Abstract
Background and Aim
The nuclear farnesoid X receptor (FXR) regulates critical pathways of hepatic metabolism, inflammation, and gut mucosal barrier. Thus, we investigated the association of FXR‐single nucleotide polymorphism (SNPs) with hepatic decompensation and liver‐related mortality in patients with advanced chronic liver disease.
Methods
Two FXR‐SNPs (rs56163822 G > T and rs35724 G > C) were genotyped in a cohort of 402 prospectively characterized patients with hepatic venous pressure gradient (HVPG) ≥ 6 mmHg.
Results
Only 19 patients (4.7%) harbored a rs56163822 T‐allele and had less pronounced liver disease as indicated by lower Child–Pugh score (CPS, 6 ± 1 vs 7 ± 2 points, P = 0.034) and higher albumin levels (38.9 ± 4.9 vs 35.9 ± 5.9 g/L, P = 0.026). In contrast, n = 267 (66.4%) patients harbored minor rs35724 allele (G/C or C/C) and had more advanced liver disease, as indicated by a higher model of end‐stage liver disease (11 ± 4 vs 10 ± 3, P = 0.016), while other baseline characteristics were similar across FXR‐SNP genotypes. In compensated CPS‐A patients, the rs35724 minor allele was independently protective for the development of ascites (adjusted hazard ratio [aHR] = 0.411, 95% confidence interval (95% CI): 0.191–0.885; P = 0.023) and tended to reduce the risk of hepatic decompensation (aHR = 0.625, 95% CI: 0.374–1.044, P = 0.072) in multivariate analyses. Of note, transplant‐free survival was longer in patients with rs35724 minor allele and HVPG ≥ 10 mmHg (at 5 years: 68.2% vs 55.8%, P = 0.047) and those with HVPG ≥ 16 mmHg (63.3% vs 44.0%, P = 0.021). After adjusting for established risk factors, the rs35724 minor allele was independently associated with reduced liver‐related mortality in the overall cohort (aHR = 0.658, 95% CI: 0.434–0.998, P = 0.049), in compensated CPS‐A patients (aHR = 0.488, 95% CI: 0.252–0.946, P = 0.034), in patients with HVPG ≥ 10 mmHg (aHR = 0.547, 95% CI: 0.346–0.864, P = 0.010), and in patients with HVPG ≥ 16 mmHg (aHR = 0.519, 95% CI: 0.307–0.878, P = 0.014).
Conclusion
The FXR‐SNP rs35724 was associated with a reduced risk for development of ascites and liver‐related mortality in patients with advanced chronic liver disease.
Keywords: advanced chronic liver disease, cirrhosis, polymorphism, rs35724, rs56163822
Introduction
The farnesoid X receptor (FXR) is a member of the nuclear receptor superfamily and main nuclear bile acid receptor that regulates the expression of key target genes in bile acid, lipid, and glucose metabolism.1, 2, 3 The receptor is highly expressed in the liver and ileum and is involved not only in metabolic pathways but also in hepatic inflammation and fibrosis.4, 5 Accordingly, FXR agonists have been shown to reduce liver fibrosis, vascular remodeling, and sinusoidal dysfunction, as well as portal hypertension in experimental studies.6, 7
Chronic liver disease progresses from fibrosis to cirrhosis, which leads to portal hypertension. Evidently, portal hypertension (hepatic venous pressure gradient [HVPG] ≥ 6 mmHg) is a major contributor to decompensating events, such as ascites, portal hypertensive bleeding, and hepatic encephalopathy (HE), which substantially increase the risk of mortality.8, 9, 10, 11
Importantly, liver disease progression shows substantial interindividual variability, and thus, research has focused on the identification of genetic factors accelerating the progression to cirrhosis and predisposing for the development of liver‐related events. Just recently, a genetic variant of the patatin‐like phospholipase domain containing 3 (PNPLA3) gene has been linked to increased risks of hepatic decompensation and mortality in patients who had already developed portal hypertension.12
The first link between genic variants of FXR and liver disease has been established by van Mill and coworkers,13 showing that the rs61755050 (C > T) single nucleotide polymorphism (SNP) was associated with a higher frequency of intrahepatic cholestasis of pregnancy, while rs56163822 (G > T) was not. Moreover, both variants were associated with reduced activity of the FXR pathway.13, 14
In other studies, rs56163822 was associated with inflammatory bowel disease, a higher cholesterol reduction under rosuvastatin, and, most interestingly, with spontaneous bacterial peritonitis (SBP) in patients with cirrhosis and ascites.15, 16, 17
Little is known about the functional relevance of rs35724 SNP (G > C). However, associations of the homozygous genotype (C/C) of this variant with cholelithiasis in male patients and of the heterozygous genotype (G/C) with body mass index were observed.18, 19
Despite the central role of FXR signaling in liver disease, information on the impact of FXR‐SNPs in (advanced) chronic liver disease is scarce. Therefore, we aimed to investigate the effect of FXR‐SNPs on (further) hepatic decompensation and mortality in patients with portal hypertension.
Methods
Patients
Four hundred two consecutive patients with portal hypertension (HVPG ≥ 6 mmHg) were tested for FXR‐SNPs (rs56163822 G > T and rs35724 G > C) between January 1, 2004, and June 30, 2014, were included in this retrospective analysis based on prospectively collected data. Patient characteristics and laboratory parameters at baseline and during clinical follow up were recorded.
Definition of hepatic decompensation and transplant‐free survival
Patients entered the survival analyses at the time of HVPG measurement. Patients' medical records were reviewed for the following events that defined (further) hepatic decompensation at baseline and during follow up: large‐volume paracentesis, SBP, portal hypertensive bleeding, severe HE (West Haven grade 3/4), and liver‐related death. Any (further) hepatic decompensation was defined by one of these events occurring during follow up.
For calculation of liver‐related transplant‐free mortality and transplant‐free survival (TFS) time, patients were censored on the day of surgery if they underwent liver transplantation, on the day of non‐liver‐related death, and at the end of follow up.
Determination of farnesoid X receptor single nucleotide polymorphisms
FXR rs56163822 and rs35724 genotyping was performed by a StepOnePlus Real‐Time PCR System (Applied Biosystems, Foster City, California, USA) using the TaqMan SNP Genotyping Assays C_25598386_10 and C_2366616_10 for rs56163822 and rs35724 (Thermo Fisher Scientific, Waltham, Massachusetts, USA).
