Abstract
To date, blood (and serum) as well as urine samples are the most commonly collected specimens for routine doping controls, which allow for the analytical coverage of an extensive set of target analytes relevant to sports drug testing programs. In the course of studies to identify potential alternative matrices to complement current testing approaches, exhaled breath (EB) has been found to offer advantageous properties especially with regard to the sample collection procedure, which is less invasive, less intrusive, and less time‐consuming when compared to conventional blood and urine testing. A yet unaddressed question has been the potential contribution of oral fluid (OF) to EB samples. The current investigation focused on characterizing an electret membrane‐based EB collection device concerning a potential introduction of OF during the sampling procedure. For that purpose, EB and OF samples collected under varying conditions from a total of 14 healthy volunteers were tested for the presence of abundant salivary proteins using bottom‐up proteomics approaches such as SDS‐PAGE followed by tryptic digestion and chromatographic‐mass spectrometric analysis. The trapping baffles integrated into the mouthpiece of the EB collection device were found to effectively retain OF introduced into the unit during sample collection as no saliva breakthrough was detectable using the established analytical approach targeting predominantly the highly abundant salivary α‐amylase. Since α‐amylase was found unaffected by storage, smoking, food intake, and exercise, it appears to be a useful marker to reveal possible OF contaminations of EB collection devices.
Keywords: complementary matrices, exhaled breath, oral fluid, sports drug testing
The investigation focused on characterizing an electret membrane‐based exhaled breath collection device concerning a potential introduction of oral fluid during the sampling procedure by using bottom‐up proteomics approaches. The trapping baffles integrated into the mouthpiece were found to effectively retain oral fluid introduced into the unit during exhaled breath sample collection. Since α‐amylase was found unaffected by storage, smoking, food intake, and exercise, it appears to be a useful marker to reveal possible oral fluid contaminations.
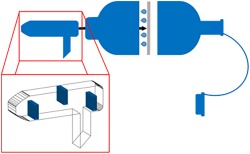
1. INTRODUCTION
To contribute to the ideal of drug‐free sport, doping controls have become an important and indispensable part of the athletics world. Currently, only blood and urine are routinely used as biological matrices for the detection of doping agents both in and out of competition. For both, the sample collection procedure is associated with several disadvantages. Blood collection is an invasive procedure and must be performed by a qualified and authorized doping control officer (Blood Collection Officer) and the provision of urine samples necessitates permanent visual control to prevent manipulation of the sample,1 which is an invasion of the athlete's privacy.2, 3 With alternative matrices such as dried blood spots (DBS), oral fluid (OF), or exhaled breath (EB), the sampling procedure is less invasive compared to blood collection or less intrusive and less time‐consuming compared to urine collection. Also from an analytical point of view, alternative matrices may be advantageous due to optimized detection windows, for example for substances banned only in competition, or a higher stability of the analytes,4, 5, 6, 7 which could compensate for the undeniable limitations of such specimens with regard to retrospectivity and/or the available analyte test spectrum.
Initially, EB has predominantly been used for the detection of volatile substances such as alcohol for estimation of blood alcohol concentration.8 Amongst others, volatile organic compounds (VOCs) of EB were found to be potential biomarkers for the diagnosis of lung diseases such as asthma, chronic obstructive pulmonary disease (COPD), or lung cancer.9 However, recent studies also demonstrated that the analysis of non‐volatile substances obtained from EB bioaerosol particles is feasible, and the first commercially available collection devices have been introduced employing either turbulent gas flow collectors or thin electret membranes.10, 11, 12, 13 The composition of EB samples as collected by means of these devices has not been studied so far, and data, especially concerning the question whether OF contributes substantially to the EB matrix, has been scarce. For scenarios where quantitative analyses are required, the definition of the matrix used is of particular importance as drug concentrations in OF and EB may not correlate.14 Therefore, the aim of this study was to establish means to investigate whether OF is present in EB sampled by means of the electret‐based device. Since saliva secreted by the salivary glands is the main component of OF,15saliva‐specific proteins were analyzed from the collection membrane and the disposable mouth piece using different proteomics approaches.
2. MATERIALS AND METHODS
2.1. EB and OF sampling
Both OF and EB samples were collected from 14 healthy volunteers (8 male, 6 female; age: 23–45) with written informed consent and approval by the local ethics committee (German Sport University Cologne, approval #107/2018). EB sampling was achieved with a SensAbues® (Stockholm, Sweden) collection device used according to the manufacturer's instructions.16, 17 OF was collected by passive drooling for approximately 1 minute and stored in polypropylene tubes.
