respiratory syncytial virus and human rhinovirus. Pediatr Allergy Immunol. 2018;29(5):555‐557. [DOI] [PubMed] [Google Scholar]
- 100. Deng H, Sun Y, Wang W, et al. The hippo pathway effector Yes‐associated protein promotes epithelial proliferation and remodeling in chronic rhinosinusitis with nasal polyps. Allergy. 2019;74(4):731‐742. [DOI] [PubMed] [Google Scholar]
- 101. Zhao L, Li YY, Li CW, et al. Increase of poorly proliferated p63(+) /Ki67(+) basal cells forming multiple layers in the aberrant remodeled epithelium in nasal polyps. Allergy. 2017;72(6):975‐984. [DOI] [PubMed] [Google Scholar]
- 102. Luukkainen A, Puan KJ, Yusof N, et al. A co‐culture model of PBMC and stem cell derived human nasal epithelium reveals rapid activation of NK and innate T cells upon influenza A virus infection of the nasal epithelium. Front Immunol. 2018;9:2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Bonertz A, Roberts G, Slater JE, et al. Allergen manufacturing and quality aspects for allergen immunotherapy in Europe and the United States: an analysis from the EAACI AIT Guidelines Project. Allergy. 2018;73(4):816‐826. [DOI] [PubMed] [Google Scholar]
- 104. Englert L, May S, Kaul S, Vieths S. The therapy allergens ordinance ("Therapieallergene‐Verordnung"). Background and effects. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2012;55(3):351‐357. [DOI] [PubMed] [Google Scholar]
- 105. German Society for Allergology and Clinical Immunology (DGAKI). http://www.dgaki.de/leitlinien/s2k-leitlinie-sit/. Accessed December 7, 2018.
- 106. Dhami S, Nurmatov U, Arasi S, et al. Allergen immunotherapy for allergic rhinoconjunctivitis: a systematic review and meta‐analysis. Allergy. 2017;72(11):1597‐1631. [DOI] [PubMed] [Google Scholar]
- 107. Pfaar O, Alvaro M, Cardona V, Hamelmann E, Mosges R, Kleine‐Tebbe J. Clinical trials in allergen immunotherapy: current concepts and future needs. Allergy. 2018;73(1):77‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Pfaar O, Demoly P, Gerth van Wijk R, et al. Recommendations for the standardization of clinical outcomes used in allergen immunotherapy trials for allergic rhinoconjunctivitis: an EAACI Position Paper. Allergy. 2014;69(7):854‐867. [DOI] [PubMed] [Google Scholar]
- 109. Roberts G, Pfaar O, Akdis CA, et al. EAACI guidelines on allergen immunotherapy: allergic rhinoconjunctivitis. Allergy. 2018;73(4):765‐798. [DOI] [PubMed] [Google Scholar]
- 110. Pfaar O, Bonini S, Cardona V, et al. Perspectives in allergen immunotherapy: 2017 and beyond. Allergy. 2018;73(suppl 104):5‐23. [DOI] [PubMed] [Google Scholar]
- 111. Noon L. Prophylactic inoculation against hay fever. Lancet. 1911;1:1572‐1573. [Google Scholar]
- 112. Eguiluz‐Gracia I, Tay TR, Hew M, et al. Recent developments and highlights in biomarkers in allergic diseases and asthma. Allergy. 2018;73(1):2290‐2305. [DOI] [PubMed] [Google Scholar]
- 113. Boyd SD, Hoh RA, Nadeau KC, Galli SJ. Immune monitoring for precision medicine in allergy and asthma. Curr Opin Immunol. 2017;48:82‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Tan HT, Sugita K, Akdis CA. Novel biologicals for the treatment of allergic diseases and asthma. Curr Allergy Asthma Rep. 2016;16(10):70. [DOI] [PubMed] [Google Scholar]
- 115. Yu H, Zhang VW, Stray‐Pedersen A, et al. Rapid molecular diagnostics of severe primary immunodeficiency determined by using targeted next‐generation sequencing. J Allergy Clin Immunol. 2016;138(4):1142‐1151. [DOI] [PubMed] [Google Scholar]
