Abstract
The molecular mechanisms driving specific regulation of neutrophils are not completely understood to date. In order to characterize fundamental granulocyte features on protein level, we analyzed changes in proteome composition as reaction to stress from cell activation processes. For this purpose, we isolated primary granulocytes from equine whole blood through density gradient centrifugation followed by sodium chloride lysis and stimulated cells for 30 min with interleukin-8 (IL8) due to its role as a chemotactic factor for neutrophils. We additionally used phorbol 12-myristate 13-acetate (PMA) and lipopolysaccharide (LPS), which are primarily associated to neutrophil extracellular trap formation and release of reactive oxygen species. From mass spectrometry analysis, we identified a total of 2,032 proteins describing the whole granulocyte proteome, including 245 proteins (12% of identified proteome) newly associated to in vivo expression in primary equine granulocytes (hypothetical proteins). We also found distinct and different changes in protein abundance (ratio ≥ 2) after short stimulation of cells with various stimuli, pointing to rapid and differentiated reaction pattern. IL8 stimulation resulted in increased protein abundance of 58 proteins (3% of proteome), whereas PMA induced changed protein abundance of 207 (10 % of proteome) and LPS of 46 proteins (2% of proteome). Enrichment analyses clearly showed fundamental differences between stimuli, with primary association of IL8 stimulation to processes in immune response, receptor signaling and signal transduction. Top enrichment for PMA on the other hand pointed to vesicle mediated transport and exocytosis. Stimulation with LPS did not result in any significant enrichment. Although we detected 43% overlap of enrichment categories for IL8 and PMA stimulation, indicating that activation of neutrophils with different stimuli partly induces some similar biological processes and pathways, hierarchical clustering showed clear differences in distribution and biological relevance of clusters between the chosen stimuli. Our studies provide novel information on the granulocyte proteome and offer insights into early, differentiated granulocyte reaction to stimuli, which contribute to a better understanding of molecular mechanisms involved in activation and recruitment of neutrophils, through inflammatory stimuli.
Keywords: innate immune cell activation, differential proteomics, interleukin 8 (IL8), PMA, LPS, neutrophil, signal transduction, biological process
Introduction
Granulocytes have initially been labeled as short-lived, terminally differentiated cells, driving innate immune response through phagocytosis, degranulation, ROS release and, as described more recently, NETosis (1, 2). However, today, neutrophil diversity and plasticity, with many different subpopulations and finely tuned functional features are evident (3–8). Still relatively little is known about specific, differentiated regulation mechanisms in early granulocyte activation involved in subsequent innate immune responses. For this reason, we investigated fundamental granulocyte features by analyzing changes in proteome composition as reaction to cell activation and allocating these changes to different biological processes and pathways in an equine model. In cells from the adaptive immune system, we previously found major differences in regulation of lymphocyte protein expression in autoimmune disease (9–12). Moreover, we detected differences in the granulocyte proteome, with Talin1 as a key player in disease pathogenesis, indicating a role of the innate immune system in lymphocyte-driven autoimmune disease (13, 14). In retrospect, the granulocytes analyzed in these studies most likely represent the subpopulation of low density neutrophils (LDN), which were recently discovered (15). In present study, LDN were excluded from analysis, due to granulocyte isolation protocol. Here, we were especially interested in the impact of initial activation on downstream innate immune response and the pathways switched on in course of activation-induced cell stress in order to provide fundamental knowledge on granulocyte activation mechanisms.
Materials and Methods
Sample Processing
The blood used in this study originated from three resident horses of the LMU equine clinic (aged 12, 20, and 21; kept in straw-embedded stalls with daily access to paddocks), which are at the student's disposal for supervised ultrasound- and health assessment training. Health status was assessed by standard clinical routine examinations. No experimental procedures were performed on these horses. Venous whole blood was collected in tubes supplemented with 25.000 I.U. heparin. After rough sedimentation of erythrocytes, PMN were isolated from plasma by density gradient centrifugation (RT, 290 × g, 25 min, brake off) using Ficoll-Paque PLUS separating solution (GE Life Sciences, Freiburg, Germany). Cells were washed gently (4 C, 400 × g, 10 min) in cold PBS (DPBS devoid of CaCl2 and MgCl2; Gibco/ThermoFisher Scientific, Germany) and remaining erythrocytes were removed by sodium chloride lysis (lysis in 0.2% NaCl, after 30 s addition of equal part 1.6% NaCl to restore isotonicity). Cells were washed (4°C, 400 × g, 10 min) and resuspended in PBS with 0.2% Glucose. From each animal used in the experiment, we prepared aliquot portions of 6 × 105 cells/500 μl. These cell aliquots were separately stimulated with recombinant equine interleukin-8 (IL8; Kingfisher Biotec; 1 ng/ml), phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich/Merck, Darmstadt, Germany; 5 μg/ml) and lipopolysaccharide (LPS; Sigma-Aldrich/Merck, Darmstadt, Germany; 5 μg/ml) for 30 min in a CO2 incubator (37°C, 5% CO2). Untreated medium control (mc) was incubated under the same conditions but without stimulating agent. After stimulation, each of the stimulated and mc aliquots was topped up to 1 ml with PBS with 0.2% Glucose and pelleted (4°C, 2,300 × g, 10 min). All Samples were stored at −20°C. Shortly before mass spectrometry analysis, cells were thawed and lysed in urea buffer (8 M urea in 0.1 M Tris/HCl pH 8.5), and protein amount was determined with Bradford protein assay (16).
Mass Spectrometry Analysis
From each sample, 10 μg total protein was digested with LysC and trypsin by filter-aided sample preparation (FASP) as previously described (17). Acidified eluted peptides were analyzed in the data-dependent mode on a Q Exactive HF mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) online coupled to a UItimate 3000 RSLC nano-HPLC (Dionex). Samples were automatically injected and loaded onto the C18 trap column, eluted after 5 min and separated on the C18 analytical column (75 μm ID × 15 cm, Acclaim PepMAP 100 C18. 100 Å/size, LC Packings, Thermo Fisher Scientific, Bremen, Germany) by a 90 min non-linear acetonitrile gradient at a flow rate of 250 nl/min. MS spectra were recorded at a resolution of 60,000. After each MS1 cycle, the 10 most abundant peptide ions were selected for fragmentation.
Data Processing
Label-free quantitative analysis was performed using Progenesis QI software (version 2.5, Non-linear Dynamics, Waters, Newcastle upon Tyne, U.K.) as described (18, 19), with raw MS spectral files imported, followed by automatic peak picking and retention time alignment and normalization of total peak intensities across all samples to minimize loading differences. All MS/MS spectra were exported from Progenesis QI software as Mascot generic files (mgf) and searched against Ensembl Horse protein database (version 3.0, http://www.ensembl.org) for peptide identification with Mascot (version 2.5.1). Search parameters used were 10 ppm peptide mass tolerance, 20 mmu fragment mass tolerance, one missed cleavage allowed, carbamidomethylation as fixed modification and methionine oxidation as well as deamidation of asparagine and glutamine as variable modifications. Mascot integrated decoy database search was set to a false discovery rate (FDR) of 1% when searching was performed on the concatenated mgf files with a percolator ion score cut-off of 13 and an appropriate significance threshold p. Identifications were re-imported into Progenesis QI and redundancies grouped following the rules of parsimony.