Hepatic venous pressure gradient measurements
The Vienna Hepatic Hemodynamic Laboratory at the Medical University of Vienna performed the HVPG measurements according to a standardized operating procedure.20, 21 HVPG measurements were performed in the absence of non‐selective beta‐blockers and nitrates. Clinically significant portal hypertension was defined by HVPG values ≥ 10 mmHg.9
Statistics
Statistical analyses were performed using IBM spss Statistics 24 (spss Inc., Armonk, New York, USA) and GraphPad Prism 6 (GraphPad Software, La Jolla, California, USA). Continuous variables were reported as mean ± standard deviation or median and interquartile range (IQR), and categorical variables were shown as numbers (n) and proportions (%) of patients. Comparisons of continuous variables were performed using Student's t‐test or Mann–Whitney U‐test, as applicable. Group comparisons of categorical variables were performed using either Pearson's χ 2 or Fisher's exact test. The incidence of any (further) hepatic decompensation, large‐volume paracentesis, SBP, portal hypertensive bleeding, HE, and TFS were assessed using the Kaplan–Meier method and compared between FXR‐SNPs (rs56163822 G/G vs G/T and rs35724 G/G vs G/C and C/C) using the log–rank test and Gehan–Breslow–Wilcoxon test. Adjusted Cox regression analyses were used to determine independent prognostic factors for any (further) hepatic decompensation, large‐volume paracentesis, and transplant‐free liver‐related mortality. A two‐sided P value ≤ 0.05 was considered as statistically significant.
Ethics
This study was approved by the ethics committee of the Medical University of Vienna (EK 1526/2017). All patients gave their written informed consent to genetic testing. The requirement of a written informed consent specific to this retrospective analysis was waived by the ethics committee.
Results
Baseline patient characteristics
In total, 402 patients with a mean age of 53.8 ± 11.2 years of predominant male gender (n = 305, 75.9%) were included. The main etiologies were viral hepatitis (n = 230, 57.2%), followed by (non‐)alcoholic fatty liver disease (n = 141, 35.1%), or other etiologies (n = 31, 7.7%). Three hundred thirteen patients (77.9%) had clinically significant portal hypertension (CSPH) with a mean HVPG of 16 ± 7 mmHg and mean model of end‐stage liver disease (MELD) of 11 ± 4 points. Two hundred twenty‐one patients (59.9%) had Child–Pugh score (CPS)‐A, 110 (29.8%) CPS‐B, and 38 (10.3%) CPS‐C, while in 33 patients, CPS could not be fully evaluated at baseline. Importantly, 298 patients (74.1%) showed compensated advanced chronic liver disease, and 104 patients (25.9%) had decompensated advanced chronic liver disease (dACLD) at baseline (Table 1).
Table 1.
Baseline patient characteristics. P values <0.05 are written in bold
| rs56163822 | rs35724 | ||||||
|---|---|---|---|---|---|---|---|
| Overall cohort, n = 402 | Wild type (G/G), n = 383 (95.3%) | Variant (G/T), n = 19 (4.7%) | P value | Wild type (G/G), n = 135 (33.6%) | Variants (G/C, C/C), n = 267 (66.4%) | P value | |
| Age, years ±SD | 53.8 ± 11.2 | 53.9 ± 11.2 | 53.6 ± 11.0 | 0.932 | 55.1 ± 11.2 | 53.2 ± 11.1 | 0.116 |
| Sex, male/female (% male) | 305/97 (75.9%) | 291/92 (76.0%) | 14/5 (73.7%) | 0.819 | 98/37 (72.6%) | 207/60 (77.5%) | 0.275 |
| Etiology | 0.067 | 0.787 | |||||
| (N)AFLD (%) | 141 (35.1%) | 134 (35.0%) | 7 (36.8%) | 50 (37.0%) | 91 (34.1%) | ||
| Viral (%) | 230 (57.2%) | 222 (58.0%) | 8 (42.1%) | 74 (54.8%) | 156 (58.4%) | ||
| Others (%) | 31 (7.7%) | 27 (7.0%) | 4 (21.1%) | 11 (8.1%) | 20 (7.5%) | ||
| Child–Pugh score,† points ± SD | 7 ± 2 | 7 ± 2 | 6 ± 1 | 0.034 | 7 ± 2 | 7 ± 2 | 0.469 |
| CPS‐A | 221 (59.9%) | 209 (59.2%) | 12 (75.0%) | 0.288 | 69 (58.0%) | 152 (60.8%) | 0.131 |
| CPS‐B | 110 (29.8%) | 106 (30.0%) | 4 (25.0%) | 42 (35.3%) | 68 (27.2%) | ||
| CPS‐C | 38 (10.3%) | 38 (10.8%) | 0 (0%) | 8 (6.7%) | 30 (12.0%) | ||
| Decompensated liver disease | 104 (25.9%) | 99 (25.8%) | 5 (26.3%) | 1.000 | 34 (25.2%) | 70 (26.2%) | 0.823 |
| MELD, points ± SD | 11 ± 4 | 11 ± 4 | 10 ± 2 | 0.158 | 10 ± 3 | 11 ± 4 | 0.016 |
| HVPG, mmHg ±SD | 16 ± 7 | 16 ± 7 | 17 ± 5 | 0.378 | 16 ± 7 | 16 ± 7 | 0.978 |
| CSPH, n (%) | 313 (77.9%) | 295 (77.0%) | 18 (94.7%) | 0.069 | 99 (73.3%) | 214 (80.1%) | 0.120 |
| Albumin, g × L−1 ± SD | 36.0 ± 5.9 | 35.9 ± 5.9 | 38.9 ± 4.9 | 0.026 | 36.5 ± 5.5 | 35.7 ± 6.0 | 0.200 |
| Bilirubin, mg × dL−1 (IQR) | 1.10 (0.74–1.87) | 1.14 (0.73–1.89) | 1.18 (0.93–1.53) | 0.973 | 1.13 (0.70–1.63) | 1.15 (0.75–2.03) | 0.301 |
| AST, U × L−1 (IQR) | 65 (45–100) | 65 (45–101) | 56 (38–86) | 0.415 | 62 (45–86) | 70 (44–102) | 0.180 |
| ALT, U × L−1 (IQR) | 52 (30–86) | 51 (30–86) | 56 (35–78) | 0.774 | 51 (31–76) | 53 (29–91) | 0.803 |
| GGT, U × L−1 (IQR) | 123 (67–212) | 123 (68–212) | 89 (47–207) | 0.472 | 118 (64–216) | 128 (67–207) | 0.564 |
P values <0.05 are written inbold.