- 116. Ponsford MJ, Klocperk A, Pulvirenti F, et al. Hyper‐IgE in the allergy clinic–when is it primary immunodeficiency? Allergy. 2018;73(11):2122‐2136. [DOI] [PubMed] [Google Scholar]
- 117. e GP. Enhancing GTEx by bridging the gaps between genotype, gene expression, and disease. Nat Genet. 2017;49(12):1664‐1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Consortium EP . An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Heeringa JJ, Rijvers L, Arends NJ, et al. IgE‐expressing memory B cells and plasmablasts are increased in blood of children with asthma, food allergy, and atopic dermatitis. Allergy. 2018;73(6):1331‐1336. [DOI] [PubMed] [Google Scholar]
- 120. Rust BJ, Wambre E. Human immune monitoring techniques during food allergen immunotherapy. Curr Allergy Asthma Rep. 2017;17(4):22. [DOI] [PubMed] [Google Scholar]
- 121. Mukai K, Gaudenzio N, Gupta S, et al. Assessing basophil activation by using flow cytometry and mass cytometry in blood stored 24 hours before analysis. J Allergy Clin Immunol. 2017;139(3):889‐899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Ryan JF, Hovde R, Glanville J, et al. Successful immunotherapy induces previously unidentified allergen‐specific CD4+ T‐cell subsets. Proc Natl Acad Sci USA. 2016;113(9):E1286‐1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Larsen LF, Juel‐Berg N, Hansen KS, et al. A comparative study on basophil activation test, histamine release assay, and passive sensitization histamine release assay in the diagnosis of peanut allergy. Allergy. 2018;73(1):137‐144. [DOI] [PubMed] [Google Scholar]
- 124. Obeso D, Mera‐Berriatua L, Rodriguez‐Coira J, et al. Multi‐omics analysis points to altered platelet functions in severe food‐associated respiratory allergy. Allergy. 2018;73(11):2137‐2149. [DOI] [PubMed] [Google Scholar]
- 125. Datema MR, van Ree R, Asero R, et al. Component‐resolved diagnosis and beyond: multivariable regression models to predict severity of hazelnut allergy. Allergy. 2018;73(3):549‐559. [DOI] [PubMed] [Google Scholar]
- 126. Papadopoulos NG, Bernstein JA, Demoly P, et al. Phenotypes and endotypes of rhinitis and their impact on management: a PRACTALL report. Allergy. 2015;70(5):474‐494. [DOI] [PubMed] [Google Scholar]
- 127. Zuberbier T, Lotvall J, Simoens S, Subramanian SV, Church MK. Economic burden of inadequate management of allergic diseases in the European Union: a GA(2) LEN review. Allergy. 2014;69(10):1275‐1279. [DOI] [PubMed] [Google Scholar]
- 128. Hellings PW, Klimek L, Cingi C, et al. Non‐allergic rhinitis: position paper of the European Academy of Allergy and Clinical Immunology. Allergy. 2017;72(11):1657‐1665. [DOI] [PubMed] [Google Scholar]
- 129. Reitsma S, Subramaniam S, Fokkens WJ, Wang DY. Recent developments and highlights in rhinitis and allergen immunotherapy. Allergy. 2018;73(12):2306‐2313. [DOI] [PubMed] [Google Scholar]
- 130. Meng Y, Lou H, Wang Y, et al. Endotypes of chronic rhinitis: a cluster analysis study. Allergy. 2019;74(4):720‐730. [DOI] [PubMed] [Google Scholar]
- 131. Rondón C, Campo P, Galindo L, et al. Prevalence and clinical relevance of local allergic rhinitis. Allergy. 2012;67(10):1282‐1288. [DOI] [PubMed] [Google Scholar]
- 132. Rondon C, Bogas G, Barrionuevo E, Blanca M, Torres MJ, Campo P. Nonallergic rhinitis and lower airway disease. Allergy. 2017;72(1):24‐34. [DOI] [PubMed] [Google Scholar]
