Abstract
Incorporating the dynamic nature of biomolecules in the modeling of their complexes is a challenge, especially when the extent and direction of the conformational changes taking place upon binding is unknown. Estimating whether the binding of a biomolecule to its partner(s) occurs in a conformational state accessible to its unbound form (“conformational selection”) and/or the binding process induces conformational changes (“induced‐fit”) is another challenge. We propose here a method combining conformational sampling using ClustENM—an elastic network‐based modeling procedure—with docking using HADDOCK, in a framework that incorporates conformational selection and induced‐fit effects upon binding. The extent of the applied deformation is estimated from its energetical costs, inspired from mechanical tensile testing on materials. We applied our pre‐ and post‐docking sampling of conformational changes to the flexible multidomain protein‐protein docking benchmark and a subset of the protein‐DNA docking benchmark. Our ClustENM‐HADDOCK approach produced acceptable to medium quality models in 7/11 and 5/6 cases for the protein‐protein and protein‐DNA complexes, respectively. The conformational selection (sampling prior to docking) has the highest impact on the quality of the docked models for the protein‐protein complexes. The induced‐fit stage of the pipeline (post‐sampling), however, improved the quality of the final models for the protein‐DNA complexes. Compared to previously described strategies to handle conformational changes, ClustENM‐HADDOCK performs better than two‐body docking in protein‐protein cases but worse than a flexible multidomain docking approach. However, it does show a better or similar performance compared to previous protein‐DNA docking approaches, which makes it a suitable alternative.
Keywords: biomolecular complexes, conformational flexibility, elastic network modeling
1. INTRODUCTION
The dynamic nature of biomolecules enables them to fulfill their functions in the cell such as catalysis, signaling, and regulation, while interacting with other bioentities. Understanding their structure and dynamics is therefore key to elucidate biological mechanisms, identify the underlying causes of diseases at molecular level and develop new therapeutical strategies. The extent of biomolecular flexibility can range from local side chain rotations of a few angstroms to global displacements like domain motions in the order of nanometers.1 Although experimental methods like X‐ray crystallography, Nuclear Magnetic Resonance (NMR) and cryo‐Electron Microscopy (EM) reveal biomolecular structures at atomic resolution, information about their mobility and flexibility is challenging to assess, especially for highly flexible biomolecules or supramolecules. Therefore, there is a need for complementary computational techniques to reveal the flexible properties and dynamics of such systems.2, 3, 4
In regard to molecular recognition mechanisms, the puzzle still exists whether a biomolecule binds to a pre‐existing conformer out of the pool of conformers existing in solution for a given partner (“conformational selection”) or rather conformational changes are induced upon binding (“induced‐fit”).5, 6 There is increasing evidence showing that different states are indeed accessible in the absence of the partner6, 7 but it is also likely that both mechanisms play a role in molecular recognition.8, 9 A study on a set of complexes with rather limited conformational changes10 has addressed the ability of current computational techniques to capture the conformational changes upon binding and concluded that current “conformational selection” methods can capture 22% of unbound‐bound transitions, “induced‐fit” methods about 57%; but backbone motions (21%) are not yet adequately covered by any technique.
Previously, ClustENM has been developed as an iterative and unbiased conformer generation technique, combining global modes from Elastic Network Model (ENM), with energy minimization and clustering.11, 12 ClustENM was applied to systems of different sizes and oligomeric states including ribosome,11, 13 highly flexible proteins like adenylate kinase and calmodulin,11, 12 and to protein‐small ligand docking.12 The generated conformers were in good agreement with experimental structures and conformations from molecular dynamics simulations. This illustrated the efficiency of ClustENM especially for large systems where molecular dynamics could become computationally time‐consuming and encounter difficulties in sampling large conformational changes. The method also has the potential to capture induced‐fit effects upon binding, as demonstrated for calmodulin where a peptide was docked onto an intermediate state between the unbound and bound forms: The conformational sampling applied to the docked complex indeed allowed to sample the bound state.12
Following this observation, we propose here a method combining molecular docking with conformer generation by ClustENM, to account both for conformational selection and induced‐fit binding mechanisms. For this, we combine conformational sampling of the individual partners (pre‐docking sampling) with that of the docked models (post‐docking sampling). As molecular docking tool we use HADDOCK,14, 15, 16 an information‐driven approach for the modeling of biomolecular complexes, which has demonstrated sustained performance in CAPRI ‐ the blind modeling experiment for the prediction of complexes,17, 18 and is a widely used method for integrative modeling of biomolecular complexes.19 HADDOCK can handle mixed systems of proteins, nucleic acids and small molecules.
To assess the performance of our ClustENM‐HADDOCK approach, we apply it to the protein‐protein flexible multidomain docking benchmark20 and to a subset of the protein‐DNA benchmark.21 Both datasets were used in previous studies focusing on modeling conformational changes, which allows us to make performance comparisons. Notably, we apply our method without prior knowledge of the extent and direction of the conformational changes.
2. MATERIALS AND METHODS
2.1. Dataset
We applied our method on two different datasets:
The flexible multidomain docking benchmark (FMD)20 (Table 1A), where the ligands (smaller partner) are mainly rigid (0.5‐1.7 å) while the receptors undergo varying degrees of conformational change (between 1.6‐19.6 å) corresponding mainly to domain‐domain motions. In this case, in order to concentrate on the conformational change aspects, we assume we have an ideal interface information.
A subset of the protein‐DNA benchmark21 for which experimental information about interfaces is available (Table 1B). Both protein and DNA (“ligand”) undergo conformational changes, but we focused here on the flexibility of the DNA.
Table 1.
List of protein‐protein (A) and protein‐DNA complexes (B) used for benchmarking
| A. Protein‐protein flexible multidomain dataset20 | ||||
|---|---|---|---|---|
| Complex ID | Receptor ID | Ligand ID | Backbone conformational changes [å] | |
| Receptor | Ligand | |||
| 1IRA37 | 1G0Y_R38 | 1ILR_139 | 19.6 | 0.7 |
| 1H1V40 | 1D0N_B41 | 1IJJ_B42 | 13.8 | 1.6 |
| 1Y6443 | 1UX5_A44 | 2FXU_A45 | 10.3 | 1.1 |
| 1F6M46 | 1CL0_A47 | 2TIR_A48 | 7.3 | 0.9 |
| 1FAK49 | 1QFK_HL50 | 1TFH_B51 | 6.3 | 1.0 |
| 1ZLI52 | 2JTO_A53 | 1KWM_A54 | 4.0 | 0.6 |
| 1E4K55 | 3AVE_AB56 | 1FNL_A57 | 2.9 | 1.7 |
| 1IBR58 | 1F59_A59 | 1QG4_A60 | 2.9 | 1.1 |
| 1KKL61 | 1JB1_A62 | 2HPR63 | 2.2 | 0.5 |
| 1NPE64 | 1KLO_A65 | 1NPE_A64 | 1.9 | ‐ |
| 1DFJ66 | 2BNH_A67 | 9RSA_B68 | 1.6 | 0.7 |
Both datasets have been previously used in benchmarking studies with HADDOCK. In the first case a multibody docking approach was followed in which the protein showing rigid‐body motion conformational changes was cut into domains with connectivity restraints between them,20 while in the second case a two steps docking strategy was followed in which DNA conformations from the first docking run were used to generate an ensemble of pre‐bent DNA conformations for a second, ensemble‐based docking stage.21
2.2. ClustENM method
ClustENM is an unbiased, iterative conformational sampling method combining ENM, energy minimization and clustering.11, 12 In ClustENM, first the initial structure is energetically minimized in implicit solvent, then a modified version of ENM (mixed resolution ENM)22 is applied to it to extract low frequency normal modes. The modified version of ENM employed in ClustENM involves placing the nodes at the residue centroids where node pairs within a cut‐off distance of 10 å are connected by elastic springs, thereby representing the residues as low‐resolution nodes. The spring constant connecting the node pairs is proportional to the total number of interacting atom pairs between the connected residues (bonded or nonbonded) within the specified cut‐off. Based on the sudden break in the smooth progression of eigenvalues in the eigenvalue spectrum, we define the number of modes “m” which are to be linearly combined using coefficients −1, 0 and 1 to obtain deformation vectors (3m vectors in total). The minimized structure is deformed in the direction of the deformation vectors with a fixed deformation RMSD d, where the deformation vector of a residue centroid is applied to all the atoms of that residue. The resulting structures are clustered based on mutual RMSD of heavy atoms with a cut‐off equal to d. The cluster containing the parent structure is discarded and a representative structure from each cluster is selected. Minimization‐ENM‐deformation‐clustering steps are applied on each representative structure for k generations.
2.2.1. Energy minimization
For proteins, the energy minimization in implicit solvent was performed using NAMD v2.1023 with the CHARMM22 force field.24 A generalized Born implicit solvent model was used with a 16 å cut‐off for non‐bonded interactions and a 14 å cut‐off for Born radius. The ion concentration was set to 0.3 M. 4000 steps of conjugate gradient were applied.