Information on Child–Pugh score is available in n = 369 (91.8%) patients.
(N)AFLD, (non)‐alcoholic fatty liver disease; ALT, alanine aminotransferase; AST, aspartate aminotransferase; CPS, Child–Pugh stage; CSPH, clinically significant portal hypertension; GGT, γ‐glutamyltransferase; HVPG, hepatic venous pressure gradient; IQR, interquartile range; MELD, model of end‐stage liver disease; SD, standard deviation.
Clinical follow up including hepatic decompensation and mortality
During a median follow up of 885 (IQR: 502–1690) days, 228 patients (56.7%) experienced any (further) hepatic decompensation. Specifically, 88 patients (21.9%) required at least one large‐volume paracentesis, 72 (17.9%) were admitted to hospital due to severe HE, and 27 (6.7%) and 25 (6.2%) had one or more events of SBP and portal hypertensive bleeding during follow up, respectively. Additionally, 30 (7.5%) underwent liver transplantation, while 109 patients (27.1%) died (Table S1).
Distribution of farnesoid X receptor single nucleotide polymorphisms and association with hepatic function
Overall, rs56163822 wild type (G/G) was present in 383 patients (95.3%), while heterozygosity for the T‐allele (G/T) was detected in 19 patients (4.7%). None of the patients was homozygous for the T‐allele. When comparing patient characteristics at baseline between these two genotypes, CPS was significantly lower in patients harboring the T‐allele (6 ± 1 vs 7 ± 2 points, P = 0.034), while albumin levels were significantly higher (38.9 ± 4.9 vs 35.9 ± 5.9 g/L, P = 0.026). Interestingly, CPS‐A was overrepresented in carriers of the rs56163822 T‐allele (75.0%), while only four (25.0%) patients had CPS‐B and no one CPS‐C. Of note, MELD score and HVPG levels were similar between C/C homozygotes and T‐allele carriers (Table 1).
Because patients without prior decompensation are of special interest when investigating the genetic influence on further disease progression, baseline characteristics of CPS‐A patients were analyzed separately with regard to FXR‐SNPs. This analysis revealed no differences in baseline characteristics between rs56163822 genotypes (Table S2).
As for rs35724, prevalence of the homozygous wild type (G/G) was 33.6% (n = 135), with heterozygosity (G/C) and homozygosity (C/C) for the minor allele being observed in 49.3% (n = 198) and 17.2% (n = 69), respectively. Apart from a significantly higher MELD score (11 ± 4 vs 10 ± 3 points, P = 0.016) at baseline, no statistically significant differences were observed between rs35724 G/G patients and patients with at least one minor allele (G/C and C/C).
A subgroup analysis of CPS‐A patients confirmed similar baseline characteristics in patients with rs35724 G/G and at least one minor allele (G/C and C/C) (Table S2).
Linkage between farnesoid X receptor single nucleotide polymorphisms and hepatic decompensation
The proportion of patients experiencing any (further) hepatic decompensation was comparable between patients with rs56163822 G/G and G/T genotypes in the overall cohort (at 5 years: 53% vs 34%, hazard ratio [HR] = 0.671, 95% confidence interval [95% CI]: 0.297–1.516, P = 0.337) and in the subgroup of CPS‐A patients (at 5 years: 38% vs 26%, HR = 0.747, 95% CI: 0.234–2.385, P = 0.623; Fig. S1) (Fig. 1; Table 2).
Figure 1.
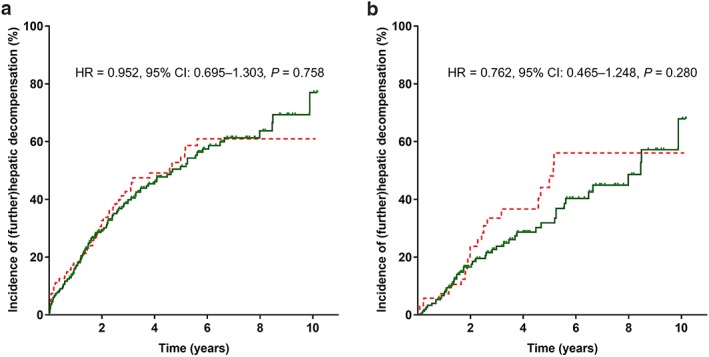
Kaplan–Meier analyses in patients with and without rs35724 minor allele (G/C or C/C) on any (further) hepatic decompensation (a) in the overall cohort and (b) first decompensation in CPS‐A patients. 95% CI, 95% confidence interval; HR, hazard ratio. (a, b)  , rs35724 G/C or C/C;
, rs35724 G/C or C/C;  , rs35724 G/G. [Color figure can be viewed at http://wileyonlinelibrary.com]
, rs35724 G/G. [Color figure can be viewed at http://wileyonlinelibrary.com]
Table 2.
Cox regression analyses on the influence of rs56163822 SNP (G/T), rs35724 (G/C or C/C), and other parameters on any (further) hepatic decompensation (A) in the overall cohort and (B) on first decompensation in CPS‐A patients during follow up. P values <0.05 are written in bold
| (A) Overall cohort, n = 402 | (B) CPS‐A patients, n = 221 | |||||
|---|---|---|---|---|---|---|
| Risk factors for hepatic decompensation | aHR | 95% CI | P value | aHR | 95% CI | P value |
| Age, per 10 years | 1.325 | 1.148–1.528 | < 0.001 | 1.214 | 0.954–1.544 | 0.115 |
| Male gender (vs female) | 1.357 | 0.947–1.944 | 0.096 | 1.840 | 0.972–3.482 | 0.061 |
| HVPG, per mmHg | 1.047 | 1.024–1.070 | < 0.001 | 1.070 | 1.021–1.123 | 0.005 |
| MELD, per point | 1.010 | 0.962–1.059 | 0.697 | 1.030 | 0.924–1.149 | 0.588 |
| Albumin, per g/dL | 0.929 | 0.903–0.955 | < 0.001 | 0.938 | 0.880–0.999 | 0.048 |
| rs56163822 SNP (G/T vs wild type) | 0.773 | 0.339–1.760 | 0.540 | 0.708 | 0.220–2.279 | 0.562 |
| rs35724 SNP (G/C or C/C vs wild type) | 0.830 | 0.602–1.145 | 0.256 | 0.625 | 0.374–1.044 | 0.072 |
P values <0.05 are written inbold.