- 133. Rondon C, Campo P, Eguiluz‐Gracia I, et al. Local allergic rhinitis is an independent rhinitis phenotype: the results of a 10‐year follow‐up study. Allergy. 2018;73(2):470‐478. [DOI] [PubMed] [Google Scholar]
- 134. Meng Y, Lou H, Wang Y, Wang C, Zhang L. The use of specific immunoglobulin E in nasal secretions for the diagnosis of allergic rhinitis. Laryngoscope. 2018;128(9):E311‐E315. [DOI] [PubMed] [Google Scholar]
- 135. She W, Yang J, Wang C, Zhang L. Diagnostic value of nasal cytology in chronic rhinosinusitis assessed by a liquid‐based cytological technique. Am J Rhinol Allergy. 2018;32(3):181‐187. [DOI] [PubMed] [Google Scholar]
- 136. Demoly P, Adkinson NF, Brockow K, et al. International consensus on drug allergy. Allergy. 2014;69(4):420‐437. [DOI] [PubMed] [Google Scholar]
- 137. Brockow K, Ardern‐Jones MR, Mockenhaupt M, et al. EAACI position paper on how to classify cutaneous manifestations of drug hypersensitivity. Allergy. 2019;74(1):14‐27. [DOI] [PubMed] [Google Scholar]
- 138. Mayorga C, Celik G, Rouzaire P, et al. In vitro tests for drug hypersensitivity reactions: an ENDA/EAACI Drug Allergy Interest Group position paper. Allergy. 2016;71(8):1103‐1134. [DOI] [PubMed] [Google Scholar]
- 139. Gomes ER, Brockow K, Kuyucu S, et al. Drug hypersensitivity in children: report from the pediatric task force of the EAACI Drug Allergy Interest Group. Allergy. 2016;71(2):149‐161. [DOI] [PubMed] [Google Scholar]
- 140. Tanno LK, Torres MJ, Castells M, Demoly P, Joint AA. What can we learn in drug allergy management from World Health Organization's international classifications? Allergy. 2018;73(5):987‐992. [DOI] [PubMed] [Google Scholar]
- 141. Pichler WJ. Immune pathomechanism and classification of drug hypersensitivity. Allergy. 2019;000:000‐000. [DOI] [PubMed] [Google Scholar]
- 142. Chiriac AM, Rerkpattanapipat T, Bousquet PJ, Molinari N, Demoly P. Optimal step doses for drug provocation tests to prove beta‐lactam hypersensitivity. Allergy. 2017;72(4):552‐561. [DOI] [PubMed] [Google Scholar]
- 143. Torres MJ, Romano A, Celik G, et al. Approach to the diagnosis of drug hypersensitivity reactions: similarities and differences between Europe and North America. Clin Transl Allergy. 2017;7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Gonzalez‐Estrada A, Archibald T, Dinsmore K, Mosier G, Campbell B, Brown S. Stability of diluted neuromuscular blocking agents utilized in perioperative hypersensitivity evaluation. Allergy. 2018;73(12):2398‐2400. [DOI] [PubMed] [Google Scholar]
- 145. Van Gasse AL, Sabato V, Uyttebroek AP, et al. Immediate moxifloxacin hypersensitivity: is there more than currently meets the eye? Allergy. 2017;72(12):2039‐2043. [DOI] [PubMed] [Google Scholar]
- 146. Salas M, Fernández‐Santamaría R, Mayorga C, et al. Use of the basophil activation test may reduce the need for drug provocation in amoxicillin‐clavulanic allergy. J Allergy Clin Immunol Pract. 2018;6(3):1010‐1018. [DOI] [PubMed] [Google Scholar]