The DNA structures and protein‐DNA complexes were minimized in implicit solvent using AMBER14 with the ff14SB force field parameters.25 The pairwise generalized Born model26, 27 was used with a 16 å cut‐off for non‐bonded interactions. We used the modified generalized Born theory‐based Debye‐Huckel limiting law for ion screening of interactions.28 The concentration of 1‐1 mobile counter‐ions in solution was set to 0.1 M. Steepest descent was applied for 500 steps followed by conjugate gradient with a maximum number of steps of 4000 and a convergence criterion of 0.01 kcal/mol/å.
2.2.2. Deformation Parameter d Estimation
The advantage of ClustENM is its computational efficiency in yielding conformers at atomic resolution and its reduction in the redundancy of the sampled conformations due to clustering. However, the degree of conformational change is usually arbitrarily defined by the user. To overcome this problem and inspired from mechanical tensile testing on material, we deformed the structures in the direction of the slow modes to assess the extent of flexibility. First, we determined the relevant low frequency modes based on the sudden break in eigenvalue spectrum; then we deformed the structure constantly in their direction and determined the energy of the deformed state by energy‐minimization. Based on the energy vs strain (deformation) curve and the reference energy state of the initial, non‐deformed structure, we assessed the maximum deformation the system could handle. The results of such energy‐deformation curves for the ribonuclease inhibitor structure (1DFJ) are shown in Figure 1. According to the consensus of four modes, the structure can handle a RMSD deformation of 1‐2 å, which actually covers the observed 1.6 å RMSD conformational change upon binding to its partner. This prediction scheme is applied to all receptors in the flexible multidomain protein docking benchmark and the DNA structures in the protein‐DNA dataset to determine the deformation RMSD d used in ClustENM. The same procedure is then applied to the docked complexes, to estimate d for post‐docking ClustENM using these complexes as starting point.
Figure 1.
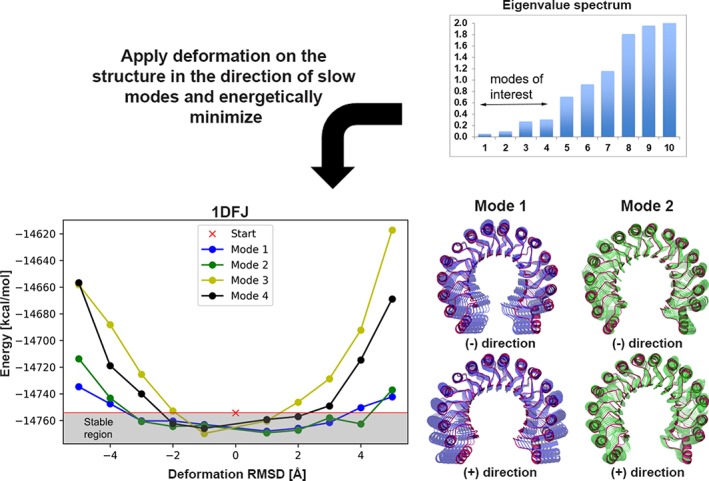
Tensile test on ribonuclease inhibitor structure along the direction of normal modes. Based on eigenvalue spectrum shown on top, the first four slowest modes are selected for application of tensile testing. The structure is first minimized (“Start”), then deformations of 1‐5 å are introduced in both negative and positive directions along each mode. To visualize the type of motions, the first and second slowest modes are also shown in the right panel. The energy of the structure increases as the deformation along an individual mode increases. The stable region corresponds to the deformations that keep the energy level below the reference state: For the first and second modes the energy starts increasing after 3 å, for the third mode after 2 å and the fourth mode after 1 å in both directions. The consensus of the four modes hints to a maximum deformation of 1‐2 å for this structure [Color figure can be viewed at http://wileyonlinelibrary.com]
The ClustENM parameters used for the conformer generation in the pre‐ and post‐sampling stages are listed in Table 2A and B for the FMD and protein‐DNA datasets, respectively.
Table 2.
ClustENM pre‐ and post‐sampling parameters
| A. Protein‐protein complexes of the FMD benchmark | ||||
|---|---|---|---|---|
| Sampling prior to docking (receptor only) | Sampling after docking (on complex structure)a | |||
| Complexes | Deformation RMSD [å] | Number of modes | Deformation RMSD [å] | Number of modes |
| 1IRA | 3 | 3 | 1 | 5 |
| 1H1V | 2 | 3 | 1 | 4 |
| 1Y64 | 4 | 3 | 2/3 | 3 |
| 1F6M | 2 | 3 | 1 | 5/4 |
| 1FAK | 2 | 3 | 1 | 3 |
| 1ZLI | 1 | 3 | 1 | 5 |
| 1E4K | 1 | 4 | 2/3 | 3 |
| 1IBR | 2 | 5 | 1 | 5 |
| 1KKL | 2 | 5 | 1/2 | 3 |
| 1NPE | 3 | 4 | 1/2 | 3 |
| 1DFJ | 2 | 4 | 1 | 4/5 |
| B. Protein‐DNA dataset | ||||
|---|---|---|---|---|
| Sampling prior to docking (DNA only) | Sampling after docking (on complex structure)a | |||
| Complexes | Deformation RMSD [å] | Number of modes | Deformation RMSD [å] | Number of modes |
| 1BY4 | 1 | 3 | 2 | 3/5 |
| 3CRO | 1 | 3 | 1 | 5 |
| 1AZP | 1 | 4 | 1 | 5 |
| 1JJ4 | 2 | 3 | 1 | 5 |
| 1A74 | 2 | 3 | 2/1 | 3 |
| 1ZME | 1 | 3 | 3/2 | 3/5 |
The presence of two different values indicates the use of different parameters for the complexes. The first value was used for the complex from the best cluster and the second value for the second‐best cluster. Single value means same parameters were used for both complex.
2.3. HADDOCK settings
We used the HADDOCK2.2 webserver16 for all dockings in this study. For the FMD benchmark, the docking was performed with “ideal” interface information, that is, the interface residues from one subunit within a 5 å distance from any atom of the partner were selected as active. No passive residues were defined. This effectively brings the interfaces together during the docking without predefining their relative orientation since no specific contacts are defined. Random removal of ambiguous interface restraints (AIR) was switched off. Since we use ensembles of conformations, the sampling was increased to 10 000/400/400 structures for rigid (it0), semi‐flexible(it1) and water stages, respectively. Clustering was performed using the fraction of common contacts (FCC)29 with a cut‐off of 0.75 and a minimum cluster size of 4.
For protein‐DNA docking, the experimental restraints described in the study by van Dijk and Bonvin, 201021 were used to drive the docking. Since we do not use the ideal interface information here, random removal of AIRs was switched on. The sampling was increased to 10 000/400/400 for the it0/it1/water stages, respectively. Because of the high charge of DNA, the value for the dielectric constant was set to 78. FCC clustering was used with 0.75 cut‐off and a minimum cluster size of 4.
Details on the data used to drive the docking for both protein‐protein and protein‐DNA systems are given in Tables S1 and S2 of the Supporting Information in Data S1.
2.4. HADDOCK scoring
The HADDOCK score was used to select the top pose of the top two cluster for the post‐docking sampling stage and also to score the models resulting from that post‐docking sampling. The HADDOCK scoring function (HS)17, 30 is a linear weighted sum of energetic terms:
where Evdw, Eelec, Edesolv, and EAIR stand for van der Waals, Coulomb electrostatics, desolvation and restraint energies, respectively. The non‐bonded components of the score (Evdw, Eelec) were calculated with the OPLS forcefield,31 the desolvation energy is a solvent accessible surface area‐dependent empirical term32 which estimates the energetic gain or penalty of burying specific sidechains upon complex formation. The restraint energy term was only used for ranking the docking poses but was excluded for the scoring of the post‐docking sampling models.
2.5. Quality assessment
The quality of the modeled complexes was assessed based on the well‐established CAPRI criteria33:
Interface RMSD (i‐RMSD): Backbone RMSD of the interface residues, defined as all residues having at least one atom within 10 å of an atom on the other partner
Ligand RMSD (l‐RMSD): Backbone RMSD of the ligand (smaller partner in the complex) after fitting on the backbone of the receptor
Fraction of native contacts (fnat): The number of correct residue‐residue contacts in the predicted complex divided by the number of residue‐residue contacts in the target complex. Two residues are defined to be in contact if they have any atom within 5 å distance to each other.