2.2. Testing of the electret filter for the presence of salivary proteins
2.2.1. Sample preparation
Myoglobin from equine heart (Merck, Darmstadt, Germany) was used as internal standard (ISTD) and 10 μL of an aqueous solution (10 μg/mL) was added to each EB collection device onto the electret filter. Where applicable, 50 μL of OF was added to the filter. Proteins were eluted from the filter by shaking the collection device with 4 mL of 8 M aqueous urea solution. To collect the eluate, the EB cartridge was placed on a plastic tube and centrifuged. The obtained eluate was concentrated by centrifugation at 4000 x g through an Amicon®Ultra‐4 Centrifugal Filter Unit with a cut‐off of 10 kDa (Merck, Darmstadt, Germany) until a final volume of approximately 100 μL was reached. After washing the retentate with 4 mL of deionized water, it was subjected to SDS‐PAGE separation (2.2.2) or in‐solution digestion (2.2.3).
2.2.2. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE) and in‐gel tryptic digestion
SDS‐PAGE separation with subsequent Coomassie staining and in‐gel tryptic digestion of selected bands was conducted as described elsewhere.18
2.2.3. In‐solution tryptic digestion with liquid chromatography–tandem mass spectrometry analysis and data evaluation
For in‐solution tryptic digestion, 25 μL of the washed retentate (2.2.1) was mixed with 25 μL of deionized water and 5 μL of a 100 mM tris(2‐carboxyethyl)phosphine solution (Roth, Karlsruhe, Germany) and incubated for 15 minutes at 60°C to accomplish reduction of disulfide bonds. Subsequently, 0.5 μL of a 2 M NH4HCO3 solution was added. After alkylation with 5.5 μL of a 250 mM iodoacetamide solution (Sigma‐Aldrich®, St Louis, MO, USA) in darkness for 30 minutes, the sample was incubated with 10 μL of a trypsin solution (40 μg/mL) and 7 μL of acetonitrile for at least 12 hours at 37°C and 400 rpm. Afterwards, 2 μL of 100% acetic acid was added.
Liquid chromatography−tandem mass spectrometry (LC−MS/MS) analyses were performed on a Vanquish UHPLC system equipped with a Poroshell 120 EC‐C8 column (3.0 x 50 mm, 2.7 μm particle size) and interfaced to a Q Exactive HF‐X mass spectrometer. The LC system was operated at a flow of 0.3 mL/min with 0.1% formic acid (solvent A) and acetonitrile with 0.1% formic acid and 1% DMSO (solvent B). The gradient started at 1% B increasing to 60% B from minute 1 to 10 followed by an increase to 90% B from minute 10 to 11 and ended with re‐equilibration at 1% B for 4 minutes. The electrospray ionization (ESI) source was operated in positive mode with an ionization voltage of 3.7 kV. Based on survey full scan data within the range m/z 200–2000 with a resolution of 120 000 FWHM (@ m/z 200), product ion mass spectra of the five most intense multiply charged molecules were generated with a resolution of 30 000 FWHM (@ m/z 200) by using nitrogen as collision gas and collision energy settings of 30% (arbitrary units).
MS data were evaluated using the Proteome Discoverer™ software as described in the literature by Walpurgis et al.18 Among others, the following search parameters were used: permitted peptide length of 6–144 amino acids, precursor mass error tolerance of 10 ppm, product ion mass error tolerance of 0.1 Da, and a minimum of two peptides per protein. The database was limited on human proteins except for confirming the ISTD.
2.3. Investigated parameters
2.3.1. Interindividual differences in OF composition and stability of salivary proteins
To probe for potential OF contributions, blank EB samples obtained from 4 male und 4 female volunteers were examined for salivary proteins. Further EB samples were fortified with 50 μL of the corresponding OF to determine interindividual differences in the protein composition of OF. In order to investigate the stability of the proteins on the electret membrane, an additional fortified EB sample per volunteer was stored over a period of 7 days at room temperature (RT). The retentates of all samples were stored at −20°C until SDS‐PAGE analysis. Following Coomassie staining, the proteins of the most abundant bands were subjected to in‐gel tryptic digestion and LC–MS/MS analysis.
2.3.2. Influence of smoking, food intake and physical activity on the OF proteome
To investigate whether smoking affects the capability of the employed approach to detect salivary proteins, an EB sample fortified with OF collected before and after smoking and a blank EB sample each were collected. The influence of food intake and sportive activities on the protein composition of OF was tested in a pilot study setting by collecting EB and OF samples at different times of the day before and after food intake as well as before and after exercise (45 minutes of running, 8 km) from one volunteer. All samples were stored at −20°C until SDS‐PAGE analysis.