Data Analysis
Differential protein abundance was determined by comparison of the mean normalized peptide abundance from the extracted ion chromatograms. Proteins were considered differentially expressed at stimulating agent/mc ratio ≥ 2.0. Bioinformatic analysis was performed on human orthologs of gene names from differentially expressed equine proteins with open source software ShinyGO v0.60: http://bioinformatics.sdstate.edu/go60/ (20) with the following settings: search species human, P-value cutoff (FDR) 0.05, number of most significant terms to show 30. P-value for enrichment analysis was calculated via hypergeometric distribution, followed by correction using FDR. Venn diagram was made with open source tool: http://bioinformatics.psb.ugent.be/webtools/Venn/.
Results
Two Thousand Thirty-Two Proteins Describing the Granulocyte Proteome
Using mass spectrometry analysis, we identified the equine whole granulocyte proteome, comprising a total of 2,032 proteins. Among the identifications, we found 245 proteins (hypothetical proteins) which have not been associated to the in vivo protein expression repertoire of equine granulocytes so far (Supplemental Table 1). These proteins represent 12% of the total granulocyte proteome identified here.
Short Stimulation Time of Only 30 min Results in Rapid and Differentiated Reactions of Cells
After stimulation with three different stimulating agents, we found distinct changes in granulocyte protein abundance compared to medium controls (ratio cut-off ≥ 2). In detail, cells stimulated with LPS showed higher expression levels of 46 proteins (2% of proteome), whereas PMA induced increased protein abundance of 207 proteins (10% of proteome). IL8 stimulation resulted in increased protein expression levels of 58 proteins (3% of proteome) (Supplemental Table 2). All of these differentially abundant proteins summed up to a total of 252, from which only 15 showed higher expression levels in all three stimulating agent groups (Figure 1, Table 1). Analysis of differentially expressed proteins per stimulation group revealed 12 unique proteins from LPS and 174 from PMA stimulated cells as well as 22 proteins with unique appearance in cells stimulated with IL8 (Figure 1, Table 1).
Figure 1.
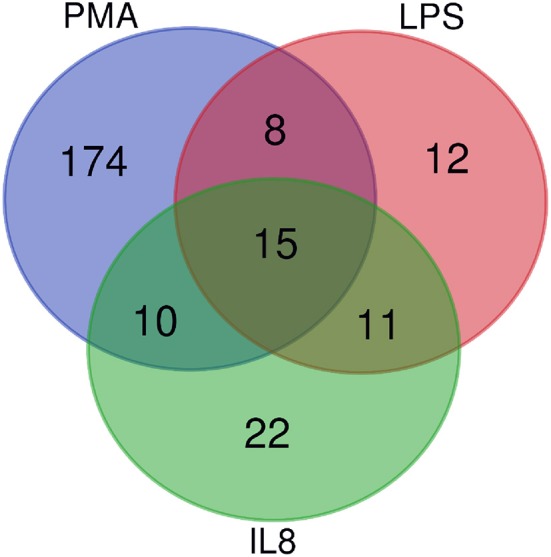
Venn Diagramm of overlapping differentially expressed proteins from IL8, PMA, and LPS stimulated cells. Fifteen proteins are differentially expressed among all stimulation groups.
Table 1.
Gene names for shared and unique proteins ≥2 from granulocyte-derived mass spectrometry list.
| Shared proteins | Unique proteins | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stimulating agent | IL8 | IL8 | IL8 | IL8 | ||||||||
| PMA | PMA | PMA | PMA | |||||||||
| LPS | LPS | LPS | LPS | |||||||||
| No. of proteins | 15 | 10 | 11 | 8 | 22 | 174 | 12 | |||||
| Gene names | AAMDC | DNM1 | ALAD | CALCOCO1 | ADAMDEC1 | A2M | COMT | GYS1 | NT5C1A | RIPOR2 | SYT5 | ACTBL2 |
| ATP2B1 | EXOSC2 | ARID1B | EIF4G2 | AIFM1 | ABHD14B | COPS6 | H2AFV | NUCKS1 | RNASEL | SYTL3 | DCUN1D1 | |
| DMTN | GMPR2 | BAX | IWS1 | CARHSP1 | ACSL4 | COX5A | HARS2 | NUDT3 | RPL12 | TACC3 | DCXR | |
| DNASE1L1 | JPT1 | CPNE6 | PROM1 | CASP1 | ADAM10 | CPSF6 | HBE1 | NUMB | RPL15 | TAF2 | DHX58 | |
| EEA1 | IMPDH2 | ECHDC1 | RHEB | CDC37 | ADD2 | CWF19L1 | HIST1H1A | NUP210 | RPL18 | TAOK3 | HIKESHI | |
| ENSA | KCNA10 | GNS | SIRPB1 | CNP | ADD3 | CXorf58 | HIST1H3A | NUP62 | RPL9 | TAPBP | RPS4X | |
| FARSB | RAP1A | PPP1R18 | TBCC | CREG1 | ADPGK | CYP2C19 | HSD17B12 | OAS3 | RPN1 | TBC1D13 | SDHB | |
| GLYR1 | SRSF4 | PSMC1 | TMEM128 | CRYZ | AGPAT2 | DCTN3 | HSPE1 | PFN | RPS6KA2 | TEDC1 | SEPT11 | |
| HCFC1 | VKORC1 | SEC23IP | DDOST | ALDH16A1 | DENND3 | HUWE1 | PGRMC1 | RPS8 | TM9SF2 | SLC47A2 | ||
| IPCEF1 | ZBTB45 | SH2D5 | DLAT | ARHGAP10 | DES | HVCN1 | PI4KA | S100A7 | TMED10 | UBE2H | ||
| LST1 | ZNF207 | HDLBP | ARL6IP1 | DHCR7 | IGSF6 | PKP1 | SARS | TPD52L2 | WASHC2A | |||
| PSIP1 | IGHG4 | ATP6AP1 | DNM1L | ILVBL | PPM1F | SEC24A | TRMT112 | WDR44 | ||||
| RPRD1B | IKBKB | ATP8A1 | DOCK10 | IMMT | PRPF8 | SELENOH | TRPC3 | |||||
| RPS4X | NAXD | B3GNT2 | DOT1L | IRAK3 | PSMA7 | SEMA3E | TUFM | |||||
| VPS37C | NPEPL1 | BAK1 | ECHS1 | IRF3 | PSMB8 | SERBP1 | UBE2M | |||||
| PAG1 | BCAP29 | EIF3H | ISG15 | PSMB9 | SF3A1 | UBR4 | ||||||
| PSMC6 | BIN1 | EIF4H | ISG20 | PSMD7 | SLC17A3 | UQCRC2 | ||||||
| PSMD12 | BMX | ENOPH1 | JAK3 | QSOX1 | SLC28A1 | USP15 | ||||||
| SARNP | BPGM | EPB41 | KARS | R3HCC1 | SMAP2 | VARS | ||||||
| SH3GLB1 | BTBD11 | ERH | KLF12 | RAB43 | SNX27 | VDAC3 | ||||||
| SIK3 | CAMKK2 | ESYT2 | LZIC | RASGRP2 | SOD2 | VPS11 | ||||||