95% CI, 95% confidence interval; aHR, adjusted hazard ratio; CPS, Child–Pugh stage; HVPG, hepatic venous pressure gradient; MELD, model of end‐stage liver disease; SNP, single nucleotide polymorphism.
Similarly, patients carrying the rs35724 minor allele had a comparable course of liver disease regarding any (further) hepatic decompensation in the overall cohort (at 5 years: 55% vs 51%, HR = 0.952, 95% CI: 0.695–1.303, P = 0.758) and the subgroup of CPS‐A patients (at 5 years: 49% vs 32%, HR = 0.762, 95% CI: 0.465–1.248, P = 0.280).
Moreover, no statistically significant differences were evident in analyses comparing the need for large‐volume paracentesis, SBP, portal hypertensive bleeding, or admission for severe HE during follow up in CPS‐A patients with or rs35724 minor allele (Fig. S2).
Neither rs56163822 T‐allele nor rs35724 minor allele increased the risk for hepatic decompensation in the overall cohort in a Cox regression analysis adjusting for age, sex, HVPG, MELD, and albumin. Nevertheless, the presence of rs35724 minor allele tended to reduce the risk of any first hepatic decompensation in patients with CPS‐A (adjusted hazard ratio [aHR] = 0.625, 95% CI: 0.374–1.044, P = 0.072) and was additionally identified as an independent protective factor for the requirement of large‐volume paracentesis during follow up (aHR = 0.411, 95% CI: 0.191–0.885, P = 0.023; Table S3).
Impact of farnesoid X receptor single nucleotide polymorphisms on transplant‐free survival and liver‐related mortality
To further investigate the influence of FXR‐SNPs on mortality in patients with portal hypertension, TFS was compared between genotypes. In line with previous analyses, no significant differences were observed between rs56163822 genotypes neither in the overall cohort (at 5 years: 67% vs 80%, HR = 0.768, 95% CI: 0.280–2.104, P = 0.608) nor in patients with CPS‐A (at 5 years: 78% vs 86%, HR = 0.760, 95% CI: 0.180–3.205, P = 0.708; Fig. S3) (Table 3; Fig. 2).
Table 3.
Cox regression analyses on the impact of rs56163822 (G/T), rs35724 (G/C or C/C), and other parameters on liver‐related mortality during follow up (A) in the overall cohort and (B) in CPS‐A patients. P values <0.05 are written in bold
| (A) Overall cohort, n = 402 | (B) CPS‐A patients, n = 221 | |||||
|---|---|---|---|---|---|---|
| Liver‐related mortality | aHR | 95% CI | P value | aHR | 95% CI | P value |
| Age, per 10 years | 1.381 | 1.134–1.681 | 0.001 | 1.224 | 0.882–1.698 | 0.227 |
| Male gender (vs female) | 1.954 | 1.165–3.277 | 0.011 | 1.978 | 0.866–4.515 | 0.106 |
| HVPG, per mmHg | 1.020 | 0.985–1.056 | 0.257 | 1.067 | 0.999–1.140 | 0.054 |
| MELD, per point | 0.974 | 0.910–1.042 | 0.439 | 0.851 | 0.712–1.016 | 0.074 |
| Albumin, per g/dL | 0.904 | 0.871–0.940 | < 0.001 | 0.932 | 0.853–1.018 | 0.119 |
| rs56163822 SNP (G/T vs wild type) | 1.027 | 0.370–2.855 | 0.959 | 0.924 | 0.215–3.970 | 0.915 |
| rs35724 SNP (G/C or C/C vs wild type) | 0.658 | 0.434–0.998 | 0.049 | 0.488 | 0.252–0.946 | 0.034 |
P values <0.05 are written inbold.
95% CI, 95% confidence interval; aHR, adjusted hazard ratio; CPS, Child–Pugh stage; HVPG, hepatic venous pressure gradient; MELD, model of end‐stage liver disease; SNP, single nucleotide polymorphism.
Figure 2.
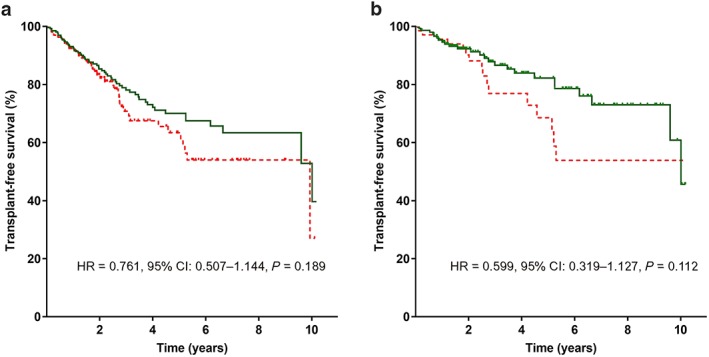
Transplant‐free survival* in patients with and without rs35724 minor allele (G/C or C/C) (a) in the overall cohort and (b) in patients with CPS‐A. *Patients were censored at the day of liver transplantation, non‐liver‐related death, or end of follow up. 95% CI, 95% confidence interval; HR, hazard ratio. (a)  , rs35724 G/C or C/C;
, rs35724 G/C or C/C;  , rs35724 G/G. (b)
, rs35724 G/G. (b)  , rs35724 G/C or C/C;
, rs35724 G/C or C/C;  , rs35724 G/G. [Color figure can be viewed at http://wileyonlinelibrary.com]
, rs35724 G/G. [Color figure can be viewed at http://wileyonlinelibrary.com]
For rs35724 SNP, TFS did not differ between homozygous wild type (G/G) patients and carriers of the C‐allele (at 5 years: 63% vs 70%, HR = 0.761, 95% CI: 0.507–1.144, P = 0.189). However, in CPS‐A patients, the presence of the minor allele was associated with a trend towards a longer TFS (at 5 years: 69% vs 82%, HR = 0.599, 95% CI: 0.319–1.127, P = 0.112).