Using i‐RMSD, l‐RMSD and fnat, the quality of models is defined as:
High (three‐stars): fnat ≥0.5 with l‐RMSD ≤1 å or i‐RMSD ≤1 å
Medium (two‐stars): fnat ≥0.3 with 1 å < l‐RMSD ≤5 å or 1 å < i‐RMSD ≤2 å
Acceptable (one‐star): fnat ≥0.1 with 5 å < l‐RMSD ≤10 å or 2 å < i‐RMSD ≤4 å
Incorrect (zero‐star): fnat <0.1
2.6. General workflow
The modeling pipeline combines conformational selection and induced‐fit mechanisms using conformational sampling prior and after complexation, respectively. Pre‐sampling was applied to the receptor in the case of protein‐protein docking, and to the DNA for the protein‐DNA dataset. The pipeline consists of the following steps (also shown in Figure 2):
The initial structure is energetically minimized and subjected to elastic network modeling to define the parameters for ClustENM. The eigenvalue spectrum is extracted to decide on the number of modes (“m”) to be used. The deformation test is applied to specify the deformation RMSD (“d”) in ClustENM.
Conformers are generated using ClustENM with the parameters defined in Step 1. For the proteins in the FMD dataset the number of generations is set to two for each system. For the DNA structures in the protein‐DNA dataset we increased the number of generations to four since DNA conformational sampling is computationally much cheaper due to the DNA's smaller size. During the generation, clusters containing the parent structures are discarded.
A maximum of 20 conformations are selected based on the minimization energy, to be used in ensemble docking. The initial structure is included in the ensemble regardless of its energy value.
Ensemble semi‐flexible docking is performed in HADDOCK.
The best scoring docked model from the best two scoring clusters are selected for the second round of ClustENM (post‐sampling).
Parameters for ClustENM are determined for each structure (same as in Step 1)
ClustENM is applied to each selected docked complex for two generations using parameters from Step 6 resulting in the final models.
The final models are scored using HADDOCK scoring and their quality is assessed.
Figure 2.
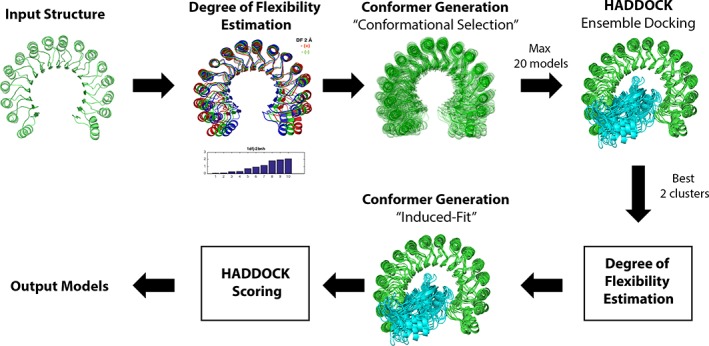
Pipeline for ClustENM‐HADDOCK [Color figure can be viewed at http://wileyonlinelibrary.com]
3. RESULTS AND DISCUSSION
3.1. Sampling prior to docking
3.1.1. Protein‐protein cases from the FMD benchmark
Starting from the unbound state of the receptors in the FMD benchmark, two generations of ClustENM sampling were applied prior to docking. The RMSDs (closest and range) of the receptor conformers to the reference bound state are given in Table 3 and the resulting ClustENM ensembles are shown in Figure 3. The RMSD range for the ensemble reflects the blind sampling of ClustENM: Some of the sampled conformations get closer to the bound state and others move further away from it. The number of conformers at the end of two generations of sampling is not so large thanks to the clustering steps in ClustENM. Two generations of ClustENM is clearly not sufficient to reach bound‐like conformations, especially for the challenging cases like 1IRA and 1H1V, where the RMSD of the closest structure to the bound state is almost the same as the unbound state (less than 1 å deviation from unbound). Figure 3 shows the generated ensemble for each case, which also reveals the expected‐to‐fail cases of 1IRA, 1H1V, 1Y64, 1FAK where the large domain motions could not be sampled in two generations. The remaining cases however do not reflect this issue.
Table 3.
RMSD values from the bound conformation for the receptors in FMD benchmark
| Complex ID | Receptor RMSD [å] unbound | Receptor RMSD [å] closest generated | Receptor RMSD [å] range in ensemble | Number of conformers inensemble |
|---|---|---|---|---|
| 1IRA | 19.6 | 19.1 | 19.1‐20.4 | 8 |
| 1H1V | 13.8 | 13.2 | 13.2‐14.0 | 11 |
| 1Y64 | 10.3 | 6.98 | 6.98‐17.0 | 16 |
| 1F6M | 7.32 | 6.40 | 6.40‐10.0 | 17 |
| 1FAK | 6.33 | 5.57 | 5.57‐7.58 | 16 |
| 1ZLI | 4.04 | 3.73 | 3.73‐5.06 | 15 |
| 1E4K | 2.91 | 2.85 | 2.85‐3.53 | 6 |
| 1IBR | 2.96 | 1.55 | 1.55‐6.26 | 20 |
| 1KKL | 2.64 | 2.72 | 2.72‐3.34 | 8 |
| 1NPE | 1.91 | 1.52 | 1.52‐4.86 | 22 |
| 1DFJ | 1.55 | 1.09 | 1.09‐4.61 | 20 |
Figure 3.
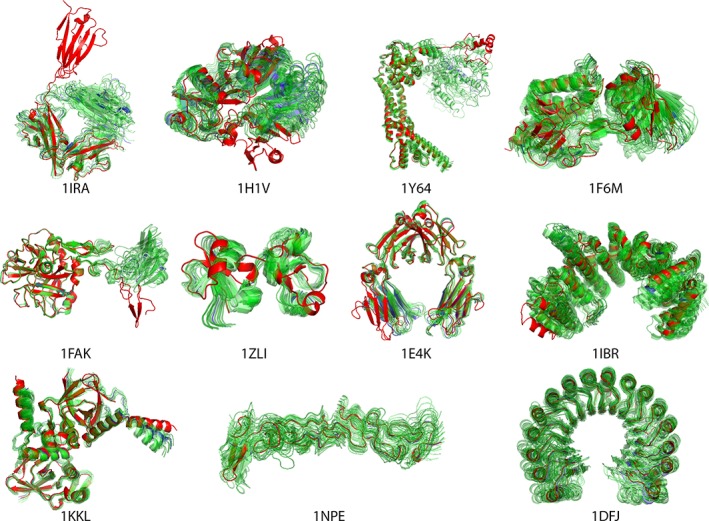
Ensembles of receptor conformers generated by ClustENM for protein‐protein docking. The starting structure is shown in blue, the bound conformation in red and the ClustENM generated conformers in green [Color figure can be viewed at http://wileyonlinelibrary.com]
3.1.2. Protein‐DNA dataset
A standard B‐DNA conformation built using our 3D‐DART webserver34 was the starting conformation for all protein‐DNA cases. We applied four generations of ClustENM on the DNA structures. The RMSD ranges of the resulting ensemble are given in Table 4. As in the case of the protein FMD benchmark, both close‐to‐bound and further‐away DNA conformations were sampled. Figure 4 shows the sampled DNA conformers for each case.
Table 4.
RMSD values from the bound conformation for the DNA structures in the protein‐DNA dataset
| Complex ID | RMSD [å]B‐DNA | RMSD [å]Closest generated | RMSD range [å]Ensemble | Number of conformers inensemble |
|---|---|---|---|---|
| 1BY4 | 1.63 | 1.71 | 1.63‐4.01 | 20 |
| 3CRO | 2.66 | 1.69 | 1.79‐4.06 | 20 |
| 1AZP | 3.95 | 2.90 | 2.90‐4.12 | 20 |
| 1JJ4 | 3.66 | 2.00 | 2.00‐7.80 | 20 |
| 1A74 | 8.34 | 4.98 | 4.98‐9.18 | 20 |
| 1ZME | 4.77 | 2.58 | 2.58‐5.82 | 20 |
Figure 4.