2.3.3. Lowest detectable OF volume
To determine the lowest OF volume detectable with the utilized SDS‐PAGE approach, 7 EB samples were fortified with 0 μL, 1 μL, 2 μL, 5 μL, 10 μL, 20 μL, 50 μL, or 100 μL of OF and analyzed as described in Sections 2.2.1 and 2.2.2.
2.3.4. Functionality of the OF traps integrated into the EB collector's mouthpiece
To investigate the functionality of the OF‐trapping baffles implemented in the mouthpiece of the SensAbues® collection device, three volunteers collected two EB samples each, one according to the manufacturer's instructions and one exhaling directly into the EB cartridge without the mouthpiece. The electret membranes were extracted as described above (2.2.1) and the used mouthpieces were rinsed with 4 mL of 8 M urea solution containing 100 ng of ISTD. Afterwards, the eluate was collected, prepared, and analyzed as described under 2.2.2.
3. RESULTS AND DISCUSSION
The aim of this study was to probe for OF potentially contributing to the EB sample matrix collected by means of a commercially available device equipped with an OF‐trapping module. Saliva‐specific proteins, particularly α‐amylase, were employed as markers for this purpose, and different aspects including inter‐individual variability, exercise, food intake, smoking, and storage were considered in a pilot study setting.
3.1. Inter‐individual differences in OF composition and stability of salivary proteins
The salivary proteome has been studied in great detail in the past and, amongst others, salivary α‐amylase has been reported as an abundant and specific constituent of this matrix.19 Especially in light of the fact that the alveolar lining fluid as the main source of the EB aerosol20 has not been shown to contain α‐amylase, this marker was considered particularly useful for the purpose of this study. As shown in Figure 1, only one band at ca. 17 kDa corresponding to the ISTD was visible in the blank EB specimens (labeled as “b”). By contrast, gel bands at apparent molecular weights (MWs) of ~ 10 kDa, ~ 14 kDa, ~ 58 kDa, ~ 62 kDa and ~ 70 kDa were visible in all OF (“pOF”) and OF‐enriched (“OF”) samples with corresponding band patterns, suggesting an effective elution of proteins spiked onto the electret membrane.
Figure 1.
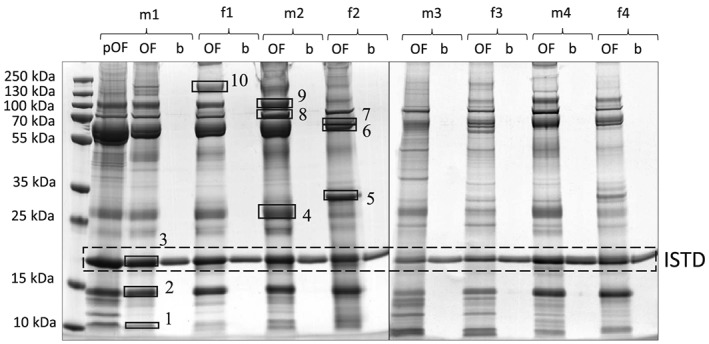
SDS‐PAGE gel stained with Coomassie of pure OF samples (pOF), fortified EB samples with OF (OF) and blank EB samples (b) obtained from 4 male (m) and 4 female (f) volunteers. 1–10: Gel bands chosen for in‐gel tryptic digestion
The protein composition of the most abundant bands was investigated by in‐gel tryptic digestion with subsequent LC–MS/MS analysis (Figure 1, Bands 1–10). As shown in Table 1, the digestive enzyme α‐amylase was identified in bands 4–10.21 This is specific for saliva and occurs in both glycosylated and non‐glycosylated forms,22 resulting in characteristic bands at MWs of ~ 58 kDa and ~ 62 kDa (Figure 1). Cystatins, amongst others, were found in bands 2 and 3 (Table 1). The protective enzymes cystatin S, cystatin SN, and cystatin SA occur primarily in saliva, which is why they are also referred to as “salivary” cystatins, but they can also be found in other tissues.23 Other proteins identified were keratins (bands 1 and 2), apolipoprotein (bands 1 and 4), 1‐antitrypsin (band 6), lactotransferrin (band 9), albumin (bands 7–10) and mucins (band 10), which occur naturally in saliva but are not specific.24 Differences between theoretical and experimental MWs may be due to posttranslational modifications such as oxidation, phosphorylation and glycosylation or to cleavage of peptide bonds during sample preparation. Posttranslational modifications may also be responsible for the interindividual differences in the band pattern after SDS‐PAGE and Coomassie staining (Figure 1).
Table 1.