Moreover, Cox regression analysis adjusted for age, sex, HVPG, MELD, and albumin revealed an independent association between presence of rs35724 minor allele and reduced liver‐related mortality both in the overall cohort (aHR = 0.658, 95% CI: 0.434–0.998, P = 0.049) and in CPS‐A patients (aHR = 0.488, 95% CI: 0.252–0.946, P = 0.034) with rs35724 minor allele being the only factor significantly associated with liver‐related mortality in CPS‐A patients.
Besides, Kaplan–Meier analysis indicated tendencies towards longer TFS in female patients (at 5 years: 56% vs 83%, HR = 0.376, 95% CI: 0.148–0.959, P = 0.041) and female patients with CPS‐A (at 5 years: 58% vs 88%, HR = 0.289, 95% CI: 0.068–1.234, P = 0.094; Fig. S4).
Subgroup analysis of patients stratified by hepatic venous pressure gradient
When analyzing patients separately according to their HVPG, the distribution of rs35724 minor alleles (G/C or C/C) was comparable between the HVPG strata: mild portal hypertension (HVPG 6–9 mmHg), HVPG 10–20 mmHg, and HVPG > 20 mmHg, P = 0.175; Table S4). While TFS did not differ between genotypes in patients with HVPG 6–9 mmHg and 10–20 mmHg, a potential survival benefit was evident in patients with CSPH (HVPG ≥ 10 mmHg; at 5 years: 68.2% vs 55.8%, HR = 0.642, 95% CI: 0.401–1.029, P = 0.047), patients with HVPG ≥ 16 mmHg (at 5 years: 63.3% vs 44.0%, HR = 0.562, 95% CI: 0.328–0.961, P = 0.021), and patients with high‐risk CSPH (HVPG > 20 mmHg, at 5 years: 62.1% vs 40.5%, HR = 0.425, 95% CI: 0.198–0.911, P = 0.036) (Table 4, Fig. 3).
Table 4.
Cox regression analyses on the influence of rs56163822 (G/T), rs35724 (G/C or C/C) genotypes, and other parameters on liver‐related transplant‐free mortality during follow up (A) in patients with clinically significant portal hypertension (CSPH; HVPG ≥ 10 mmHg) and (B) in patients with HVPG ≥ 16 mmHg. P values <0.05 are written in bold
| (A) CSPH, n = 313 | (B) HVPG ≥ 16 mmHg, n = 198 | |||||
|---|---|---|---|---|---|---|
| Liver‐related mortality | aHR | 95% CI | P value | aHR | 95% CI | P value |
| Age, per 10 years | 1.379 | 1.113–1.709 | 0.003 | 1.355 | 1.050–1.748 | 0.019 |
| Male gender (vs female) | 2.463 | 1.374–4.414 | 0.002 | 2.240 | 1.144–4.383 | 0.019 |
| HVPG, per mmHg | 1.017 | 0.974–1.062 | 0.443 | 1.003 | 0.940–1.070 | 0.939 |
| MELD, per point | 0.963 | 0.895–1.037 | 0.320 | .917 | 0.837–1.005 | 0.064 |
| Albumin, per g/dL | 0.900 | 0.865–0.937 | < 0.001 | 0.902 | 0.855–0.951 | < 0.001 |
| rs56163822 SNP (G/T vs wild type) | 1.044 | 0.374–2.915 | 0.935 | 1.500 | 0.514–4.378 | 0.459 |
| rs35724 SNP (G/C or C/C vs wild type) | 0.547 | 0.346–0.864 | 0.010 | 0.519 | 0.307–0.878 | 0.014 |
P values <0.05 are written inbold.
95% CI, 95% confidence interval; aHR, adjusted hazard ratio; CSPH, clinically significant portal hypertension; HVPG, hepatic venous pressure gradient; HVPG, hepatic venous pressure gradient; MELD, model of end‐stage liver disease; SNP, single nucleotide polymorphism.
Figure 3.
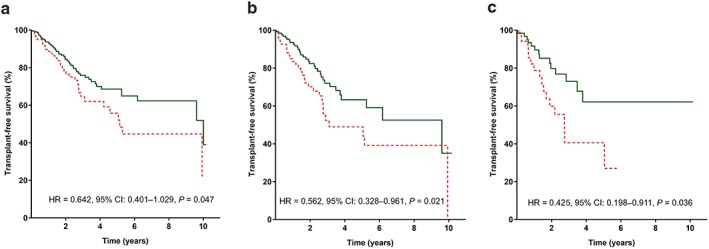
Transplant‐free survival* in patients with and without rs35724 minor allele (G/C or C/C) (a) in patients with clinically significant portal hypertension (CSPH; HVPG ≥ 10 mmHg), (b) in patients with HVPG ≥ 16 mmHg, and (c) in patients with high‐risk CSPH (HVPG > 20 mmHg). *Patients were censored at the day of liver transplantation, non‐liver‐related death, or end of follow up. 95% CI, 95% confidence interval; CSPH, clinically significant portal hypertension; HR, hazard ratio; HVPG, hepatic venous pressure gradient. (a–c)  , rs35724 G/C or C/C;
, rs35724 G/C or C/C;  , rs35724 G/G. [Color figure can be viewed at http://wileyonlinelibrary.com]
, rs35724 G/G. [Color figure can be viewed at http://wileyonlinelibrary.com]
However, Cox regression analysis adjusted for age, sex, HVPG, MELD, and albumin revealed an independent association between presence of rs35724 minor allele and reduced liver‐related transplant‐free mortality in patient with CSPH (HR = 0.425, 95% CI: 0.198–0.911), P = 0.036) and in patients with HVPG ≥ 16 mmHg (aHR = 0.519, 95% CI: 0.307–0.878, P = 0.014).
The risk of (further) hepatic decompensation did not differ between genotypes in all HVPG subcategories (Fig. S5). The hemodynamic response rate to non‐selective beta‐blocker therapy was comparable between rs35724 variants (58.5% of patients with G/G and 59.3% with G/C or C/C, P = 0.935). Noteworthy, the relative change of HVPG was also similar with regard to the rs35724 SNPs, G/G: median −17.0% (IQR: −23.8–[−3.0%]) versus minor alleles: −15.8% (IQR: −31.8–[−3.4%]), P = 0.976.