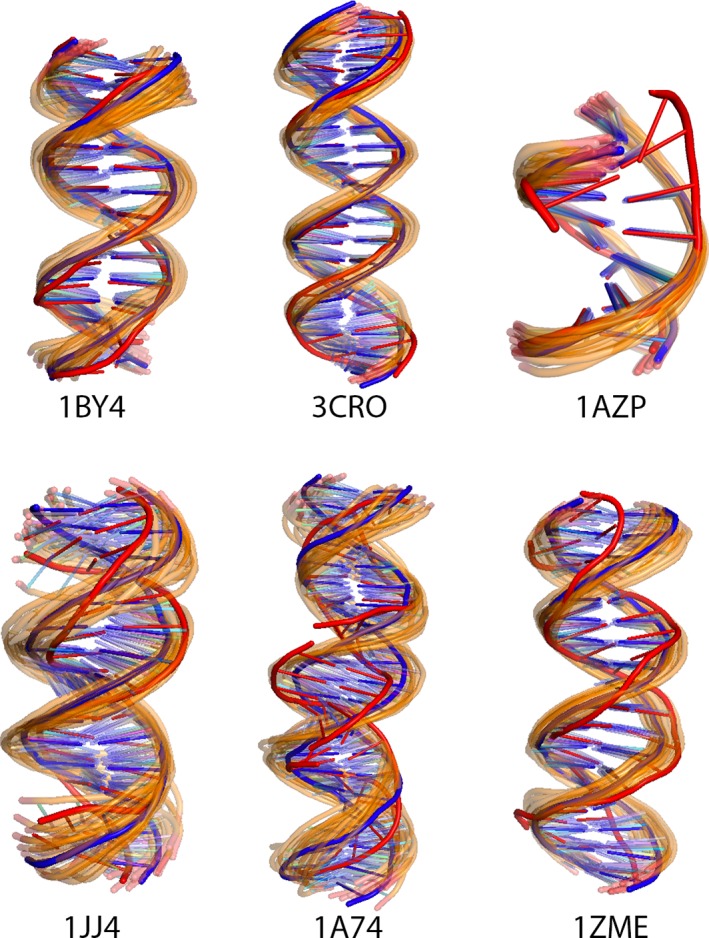
Ensembles of DNA conformers generated by ClustENM for protein‐DNA dockings. Starting structures are shown in blue, bound structure in red and the generated conformers in orange [Color figure can be viewed at http://wileyonlinelibrary.com]
3.2. Docking
After the pre‐docking conformational sampling, the resulting ensembles of conformers were used for ensemble docking with HADDOCK following the settings specified in Materials and Methods. This docking phase corresponds to the “conformational selection” part of our pipeline (with some induced fit as well since the flexible stage of HADDOCK allows for limited conformational changes). In this stage, ideally, HADDOCK should select the best conformers in its docking workflow from rigid body to final flexible refinement in explicit solvent. The best scoring docking pose from the top 2 ranked clusters were subsequently selected for the post‐docking induced‐fit part of the pipeline. The selected poses and their i‐RMSD, l‐RMSD, fnat and CAPRI quality metrics are shown in Figures 5 and 6 for the FMD and protein‐DNA docking, respectively.
Figure 5.
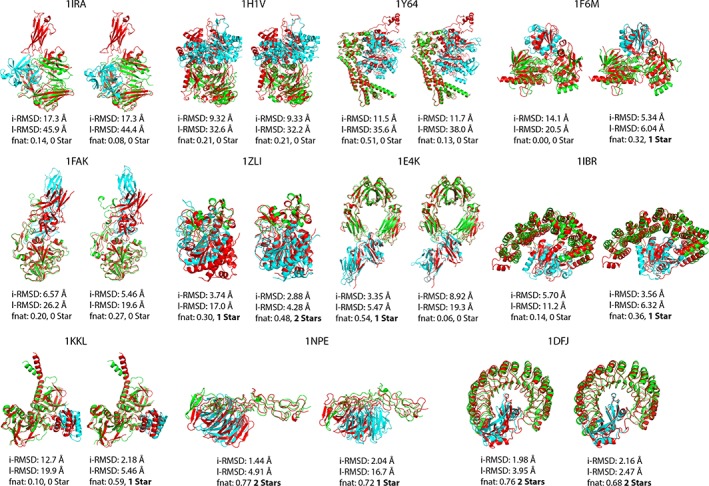
Protein‐protein docking top pose from the best two clusters of HADDOCK (green receptor/blue ligand) superimposed onto bound structure (red) [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 6.
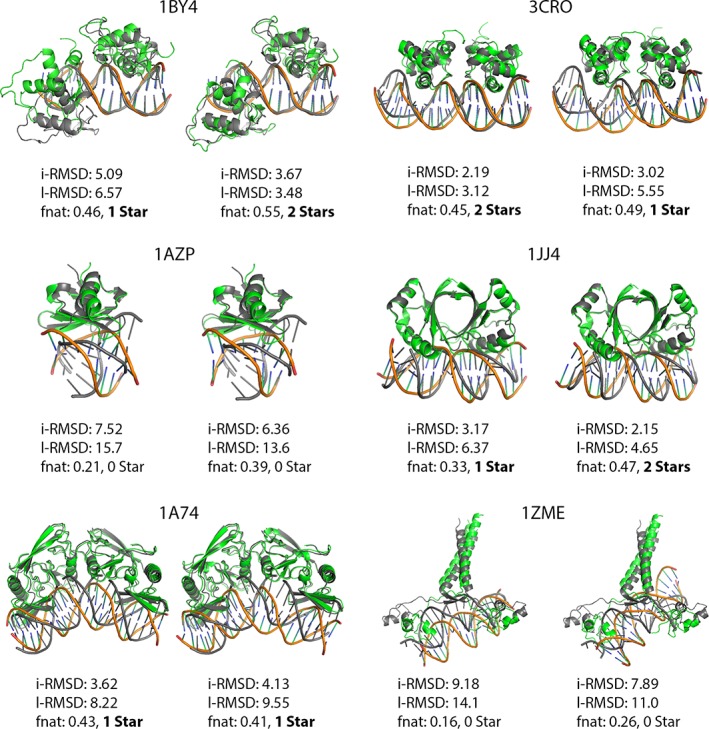
Protein‐DNA docking top pose from the best two clusters of HADDOCK (green receptor/orange DNA) superimposed on bound structure (gray) [Color figure can be viewed at http://wileyonlinelibrary.com]
In the FMD dataset, we were able to obtain models of at least acceptable quality in 7/11 cases. The failed cases are, as expected, 1IRA, 1H1V, 1Y64, and 1FAK, which all undergo large domain motions. As already explained in the “Sampling prior docking” section, the conformational sampling using only two generations was clearly not sufficient to model such large conformational change in these cases.
As for the protein‐DNA complexes, HADDOCK was able to obtain at least acceptable quality structures in 4/6 cases as shown in Figure 6. The binding mode in 1AZP is reverted (180° rotation), which already nullifies its chance to be a successful case at the end of the pipeline. In this particular case, however, near native poses were generated during docking but did not end up in the top 2 clusters.
3.3. Post‐docking sampling
We applied a second round of ClustENM to the best ranking model of the top 2 ranked clusters obtained from docking to perform the “induced‐fit” stage of our pipeline. The resulting models were ranked using the HADDOCK score calculated after a short energy minimization of each model.17 The results for protein‐protein and protein‐DNA systems are presented in the following sections.
3.3.1. Protein‐protein cases from the FMD benchmark
The quality of the starting and final conformers is summarized in Table 5, together with the rank of the best generated conformer. Additionally, we show the change in i‐RMSD, l‐RMSD and fnat through the “generations” of post‐sampling ClustENM in Figures S1–S3 in Data S1, respectively. We were able to obtain acceptable/medium quality (1‐2‐star) poses ranked within the top 20 in 7/11 cases, 4 of which were ranked as top 1. Unsurprisingly, the failed cases are no other than 1IRA, 1H1V, 1Y64 and 1FAK. Figures S1–S3 in Data S1 indicate that the quality of the starting docked model (which is the result of pre‐sampling followed by docking) is the limiting factor for the final complex structures. Indeed the “induced‐fit” sampling phase does not dramatically change the quality of the final conformers for the FMD benchmark. Conformational sampling prior to docking is thus contributing more to the quality of the final complexes than post‐sampling in the case of the protein‐protein FMD benchmark. The final complex structures are shown in Figure 7.
Table 5.
Quality and rank of the sampled conformers after the docking—FMD benchmark
| Quality—Number of conformers | Quality/Rank | |||
|---|---|---|---|---|
| Complex ID | Starting docked models | All ‐post‐docking ClustENM generated complex models | Best conformer | First acceptable or better model |
| 1IRA | 0*,0**,0***—2 | 0*,0**,0***—58 | — | — |
| 1H1V | 0*,0**,0***—2 | 0*,0**,0***—17 | — | — |
| 1Y64 | 0*,0**,0***—2 | 0*,0**,0***—31 | — | — |
| 1F6M | 1*,0**,0***—2 | 22*,0**,0***—72 | */15 | */15 |
| 1FAK | 0*,0**,0***—2 | 0*,0**,0***—30 | ‐ | ‐ |
| 1ZLI | 1*,1**,0***—2 | 20*,7**,0***—30 | **/1 | **/1 |
| 1E4K | 1*,0**,0***—2 | 13*,0**,0***—28 | */1 | */1 |
| 1IBR | 1*,0**,0***—2 | 27*,0**,0***—63 | */1 | */1 |
| 1KKL | 0*,1**,0***—2 | 12*,3**,0***—30 | **/20 | */16 |
| 1NPE | 1*,1*,0***—2 | 17*,17**,0***—34 | **/15 | */1 |
| 1DFJ | 0*,2**,0***—2 | 0*,18**,0***—18 | **/1 | **/1 |
Figure 7.