In‐gel tryptic digestion with subsequent LC–MS/MS analysis evaluated by using proteome discoverer™ software of the most abundant bands (Figure 1); three proteins per excised band with the highest scores are listed
| Band | Protein | Accession # | # of Identified Peptides/Sequence Coverage | Score | MW [kDa] (theor.) | MW [kDa] (exp.) |
|---|---|---|---|---|---|---|
| 1 | Keratin, type II cytoskeletal | P04264 | 14/22.98% | 40.99 | 66.0 | ~ 10 |
| Keratin, type I cytoskeletal | P35527 | 11/19.10% | 27.90 | 62.0 | ||
| Apolipoprotein A‐II | P02652 | 3/21.00% | 6.46 | 11.2 | ||
| 2 | Keratin, type II cytoskeletal 1 | P04264 | 20/25.47% | 41.62 | 66.0 | ~ 14 |
| Keratin, type I cytoskeletal 10 | P13645 | 15/22.6% | 35.28 | 58.8 | ||
| Cystatin‐S | P01036 | 6/46.1% | 33.09 | 16.2 | ||
| 3 | Myoglobin OS = Equus burchelli | P68083 | 15/81.82% | 198.62 | 17.1 | ~ 17 |
| Lipocalin‐1 | P31025 | 5/28.41% | 17.42 | 19.2 | ||
| Cystatin‐S | P01036 | 3/24.82% | 8.31 | 16.2 | ||
| 4 | Alpha‐amylase 1 | P04745 | 12/27.59% | 42.56 | 57.7 | ~ 26 |
| Apolipoprotein A‐I | P02647 | 14/56.93% | 38.71 | 30.8 | ||
| Zymogen granule protein 16 homolog B | Q96DA0 | 6/26.92% | 34.96 | 22.7 | ||
| 5 | Alpha‐amylase 1 | P04745 | 9/21.72% | 23.60 | 57.7 | ~ 31 |
| Zymogen granule protein 16 homolog B | Q96DA0 | 3/16.83% | 11.64 | 22.7 | ||
| BPI fold‐containing family A member 2 | Q96DR5 | 4/18.47% | 11.55 | 27.0 | ||
| 6 | Alpha‐amylase 1 | P04745 | 18/37.18% | 112.98 | 57.7 | ~ 58 |
| BPI fold‐containing family B member 1 | Q8TDL5 | 10/26.65% | 34.35 | 52.4 | ||
| Alpha‐1‐antitrypsin | P01009 | 5/15.07% | 17.82 | 46.7 | ||
| 7 | Alpha‐amylase 1 | P04745 | 18/36.79% | 108.30 | 57.7 | ~ 62 |
| BPI fold‐containing family B member 1 | Q8TDL5 | 11/32.44% | 48.75 | 52.4 | ||
| Serum albumin | P02768 | 12/16.42% | 43.12 | 69.3 | ||
| 8 | Serum albumin | P02768 | 30/38.59% | 127.58 | 69.3 | ~ 70 |
| Complement C3 | P01024 | 15/10.52% | 43.89 | 187.0 | ||
| Alpha‐amylase 1 | P04745 | 11/24.46% | 43.27 | 57.7 | ||
| 9 | Lactotransferrin | P02788 | 20/26.48% | 78.84 | 78.1 | ~ 100 |
| Alpha‐amylase 1 | P04745 | 14/31.51% | 76.78 | 57.7 | ||
| Serum albumin | P02768 | 15/20.2% | 55.36 | 69.3 | ||
| 10 | Mucin‐5B | Q9HC84 | 12/2.52% | 31.86 | 596.0 | ~ 130 |
| Serum albumin | P02768 | 6/9.03% | 23.18 | 69.3 | ||
| Alpha‐amylase 1 | P04745 | 6/16.05% | 17.41 | 57.7 |
Another aspect relevant to doping controls is analyte stability. As the duration of doping control sample transport from the collection site to the laboratory might vary from hours to days, the stability of salivary proteins on the collection membrane was investigated over a storage period of one week at RT. Band patterns differing between fresh and stored samples were observed, with several additional bands at lower MWs detected in stored EB specimens, which proved to contain fragments of cystatins. No difference however was detected concerning the intense bands at ~ 58 kDa and ~ 62 kDa previously identified as α‐amylase (Figure S1, Table S1 in the Supporting Information).
3.2. In‐solution tryptic digestion
Complementary to SDS‐PAGE with Coomassie staining, the option to analyze potential salivary markers by in‐solution tryptic digestion was assessed. Also here, α‐amylase and other saliva‐specific proteins were identified only in samples fortified with OF (Table 2) but not in ordinarily collected EB samples. The ISTD was detected in all samples, thus confirming that the presence of OF would be revealed by in‐solution tryptic digestion and LC–MS/MS analysis.