Discussion
For a better understanding of modulators of liver disease progression, genetic factors have been increasingly investigated: For example, the PNPLA3 rs738409 (C > G) SNP has been associated with an increased risk for hepatic decompensation and liver‐related mortality.12, 22 Additionally, the G‐allele is associated with steatosis, liver fibrosis, and disease activity in non‐alcoholic fatty liver disease23 and viral hepatitis.24, 25
In the present study, we aimed to explore the potential impact of two FXR‐SNPs on hepatic decompensation and mortality in patients with portal hypertension. Interestingly, the presence of rs35724 minor allele was independently associated with reduced risk for liver‐related mortality in the overall cohort, in the subgroup of CPS‐A patients, and in patients with CSPH, severe portal hypertension (HVPG ≥ 16 mmHg), and high‐risk portal hypertension (HVPG > 20 mmHg). Finally, patients harboring the rs35724 minor allele had a reduced requirement for large‐volume paracentesis.
The FXR‐SNP rs56163822 G/T is prevalent in 2.5% of European population and 12.1% of Chinese patients and associated with reduced activity of FXR, but here, we did not find a significant clinical impact of this SNP in our cohort with a prevalence of 4.7% of the minor allele.13, 14 However, because FXR agonists have demonstrated anti‐fibrotic effects and a reduction of portal pressure in animal models, one may hypothesize that genotypes leading to a downregulation of FXR (such as rs56163822 G/T) increase the risks of hepatic decompensation and mortality.6, 7, 13 Although we could confirm the prevalence of 4.7% in a Caucasian population, the low number of patients with heterozygous genotype substantially limits the conclusions that can be drawn from this study. However, a significantly lower CPS hints towards a protective effect on disease progression. This is in contrast to the cross‐sectional study of Lutz et al. reporting a higher prevalence of rs56163822 G/T in patients with SBP compared with those without (including a total of 11 patients with this genotype).17 Although we were able to include a higher number of thoroughly characterized patients in our longitudinal study, a definite statement regarding the impact of this SNP on SBP incidence cannot be made based on our cohort.
The rs35724 G/G, G/C, and C/C genotypes were reported to be prevalent in 37%/45%/17% in European patients, which was confirmed by our study.15 While the functional effect of the FXR rs35724 SNP is currently unknown, one study reported a higher prevalence of the C/C genotype in male gallstone carriers, although there was no impact on the expression of genes involved in bile synthesis and transport.18 Our study uncovered a significant association of rs35724 minor allele and improved survival in the overall cohort and subgroups of different disease severity. These findings highlight the potential “hepatoprotective” effect of the rs35724 SNP on disease progression leading to a less pronounced course of liver disease as compared with patients with the homozygous wild type. In line with our observations, Mueller et al. (2017) reported a better liver function and improved survival in 341 patients with dACLD and ascites harboring rs35724 minor allele.26 Just recently, Grimaudo et al. reported significantly higher hepatic FXR mRNA levels associated with the rs35724 minor alleles—together with higher levels of circulating cholesterol and lower carotid artery intima‐media thickness of the common carotid arteries in a cohort of 124 patients biopsy‐proven non‐alcoholic steatohepatitis (NASH). Moreover, this variant was protective against severity of steatohepatitis and fibrosis.27
These reports support our hypothesis of a protective role of rs35724 minor allele on liver disease progression and suggest an upregulation/activation of FXR signaling by this genotype, thus being a “gain‐of‐function” mutation.
Of note, the influence of the genetic background may be easier to detect in CPS‐A patients than in the overall cohort because these patients represent a rather homogenous cohort of patients without prior decompensation, in contrast to patients with dACLD, in whom non‐genetic, extrahepatic factors may have a more profound impact on the course of liver disease.28, 29, 30 Accordingly, less consistent results in the overall cohort seem reasonable. Despite the association between longer TFS and rs35724, minor allele was more pronounced in female patients. The results of our study regarding female subgroup must be interpreted with caution due to the limited sample size. Because no gender‐specific studies evaluating different expression of FXR in male and female have been carried out so far, further gender‐specific studies are warranted to investigate the specific functional consequences of this variant in female versus male patients.
Some limitations need to be acknowledged when interpreting the results of this study: Despite the retrospective design, patients were prospectively characterized at the timepoint of HVPG measurement, and clinical events during follow up were carefully assessed during scheduled clinical visits. Second, the statistical power to detect a clinically meaningful impact of rs56163822 genotype was limited, because the number of patients harboring the rs56163822 G/T genotype was small. Similarly, the results of subgroup analyses need to be interpreted with caution due to limited sample size in some analyses.
In conclusion, we are the first to investigate the impact of two FXR‐SNP (rs56163822 and rs35724) on hepatic decompensation and liver‐related mortality in patients with portal hypertension. While we found prognostic value of the rs35724 genotype for hepatic decompensation and mortality, further studies are needed to explore the underlying mechanisms and to confirm these results. However, the influence of this variant should be taken into account in future studies evaluating the effect of FXR agonists in humans.
Supporting information
Table S1. Comparison of relative incidence of hepatic decompensation between FXR‐SNPs.
Table S2. Baseline characteristics and follow‐up in CPS‐A patients.
Table S3. Cox regression analysis on the influence of rs56163822 SNP (G/T) and rs35724 SNP (G/C or C/C), as well as other factors for the requirement of large‐volume paracentesis in CPS‐A patients.
Table S4. Distribution of rs35724 variants among patients stratified by HVPG levels.
Figure S1. Kaplan‐Meier analyses on any (further) hepatic decompensation A in patients with and without rs56163822 SNP in the overall cohort and B in patients with CPS‐A.
Figure S2. Kaplan‐Meier analyses on the incidence of A large‐volume paracentesis, B hepatic encephalopathy, C spontaneous bacterial peritonitis and D portal hypertensive bleeding in patients with CPS‐A and rs35724 minor allele.
Figure S3. Transplant‐free survival* in patients with and without rs56163822 SNP A in the overall cohort and B in patients with CPS‐A.
Figure S4. Kaplan Meier analyses on transplant‐free survival A in female patients and B in female CPS‐A patients with rs35724 SNP minor allele.