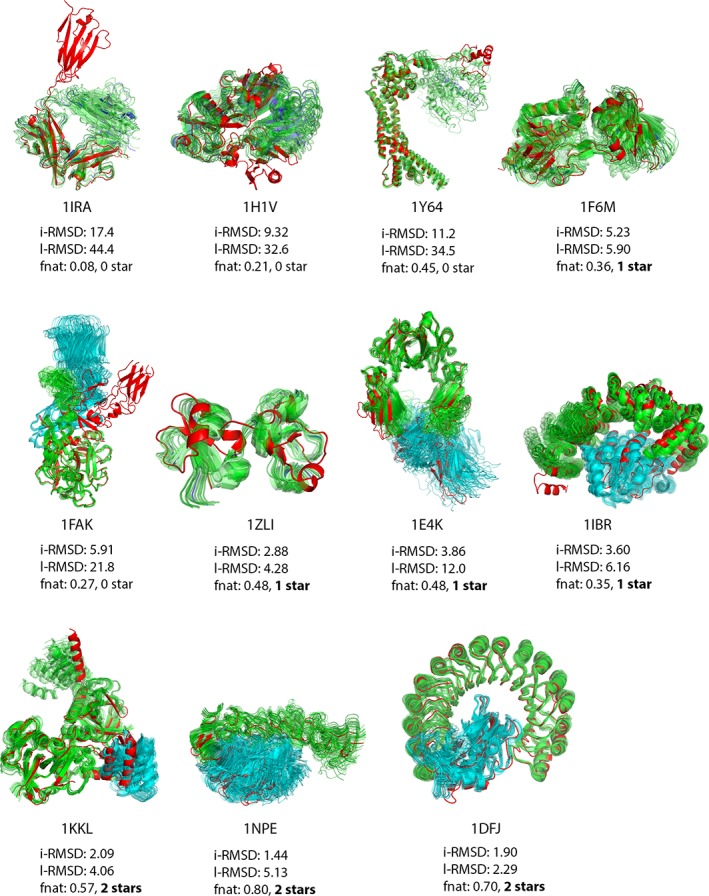
Final protein‐protein complexes for the protein‐protein FMD benchmark. The receptor is shown in green, the ligand in blue and the reference bound structure in red. The quality of the best conformer (listed in Table 5) together with its i‐RMSD [å], l‐RMSD [å] and fnat is given for each protein‐protein complex. In cases where no acceptable or better model is present, the quality of best scoring complex is given [Color figure can be viewed at http://wileyonlinelibrary.com]
3.3.2. Protein‐DNA cases
Table 6 shows the quality of the starting and final conformers with the rank of the best conformer for the protein‐DNA dataset. The change in i‐RMSD, l‐RMSD and fnat through the ClustENM generations are shown in Figures S4–S6 in Data S1, respectively. We obtained 1‐2 star poses for 5 out of 6 cases, 3 of which were ranked as top 1 and one as top 2 and the last one as 21st. As expected, there is no successful conformation generated for 1AZP.
Table 6.
Quality and rank of the sampled conformers after the docking—protein‐DNA dataset
| Quality—Number of conformers | Quality/Rank | |||
|---|---|---|---|---|
| Complex ID | Starting docked models | All post‐docking ClusENM generated complex models | Best conformer | First acceptable or better model |
| 1BY4 | 1*, 1**, 0***—2 | 59*,15**,0***—75 | **/1 | **/1 |
| 3CRO | 1*,1**,0***—2 | 84*,74**,0***—158 | **/1 | **/1 |
| 1AZP | 0*,0**,0***—2 | 0*,0**,0***—174 | — | — |
| 1JJ4 | 1*,1**,0***—2 | 235*,52**,0***—287 | **/2 | */1 |
| 1A74 | 2*,0**,0***—2 | 34*,0**,0***—53 | */1 | */1 |
| 1ZME | 0*,0**,0***—2 | 5*,0**,0***—91 | */21 | */21 |
Different from the protein‐protein FMD benchmark, sampling after docking for induced‐fit increases the quality of the models for the protein‐DNA cases as shown in Figures S4–S6 in Data S1. For example, in the case of 1ZME, we were able to obtain 1‐star quality structures by starting from zero quality docking poses. Nevertheless, the quality of the starting structures has again the most impact on the quality of the final conformers (shown in Figure 8).
Figure 8.
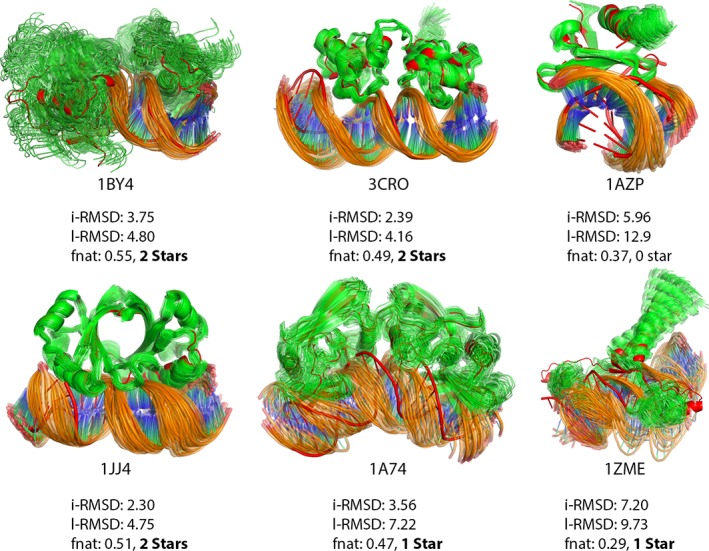
Final ClustENM‐HADDOCK models for protein‐DNA complexes (green protein/orange DNA), aligned on bound structure (red). The quality of the best conformer (listed in Table 6) together with its i‐RMSD [å], l‐RMSD [å] and fnat is given for each protein‐DNA complex
3.4. Comparison with previous studies
3.4.1. FMD benchmark: ClustENM‐HADDOCK vs Multidomain Flexible docking and Two‐Body Docking
We compared the performance of our pipeline with that of the multidomain flexible docking and two‐body docking approaches reported in the study of Karaca and Bonvin20 (Table 7). For multidomain flexible docking, the possible hinge regions were predicted using HingeProt35; the proteins were cut at those hinges and connectivity restraints were defined between the domains, allowing rigid‐body motion conformational changes during docking. The two‐body docking was only allowing for limited conformational changes in the interface. The results indicate that the ClustENM‐HADDOCK approach performs much better than two‐body docking but worse than the multidomain flexible docking approach of Karaca and Bonvin.20 The latter is especially advantageous to effectively incorporate rigid‐body hinge motions compared to our elastic network‐based conformational sampling, where two generations of ClustENM are clearly insufficient for the systems undergoing extremely large conformational changes (e.g. 1IRA, 1H1V, 1Y64). This might however depend on the type of motions since, in a previous study,11 for the adenylate kinase system undergoing conformational change of 7 å for example, we were able to observe full closing and opening of the domains when we applied ClustENM for 5 generations.
Table 7.
Comparison of ClustENM‐HADDOCK with previous protocols for FMD benchmark
| ClustENM‐HADDOCK | Flexible Multidomain Docking results20 | 2‐Body Docking results20 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Complex | Quality/Rank | i‐RMSD [å] | Fnat | Quality/Rank | i‐RMSD [å] | Fnat | Quality/Rank | i‐RMSD [å] | Fnat |
| 1IRA | — | 17.5 | 0.08 | */1 | 3.9 | 0.55 | — | 17.5 | 0.04 |
| 1H1V | — | 9.3 | 0.21 | */11 | 4.6 | 0.49 | — | 11.9 | 0.08 |
| 1Y64 | — | 11.1 | 0.45 | */5 | 3.9 | 0.48 | — | 10.3 | 0.07 |
| 1F6M | */15 | 5.2 | 0.36 | */1 | 3.5 | 0.69 | — | 14.1 | 0.00 |
| 1FAK | — | 5.9 | 0.27 | **/37 | 2.8 | 0.55 | — | 11.4 | 0.01 |
| 1ZLI | **/1 | 2.9 | 0.48 | **/1 | 2.1 | 0.74 | — | 14.8 | 0.02 |
| 1E4K | */1 | 3.9 | 0.48 | **/5 | 2.3 | 0.70 | */1 | 4.1 | 0.58 |
| 1IBR | */1 | 3.6 | 0.35 | **/1 | 2.3 | 0.63 | — | 9.6 | 0.11 |
| 1KKL | **/20 | 2.1 | 0.57 | **/1 | 2.2 | 0.67 | */1 | 3.1 | 0.56 |
| 1NPE | **/15 | 1.4 | 0.80 | **/16 | 1.2 | 0.95 | **/1 | 1.7 | 0.85 |
| 1DFJ | **/1 | 1.9 | 0.69 | **/5 | 2.0 | 0.68 | **/116 | 1.8 | 0.63 |
3.4.2. Protein‐DNA dataset: ClustENM‐HADDOCK vs unbound‐unbound docking using canonical B‐DNA models and unbound‐unbound docking using custom‐build B‐DNA models
We compared our protein‐DNA results with those reported by van Dijk and Bonvin21 to assess our performance against (i) one round of unbound docking starting from canonical B‐DNA models (“Unbound flex”) and (ii) two‐rounds of unbound docking where the second round uses an ensemble of custom‐built bent B‐DNA models obtained from an analysis of the conformations obtained during the first docking stage (“DNA lib”). The results in Table 8 indicate that ClustENM‐HADDOCK approach performs better than “Unbound flex” in 4/6 cases, the same in 1 case (1AZP) and worse in one case (1ZME) when the top 10 best scoring structures are considered. Generated acceptable quality solutions for 1ZME from ClustENM‐HADDOCK were not scored in the top 10 unfortunately (rank of first acceptable model is 21st). The “DNA lib” approach on the other hand was able to obtain at least acceptable quality models in all cases. One thing to note is that for both “Unbound flex” and “DNA lib” approaches, a multidomain flexible docking method (ie, cutting from the hinge(s) and using connectivity restraints) was used for the protein in the 1ZME case, which prevents a one‐to‐one comparison.