Table 2.
In‐solution tryptic digestion with subsequent LC–MS/MS analysis evaluated by using Proteome Discoverer™ software of fortified EB samples with OF (OF) and blank EB samples (b) obtained from 3 male (m) and 2 female (f) volunteers; listed are four proteins each with the highest scores for fortified samples and each identified protein for blank samples
| Sample | Protein | Accession # | # of Identified Peptides/Sequence Coverage | Score | MW [kDa] |
|---|---|---|---|---|---|
| m1 (OF) | Alpha‐amylase 1 | P04745 | 17/30.72% | 64.44 | 57.7 |
| Lysozyme C | P61626 | 8/35.14% | 36.87 | 16.5 | |
| Serum albumin | P02768 | 8/16.09% | 35.18 | 69.3 | |
| Zinc‐alpha‐2‐glycoprotein | P25311 | 12/34.90% | 34.77 | 34.2 | |
| Myoglobin OS = Equus burchelli | P68083 | 18/75.32% | 101.98 | 17.1 | |
| m1 (b) | Keratin, type II cytoskeletal 1 | P04264 | 16/25.00% | 51.96 | 66.0 |
| Serum albumin | P02768 | 12/19.54% | 43.10 | 69.3 | |
| Keratin, type I cytoskeletal 10 | P13645 | 11/21.23% | 31.06 | 58.8 | |
| Myoglobin OS = Equus burchelli | P68083 | 21/88.96% | 167.44 | 17.1 | |
| f1 (OF) | Zinc‐alpha‐2‐glycoprotein | P25311 | 11/34.90% | 30.09 | 34.2 |
| Cystatin‐B | P04080 | 6/54.08% | 29.24 | 11.1 | |
| Ig alpha‐1 chain C region | P01876 | 9/19.55% | 26.48 | 37.6 | |
| Alpha‐amylase 1 | P04745 | 8/13.50% | 25.87 | 57.7 | |
| Myoglobin OS = Equus burchelli | P68083 | 18/64.94% | 105.09 | 17.1 | |
| f1 (b) | Triosephosphate isomerase | P60174 | 3/13.29% | 7.65 | 30.8 |
| Myoglobin OS = Equus burchelli | P68083 | 21/88.96% | 138.25 | 17.1 | |
| m2 (OF) | Alpha‐amylase 1 | P04745 | 19/38.75% | 72.03 | 57.7 |
| Cystatin‐B | P04080 | 7/54.08% | 31.54 | 11.1 | |
| Zinc‐alpha‐2‐glycoprotein | P25311 | 10/30.54% | 27.81 | 34.2 | |
| Ig alpha‐1 chain C region | P01876 | 8/19.55% | 22.04 | 37.6 | |
| Myoglobin OS = Equus burchelli | P68083 | 19/88.96% | 99.29 | 17.1 | |
| m2 (b) | Myoglobin OS = Equus burchelli | P68083 | 20/88.96% | 113.87 | 17.1 |
| f2 (OF) | Alpha‐amylase 1 | P04745 | 15/28.96% | 53.07 | 57.7 |
| Cystatin‐SN | P01037 | 6/30.50% | 27.05 | 16.4 | |
| Zinc‐alpha‐2‐glycoprotein | P25311 | 8/28.19% | 23.84 | 34.2 | |
| Ig alpha‐1 chain C region | P01876 | 8/19.55% | 21.54 | 37.6 | |
| Myoglobin OS = Equus burchelli | P68083 | 21/88.96% | 166.15 | 17.1 | |
| f2 (b) | Myoglobin OS = Equus burchelli | P68083 | 21/85.06% | 161.18 | 17.1 |
| m3 (OF) | Lysozyme C | P61626 | 10/35.81% | 53.55 | 16.5 |
| Alpha‐amylase 1 | P04745 | 13/19.77% | 42.83 | 57.7 | |
| Serum albumin | P02768 | 10/17.24% | 37.29 | 69.3 | |
| Cystatin‐SN | P01037 | 8/30.50% | 35.32 | 16.4 | |
| Myoglobin OS = Equus burchelli | P68083 | 18/78.57% | 120.28 | 17.1 | |
| m3 (b) | Myoglobin OS = Equus burchelli | P68083 | 23/88.96% | 158.61 | 17.1 |
3.3. Influence of smoking, food intake, and physical activity on the salivary proteome
Various factors might affect the composition of the salivary proteome.25 To investigate the robustness of potential markers indicative for the presence of OF in EB samples, specimens collected under varying conditions were analyzed. Whilst the number of individuals/samples per scenario cannot be considered as statistically significant, it was noted that in this pilot study dataset neither smoking, nor food intake or exercise were found to have a visible influence on the detected protein band pattern in SDS‐PAGE analyses (Figures S2 and S3). As the interest in salivary proteins traceable in EB samples was of purely qualitative nature in the present study, no additional tests were conducted but might become relevant if EB specimens would be routinely analyzed for contributions caused by OF.