- 147. Trubiano JA, Thursky KA, Stewardson AJ, et al. Impact of an integrated antibiotic allergy testing program on antimicrobial stewardship: a multicenter evaluation. Clin Infect Dis. 2017;65(1):166‐174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Doña I, Caubet JC, Brockow K, et al. An EAACI task force report: recognising the potential of the primary care physician in the diagnosis and management of drug hypersensitivity. Clin Transl Allergy. 2018;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Madrigal‐Burgaleta R, Bernal‐Rubio L, Berges‐Gimeno MP, et al. A large single‐hospital experience using drug provocation testing and rapid drug desensitization in hypersensitivity to antineoplastic and biological agents. J Allergy Clin Immunol Pract. 2019;7(2):618‐632. [DOI] [PubMed] [Google Scholar]
- 150. Castells M. Drug hypersensitivity and anaphylaxis in cancer and chronic inflammatory diseases: the role of desensitizations. Front Immunol. 2017;8:1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Cortellini G, Romano A, Santucci A, et al. Clinical approach on challenge and desensitization procedures with aspirin in patients with ischemic heart disease and nonsteroidal anti‐inflammatory drug hypersensitivity. Allergy. 2017;72(3):498‐506. [DOI] [PubMed] [Google Scholar]
- 152. Investigators PGoC , Vickery BP, Vereda A, et al. AR101 oral immunotherapy for peanut allergy. N Engl J Med. 2018;379(21):1991‐2001. [DOI] [PubMed] [Google Scholar]
- 153. Sampson HA, Shreffler WG, Yang WH, et al. Effect of varying doses of epicutaneous immunotherapy vs placebo on reaction to peanut protein exposure among patients with peanut sensitivity: a randomized clinical trial. JAMA. 2017;318(18):1798‐1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Matthews JG, Zawadzki R, Haselkorn T, Rosen K. Clarification of epicutaneous immunotherapy trial phase 3 results and methods for qualitative survey design. Ann Allergy Asthma Immunol. 2018;121(5):641‐642. [DOI] [PubMed] [Google Scholar]
- 155. Pajno GB, Fernandez‐Rivas M, Arasi S, et al. EAACI Guidelines on allergen immunotherapy: IgE‐mediated food allergy. Allergy. 2018;73(4):799‐815. [DOI] [PubMed] [Google Scholar]
- 156. Nurmatov U, Dhami S, Arasi S, et al. Allergen immunotherapy for IgE‐mediated food allergy: a systematic review and meta‐analysis. Allergy. 2017;72(8):1133‐1147. [DOI] [PubMed] [Google Scholar]
- 157. Bluemchen K, Eiwegger T. Oral peanut immunotherapy ‐ how much is too much? How much is enough? Allergy. 2019;74(2):220‐222. [DOI] [PubMed] [Google Scholar]
- 158. Jappe U, Breiteneder H. Peanut allergy ‐ individual molecules as a key to precision medicine. Allergy. 2019;74(2):216‐219. [DOI] [PubMed] [Google Scholar]
- 159. Cook QS, Burks AW. Peptide and recombinant allergen vaccines for food allergy. Clin Rev Allergy Immunol. 2018;55(2):162‐171. [DOI] [PubMed] [Google Scholar]
- 160. Saidova A, Hershkop AM, Ponce M, Eiwegger T. Allergen‐specific T cells in IgE‐mediated food allergy. Arch Immunol Ther Exp. 2018;66(3):161‐170. [DOI] [PubMed] [Google Scholar]
- 161. O'Hehir RE, Prickett SR, Rolland JM. T cell epitope peptide therapy for allergic diseases. Curr Allergy Asthma Rep. 2016;16(2):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. van der Kleij H, Warmenhoven H, van Ree R, et al. Chemically modified peanut extract shows increased safety while maintaining immunogenicity. Allergy. 2018. 10.1111/all.13687 [DOI] [PubMed] [Google Scholar]
- 163. Upton J, Nowak‐Wegrzyn A. The impact of baked egg and baked milk diets on IgE‐ and non‐IgE‐mediated allergy. Clin Rev Allergy Immunol. 2018;55(2):118‐138. [DOI] [PubMed] [Google Scholar]
- 164. Bublin M, Kostadinova M, Radauer C, et al. Engineering of structural variants of the major peanut allergens Ara h 2 and Ara h 6 for allergen‐specific immunotherapy. J Allergy Clin Immunol. 2019;143(3):1226‐1229. [DOI] [PubMed] [Google Scholar]