Table 8.
Comparison of the performance of ClustENM‐HADDOCK with the previous protocols by van Dijk and Bonvin21 for the protein‐DNA dataset
| ClustENM‐HADDOCK | Unbound flex21 | DNA lib21 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Complex | Quality | i‐RMSD [å] | Fnat | Quality | i‐RMSD [å] | Fnat | Quality | i‐RMSD [å] | Fnat |
| 1BY4 | 7*,3**,0*** | 4.55 (0.70) | 0.47 (0.05) | 5*,0**,0*** | 5.87 (1.71) | 0.17 (0.05) | 4*,3**,0*** | 4.91 (2.32) | 0.27 (0.09) |
| 3CRO | 8*,2**,0*** | 3.19 (0.60) | 0.50 (0.01) | 6*,2**,0*** | 3.29 (0.68) | 0.27 (0.07) | 3*,7**,0*** | 2.62 (0.73) | 0.40 (0.06) |
| 1AZP | 0*,0**,0*** | 6.25 (0.52) | 0.37 (0.06) | 0*,0**,0*** | 6.68 (2.26) | 0.04 (0.04) | 5*,0**,0*** | 4.00 (0.45) | 0.10 (0.04) |
| 1JJ4 | 5*,5**,0*** | 2.39 (0.18) | 0.51 (0.03) | 6*,0**,0*** | 4.55 (0.58) | 0.16 (0.07) | 9*,1**,0*** | 3.62 (0.38) | 0.21 (0.07) |
| 1A74 | 10*,0**,0*** | 3.81 (0.40) | 0.45 (0.02) | 8*,0**,0*** | 6.30 (0.46) | 0.14 (0.04) | 9*,1**,0*** | 3.37 (0.37) | 0.24 (0.05) |
| 1ZME | 0*,0**,0*** | 8.08 (0.70) | 0.50 (0.01) | 4*,0**,0*** | 5.29 (0.59) | 0.12 (0.06) | 8*,0**,0*** | 4.63 (0.80) | 0.15 (0.04) |
Note: Values between parenthesis correspond to standard deviations.
Based on the quality of the modeled complexes generated by our ClustENM‐HADDOCK approach, our pipeline offers a nice alternative for protein‐DNA docking cases.
4. CONCLUSIONS
We applied our ClustENM‐HADDOCK pre‐ and post‐sampling method to protein‐protein and protein‐DNA complexes to incorporate dynamics and simulate conformational selection and induced‐fit effects upon binding. The method does not use any prior knowledge on the direction and extent of the conformational changes. The extent of the applied deformation is estimated from its energetical costs, inspired from mechanical tensile testing on material. Our pre‐sampling/docking/post‐sampling approach was able to produce acceptable to medium quality models in 7 out of 11 cases for the protein‐protein complexes and 5 out of 6 for the protein‐DNA complexes. We observed that the conformational selection (“pre‐sampling” and also docking) has the highest impact on the quality of the final models especially in the case of protein‐protein complexes. For protein‐DNA complexes, in contrast, the induced‐fit stage of the pipeline (post‐sampling) significantly improves the quality of the final models.
Comparison of our method with previous studies showed that the previously proposed flexible multidomain docking remains the best performing approach, provided that reliable hinge region information is available. In the absence of such information, our approach could however be a good alternative to incorporate flexibility. As for the protein‐DNA cases, our ClustENM‐HADDOCK approach does perform better than standard flexible docking and shows a comparable performance with a two‐stage docking approach in which a library of pre‐bent DNA conformation is used in a second docking stage. This makes ClustENM‐HADDOCK a suitable alternative for modeling conformational changes in protein‐DNA binding.
We also observed that 2‐generations of ClustENM for pre‐sampling was not sufficient in the cases of large conformational changes (eg, 1IRA, 1H1V, 1Y64). For these cases, the number of ClustENM generations could be increased further to obtain large domain motions. Additionally, here we considered only the best two scoring clusters from the docking as starting points for post‐sampling. Instead of two, we could select more clusters from docking and apply post‐sampling. This would have increased our chance of success for the 1AZP case for example (although near native poses were generated during the docking, these did not end up in the top 2 clusters).
Supporting information
Data S1. Supporting Information
ACKNOWLEDGMENTS
We would like to thank Dr. Rodrigo Vargas Honorato for his help with the scoring of protein‐DNA post‐sampling models. This work was supported by European H2020 e‐Infrastructure grants (INDIGO‐DataCloud, grant no. 653549 and BioExcel grants no. 675728 and 823830).
Kurkcuoglu Z, Bonvin AMJJ. Pre‐ and post‐docking sampling of conformational changes using ClustENM and HADDOCK for protein‐protein and protein‐DNA systems. Proteins. 2020;88:292–306. 10.1002/prot.25802
Funding information BioExcel, Grant/Award Numbers: 823830, 675728; INDIGO‐DataCloud, Grant/Award Number: 653549; European H2020 e‐Infrastructure
Contributor Information
Zeynep Kurkcuoglu, Email: zeynepkurkcuoglu@gmail.com.
Alexandre M. J. J. Bonvin, Email: a.m.j.j.bonvin@uu.nl.
DATA ACCESSIBILITY
The information (AIRs) used to drive the docking together with i‐RMSD, l‐RMSD and fnat figures for the protein‐protein, protein‐DNA systems in post‐sampling are available as supplementary material (Data S1). All models generated for the various complexes together with their statistics are available for download from the SBGrid data repository36 (https://data.sbgrid.org/labs/32/, https://doi.org/10.15785/SBGRID/707).
REFERENCES
- 1. Palamini M, Canciani A, Forneris F. Identifying and visualizing macromolecular flexibility in structural biology. Front Mol Biosci. 2016;3:47 10.3389/fmolb.2016.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Karaca E, Bonvin AMJJ. Advances in integrative modeling of biomolecular complexes. Methods. 2013;59(3):372‐381. 10.1016/j.ymeth.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 3. Rodrigues JPGLM, Bonvin AMJJ. Integrative computational modeling of protein interactions. FEBS J. 2014;281(8):1988‐2003. 10.1111/febs.12771. [DOI] [PubMed] [Google Scholar]
- 4. Johansson KE, Lindorff‐Larsen K. Structural heterogeneity and dynamics in protein evolution and design. Curr Opin Struct Biol. 2018;48:157‐163. 10.1016/j.sbi.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 5. Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5(11):789‐796. 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Michel D. Conformational selection or induced fit? New insights from old principles. Biochimie. 2016;128–129:48‐54. 10.1016/j.biochi.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 7. Haliloglu T, Bahar I. Adaptability of protein structures to enable functional interactions and evolutionary implications. Curr Opin Struct Biol. 2015;35:17‐23. 10.1016/j.sbi.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Csermely P, Palotai R, Nussinov R. Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem Sci. 2010;35(10):539‐546. 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu F, Chu X, Lu HP, Wang J. Molecular mechanism of multispecific recognition of Calmodulin through conformational changes. Proc Natl Acad Sci USA. 2017;114(20):E3927‐E3934. 10.1073/pnas.1615949114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kuroda D, Gray JJ. Pushing the backbone in protein‐protein docking. Structure. 2016;24(10):1821‐1829. 10.1016/j.str.2016.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kurkcuoglu Z, Bahar I, Doruker P. ClustENM: ENM‐based sampling of essential conformational space at full atomic resolution. J Chem Theory Comput. 2016;12(9):4549‐4562. 10.1021/acs.jctc.6b00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kurkcuoglu Z, Doruker P. Ligand docking to intermediate and close‐to‐bound conformers generated by an elastic network model based algorithm for highly flexible proteins. PLoS One. 2016;11(6):e0158063 10.1371/journal.pone.0158063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Can MT, Kurkcuoglu Z, Ezeroglu G, Uyar A, Kurkcuoglu O, Doruker P. Conformational dynamics of bacterial trigger factor in apo and ribosome‐bound states. PLoS One. 2017;12(4):e0176262. 10.1371/journal.pone.0176262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dominguez C, Boelens R, Bonvin AMJJ. HADDOCK: a protein−protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731‐1737. 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 15. De Vries SJ, van Dijk M, Bonvin AMJJ. The HADDOCK web server for data‐driven biomolecular docking. Nat Protoc. 2010;5:883‐897. [DOI] [PubMed] [Google Scholar]