3.4. Lowest detectable OF volume
Both the intensity and the number of protein bands increased consistently with the added volume of OF to EB samples. Using Coomassie staining, the most intense bands at 58 kDa and 62 kDa (representing α‐amylase) were visible at a volume of 2 μL and above (Figure 2), demonstrating that if OF passed the mouthpiece baffles and contributed to the EB matrix collected on the electret membrane, its volume would be below 2 μL.
Figure 2.
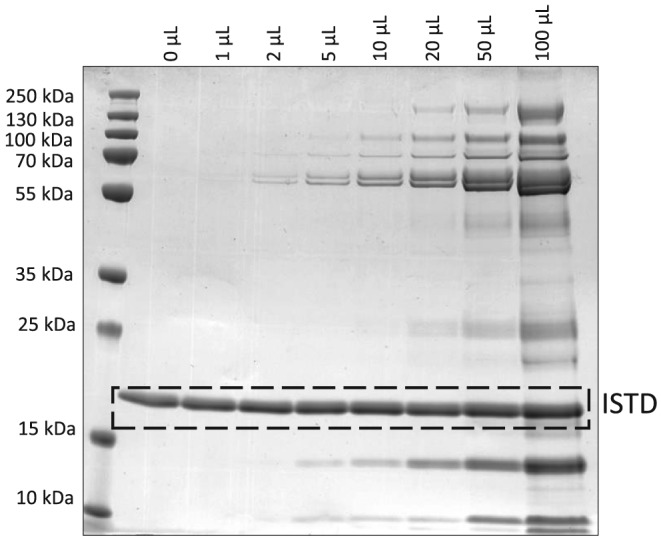
SDS‐PAGE gel stained with Coomassie of EB samples fortified with different volumes of OF
3.5. Functionality of the OF trapping baffles integrated into the mouthpiece
The main purpose of the OF trapping baffles integrated into the mouthpiece is preventing OF from entering the main EB collection container. Whether or not the disposable/removable mouthpiece in fact retains OF was tested by analyzing used OF‐traps and EB samples collected without the mouthpiece mounted to the EB collection container for saliva‐specific proteins. As shown in Figure 3, after elution of the mouthpiece (Figure 3, mp) a characteristic pattern with typical α‐amylase bands at ~ 58 kDa and ~ 62 kDa as observed previously in OF samples was visible. Moreover, in 2 out of 3 EB samples generated by breathing into the collection container without installed mouthpiece (Figure 3; f1, m2) resulted in a similar SDS‐PAGE band pattern, corroborating that the mouthpiece indeed retains significant amounts of OF during the sampling process.
Figure 3.
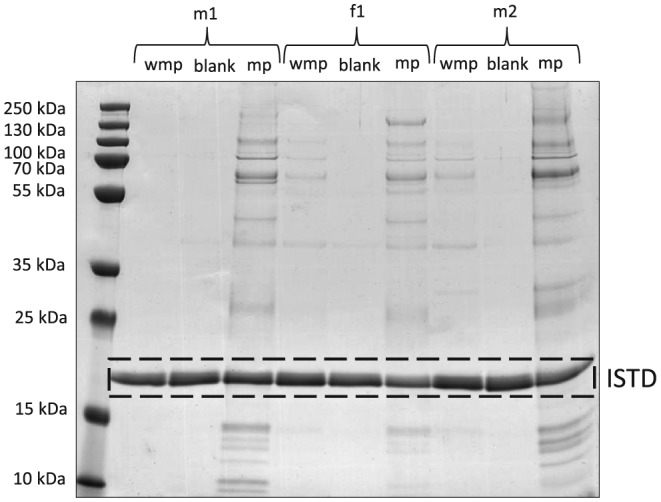
SDS‐PAGE gel stained with Coomassie of EB samples collected without mouthpiece (wmp), EB blank samples (b) and eluates of the mouthpiece (mp) from male (m) and female (f) volunteers
4. CONCLUSION
The question whether OF contributes substantially to EB samples and thus potentially complicates the interpretation of results has been controversially discussed in previous studies.14, 26 Within this study, it could be demonstrated that the introduction of OF into EB samples during conventional collection procedures is possible, and that geometries forming OF‐traps are useful means, if the presence of OF in such specimens needs to be minimized. For tests aiming merely at qualitative results, ie, probing for the presence or absence of a drug, as for instance in abstinence controls, contributions of OF to the EB sample matrix might not be critical. If investigations focus on exhaled breath only, the use of collection devices offering OF‐traps is advisable. The saliva‐specific protein α‐amylase was found to be a useful marker to uncover OF contamination. It can be detected either by SDS‐PAGE followed by Coomassie staining by two characteristic bands at ~ 58 kDa and ~ 62 kDa or by less time‐consuming in‐solution tryptic digestion and subsequent LC–MS/MS analysis.