| VAV2 | CARMIL2 | FABP5 | MCFD2 | RBBP4 | SPCS2 | VPS26A | ||||||
| CASP14 | GHDC | MCU | RBBP7 | SRSF6 | VPS28 | |||||||
| CASS4 | GLOD4 | MOGS | RBM8A | SSR1 | VTI1A | |||||||
| CD109 | GM2A | MPDU1 | RDH16 | STK38 | WDR5 | |||||||
| CD300LF | GMFG | MTCH2 | RECQL | STRN | WFIKKN1 | |||||||
| CES2 | GNG12 | MYADM | RENBP | SYNE1 | YARS | |||||||
| CHMP3 | GRHPR | MYO1E | REXO2 | SYNE2 | YIF1B | |||||||
| COL4A3BP | GRN | NAF1 | RHAG | SYPL1 | ZSCAN4 | |||||||
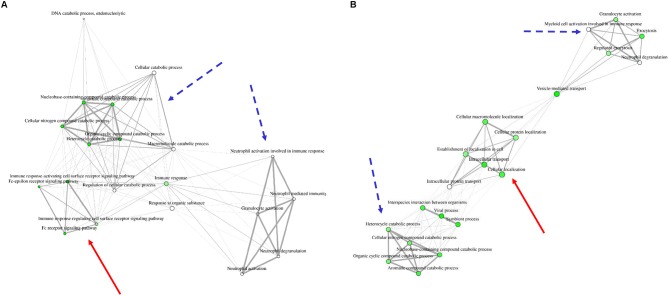
Reaction of Innate Immune Cells to Different Stimuli Are Respectively Clustered in Three Distinct Networks
In order to understand the association of the differentially expressed proteins to biological processes and their known role in granulocyte activation pathways, we analyzed the data from the 15 proteins present in all groups (Figure 1, Table 1, Supplemental Figure 1, Supplemental Table 3) as well as LPS, PMA, and IL8 groups with open source software ShinyGO. LPS stimulation data did not result in any significant enrichment and clustering of the differentially expressed proteins in these cells. Therefore, we looked into GO category assignment for these proteins and found eight high level categories mainly connected to cell metabolism, intracellular transport and response to stress (Supplemental Table 4). Data from IL8 and PMA stimulated cells, however, revealed three distinct clusters. Comparison of these enrichment category clusters showed a 43% overlap between IL8 and PMA stimulation groups, with neutrophil activation and cellular catabolic processes as the two major shared functional categories (Figure 3, Table 3). The unique clusters for each stimulant, however, showed a clear difference in reaction of cells to stimuli: PMA stimulated innate immune cells reacted with processes involved in intracellular transportation processes, whereas IL8 stimulated cells showed involvement in signal transduction pathways (Figure 3, Table 3).
Unique Reaction of Cells to IL8 Stimulation Associates to Receptor Signaling, Signal Transduction, and Immune Response
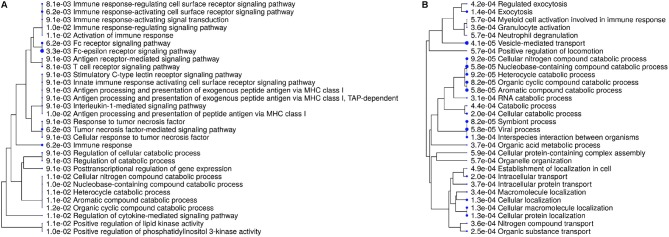
For more detailed analysis, we subsequently focused on proteins which were differentially abundant in either IL8 or PMA stimulated cells and therefore described as unique for respective stimulus. Hierarchical clustering of enrichment analysis data from unique proteins expressed after PMA stimulation pointed to primary involvement in vesicle-mediated and intracellular transport as well as exocytosis on the one and metabolic processes on the other hand (Figure 2, Table 2). Results from IL8 stimulated cells showed primary association of uniquely expressed proteins to receptor signaling, signal transduction, and immune response with top enrichment for Fc-epsilon receptor and Tumor necrosis factor (TNF) mediated signaling pathways (Figure 2, Table 2). Also, enrichment of PI 3-kinase activity pointed to processes in cytoskeleton dynamics (Figure 2, Table 2). Interestingly, the protein proteasome 26S subunit, ATPase 6 (PSMC6), was allocated to the majority of functional enrichment categories from the IL8 stimulation group (Table 2).
Figure 2.
Enrichment tree showing 30 most significant functional categories from biological processes generated from gene names of differentially expressed proteins after stimulation with either IL8 (A) or PMA (B). Size of the solid circles corresponds to the enrichment FDR. Proteins used for calculation of enrichment were uniquely present in respective stimulation group.
Table 2.
Enrichment of functional categories describing biological proccesses generated from proteins with differential expression after IL8 and PMA stimulation.
| Enrichment FDR | Genes in list | Total genes | Functional category | Genes |
|---|---|---|---|---|
| Biological proccesses from unique Proteins with ratio IL8/mc ≥ 2 | ||||
| 0.0033 | 4 | 133 | Fc-epsilon receptor signaling pathway | PSMC6 IKBKB VAV2 PSMD12 |
| 0.0062 | 10 | 2,602 | Immune response | PAG1 CDC37 CASP1 ADAMDEC1 PSMC6 IKBKB CREG1 VAV2 PSMD12 DDOST |
| 0.0062 | 4 | 210 | Tumor necrosis factor-mediated signaling pathway | IKBKB CASP1 PSMC6 PSMD12 |
| 0.0062 | 4 | 203 | Fc receptor signaling pathway | PSMC6 IKBKB VAV2 PSMD12 |
| 0.0062 | 5 | 466 | Immune response-activating cell surface receptor signaling pathway | PAG1 PSMC6 IKBKB VAV2 PSMD12 |
| 0.0081 | 4 | 275 | T cell receptor signaling pathway | PAG1 PSMC6 IKBKB PSMD12 |
| 0.0081 | 5 | 520 | Immune response-regulating cell surface receptor signaling pathway | PAG1 PSMC6 IKBKB VAV2 PSMD12 |
| 0.0091 | 6 | 1,050 | Regulation of catabolic process | CARHSP1 SH3GLB1 PSMC6 CDC37 AIFM1 PSMD12 |
| 0.0091 | 5 | 662 | Posttranscriptional regulation of gene expression | CARHSP1 CDC37 SARNP PSMC6 PSMD12 |