- 165. Srivastava KD, Siefert A, Fahmy TM, Caplan MJ, Li XM, Sampson HA. Investigation of peanut oral immunotherapy with CpG/peanut nanoparticles in a murine model of peanut allergy. J Allergy Clin Immunol. 2016;138(2):536‐543. [DOI] [PubMed] [Google Scholar]
- 166. Dunn Galvin A, McMahon S, Ponsonby AL, Hsiao KC, Tang M, Team Ps . The longitudinal impact of probiotic and peanut oral immunotherapy on health‐related quality of life. Allergy. 2018;73(3):560‐568. [DOI] [PubMed] [Google Scholar]
- 167. Rial MJ, Barroso B, Sastre J. Dupilumab for treatment of food allergy. J Allergy Clin Immunol Pract. 2019;7(2):673‐674. [DOI] [PubMed] [Google Scholar]
- 168. Brandstrom J, Vetander M, Lilja G, et al. Individually dosed omalizumab: an effective treatment for severe peanut allergy. Clin Exp Allergy. 2017;47(4):540‐550. [DOI] [PubMed] [Google Scholar]
- 169. Andorf S, Purington N, Block WM, et al. Anti‐IgE treatment with oral immunotherapy in multifood allergic participants: a double‐blind, randomised, controlled trial. Lancet Gastroenterol Hepatol. 2018;3(2):85‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Smaldini PL, Trejo F, Cohen JL, Piaggio E, Docena GH. Systemic IL‐2/anti‐IL‐2Ab complex combined with sublingual immunotherapy suppresses experimental food allergy in mice through induction of mucosal regulatory T cells. Allergy. 2018;73(4):885‐895. [DOI] [PubMed] [Google Scholar]
- 171. Blumchen K, Trendelenburg V, Ahrens F, et al. Efficacy, safety, and quality of life in a multi‐center, randomized, placebo‐controlled trial of low‐dose peanut oral immunotherapy in peanut allergic children. J Allergy Clin Immunol Pract. 2019;7(2):479‐479. [DOI] [PubMed] [Google Scholar]
- 172. Nagakura KI, Yanagida N, Sato S, et al. Low‐dose oral immunotherapy for children with anaphylactic peanut allergy in Japan. Pediatr Allergy Immunol. 2018;29(5):512‐518. [DOI] [PubMed] [Google Scholar]
- 173. Croote D, Darmanis S, Nadeau KC, Quake SR. High‐affinity allergen‐specific human antibodies cloned from single IgE B cell transcriptomes. Science. 2018;362(6420):1306‐1309. [DOI] [PubMed] [Google Scholar]
- 174. Trischler J, Lieb A, Arnold M, et al. Omalizumab effectively protects against early and late allergic responses in asthma after 4 weeks. Allergy. 2017;72(12):1912‐1915. [DOI] [PubMed] [Google Scholar]
- 175. Zuberbier T, Aberer W, Asero R, et al. The EAACI/GA(2)LEN/EDF/WAO guideline for the definition, classification, diagnosis and management of urticaria. Allergy. 2018;73(7):1393‐1414. [DOI] [PubMed] [Google Scholar]
- 176. Castro M, Corren J, Pavord ID, et al. Dupilumab efficacy and safety in moderate‐to‐severe uncontrolled asthma. N Engl J Med. 2018;378(26):2486‐2496. [DOI] [PubMed] [Google Scholar]
- 177. Rabe KF, Nair P, Brusselle G, et al. Efficacy and safety of dupilumab in glucocorticoid‐dependent severe asthma. N Engl J Med. 2018;378(26):2475‐2485. [DOI] [PubMed] [Google Scholar]
- 178. Bachert C, Sousa AR, Lund VJ, et al. Reduced need for surgery in severe nasal polyposis with mepolizumab: randomized trial. J Allergy Clin Immunol. 2017;140(4):1024‐1031. [DOI] [PubMed] [Google Scholar]
- 179. Simpson EL, Bieber T, Guttman‐Yassky E, et al. Two phase 3 trials of dupilumab versus placebo in atopic dermatitis. N Engl J Med. 2016;375(24):2335‐2348. [DOI] [PubMed] [Google Scholar]