Supporting information
Figure S1:
SDS‐PAGE gel stained with Coomassie of fortified EB samples with OF prepared to retentate on day 1 (d1) or day 8 (d8) after storage for 7 days at room temperature and blank EB samples (b) obtained from 4 male (m) and 4 female (f) volunteers; 1–10: Gel spots chosen for in‐gel tryptic digestion
Table S1: In‐gel tryptic digestion with subsequent LC–MS/MS analysis evaluated by using Proteome Discoverer™ Software of the most abundant bands (Supplementary Figure 1); three proteins per excised band with the highest scores are listed.
Figure S2: SDS‐PAGE gel stained with Coomassie of fortified EB samples with OF (OF) and blank EB samples (b) obtained from 1 male volunteer before and after smoking
Figure S3: SDS‐PAGE Gel stained with Coomassie of fortified EB samples with OF obtained at different times of the day before and after food intake (fi) or after exercise (45 min of running, 8 km)
ACKNOWLEDGEMENTS
This project was supported by funding from the Partnership for Clean Competition Research Collaborative (#110638TRF18). The content of this publication does not necessarily reflect the views or policies of the Research Collaborative. The authors also acknowledge the constructive collaboration with SensAbues AB (Stockholm, Sweden).
Garzinsky A‐M, Walpurgis K, Krug O, Thevis M. Does oral fluid contribute to exhaled breath samples collected by means of an electret membrane? Drug Test Anal. 2019;11:1764–1770. 10.1002/dta.2597
REFERENCES
- 1. Thevis M, Geyer H, Mareck U, et al. Detection of manipulation in doping control urine sample collection: a multidisciplinary approach to determine identical urine samples. Anal Bioanal Chem. 2007;388(7):1539‐1543. [DOI] [PubMed] [Google Scholar]
- 2. World Anti‐Doping Agency . International Standard for Laboratories. https://www.wada-ama.org/sites/default/files/resources/files/isl_june_2016.pdf. Accessed 02.01.2019.
- 3. World Anti‐Doping Agency . International standard for testing and investigations. https://www.wada-ama.org/sites/default/files/resources/files/2016-09-30_-_isti_final_january_2017.pdf. Accessed 02.01.2019.
- 4. Henion J, Oliveira RV, Chace DH. Microsample analyses via DBS: challenges and opportunities. Bioanalysis. 2013;5(20):2547‐2565. [DOI] [PubMed] [Google Scholar]
- 5. Anizan S, Huestis MA. The potential role of oral fluid in antidoping testing. Clin Chem. 2014;60(2):307‐322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thevis M, Geyer H, Tretzel L, Schänzer W. Sports drug testing using complementary matrices: advantages and limitations. J Pharm Biomed Anal. 2016;130:220‐230. [DOI] [PubMed] [Google Scholar]
- 7. Thevis M, Krug O, Geyer H, Schänzer W. Expanding analytical options in sports drug testing: mass spectrometric detection of prohibited substances in exhaled breath. Rapid Commun Mass Spectrom. 2017;31(15):11290‐1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lubkin SR, Gullberg RG, Logan BK, Maini PK, Murray JD. Simple versus sophisticated models of breath alcohol exhalation profiles. Alcohol Alcohol. 1996;31(1):61‐67. [DOI] [PubMed] [Google Scholar]
- 9. Chan HP, Lewis C, Thomas PS. Exhaled breath analysis: novel approach for early detection of lung cancer. Lung Cancer. 2009;63(2):164‐168. [DOI] [PubMed] [Google Scholar]
- 10. Seferaj S, Ullah S, Tinglev A, et al. Evaluation of a new simple collection device for sampling of microparticles in exhaled breath. J Breath Res. 2018;12(3):036005. [DOI] [PubMed] [Google Scholar]
- 11. Ullah S, Sandqvist S, Beck O. A liquid chromatography and tandem mass spectrometry method to determine 28 non‐volatile drugs of abuse in exhaled breath. J Pharm Biomed Anal. 2018;148:251‐258. [DOI] [PubMed] [Google Scholar]
- 12. Coucke L, Massarini E, Ostijn Z, Beck O, Verstraete AG. Delta(9)‐tetrahydrocannabinol concentrations in exhaled breath and physiological effects following cannabis intake ‐ a pilot study using illicit cannabis. Clin Biochem. 2016;49(13–14):1072‐1077. [DOI] [PubMed] [Google Scholar]
- 13. Wang C‐s, Otani Y. Removal of nanoparticles from gas streams by fibrous filters: a review. Ind Eng Chem Res. 2013;52(1):5‐17. [Google Scholar]
- 14. Meyer MR, Rosenborg S, Stenberg M, Beck O. First report on the pharmacokinetics of tramadol and O‐desmethyltramadol in exhaled breath compared to plasma and oral fluid after a single oral dose. Biochem Pharmacol. 2015;98(3):502‐510. [DOI] [PubMed] [Google Scholar]
- 15. de Almeida PDV, Grégio AMT, Machado MÂN, de Lima AAS, Azevedo LR. Saliva composition and functions: a comprehensive review. J Contemp Dent Pract. 2008;9(3):72‐80. [PubMed] [Google Scholar]
- 16. SensAbues . Sensabues‐user‐instructions. http://sensabues.com/wp-content/uploads/2016/11/Sensabues-user-instructions.pdf. Accessed 02.01.2019.