| 0.0091 | 6 | 928 | Regulation of cellular catabolic process | CARHSP1 SH3GLB1 PSMC6 CDC37 AIFM1 PSMD12 |
| 0.0091 | 3 | 141 | Innate immune response activating cell surface receptor signaling pathway | PSMC6 IKBKB PSMD12 |
| 0.0091 | 3 | 135 | Stimulatory C-type lectin receptor signaling pathway | PSMC6 IKBKB PSMD12 |
| 0.0091 | 4 | 368 | Response to tumor necrosis factor | IKBKB CASP1 PSMC6 PSMD12 |
| 0.0091 | 3 | 147 | Antigen processing and presentation of exogenous peptide antigen via MHC class I | PSMC6 IKBKB PSMD12 |
| 0.0091 | 3 | 143 | Antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | PSMC6 IKBKB PSMD12 |
| 0.0091 | 4 | 323 | Antigen receptor-mediated signaling pathway | PAG1 PSMC6 IKBKB PSMD12 |
| 0.0091 | 5 | 662 | Immune response-activating signal transduction | PAG1 PSMC6 IKBKB VAV2 PSMD12 |
| 0.0091 | 3 | 118 | Interleukin-1-mediated signaling pathway | IKBKB PSMC6 PSMD12 |
| 0.0091 | 4 | 344 | Cellular response to tumor necrosis factor | IKBKB CASP1 PSMC6 PSMD12 |
| 0.0102 | 5 | 712 | Nucleobase-containing compound catabolic process | CARHSP1 AIFM1 CNP PSMC6 PSMD12 |
| 0.0102 | 2 | 34 | Positive regulation of phosphatidylinositol 3-kinase activity | SH3GLB1 VAV2 |
| 0.0102 | 5 | 714 | Immune response-regulating signaling pathway | PAG1 PSMC6 IKBKB VAV2 PSMD12 |
| 0.0104 | 3 | 167 | Antigen processing and presentation of peptide antigen via MHC class I | PSMC6 IKBKB PSMD12 |
| 0.0110 | 2 | 38 | Positive regulation of lipid kinase activity | SH3GLB1 VAV2 |
| 0.0110 | 3 | 182 | Regulation of cytokine-mediated signaling pathway | CDC37 CASP1 IKBKB |
| 0.0110 | 5 | 763 | Activation of immune response | PAG1 PSMC6 IKBKB VAV2 PSMD12 |
| 0.0110 | 5 | 765 | Cellular nitrogen compound catabolic process | CARHSP1 AIFM1 CNP PSMC6 PSMD12 |
| 0.0110 | 5 | 764 | Heterocycle catabolic process | CARHSP1 AIFM1 CNP PSMC6 PSMD12 |
| 0.0114 | 5 | 778 | Aromatic compound catabolic process | CARHSP1 AIFM1 CNP PSMC6 PSMD12 |
| 0.0122 | 5 | 813 | Organic cyclic compound catabolic process | CARHSP1 AIFM1 CNP PSMC6 PSMD12 |
| Biological proccesses from unique proteins with ratio PMA/mc ≥ 2 | ||||
| 4.15E-05 | 42 | 2,220 | Vesicle-mediated transport | BCAP29 CHMP3 VPS26A HIST1H1A SYT5 SNX27 VTI1A VPS11 VPS28 TMED10 RAB43 SEC24A ATP6AP1 DNM1L DENND3 ESYT2 NUMB BIN1 SYTL3 MCFD2 MYO1E CD300LF TAPBP GRN PKP1 HUWE1 PGRMC1 PSMD7 QSOX1 HVCN1 ATP8A1 UBR4 GMFG DCTN3 ADAM10 S100A7 CD109 FABP5 GHDC AGPAT2 A2M GM2A |
| 5.76E-05 | 22 | 778 | Aromatic compound catabolic process | COMT RBM8A NUDT3 ISG20 NT5C1A RNASEL ADPGK BPGM PKP1 NAF1 SERBP1 RPL18 PSMA7 PSMD7 NUP210 RPS8 RPL9 RPL15 RPL12 PSMB8 NUP62 PSMB9 |
| 5.76E-05 | 21 | 712 | Nucleobase-containing compound catabolic process | RBM8A NUDT3 ISG20 NT5C1A RNASEL ADPGK BPGM PKP1 NAF1 SERBP1 RPL18 PSMA7 PSMD7 NUP210 RPS8 RPL9 RPL15 RPL12 PSMB8 NUP62 PSMB9 |
| 5.76E-05 | 25 | 951 | Viral process | OAS3 CHMP3 RNASEL KARS PSMA7 EIF4H IRF3 UBR4 BIN1 COPS6 ISG15 PSMB8 NUP62 PSMB9 NUCKS1 ISG20 RAB43 PI4KA RPL18 NUP210 RPS8 VPS28 RPL9 RPL15 RPL12 |
| 8.17E-05 | 22 | 813 | Organic cyclic compound catabolic process | COMT RBM8A NUDT3 ISG20 NT5C1A RNASEL ADPGK BPGM PKP1 NAF1 SERBP1 RPL18 PSMA7 PSMD7 NUP210 RPS8 RPL9 RPL15 RPL12 PSMB8 NUP62 PSMB9 |
| 8.17E-05 | 25 | 1,024 | Symbiont process | OAS3 CHMP3 RNASEL KARS PSMA7 EIF4H IRF3 UBR4 BIN1 COPS6 ISG15 PSMB8 NUP62 PSMB9 NUCKS1 ISG20 RAB43 PI4KA RPL18 NUP210 RPS8 VPS28 RPL9 RPL15 RPL12 |
| 9.22E-05 | 21 | 765 | Cellular nitrogen compound catabolic process | RBM8A NUDT3 ISG20 NT5C1A RNASEL ADPGK BPGM PKP1 NAF1 SERBP1 RPL18 PSMA7 PSMD7 NUP210 RPS8 RPL9 RPL15 RPL12 PSMB8 NUP62 PSMB9 |
| 9.22E-05 | 21 | 764 | Heterocycle catabolic process | RBM8A NUDT3 ISG20 NT5C1A RNASEL ADPGK BPGM PKP1 NAF1 SERBP1 RPL18 PSMA7 PSMD7 NUP210 RPS8 RPL9 RPL15 RPL12 PSMB8 NUP62 PSMB9 |
| 0.0001 | 36 | 1,981 | Cellular protein localization | HUWE1 VPS11 BCAP29 TBC1D13 SPCS2 VPS26A TM9SF2 UQCRC2 SNX27 VTI1A VPS28 TMED10 ARL6IP1 RAB43 NUP62 SYNE2 SYNE1 DNM1L RIPOR2 SEC24A ADAM10 SYTL3 PPM1F MTCH2 NUMB EPB41 MYADM RPL18 SRSF6 SSR1 NUP210 RPS8 RPL9 RPL15 RPL12 RBM8A |
| 0.0001 | 48 | 3,087 | Cellular localization | HUWE1 VPS11 BCAP29 DNM1L TBC1D13 COL4A3BP CHMP3 SPCS2 VPS26A TM9SF2 SYT5 UQCRC2 SNX27 VTI1A VPS28 TMED10 ARL6IP1 RAB43 NUP62 SYNE2 SEC24A SYNE1 TRPC3 BAK1 ATP6AP1 DENND3 RIPOR2 ESYT2 NUMB BIN1 ADAM10 SYTL3 PPM1F MTCH2 CPSF6 EPB41 MYADM RPL18 SRSF6 SSR1 NUP210 DCTN3 RPS8 RPL9 RPL15 MCFD2 RPL12 RBM8A |
| 0.0001 | 25 | 1,084 | Interspecies interaction between organisms | OAS3 CHMP3 RNASEL KARS PSMA7 EIF4H IRF3 UBR4 BIN1 COPS6 ISG15 PSMB8 NUP62 PSMB9 NUCKS1 ISG20 RAB43 PI4KA RPL18 NUP210 RPS8 VPS28 RPL9 RPL15 RPL12 |