- 180. Ortega HG, Yancey SW, Mayer B, et al. Severe eosinophilic asthma treated with mepolizumab stratified by baseline eosinophil thresholds: a secondary analysis of the DREAM and MENSA studies. Lancet Respir Med. 2016;4(7):549‐556. [DOI] [PubMed] [Google Scholar]
- 181. Castro M, Zangrilli J, Wechsler ME, et al. Reslizumab for inadequately controlled asthma with elevated blood eosinophil counts: results from two multicentre, parallel, double‐blind, randomised, placebo‐controlled, phase 3 trials. Lancet Respir Med. 2015;3(5):355‐366. [DOI] [PubMed] [Google Scholar]
- 182. Bleecker ER, FitzGerald JM, Chanez P, et al. Efficacy and safety of benralizumab for patients with severe asthma uncontrolled with high‐dosage inhaled corticosteroids and long‐acting beta2‐agonists (SIROCCO): a randomised, multicentre, placebo‐controlled phase 3 trial. Lancet. 2016;388(10056):2115‐2127. [DOI] [PubMed] [Google Scholar]
- 183. FitzGerald JM, Bleecker ER, Nair P, et al. Benralizumab, an anti‐interleukin‐5 receptor alpha monoclonal antibody, as add‐on treatment for patients with severe, uncontrolled, eosinophilic asthma (CALIMA): a randomised, double‐blind, placebo‐controlled phase 3 trial. Lancet. 2016;388(10056):2128‐2141. [DOI] [PubMed] [Google Scholar]
- 184. Tsetsos N, Goudakos JK, Daskalakis D, Konstantinidis I, Markou K. Monoclonal antibodies for the treatment of chronic rhinosinusitis with nasal polyposis: a systematic review. Rhinology J. 2018;56(1):11‐21. [DOI] [PubMed] [Google Scholar]
- 185. Wollenberg A, Barbarot S, Bieber T, et al. Consensus‐based European guidelines for treatment of atopic eczema (atopic dermatitis) in adults and children: part II. J Eur Acad Dermatol Venereol. 2018;32(6):850‐878. [DOI] [PubMed] [Google Scholar]
- 186. Wollenberg A, Barbarot S, Bieber T, et al. Consensus‐based European guidelines for treatment of atopic eczema (atopic dermatitis) in adults and children: part I. J Eur Acad Dermatol Venereol. 2018;32(5):657‐682. [DOI] [PubMed] [Google Scholar]
- 187. Bachert C, Mannent L, Naclerio RM, et al. Effect of subcutaneous dupilumab on nasal polyp burden in patients with chronic sinusitis and nasal polyposis: a randomized clinical trial. JAMA. 2016;315(5):469‐479. [DOI] [PubMed] [Google Scholar]
- 188. Jonstam K, Swanson BN, Mannent LP, et al. Dupilumab reduces local type 2 pro‐inflammatory biomarkers in chronic rhinosinusitis with nasal polyposis. Allergy. 2019;74(4):743‐752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Broesby‐Olsen S, Vestergaard H, Mortz CG, et al. Omalizumab prevents anaphylaxis and improves symptoms in systemic mastocytosis: efficacy and safety observations. Allergy. 2018;73(1):230‐238. [DOI] [PubMed] [Google Scholar]
- 190. Corren J, Parnes JR, Wang L, et al. Tezepelumab in adults with uncontrolled asthma. N Engl J Med. 2017;377(10):936‐946. [DOI] [PubMed] [Google Scholar]
- 191. Furue M, Yamamura K, Kido‐Nakahara M, Nakahara T, Fukui Y. Emerging role of interleukin‐31 and interleukin‐31 receptor in pruritus in atopic dermatitis. Allergy. 2018;73(1):29‐36. [DOI] [PubMed] [Google Scholar]
- 192. Uchida M, Anderson EL, Squillace DL, et al. Oxidative stress serves as a key checkpoint for IL‐33 release by airway epithelium. Allergy. 2017;72(10):1521‐1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Colás C, Brosa M, Antón E, et al. Estimate of the total costs of allergic rhinitis in specialized care based on real‐world data: the FERIN Study. Allergy. 2017;72(6):959‐966. [DOI] [PubMed] [Google Scholar]