- 17. SensAbues . SensAbues ‐ next generation drug detection and health monitoring. http://sensabues.com/product. Accessed 02.01.2019.
- 18. Walpurgis K, Krug O, Thomas A, Laussmann T, Schänzer W, Thevis M. Detection of an unknown fusion protein in confiscated black market products. Drug Test Anal. 2014;6(11–12):1117‐1124. [DOI] [PubMed] [Google Scholar]
- 19. Schenkels LC, Veerman EC, Nieuw Amerongen AV. Biochemical composition of human saliva in relation to other mucosal fluids. Crit Rev Oral Biol Med. 1995;6(2):161‐175. [DOI] [PubMed] [Google Scholar]
- 20. Johnson GR, Morawska L. The mechanism of breath aerosol formation. J Aerosol Med Pulm Drug Deliv. 2009;22(3):229‐237. [DOI] [PubMed] [Google Scholar]
- 21. European bioinformatics institute (EMBL‐EBI) SSIoB, protein information resource (PIR) P04745 (AMY1_HUMAN). https://www.uniprot.org/uniprot/P04745. Accessed 17.01.2019.
- 22. Yang ZM, Lin J, Chen LH, Zhang M, Chen WW, Yang XR. The roles of AMY1 copies and protein expression in human salivary alpha‐amylase activity. Physiol Behav. 2015;138:173‐178. [DOI] [PubMed] [Google Scholar]
- 23. Dickinson DP, Thiesse M, Hicks MJ. Expression of type 2 cystatin genes CST1‐CST5 in adult human tissues and the developing submandibular gland. DNA Cell Biol. 2002;21(1):47‐65. [DOI] [PubMed] [Google Scholar]
- 24. Huang CM. Comparative proteomic analysis of human whole saliva. Arch Oral Biol. 2004;49(12):951‐962. [DOI] [PubMed] [Google Scholar]
- 25. Siqueira WL, Dawes C. The salivary proteome: challenges and perspectives. Proteomics Clin Appl. 2011;5(11–12):575‐579. [DOI] [PubMed] [Google Scholar]
- 26. Ellefsen KN, Concheiro M, Beck O, Gorelick DA, Pirard S, Huestis MA. Quantification of cocaine and metabolites in exhaled breath by liquid chromatography‐high‐resolution mass spectrometry following controlled administration of intravenous cocaine. Anal Bioanal Chem. 2014;406(25):6213‐6223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1:
SDS‐PAGE gel stained with Coomassie of fortified EB samples with OF prepared to retentate on day 1 (d1) or day 8 (d8) after storage for 7 days at room temperature and blank EB samples (b) obtained from 4 male (m) and 4 female (f) volunteers; 1–10: Gel spots chosen for in‐gel tryptic digestion
Table S1: In‐gel tryptic digestion with subsequent LC–MS/MS analysis evaluated by using Proteome Discoverer™ Software of the most abundant bands (Supplementary Figure 1); three proteins per excised band with the highest scores are listed.
Figure S2: SDS‐PAGE gel stained with Coomassie of fortified EB samples with OF (OF) and blank EB samples (b) obtained from 1 male volunteer before and after smoking
Figure S3: SDS‐PAGE Gel stained with Coomassie of fortified EB samples with OF obtained at different times of the day before and after food intake (fi) or after exercise (45 min of running, 8 km)