- 16. van GCP Z, JPGLM R, Trellet M, et al. The HADDOCK2.2 web server: user‐friendly integrative modeling of biomolecular complexes. J Mol Biol. 2016;428(4):720‐725. 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 17. Vangone A, Rodrigues JPGLM, Xue LC, et al. Sense and simplicity in HADDOCK scoring: lessons from CASP‐CAPRI round 1. Proteins. 2017;85(3):417‐423. 10.1002/prot.25198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lensink MF, Velankar S, Kryshtafovych A, et al. Prediction of homoprotein and heteroprotein complexes by protein docking and template‐based modeling: a CASP‐CAPRI experiment. Proteins Struct Funct Bioinforma. 2016;84(S1):323‐348. 10.1002/prot.25007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moreira IS, Fernandes PA, Ramos MJ. Protein‐protein docking dealing with the unknown. J Comput Chem. 2010;31(2):317‐342. 10.1002/jcc.21276. [DOI] [PubMed] [Google Scholar]
- 20. Karaca E, Bonvin AMJJ. A multidomain flexible docking approach to Deal with large conformational changes in the modeling of biomolecular complexes. Structure. 2011;19(4):555‐565. 10.1016/j.str.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 21. van Dijk M, Bonvin AMJJ. Pushing the limits of what is achievable in protein–DNA docking: benchmarking HADDOCK's performance. Nucleic Acids Res. 2010;38(17):5634‐5647. 10.1093/nar/gkq222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kurkcuoglu O, Turgut OT, Cansu S, Jernigan RL, Doruker P. Focused functional dynamics of supramolecules by use of a mixed‐resolution elastic network model. Biophys J. 2009;97:1178‐1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Phillips JC, Braun R, Wang W, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781‐1802. 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. MacKerell AD, Bashford D, Bellott M, et al. All‐atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102(18):3586‐3616. 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 25. Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11(8):3696‐3713. 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hawkins GD, Cramer CJ, Truhlar DG. Pairwise solute descreening of solute charges from a dielectric medium. Chem Phys Lett. 1995;246:122‐129. [Google Scholar]
- 27. Hawkins GD, Cramer CJ, Truhlar DG. Parametrized models of aqueous free energies of solvation based on pairwise descreening of solute atomic charges from a dielectric medium. J Phys Chem. 1996;100:19824‐19839. [Google Scholar]
- 28. Srinivasan J, Trevathan MW, Beroza P, Case DA. Application of a pairwise generalized born model to proteins and nucleic acids: inclusion of salt effects. Theor Chem Acc. 1999;101:426‐434. [Google Scholar]
- 29. Rodrigues JPGLM, Trellet M, Schmitz C, et al. Clustering biomolecular complexes by residue contacts similarity. Proteins Struct Funct Bioinforma. 2012;80(7):1810‐1817. 10.1002/prot.24078. [DOI] [PubMed] [Google Scholar]
- 30. Vangone A, Rodrigues JPGLM, Xue LC, et al. Sense and simplicity in HADDOCK scoring: lessons from CASP‐CAPRI (page 418). Proteins. 2017;85(8):1589‐1590. 10.1002/prot.25339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jorgensen WL, Tirado‐Rives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc. 1988;110(6):1657‐1666. 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 32. Fernandez‐Recio J, Totrov M, Abagyan R. Identification of protein‐protein interaction sites from docking energy landscapes. J Mol Biol. 2004;335(3):843‐865. [DOI] [PubMed] [Google Scholar]
- 33. Mendez R, Leplae R, De Maria L, Wodak SJ. Assessment of blind predictions of protein‐protein interactions: current status of docking methods. Proteins. 2003;52(1):51‐67. 10.1002/prot.10393. [DOI] [PubMed] [Google Scholar]
- 34. van Dijk M, Bonvin AMJJ. 3D‐DART: a DNA structure modelling server. Nucleic Acids Res. 2009;37(Web Server issue:W235‐9 10.1093/nar/gkp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Emekli U, Schneidman‐Duhovny D, Wolfson HJ, Nussinov R, Haliloglu T. HingeProt: automated prediction of hinges in protein structures. Proteins. 2008;70(4):1219‐1227. 10.1002/prot.21613. [DOI] [PubMed] [Google Scholar]
- 36. Meyer PA, Socias S, Key J, et al. Data publication with the structural biology data grid supports live analysis. Nat Commun. 2016;7:10882 10.1038/ncomms10882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schreuder H, Tardif C, Trump‐Kallmeyer S, et al. A new cytokine‐receptor binding mode revealed by the crystal structure of the IL‐1 receptor with an antagonist. Nature. 1997;386:194‐200. 10.2210/PDB1IRA/PDB. [DOI] [PubMed] [Google Scholar]
- 38. Vigers GP, Dripps DJ, Edwards CK III, Brandhuber BJ. X‐ray crystal structure of a small antagonist peptide bound to interleukin‐1 receptor type 1. JBiolChem. 2000;275:36927‐36933. 10.2210/PDB1G0Y/PDB. [DOI] [PubMed] [Google Scholar]
- 39. Schreuder HA, Rondeau JM, Tardif C, et al. Refined crystal structure of the interleukin‐1 receptor antagonist. Presence of a disulfide link and a cis‐proline. EurJBiochem. 1995;227:838‐847. 10.2210/PDB1ILR/PDB. [DOI] [PubMed] [Google Scholar]
- 40. Choe H, Burtnick LD, Mejillano M, Yin HL, Robinson RC, Choe S. The calcium activation of gelsolin: insights from the 3A structure of the G4‐G6/Actin complex. J Mol Biol. 2002;324:691 10.2210/PDB1H1V/PDB. [DOI] [PubMed] [Google Scholar]
- 41. Burtnick LD, Koepf EK, Grimes J, et al. The crystal structure of plasma gelsolin: implications for Actin severing, capping, and nucleation. Cell. 1997;90:661‐670. 10.2210/PDB1D0N/PDB. [DOI] [PubMed] [Google Scholar]
- 42. Bubb MR, Govindasamy L, Yarmola EG, et al. Polylysine induces an antiparallel Actin dimer that nucleates filament assembly: crystal structure at 3.5‐a resolution. J Biol Chem. 2002;277:20999‐21006. 10.2210/PDB1IJJ/PDB. [DOI] [PubMed] [Google Scholar]
- 43. Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of Actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488‐494. 10.2210/PDB1Y64/PDB. [DOI] [PubMed] [Google Scholar]
- 44. Xu Y, Moseley JB, Sagot I, et al. Crystal structures of a formin homology‐2 domain reveal a tethered dimer architecture. Cell. 2004;116:711‐723. 10.2210/PDB1UX5/PDB. [DOI] [PubMed] [Google Scholar]
- 45. Rizvi SA, Tereshko V, Kossiakoff AA, Kozmin SA. Structure of bistramide a‐Actin complex at a 1.35 a resolution. J Am Chem Soc. 2006;128:3882‐3883. 10.2210/PDB2FXU/PDB. [DOI] [PubMed] [Google Scholar]
- 46. Lennon BW, Williams CH Jr, Ludwig ML. Twists in catalysis: alternating conformations of Escherichia coli thioredoxin reductase. Science (80‐ ). 2000;289:1190‐1194. 10.2210/PDB1F6M/PDB. [DOI] [PubMed] [Google Scholar]
- 47. Lennon BW, Williams CH Jr, Ludwig ML. Crystal structure of reduced thioredoxin reductase from Escherichia coli: structural flexibility in the isoalloxazine ring of the flavin adenine dinucleotide cofactor. Protein Sci. 1999;8:2366‐2379. 10.2210/PDB1CL0/PDB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nikkola M, Gleason FK, Fuchs JA, Eklund H. Crystal structure analysis of a mutant Escherichia coli thioredoxin in which lysine 36 is replaced by glutamic acid. Biochemistry. 1993;32:5093‐5098. 10.2210/PDB2TIR/PDB. [DOI] [PubMed] [Google Scholar]
- 49. Zhang E, St Charles R, Tulinsky A. Structure of extracellular tissue factor complexed with factor VIIa inhibited with a BPTI mutant. J Mol Biol. 1999;285:2089‐2104. 10.2210/PDB1FAK/PDB. [DOI] [PubMed] [Google Scholar]