Figure S5. Kaplan‐Meier analyses on (further) hepatic decompensation A in patients with mild portal hypertension (HVPG 6‐9mmHg), B in patients with HVPG 10‐20mmHg, C in patients with high risk CSPH (HVPG >20mmHg) as well as D in patients with clinically significant portal hypertension (CPSH, HVPG ≥10mmHg) and E HVPG ≥16mmHg, stratified according to presence of rs35724 minor allele.
Acknowledgment
We especially want to thank Claudia Willheim for assistance with FXR genotyping.
Semmler, G. , Simbrunner, B. , Scheiner, B. , Schwabl, P. , Paternostro, R. , Bucsics, T. , Stättermayer, A. F. , Bauer, D. , Pinter, M. , Ferenci, P. , Trauner, M. , Mandorfer, M. , and Reiberger, T. (2019) Impact of farnesoid X receptor single nucleotide polymorphisms on hepatic decompensation and mortality in cirrhotic patients with portal hypertension. Journal of Gastroenterology and Hepatology, 34: 2164–2172. 10.1111/jgh.14700.
Declaration of conflict of interest: The authors have nothing to disclose regarding the work under consideration for publication. The following authors disclose conflicts of interests outside the submitted work: B. S. received travel support from AbbVie and Gilead. P. S. received speaker fees from Boehringer Ingelheim and Roche and travel support from Boehringer Ingelheim, Gilead, and Roche. T. B. received travel support from AbbVie, BMS, and Medis; speaker fees from BMS; and grant support from Medis. P. F. received unrestricted grant support from Gilead and has served as a speaker and advisory board member for MSD and AbbVie. M. T. received grant support from Cymabay, Falk, Gilead, Intercept, MSD, and Takeda; honoraria for consulting from AbbVie, Gilead, Intercept, Janssen, MSD, and Regulus; speaker fees from Falk, Gilead, and MSD; and travel support from AbbVie and Gilead. M. P. received speaker fees from Bayer and BMS; travel support from Bayer; and he is an investigator for Bayer, BMS, and Lilly and served as advisory board member for Bayer, BMS, and Eisai. M. M. has served as a speaker and consultant for AbbVie, BMS, Gilead, Gore, and Janssen. T. R. received grant support from AbbVie, Boehringer Ingelheim, Gilead, MSD, Philips Healthcare, Gore; speaking honoraria from Gilead, Gore, Intercept, Roche, MSD; consulting/advisory board fee from AbbVie, Boehringer Ingelheim, Gilead, MSD; and travel support from Boehringer Ingelheim, Gilead, and Roche. G. S, B. Si., R. P., A. F. S., and D. B. have nothing to disclose.
Author contribution: G. S., M. M., and T. R. did the concept of the study. G. S., B. Si., B. S., and M. M. performed the extraction of clinical data. A. S. and P. F. did the genotyping. Statistical analysis was carried out by G. S., M. M., and T. R. G. S., B. S., M. M., and T. R. drafted the manuscript. G. S., B. S., M. M., and T. R. did the writing of the manuscript. All authors revised the study for important intellectual content and approved the final version of this manuscript.
References
- 1. Makishima M, Okamoto AY, Repa JJ et al Identification of a nuclear receptor for bile acids. Science (New York, N.Y.) 1999; 284: 1362–1365. [DOI] [PubMed] [Google Scholar]
- 2. Parks DJ, Blanchard SG, Bledsoe RK et al Bile acids: natural ligands for an orphan nuclear receptor. Science (New York, N.Y.) 1999; 284: 1365–1368. [DOI] [PubMed] [Google Scholar]
- 3. Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell 1999; 3: 543–553. [DOI] [PubMed] [Google Scholar]
- 4. Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 2006; 126: 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trauner M, Halilbasic E. Nuclear receptors as new perspective for the management of liver diseases. Gastroenterology 2011; 140: 1120–1125.e1‐12. [DOI] [PubMed] [Google Scholar]
- 6. Schwabl P, Hambruch E, Seeland BA et al The FXR agonist PX20606 ameliorates portal hypertension by targeting vascular remodelling and sinusoidal dysfunction. J. Hepatol. 2017; 66: 724–733. [DOI] [PubMed] [Google Scholar]
- 7. Verbeke L, Farre R, Trebicka J et al Obeticholic acid, a farnesoid X receptor agonist, improves portal hypertension by two distinct pathways in cirrhotic rats. Hepatol. (Baltimore, Md) 2014; 59: 2286–2298. [DOI] [PubMed] [Google Scholar]
- 8. Pfisterer N, Riedl F, Pachofszky T et al Outcomes after placement of a SX‐ELLA oesophageal stent for refractory variceal bleeding—a national multicentre study. Liver Int. Off. J. Int. Assoc. Study Liver 2019; 39(2): 290‐8. 10.1111/liv.13971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reiberger T, Puspok A, Schoder M et al Austrian consensus guidelines on the management and treatment of portal hypertension (Billroth III). Wien. Klin. Wochenschr. 2017; 129: 135–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schwabl P, Bucsics T, Soucek K et al Risk factors for development of spontaneous bacterial peritonitis and subsequent mortality in cirrhotic patients with ascites. Liver Int. Off. J. Int. Assoc. Study Liver 2015; 35: 2121–2128. [DOI] [PubMed] [Google Scholar]
- 11. Mandorfer M, Bota S, Schwabl P et al Nonselective beta blockers increase risk for hepatorenal syndrome and death in patients with cirrhosis and spontaneous bacterial peritonitis. Gastroenterology 2014; 146: 1680–1690.e1. [DOI] [PubMed] [Google Scholar]
- 12. Mandorfer M, Scheiner B, Stättermayer AF et al Impact of patatin‐like phospholipase domain containing 3 rs738409 G/G genotype on hepatic decompensation and mortality in patients with portal hypertension. Aliment. Pharmacol. Ther. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Mil SW, Milona A, Dixon PH et al Functional variants of the central bile acid sensor FXR identified in intrahepatic cholestasis of pregnancy. Gastroenterology 2007; 133: 507–516. [DOI] [PubMed] [Google Scholar]