- 194. Tavakoli H, FitzGerald JM, Chen W, et al. Ten‐year trends in direct costs of asthma: a population‐based study. Allergy. 2017;72(2):291‐299. [DOI] [PubMed] [Google Scholar]
- 195. Thyssen JP, Hamann CR, Linneberg A, et al. Atopic dermatitis is associated with anxiety, depression, and suicidal ideation, but not with psychiatric hospitalization or suicide. Allergy. 2018;73(1):214‐220. [DOI] [PubMed] [Google Scholar]
- 196. Hilvering B, Vijverberg SJH, Jansen J, et al. Diagnosing eosinophilic asthma using a multivariate prediction model based on blood granulocyte responsiveness. Allergy. 2017;72(8):1202‐1211. [DOI] [PubMed] [Google Scholar]
- 197. Roth‐Walter F, Adcock IM, Benito‐Villalvilla C, et al. Comparing biologicals and small molecule drug therapies for chronic respiratory diseases. Allergy. 2019;74(3):432‐448. [DOI] [PubMed] [Google Scholar]
- 198. Diamant Z, Mantzouranis E, Bjermer L. Montelukast in the treatment of asthma and beyond. Expert Rev Clin Immunol. 2009;5(6):639‐658. [DOI] [PubMed] [Google Scholar]
- 199. Chauhan BF, Jeyaraman MM, Singh Mann A, et al. Addition of anti‐leukotriene agents to inhaled corticosteroids for adults and adolescents with persistent asthma. Cochrane Database Syst Rev. 2017;3:CD010347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Diamant Z, Aalders W, Parulekar A, Bjermer L, Hanania NA. Targeting lipid mediators in asthma: time for reappraisal. Curr Opin Pulm Med. 2019;25(1):121‐127. [DOI] [PubMed] [Google Scholar]
- 201. Domingo C, Palomares O, Sandham DA, Erpenbeck VJ, Altman P. The prostaglandin D2 receptor 2 pathway in asthma: a key player in airway inflammation. Respir Res. 2018;19(1):189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Singh D, Cadden P, Hunter M, et al. Inhibition of the asthmatic allergen challenge response by the CRTH2 antagonist OC000459. Eur Respir J. 2013;41(1):46‐52. [DOI] [PubMed] [Google Scholar]
- 203. Diamant Z, Sidharta PN, Singh D, et al. Setipiprant, a selective CRTH2 antagonist, reduces allergen‐induced airway responses in allergic asthmatics. Clin Exp Allergy. 2014;44(8):1044‐1052. [DOI] [PubMed] [Google Scholar]
- 204. Fajt ML, Gelhaus SL, Freeman B, et al. Prostaglandin D(2) pathway upregulation: relation to asthma severity, control, and TH2 inflammation. J Allergy Clin Immunol. 2013;131(6):1504‐1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Kuna P, Bjermer L, Tornling G. Two Phase II randomized trials on the CRTh2 antagonist AZD1981 in adults with asthma. Drug Des Devel Ther. 2016;10:2759‐2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Hall IP, Fowler AV, Gupta A, et al. Efficacy of BI 671800, an oral CRTH2 antagonist, in poorly controlled asthma as sole controller and in the presence of inhaled corticosteroid treatment. Pulm Pharmacol Ther. 2015;32:37‐44. [DOI] [PubMed] [Google Scholar]
- 207. Bateman ED, Guerreros AG, Brockhaus F, et al. Fevipiprant, an oral prostaglandin DP2 receptor (CRTh2) antagonist, in allergic asthma uncontrolled on low‐dose inhaled corticosteroids. Eur Respir J. 2017;50(2). 10.1183/13993003.00670-2017 [DOI] [PubMed] [Google Scholar]
- 208. Gonem S, Berair R, Singapuri A, et al. Fevipiprant, a prostaglandin D2 receptor 2 antagonist, in patients with persistent eosinophilic asthma: a single‐centre, randomised, double‐blind, parallel‐group, placebo‐controlled trial. Lancet Respir Med. 2016;4(9):699‐707. [DOI] [PubMed] [Google Scholar]