- 50. Pike AC, Brzozowski AM, Roberts SM, Olsen OH, Persson E. Structure of human factor VIIa and its implications for the triggering of blood coagulation. Proc Natl Acad Sci USA. 1999;96:8925‐8930. 10.2210/PDB1QFK/PDB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huang M, Syed R, Stura EA, et al. The mechanism of an inhibitory antibody on TF‐initiated blood coagulation revealed by the crystal structures of human tissue factor, fab 5G9 and TF.5G9 complex. J Mol Biol. 1998;275:873‐894. 10.2210/PDB1TFH/PDB. [DOI] [PubMed] [Google Scholar]
- 52. Arolas JL, Popowicz GM, Lorenzo J, et al. The three‐dimensional structures of tick Carboxypeptidase inhibitor in complex with a/B Carboxypeptidases reveal a novel double‐headed binding mode. J Mol Biol. 2005;350:489‐498. 10.2210/PDB1ZLI/PDB. [DOI] [PubMed] [Google Scholar]
- 53. Pantoja‐Uceda D, Arolas JL, Garcia P, et al. The NMR structure and dynamics of the two‐domain tick carboxypeptidase inhibitor reveal flexibility in its free form and stiffness upon binding to human carboxypeptidase B. Biochemistry. 2008;47:7066‐7078. 10.2210/PDB2JTO/PDB. [DOI] [PubMed] [Google Scholar]
- 54. Barbosa Pereira PJ, Segura‐Martin S, Oliva B, et al. Human procarboxypeptidase B: three‐dimensional structure and implications for thrombin‐activatable fibrinolysis inhibitor (TAFI). J Mol Biol. 2002;321:537‐547. 10.2210/PDB1KWM/PDB. [DOI] [PubMed] [Google Scholar]
- 55. Sondermann P, Huber R, Oosthuizen V, Jacob U. The 3.2A crystal structure of the human Igg1 fc fragment‐fc‐gamma‐Riii complex. Nature. 2000;406:267‐273. 10.2210/PDB1E4K/PDB. [DOI] [PubMed] [Google Scholar]
- 56. Matsumiya S, Yamaguchi Y, Saito J, et al. Corrigendum to " structural comparison of fucosylated and nonfucosylated Fc fragments of human immunoglobulin G1" [J. Mol. Biol. 386/3 (2007) 767–779]. J Mol Biol. 2011;408:1001‐1001. 10.2210/PDB3AVE/PDB. [DOI] [PubMed] [Google Scholar]
- 57. Zhang Y, Boesen CC, Radaev S, et al. Crystal structure of the extracellular domain of a human fc gamma RIII. Immunity. 2000;13:387‐395. 10.2210/PDB1FNL/PDB. [DOI] [PubMed] [Google Scholar]
- 58. Vetter IR, Arndt A, Kutay U, Gorlich D, Wittinghofer A. Structural view of the ran‐importin beta interaction at 2.3 A resolution. Cell. 1999;97:635‐646. 10.2210/PDB1IBR/PDB. [DOI] [PubMed] [Google Scholar]
- 59. Bayliss R, Littlewood T, Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin‐beta in nuclear trafficking. Cell. 2000;102:99‐108. 10.2210/PDB1F59/PDB. [DOI] [PubMed] [Google Scholar]
- 60. Kent HM, Moore MS, Quimby BB, et al. Engineered mutants in the switch II loop of Ran define the contribution made by key residues to the interaction with nuclear transport factor 2 (NTF2) and the role of this interaction in nuclear protein import. J Mol Biol. 1999;289:565‐577. 10.2210/PDB1QG4/PDB. [DOI] [PubMed] [Google Scholar]
- 61. Fieulaine S, Morera S, Poncet S, et al. X‐ray structure of a bifunctional protein kinase in complex with its protein substrate HPr. Proc Natl Acad Sci USA. 2002;99:13437‐13441. 10.2210/PDB1KKL/PDB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fieulaine S, Morera S, Poncet S, et al. X‐ray structure of HPr kinase: a bacterial protein kinase with a P‐loop nucleotide‐binding domain. EMBO J. 2001;20:3917‐3927. 10.2210/PDB1JB1/PDB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liao DI, Herzberg O. Refined structures of the active Ser83‐‐>Cys and impaired Ser46‐‐>Asp histidine‐containing phosphocarrier proteins. Structure. 1994;2:1203‐1216. 10.2210/PDB2HPR/PDB. [DOI] [PubMed] [Google Scholar]
- 64. Takagi J, Yang YT, Liu J‐H, Wang J‐H, Springer TA. Complex between nidogen and laminin fragments reveals a paradigmatic beta‐propeller interface. Nature. 2003;424:969‐974. 10.2210/PDB1NPE/PDB. [DOI] [PubMed] [Google Scholar]
- 65. Stetefeld J, Mayer U, Timpl R, Huber R. Crystal structure of three consecutive laminin‐type epidermal growth factor‐like (LE) modules of laminin gamma1 chain harboring the nidogen binding site. JMolBiol. 1996;257:644‐657. 10.2210/PDB1KLO/PDB. [DOI] [PubMed] [Google Scholar]
- 66. Kobe B, Deisenhofer J. A structural basis of the interactions between leucine‐rich repeats and protein ligands. Nature. 1995;374:183‐186. 10.2210/PDB1DFJ/PDB. [DOI] [PubMed] [Google Scholar]
- 67. Kobe B, Deisenhofer J. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease a. JMolBiol. 1996;264:1028‐1043. 10.2210/PDB2BNH/PDB. [DOI] [PubMed] [Google Scholar]
- 68. Nachman J, Miller M, Gilliland GL, Carty R, Pincus M, Wlodawer A. Crystal structure of two covalent nucleoside derivatives of ribonuclease a. Biochemistry. 1990;29:928‐937. 10.2210/PDB9RSA/PDB. [DOI] [PubMed] [Google Scholar]
- 69. Zhao Q, Chasse SA, Devarakonda S, Sierk ML, Ahvazi B, Rastinejad F. Structural basis of RXR‐DNA interactions. JMolBiol. 2000;296:509‐520. 10.2210/PDB1BY4/PDB. [DOI] [PubMed] [Google Scholar]
- 70. Holmbeck SM, Foster MP, Casimiro DR, Sem DS, Dyson HJ, Wright PE. High‐resolution solution structure of the retinoid X receptor DNA‐binding domain. JMolBiol. 1998;281:271‐284. 10.2210/PDB1RXR/PDB. [DOI] [PubMed] [Google Scholar]
- 71. Mondragon A, Harrison SC. The phage 434 Cro/OR1 complex at 2.5 A resolution. JMolBiol. 1991;219:321‐334. 10.2210/PDB3CRO/PDB. [DOI] [PubMed] [Google Scholar]
- 72. Padmanabhan S, Jimenez MA, Gonzalez C, Sanz JM, Gimenez‐Gallego G, Rico M. Three‐dimensional solution structure and stability of phage 434 Cro protein. Biochemistry. 1997;36:6424‐6436. 10.2210/PDB1ZUG/PDB. [DOI] [PubMed] [Google Scholar]
- 73. Robinson H, Gao YG, McCrary BS, Edmondson SP, Shriver JW, Wang AH. The hyperthermophile chromosomal protein Sac7d sharply kinks DNA. Nature. 1998;392:202‐205. 10.2210/PDB1AZP/PDB. [DOI] [PubMed] [Google Scholar]
- 74. Edmondson SP, Qiu L, Shriver JW. Solution structure of the DNA‐binding protein Sac7d from the hyperthermophile Sulfolobus acidocaldarius. Biochemistry. 1995;34:13289‐13304. 10.2210/PDB1SAP/PDB. [DOI] [PubMed] [Google Scholar]
- 75. Kim SS, Tam JK, Wang AF, Hegde RS. The structural basis of DNA target discrimination by papillomavirus E2 proteins. JBiolChem. 2000;275:31245‐31254. 10.2210/PDB1JJ4/PDB. [DOI] [PubMed] [Google Scholar]
- 76. Flick KE, Jurica MS, Monnat RJ Jr, Stoddard BL. DNA binding and cleavage by the nuclear intron‐encoded homing endonuclease I‐PpoI. Nature. 1998;394:96‐101. 10.2210/PDB1A74/PDB. [DOI] [PubMed] [Google Scholar]
- 77. Galburt EA, Chadsey MS, Jurica MS, et al. Conformational changes and cleavage by the homing endonuclease I‐PpoI: a critical role for a leucine residue in the active site. JMolBiol. 2000;300:877‐887. 10.2210/PDB1EVX/PDB. [DOI] [PubMed] [Google Scholar]
- 78. Swaminathan K, Flynn P, Reece RJ, Marmorstein R. Crystal structure of a PUT3‐DNA complex reveals a novel mechanism for DNA recognition by a protein containing a Zn2Cys6 binuclear cluster. NatStructMolBiol. 1997;4:751‐759. 10.2210/PDB1ZME/PDB. [DOI] [PubMed] [Google Scholar]
- 79. Walters KJ, Dayie KT, Reece RJ, Ptashne M, Wagner G. Structure and mobility of the PUT3 dimer. NatStructMolBiol. 1997;4:744‐750. 10.2210/PDB1AJY/PDB. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supporting Information
Data Availability Statement
The information (AIRs) used to drive the docking together with i‐RMSD, l‐RMSD and fnat figures for the protein‐protein, protein‐DNA systems in post‐sampling are available as supplementary material (Data S1). All models generated for the various complexes together with their statistics are available for download from the SBGrid data repository36 (https://data.sbgrid.org/labs/32/, https://doi.org/10.15785/SBGRID/707).