| 0.0001 | 36 | 1,993 | Cellular macromolecule localization | HUWE1 VPS11 BCAP29 TBC1D13 SPCS2 VPS26A TM9SF2 UQCRC2 SNX27 VTI1A VPS28 TMED10 ARL6IP1 RAB43 NUP62 SYNE2 SYNE1 DNM1L RIPOR2 SEC24A ADAM10 SYTL3 PPM1F MTCH2 NUMB EPB41 MYADM RPL18 SRSF6 SSR1 NUP210 RPS8 RPL9 RPL15 RPL12 RBM8A |
| 0.0001 | 24 | 1,023 | Exocytosis | SYT5 VPS11 ATP6AP1 DNM1L SYTL3 TMED10 GRN PKP1 HUWE1 PGRMC1 PSMD7 QSOX1 HVCN1 ATP8A1 UBR4 GMFG ADAM10 S100A7 CD109 FABP5 GHDC AGPAT2 A2M GM2A |
| 0.0002 | 35 | 1,959 | Intracellular transport | HUWE1 VPS11 BCAP29 TBC1D13 COL4A3BP CHMP3 SPCS2 VPS26A SYT5 UQCRC2 SNX27 VTI1A VPS28 TMED10 ARL6IP1 RAB43 NUP62 SEC24A SYNE2 DNM1L DENND3 SYTL3 CPSF6 BIN1 RPL18 SRSF6 SSR1 NUP210 DCTN3 RPS8 RPL9 RPL15 MCFD2 RPL12 RBM8A |
| 0.0002 | 41 | 2,500 | Cellular catabolic process | HUWE1 DNM1L COMT PSMA7 PSMD7 UBR4 ECHS1 VPS28 CYP2C19 PSMB8 HBE1 PSMB9 RBM8A NUDT3 CAMKK2 ISG20 RENBP DENND3 NT5C1A USP15 RNASEL ADPGK VPS11 BPGM ISG15 GM2A PKP1 QSOX1 NAF1 VTI1A SERBP1 RPL18 CHMP3 VPS26A NUP210 RPS8 RPL9 FABP5 RPL15 RPL12 NUP62 |
| 0.0003 | 46 | 3,011 | Organic substance transport | HUWE1 VPS11 TBC1D13 COL4A3BP CHMP3 SPCS2 VPS26A ATP8A1 SLC17A3 SYT5 UQCRC2 SNX27 VTI1A SLC28A1 VPS28 TMED10 ARL6IP1 RAB43 NUP62 KARS DNM1L RHAG ACSL4 BCAP29 SEC24A ESYT2 IRF3 NUP210 RNASEL SYTL3 FABP5 MCFD2 GM2A RBM8A PPM1F CPSF6 MCU RPL18 ATP6AP1 VDAC3 SRSF6 SSR1 RPS8 RPL9 RPL15 RPL12 |
| 0.0003 | 15 | 465 | RNA catabolic process | RBM8A ISG20 RNASEL PKP1 NAF1 SERBP1 RPL18 PSMA7 PSMD7 RPS8 RPL9 RPL15 RPL12 PSMB8 PSMB9 |
| 0.0003 | 49 | 3,354 | Macromolecule localization | HUWE1 NAF1 VPS11 BCAP29 TBC1D13 COL4A3BP CHMP3 SPCS2 VPS26A ATP8A1 TM9SF2 UQCRC2 SNX27 VTI1A VPS28 TMED10 ARL6IP1 RAB43 NUP62 SYNE2 KARS DNM1L SYNE1 ACSL4 RIPOR2 SEC24A ESYT2 IRF3 NUP210 ADAM10 SYTL3 MCFD2 GM2A RBM8A PPM1F MTCH2 CPSF6 NUMB MCU EPB41 MYADM RPL18 ATP6AP1 SRSF6 SSR1 RPS8 RPL9 RPL15 RPL12 |
| 0.0004 | 41 | 2,592 | Nitrogen compound transport | HUWE1 VPS11 TBC1D13 RHAG COL4A3BP CHMP3 SPCS2 VPS26A SLC17A3 SYT5 UQCRC2 SNX27 VTI1A SLC28A1 VPS28 TMED10 ARL6IP1 RAB43 NUP62 KARS DNM1L BCAP29 TAPBP SEC24A IRF3 NUP210 SYTL3 MCFD2 RBM8A PPM1F CPSF6 MCU RPL18 ATP6AP1 VDAC3 SRSF6 SSR1 RPS8 RPL9 RPL15 RPL12 |
| 0.0004 | 17 | 603 | Granulocyte activation | KARS GRN PKP1 HUWE1 PGRMC1 PSMD7 QSOX1 HVCN1 ATP8A1 UBR4 GMFG ADAM10 S100A7 FABP5 GHDC AGPAT2 GM2A |
| 0.0004 | 25 | 1,194 | Intracellular protein transport | HUWE1 VPS11 TBC1D13 SPCS2 VPS26A UQCRC2 SNX27 VTI1A VPS28 TMED10 ARL6IP1 RAB43 NUP62 BCAP29 SEC24A SYTL3 RPL18 SRSF6 SSR1 NUP210 RPS8 RPL9 RPL15 RPL12 RBM8A |
| 0.0004 | 26 | 1,276 | Organic acid metabolic process | SARS KARS VARS HARS2 ECHS1 YARS ENOPH1 CYP2C19 GRHPR CES2 ACSL4 COMT RENBP HSD17B12 ADPGK FABP5 BPGM SLC17A3 PSMA7 PSMD7 ABHD14B NUP210 B3GNT2 PSMB8 NUP62 PSMB9 |
| 0.0004 | 21 | 901 | Regulated exocytosis | SYT5 DNM1L TMED10 GRN PKP1 HUWE1 PGRMC1 PSMD7 QSOX1 HVCN1 ATP8A1 UBR4 GMFG ADAM10 S100A7 CD109 FABP5 GHDC AGPAT2 A2M GM2A |
| 0.0004 | 43 | 2,825 | Catabolic process | HUWE1 DNM1L COMT PSMA7 PSMD7 UBR4 ECHS1 VPS28 CYP2C19 PSMB8 HBE1 PSMB9 RBM8A NUDT3 CAMKK2 ISG20 IRAK3 RENBP DENND3 NT5C1A USP15 RNASEL ADPGK VPS11 BPGM ISG15 GM2A PKP1 QSOX1 NAF1 VTI1A SERBP1 RPL18 CHMP3 VPS26A NUP210 RPS8 RPL9 FABP5 CES2 RPL15 RPL12 NUP62 |
| 0.0005 | 38 | 2,364 | Establishment of localization in cell | HUWE1 VPS11 BCAP29 TBC1D13 COL4A3BP CHMP3 SPCS2 VPS26A SYT5 UQCRC2 SNX27 VTI1A VPS28 TMED10 ARL6IP1 RAB43 NUP62 SEC24A TRPC3 BAK1 SYNE2 DNM1L DENND3 NUMB SYTL3 CPSF6 BIN1 RPL18 SRSF6 SSR1 NUP210 DCTN3 RPS8 RPL9 RPL15 MCFD2 RPL12 RBM8A |
| 0.0006 | 16 | 577 | Neutrophil degranulation | GRN PKP1 HUWE1 PGRMC1 PSMD7 QSOX1 HVCN1 ATP8A1 UBR4 GMFG ADAM10 S100A7 FABP5 GHDC AGPAT2 GM2A |
| 0.0006 | 17 | 640 | Myeloid cell activation involved in immune response | KARS GRN PKP1 HUWE1 PGRMC1 PSMD7 QSOX1 HVCN1 ATP8A1 UBR4 GMFG ADAM10 S100A7 FABP5 GHDC AGPAT2 GM2A |
| 0.0006 | 16 | 576 | Positive regulation of locomotion | SEMA3E KARS S100A7 CARMIL2 SYNE2 RIPOR2 ATP8A1 PPM1F CHMP3 NUMB GRN CASS4 PFN1 SOD2 ADAM10 MYADM |
| 0.0006 | 55 | 4,098 | Organelle organization | RBBP4 HUWE1 VPS11 RECQL TACC3 SYNE2 ARHGAP10 ADD2 PKP1 DNM1L CASS4 DOT1L H2AFV PFN1 HIST1H1A GMFG UQCRC2 ADD3 VTI1A EPB41 TMED10 WDR5 RPL12 CAMKK2 BAK1 GRN PPM1F SEC24A SYNE1 USP15 SERBP1 CARMIL2 ARL6IP1 HIST1H3A NUCKS1 RPS6KA2 VDAC3 RBBP7 COL4A3BP BIN1 SEMA3E CHMP3 IMMT NAF1 RAB43 DES MYADM NUP62 SOD2 ZSCAN4 TAPBP DCTN3 VPS28 COPS6 MCFD2 |
| 0.0006 | 25 | 1,253 | Cellular protein-containing complex assembly | ADD2 DNM1L EIF4H PFN1 SRSF6 GMFG NAF1 ADD3 VPS11 PRPF8 RPL12 SF3A1 CPSF6 SEC24A CHMP3 CARMIL2 RBBP4 HIST1H3A HIST1H1A BIN1 EIF3H TAPBP MYADM RBBP7 TMED10 |
Proteins used were uniquely expressed in each stimulation group.