- 14. Marzolini C, Tirona RG, Gervasini G et al A common polymorphism in the bile acid receptor farnesoid X receptor is associated with decreased hepatic target gene expression. Mol. Endocrinol. (Baltimore, MD) 2007; 21: 1769–1780. [DOI] [PubMed] [Google Scholar]
- 15. Attinkara R, Mwinyi J, Truninger K et al Association of genetic variation in the NR1H4 gene, encoding the nuclear bile acid receptor FXR, with inflammatory bowel disease. BMC. Res. Notes 2012; 5: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu M, Lui SS, Tam LS, Li EK, Tomlinson B. The farnesoid X receptor‐1G>T polymorphism influences the lipid response to rosuvastatin. J. Lipid Res. 2012; 53: 1384–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lutz P, Berger C, Langhans B et al A farnesoid X receptor polymorphism predisposes to spontaneous bacterial peritonitis. Digest. Liver Disease Off. J. Italian Soc. Gastroenterol. Italian Assoc. Study Liver 2014; 46: 1047–1050. [DOI] [PubMed] [Google Scholar]
- 18. Hirobe‐Jahn S, Harsch S, Renner O, Richter D, Muller O, Stange EF. Association of FXR gene variants with cholelithiasis. Clin. Res. Hepatol. Gastroenterol. 2015; 39: 68–79. [DOI] [PubMed] [Google Scholar]
- 19. van den Berg SW, Dolle ME, Imholz S et al Genetic variations in regulatory pathways of fatty acid and glucose metabolism are associated with obesity phenotypes: a population‐based cohort study. Int. J. Obes. (2005 2009; 33: 1143–1152. [DOI] [PubMed] [Google Scholar]
- 20. Ferlitsch A, Bota S, Paternostro R et al Evaluation of a new balloon occlusion catheter specifically designed for measurement of hepatic venous pressure gradient. Liver international: official journal of the International Association for the Study of the Liver. Liver International 2015; 35: 2115–2120. [DOI] [PubMed] [Google Scholar]
- 21. Schwarzer R, Kivaranovic D, Mandorfer M et al Randomised clinical study: the effects of oral taurine 6g/day vs placebo on portal hypertension. Aliment. Pharmacol. Ther. 2018; 47: 86–94. [DOI] [PubMed] [Google Scholar]
- 22. Scheiner B, Mandorfer M, Schwabl P et al The impact of PNPLA3 rs738409 SNP on liver fibrosis progression, portal hypertension and hepatic steatosis in HIV/HCV coinfection. PLoS ONE 2015; 10: e0143429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stättermayer AF, Traussnigg S, Aigner E et al Low hepatic copper content and PNPLA3 polymorphism in non‐alcoholic fatty liver disease in patients without metabolic syndrome. J. Trace Elem. Med. Biol. Organ Soc. Min. Trace Elem. (GMS) 2017; 39: 100–107. [DOI] [PubMed] [Google Scholar]
- 24. Stättermayer AF, Rutter K, Beinhardt S et al Role of FDFT1 polymorphism for fibrosis progression in patients with chronic hepatitis C. Liver Int. Off. J. Int. Assoc. Study Liver 2014; 34: 388–395. [DOI] [PubMed] [Google Scholar]
- 25. Stättermayer AF, Rutter K, Beinhardt S et al Association of the IL28B genotype with insulin resistance in patients with chronic hepatitis C. J. Hepatol. 2012; 57: 492–498. [DOI] [PubMed] [Google Scholar]
- 26. Müller N, Fischer J, Krohn S et al Farnesoid‐X receptor single nucleotide polymorphism rs35724 is potentially associated with patients' outcome in decompensated liver cirrhosis. J. Hepatol. 2017: S561. [Google Scholar]
- 27. Grimaudo S, Dongiovanni P, Pipitone RM et al FXR rs35724 G>C variant modulates cholesterol levels, carotid atherosclerosis and liver damage in non‐alcoholic fatty liver. Dig. Liver Dis. 2019; 51: e26. [Google Scholar]
- 28. Reiberger T, Ulbrich G, Ferlitsch A et al Carvedilol for primary prophylaxis of variceal bleeding in cirrhotic patients with haemodynamic non‐response to propranolol. Gut 2013; 62: 1634–1641. [DOI] [PubMed] [Google Scholar]
- 29. Bucsics T, Mandorfer M, Schwabl P et al Impact of acute kidney injury on prognosis of patients with liver cirrhosis and ascites: a retrospective cohort study. J. Gastroenterol. Hepatol. 2015; 30: 1657–1665. [DOI] [PubMed] [Google Scholar]
- 30. Scheiner B, Steininger L, Semmler G et al Controlled attenuation parameter (CAP) does not predict hepatic decompensation in patients with advanced chronic liver disease. Liver Int. Off. J. Int. Assoc. Study Liver 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Comparison of relative incidence of hepatic decompensation between FXR‐SNPs.
Table S2. Baseline characteristics and follow‐up in CPS‐A patients.
Table S3. Cox regression analysis on the influence of rs56163822 SNP (G/T) and rs35724 SNP (G/C or C/C), as well as other factors for the requirement of large‐volume paracentesis in CPS‐A patients.
Table S4. Distribution of rs35724 variants among patients stratified by HVPG levels.
Figure S1. Kaplan‐Meier analyses on any (further) hepatic decompensation A in patients with and without rs56163822 SNP in the overall cohort and B in patients with CPS‐A.
Figure S2. Kaplan‐Meier analyses on the incidence of A large‐volume paracentesis, B hepatic encephalopathy, C spontaneous bacterial peritonitis and D portal hypertensive bleeding in patients with CPS‐A and rs35724 minor allele.
Figure S3. Transplant‐free survival* in patients with and without rs56163822 SNP A in the overall cohort and B in patients with CPS‐A.
Figure S4. Kaplan Meier analyses on transplant‐free survival A in female patients and B in female CPS‐A patients with rs35724 SNP minor allele.
Figure S5. Kaplan‐Meier analyses on (further) hepatic decompensation A in patients with mild portal hypertension (HVPG 6‐9mmHg), B in patients with HVPG 10‐20mmHg, C in patients with high risk CSPH (HVPG >20mmHg) as well as D in patients with clinically significant portal hypertension (CPSH, HVPG ≥10mmHg) and E HVPG ≥16mmHg, stratified according to presence of rs35724 minor allele.