- 209. Pettipher R, Hunter MG, Perkins CM, et al. Heightened response of eosinophilic asthmatic patients to the CRTH2 antagonist OC000459. Allergy. 2014;69(9):1223‐1232. [DOI] [PubMed] [Google Scholar]
- 210. Dhami S, Kakourou A, Asamoah F, et al. Allergen immunotherapy for allergic asthma: a systematic review and meta‐analysis. Allergy. 2017;72(12):1825‐1848. [DOI] [PubMed] [Google Scholar]
- 211. Cardona V, Luengo O, Labrador‐Horrillo M. Immunotherapy in allergic rhinitis and lower airway outcomes. Allergy. 2017;72(1):35‐42. [DOI] [PubMed] [Google Scholar]
- 212. Di Bona D, Plaia A, Leto‐Barone MS, La Piana S, Macchia L, Di Lorenzo G. Efficacy of allergen immunotherapy in reducing the likelihood of developing new allergen sensitizations: a systematic review. Allergy. 2017;72(5):691‐704. [DOI] [PubMed] [Google Scholar]
- 213. Ponce M, Schroeder F, Bannert C, et al. Preventive sublingual immunotherapy with House Dust Mite extract modulates epitope diversity in pre‐school children. Allergy. 2019;74(4):780‐787. [DOI] [PubMed] [Google Scholar]
- 214. Asaria M, Dhami S, van Ree R, et al. Health economic analysis of allergen immunotherapy for the management of allergic rhinitis, asthma, food allergy and venom allergy: a systematic overview. Allergy. 2018;73(2):269‐283. [DOI] [PubMed] [Google Scholar]
- 215. Hesse L, van Ieperen N, Habraken C, et al. Subcutaneous immunotherapy with purified Der p1 and 2 suppresses type 2 immunity in a murine asthma model. Allergy. 2018;73(4):862‐874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Huang Y, Wang C, Wang X, Zhang L, Lou H. Efficacy and safety of subcutaneous immunotherapy with house dust mite for allergic rhinitis: a meta‐analysis of randomized controlled trials. Allergy. 2019;74(1):189‐192. [DOI] [PubMed] [Google Scholar]
- 217. Couroux P, Ipsen H, Stage BS, et al. A birch sublingual allergy immunotherapy tablet reduces rhinoconjunctivitis symptoms when exposed to birch and oak and induces IgG4 to allergens from all trees in the birch homologous group. Allergy. 2019;74(2):361‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Ihara F, Sakurai D, Yonekura S, et al. Identification of specifically reduced Th2 cell subsets in allergic rhinitis patients after sublingual immunotherapy. Allergy. 2018;73(9):1823‐1832. [DOI] [PubMed] [Google Scholar]
- 219. Masuyama K, Okamoto Y, Okamiya K, et al. Efficacy and safety of SQ house dust mite sublingual immunotherapy‐tablet in Japanese children. Allergy. 2018;73(12):2352‐2363. [DOI] [PubMed] [Google Scholar]
- 220. Hoffmann HJ, Valovirta E, Pfaar O, et al. Novel approaches and perspectives in allergen immunotherapy. Allergy. 2017;72(7):1022‐1034. [DOI] [PubMed] [Google Scholar]
- 221. Pohlit H, Bellinghausen I, Frey H, Saloga J. Recent advances in the use of nanoparticles for allergen‐specific immunotherapy. Allergy. 2017;72(10):1461‐1474. [DOI] [PubMed] [Google Scholar]
- 222. Augé J, Vent J, Agache I, et al. EAACI Position paper on the standardization of nasal allergen challenges. Allergy. 2018;73(8):1597‐1608. [DOI] [PubMed] [Google Scholar]
- 223. Larenas‐Linnemann DES, Antolín‐Amérigo D, Parisi C, et al. National clinical practice guidelines for allergen immunotherapy: an international assessment applying AGREE‐II. Allergy. 2018;73(3):664‐672. [DOI] [PubMed] [Google Scholar]