Discussion
Knowledge about the molecular mechanisms involved in specific granulocyte activation and subsequent choice of pathways depending on different stressors is still incomplete to date. Moreover, in the past, granulocytes have frequently been underestimated in their ability to execute distinct heterogenic reactions rather than uniform response cascades to any, mainly pathogen-induced stimulus. The past decade has yielded more details on granulocyte heterogeneity and function, not only for processes in the innate immune system but also for regulatory involvement in adaptive immune responses (4, 21). Nevertheless, there are still many signaling processes in granulocyte activation, which need clarification. To gain deeper insight into these processes and to find possible downstream reaction differences between initiating stimuli, we performed a short stimulation assay with freshly obtained, primary equine granulocytes. PMA and LPS, were used as universal stimuli. PMA induces exocytosis, ROS release and NET formation through direct activation of protein kinase C (PKC) and subsequent signal transduction cascade (22). LPS triggers similar responses by binding to TLR4 on neutrophils (23). For specific activation of granulocytes, we used IL8. This cytokine is expressed by a variety of cells, such as monocytes, fibroblasts and endothelial cells and acts as a potent chemoattractant for granulocytes, inducing neutrophil recruitment and chemotaxis via chemokine receptors CXCR1 and CXCR2 (24). Granulocyte activation and identification of resulting differences in behavior, gene regulation and protein expression have previously been performed in other uncommon models such as cattle (25) and pigs (26). Predominantly, studies focus on human granulocytes, however, most of these studies concentrate on one particular morphologic (granules, membrane proteins) (27–29) or functional (NET formation) (30, 31) feature of granulocytes. Few studies describe stimulatory experiments and their effect on the whole granulocyte proteome (32, 33). Compared to these studies, we chose a very short stimulation time in order to detect early proteome changes with possibly transient character. Also, we did not separate the proteome via 2D prior to mass spectrometry analysis.
From our initial proteomics experiment, we unraveled the equine granulocyte proteome, detecting 2,032 proteins (Supplemental Table 1). Similar proteomic based studies have been performed on human granulocytes (34), granulocytes from other species such as cattle (35) and rats (36), as well as neutrophil-associated BALF proteins in horses (37). However, to our knowledge, the full equine neutrophil proteome has not been described to date. Interestingly, 12% of the identified proteins in our study were classified as “hypothetical proteins,” whose existence is predicted, but experimental evidence for in vivo expression is lacking. With our studies, we could confirm actual in vivo expression of these proteins, associating them to primary granulocyte proteome in horses. We chose equine granulocytes to conduct our experiments, because the equine and human immune system share a wide range of similarities both in granulocyte-lymphocyte ratio, composition and function (38–40). Furthermore, the horse is prone to allergies and autoimmune diseases, which are similarly found in humans (41–45) and adaptive as well as innate immune cells from horses have proven to be valuable tools for studying human diseases (37, 42, 44, 46). Despite certain differences between human and horse neutrophils (47, 48), the horse is still a very promising model, especially for processes and diseases which are not adequately addressed by rodent models. However, more investigations are needed to determine its exact and true translational value, which we provide a basis for with our studies.
Among all identified proteins, we found a total of 252 differentially abundant proteins after cell stimulation with different stimuli (Supplemental Table 2). Fifteen of these proteins showed higher expression levels in all three stimulating agent groups (Figure 1, Table 1, Supplemental Figure 1, Supplemental Table 3), indicating onset of some mutual reactions to the different stimuli. A larger number of unique proteins with differential expression per stimulant, however, pointed to predominantly differentiated reactions to the different stimuli (Figure 1, Table 1). Further assessment of all differentially abundant proteins from PMA and IL8 samples with ShinyGO enrichment analysis revealed 57% unique network clustering for each stimulant, respectively (Figure 3, Table 3). This shows the ability of granulocytes to distinguish between stimuli and regulate specific pathways in response to selective cell-stressors, although partial immune response is executed independent of stimulation type. Subsequent analysis of solely those proteins that changed abundance uniquely after either IL8 or PMA stimulation highlighted their association to stimulant-characteristic reactions, such as exocytosis and degranulation after PMA stimulation (22), and cytoskeleton dynamics after stimulation with IL8 (49, 50) (Figure 2, Table 2).
Figure 3.
Network clustering for biological processes to which differentially expressed proteins from IL8 (A) and PMA (B) stimulation were appointed. Three distinct clusters are visible in each stimulation group. Two clusters show similarities between stimulants (dotted blue arrow: granulocyte activation and metabolic processes) whereas one cluster is unique for each group [red arrow: immune response signaling pathways in IL8 stimulated cells (A) and cellular protein localization in PMA stimulated cells (B)]. For a more clear presentation of clusters, we searched 20 most significant categories instead of 30.
Table 3.
Shared and unique functional categories generated from differentially expressed proteins after IL8 and PMA stimulation.
| Functional categories from biological process IL8 and PMA | |||
|---|---|---|---|
| Stimulating agent | Unique | Shared | Unique |
| IL8 | IL8 /PMA | PMA | |
| No. of categories | 17 | 13 | 17 |
| Functional categories | Antigen receptor-mediated signaling pathway | Aromatic compound catabolic process | Cellular localization |
| Cell activation | Cellular catabolic process | Cellular macromolecule localization | |
| Cellular response to mineralocorticoid stimulus | Cellular nitrogen compound catabolic process | Cellular protein localization | |
| DNA catabolic process | Granulocyte activation | Establishment of localization in cell | |
| DNA catabolic process, endonucleolytic | Heterocycle catabolic process | Exocytosis | |
| Fc receptor signaling pathway | Leukocyte degranulation | Interspecies interaction between organisms | |
| Fc-epsilon receptor signaling pathway | Myeloid cell activation involved in immune response | Intracellular protein transport | |
| Immune response | Neutrophil activation | Intracellular transport | |
| Immune response-activating cell surface receptor signaling pathway | Neutrophil activation involved in immune response | Macromolecule localization | |
| Immune response-regulating cell surface receptor signaling pathway | Neutrophil degranulation | Nitrogen compound transport | |
| Macromolecule catabolic process | Neutrophil mediated immunity | Organic substance transport | |
| MRNA metabolic process | Nucleobase-containing compound catabolic process | Regulated exocytosis | |
| Regulation of cellular catabolic process | Organic cyclic compound catabolic process | Regulation of biological quality | |
| Regulation of mRNA stability | RNA catabolic process | ||
| Regulation of RNA stability | Symbiont process | ||
| Response to organic substance | Vesicle-mediated transport | ||
| Tumor necrosis factor-mediated signaling pathway | Viral process | ||
IL8 stimulation yielded the identification of proteasome 26S subunit, ATPase 6 (PSMC6), which showed higher abundance unique to this stimulant (IL8/mc ratio 2.1; p < 0.001). PSMC6 is an ATP-dependent proteolytic complex responsible for ubiquitin-dependent protein degradation (51, 52), which is an important regulator of the majority of cellular activity and homeostasis (53). Divergent levels of proteasome activity have a strong impact on disease pathogenesis of several diseases and are used as drug targets in disease treatment (51, 54–56). Thus, the higher abundance of PSMC6 in IL8 stimulated cells might indicate activation of the proteasome in granulocytes with functional importance in downstream regulation of immune response to stress. Subsequent analysis of proteomic data from IL8 stimulated cells revealed that PSMC6 was present in the majority of functional enrichment clusters from biological processes, including the top enriched functional categories tumor necrosis factor (TNF) mediated signaling and Fc-epsilon receptor pathways (Table 2). These two pathways are essential for signal transduction in cells, with a wide functional variety of downstream responses such as apoptosis but also immune and inflammatory responses as well as cell survival, activation and differentiation (57, 58). Interestingly, occurrence of Fc receptors on granulocytes have initially been described as a marker of neutrophil heterogeneity rather than a necessity for optimal neutrophil aggregation and adhesion (59). Especially Fc-epsilon receptor signaling is only present in neutrophils under certain conditions and their exact role is still discussed among experts, whereas other Fc receptor types, such as low-affinity Fc-gamma receptors, are commonly expressed on granulocytes playing an important role in immune complex mediated activation of neutrophils through their downstream pathways (58). Furthermore, Fc receptors are unlikely to mediate PMA-induced cell activation (59), which is consistent with our findings on PMA-stimulated granulocytes, where we found no allocation of uniquely expressed proteins to Fc receptor signaling pathways (Figures 2, 3, Tables 2, 3).
Our findings undermine the ongoing appreciation of granulocyte function toward finely tuned, heterogeneous, specific reactions of more than one subpopulation of neutrophils (4, 7, 8, 60–63). Furthermore, our data shows that a stimulation time of only 30 min is sufficient to initiate substantial and specific changes in granulocyte proteome as reaction to individual stimulating agents (Figure 1, Table 1, Supplemental Table 2). These rapid changes most likely occur due to gene induction of early responding genes but may also be the result of posttranslational modifications mediated by proteins that are activated early in neutrophil responses to stimuli, such as phosphatidylinositol 3-kinase (PI 3-kinase). Interestingly, PI 3-kinase activity appears as functional category from enrichment analysis of our IL8 data (Figure 2A, Table 2), supporting the involvement of PI 3-kinase in IL8-induced protein changes. With its ability to phosphorylate molecules acting as second messengers and thereby switch on downstream intracellular signaling (64), and its involvement in neutrophil chemokinesis and phagosome formation (49, 50, 65), PI 3-kinase merits further investigations in future functional studies. No matter the origin of the changed granulocyte protein repertoire described in our data, it gives insight into early onset of granulocyte activation on protein level, which may be useful to modulate granulocyte mediated pathological processes in future functional experiments. However, more experiments are needed, not only for determination of minimal stimulation times triggering regulation of protein expression levels in granulocytes, but also for analysis of expression kinetics in course of longer stimulation assays. From other comprehensive studies on equine neutrophils we know, that neutrophil extracellular traps (NETs) readily occur in response to adequate stimuli (66) as opposed to cells from other animal models (67). Our protein data however, lack association to this process (Figure 2, Table 2). We assume that the expression differences of proteins associated to NET formation occur after longer stimulation time, as recently described (66). Therefore, increasing the stimulation time in these assays could address protein repertoire changes associated to NET-relevant biological processes such as DNA decondensation, histone citrullination, and related signal transduction. Also, we would expect more prominent clustering of IL8 induced protein changes to cytoskeleton dynamics involved in chemotaxis and phagosome formation, as functional answers of cells to stimuli fluctuate over time (68). Keeping in mind the dynamic character of protein expression patterns in course of cell activation, our data put a spotlight merely on the first reaction to stimuli. This is a very interesting time point in our opinion, because it shows the initiating functional answers in activated cells, which are potentially accessible to experimental modulation. Adding proteomic data from more stimulation times would give a more precise insight into dynamic whole-cell proteome changes throughout the activation process of granulocytes, similar to previous analysis of pre-determined cytokines and degranulation markers by kinetic flow cytometry (69), which needs to be addressed in future studies.
Conclusion
With our data we provide a fundamental study on activation of primary granulocytes and regulation of downstream immune response by showing that different stimuli provoke divergent and rapid downstream responses through regulation of protein expression in these cells. These expression differences show involvement in various different pathways and biological processes which, among some similarities, differ between stimuli and support knowledge on heterogeneity of granulocytes and their highly selective response to stimuli. The presented data may therefore act as a guide for further, in-depth research on granulocyte response patterns and behavior in health and disease.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (70) partner repository with the dataset identifier PXD013648.
Ethics Statement
Collection of blood was permitted by the local authority, Regierung von Oberbayern (Permit number: ROB-55.2Vet-2532.Vet_03-17-88).
Author Contributions
CD conceived and designed the experiments. RD and SH performed the experiments. RD, SH, MW, and CD analyzed the data. RD wrote the manuscript. All authors critically read the manuscript and approved the final version to be published.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Tanja Witte, Kirsten Hahn, and Yvette Ballauf for support in blood withdrawal from the horses.
Footnotes
Funding. This work was supported by grants from the Deutsche Forschungsgemeinschaft DFG DE 719/4-3 (to CD).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.03064/full#supplementary-material
References
- 1.van der Linden M, Meyaard L. Fine-tuning neutrophil activation: strategies and consequences. Immunol Lett. (2016) 178:3–9. 10.1016/j.imlet.2016.05.015 [DOI] [PubMed] [Google Scholar]
- 2.de Buhr N, Bonilla MC, Jimenez-Soto M, von Kockritz-Blickwede M, Dolz G. Extracellular trap formation in response to Trypanosoma cruzi infection in granulocytes isolated from dogs and common opossums, natural reservoir hosts. Front Microbiol. (2018) 9:966. 10.3389/fmicb.2018.00966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakravarti A, Rusu D, Flamand N, Borgeat P, Poubelle PE. Reprogramming of a subpopulation of human blood neutrophils by prolonged exposure to cytokines. Lab Invest. (2009) 89:1084–99. 10.1038/labinvest.2009.74 [DOI] [PubMed] [Google Scholar]
- 4.Eruslanov EB, Singhal S, Albelda SM. Mouse versus human neutrophils in cancer: a major knowledge gap. Trends Cancer. (2017) 3:149–60. 10.1016/j.trecan.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deniset JF, Kubes P. Neutrophil heterogeneity: bona fide subsets or polarization states? J Leukoc Biol. (2018) 103:829–38. 10.1002/JLB.3RI0917-361R [DOI] [PubMed] [Google Scholar]
- 6.Grieshaber-Bouyer R, Nigrovic PA. Neutrophil heterogeneity as therapeutic opportunity in immune-mediated disease. Front Immunol. (2019) 10:346. 10.3389/fimmu.2019.00346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng LG, Ostuni R, Hidalgo A. Heterogeneity of neutrophils. Nat Rev Immunol. (2019) 19:255–65. 10.1038/s41577-019-0141-8 [DOI] [PubMed] [Google Scholar]
- 8.Kubes P. The enigmatic neutrophil: what we do not know. Cell Tissue Res. (2018) 371:399–406. 10.1007/s00441-018-2790-5 [DOI] [PubMed] [Google Scholar]
- 9.Degroote RL, Hauck SM, Amann B, Hirmer S, Ueffing M, Deeg CA. Unraveling the equine lymphocyte proteome: differential septin 7 expression associates with immune cells in equine recurrent uveitis. PLoS ONE. (2014) 9:e91684. 10.1371/journal.pone.0091684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Degroote RL, Helm S, Klein U, Schmitt R, Ueffing M, Hauck SM, et al. The equine CD4+ lymphocyte proteome. Dataset Pap Sci. (2014) 2014:4 10.1155/2014/105312 [DOI] [Google Scholar]
- 11.Degroote RL, Uhl PB, Amann B, Krackhardt AM, Ueffing M, Hauck SM, et al. Formin like 1 expression is increased on CD4+ T lymphocytes in spontaneous autoimmune uveitis. J Proteomics. (2017) 154:102–8. 10.1016/j.jprot.2016.12.015 [DOI] [PubMed] [Google Scholar]
- 12.Schauer M, Kleinwort KJH, Degroote RL, Wiedemann C, Kremmer E, Hauck SM, et al. Interaction of septin 7 and DOCK8 in equine lymphocytes reveals novel insights into signaling pathways associated with autoimmunity. Sci Rep. (2018) 8:12332. 10.1038/s41598-018-30753-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Degroote RL, Hauck SM, Kremmer E, Amann B, Ueffing M, Deeg CA. Altered expression of talin 1 in peripheral immune cells points to a significant role of the innate immune system in spontaneous autoimmune uveitis. J Proteomics. (2012) 75:4536–44. 10.1016/j.jprot.2012.01.023 [DOI] [PubMed] [Google Scholar]
- 14.Degroote RL, Hauck SM, Treutlein G, Amann B, Frohlich KJ, Kremmer E, et al. Expression changes and novel interaction partners of Talin 1 in effector cells of autoimmune uveitis. J Proteome Res. (2013) 12:5812–9. 10.1021/pr400837f [DOI] [PubMed] [Google Scholar]
- 15.Kanamaru R, Ohzawa H, Miyato H, Matsumoto S, Haruta H, Kurashina K, et al. Low density neutrophils (LDN) in postoperative abdominal cavity assist the peritoneal recurrence through the production of neutrophil extracellular traps (NETs). Sci Rep. (2018) 8:632. 10.1038/s41598-017-19091-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. (1976) 72:248–54. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 17.Grosche A, Hauser A, Lepper MF, Mayo R, von Toerne C, Merl-Pham J, et al. The proteome of native adult muller glial cells from murine retina. Mol Cell Proteomics. (2016) 15:462–80. 10.1074/mcp.M115.052183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauck SM, Hofmaier F, Dietter J, Swadzba ME, Blindert M, Amann B, et al. Label-free LC-MSMS analysis of vitreous from autoimmune uveitis reveals a significant decrease in secreted Wnt signalling inhibitors DKK3 and SFRP2. J Proteomics. (2012) 75:4545–54. 10.1016/j.jprot.2012.04.052 [DOI] [PubMed] [Google Scholar]
- 19.Hauck SM, Lepper MF, Hertl M, Sekundo W, Deeg CA. Proteome dynamics in biobanked horse peripheral blood derived lymphocytes (PBL) with induced autoimmune uveitis. Proteomics. (2017) 17:1700013. 10.1002/pmic.201700013 [DOI] [PubMed] [Google Scholar]
- 20.Ge S, Jung D. ShinyGO: a graphical enrichment tool for animals and plants. bioRxiv. (2018) 315150. 10.1101/315150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Wang W, Yang F, Xu Y, Feng C, Zhao Y. The regulatory roles of neutrophils in adaptive immunity. Cell Commun Signal. (2019) 17:147. 10.1186/s12964-019-0471-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito T, Takahashi H, Doken H, Koyama H, Aratani Y. Phorbol myristate acetate induces neutrophil death through activation of p38 mitogen-activated protein kinase that requires endogenous reactive oxygen species other than HOCl. Biosci Biotechnol Biochem. (2005) 69:2207–12. 10.1271/bbb.69.2207 [DOI] [PubMed] [Google Scholar]
- 23.Yipp BG, Kim JH, Lima R, Zbytnuik LD, Petri B, Swanlund N, et al. (2017). The lung is a host defense niche for immediate neutrophil-mediated vascular protection. Sci Immunol. 2:eaam8929. 10.1126/sciimmunol.aam8929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.French SW, Mendoza AS, Afifiyan N, Tillman B, Vitocruz E, French BA. The role of the IL-8 signaling pathway in the infiltration of granulocytes into the livers of patients with alcoholic hepatitis. Exp Mol Pathol. (2017) 103:137–40. 10.1016/j.yexmp.2017.08.005 [DOI] [PubMed] [Google Scholar]
- 25.Bassel LL, Caswell JL. Bovine neutrophils in health and disease. Cell Tissue Res. (2018) 371:617–37. 10.1007/s00441-018-2789-y [DOI] [PubMed] [Google Scholar]
- 26.de Buhr N, Reuner F, Neumann A, Stump-Guthier C, Tenenbaum T, Schroten H, et al. Neutrophil extracellular trap formation in the Streptococcus suis-infected cerebrospinal fluid compartment. Cell Microbiol. (2017) 19:e12649. 10.1111/cmi.12649 [DOI] [PubMed] [Google Scholar]
- 27.Lominadze G, Powell DW, Luerman GC, Link AJ, Ward RA, McLeish KR. Proteomic analysis of human neutrophil granules. Mol Cell Proteomics. (2005) 4:1503–21. 10.1074/mcp.M500143-MCP200 [DOI] [PubMed] [Google Scholar]
- 28.Xu P, Crawford M, Way M, Godovac-Zimmermann J, Segal AW, Radulovic M. Subproteome analysis of the neutrophil cytoskeleton. Proteomics. (2009) 9:2037–49. 10.1002/pmic.200800674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rorvig S, Ostergaard O, Heegaard NH, Borregaard N. Proteome profiling of human neutrophil granule subsets, secretory vesicles, and cell membrane: correlation with transcriptome profiling of neutrophil precursors. J Leukoc Biol. (2013) 94:711–21. 10.1189/jlb.1212619 [DOI] [PubMed] [Google Scholar]
- 30.Chapman EA, Lyon M, Simpson D, Mason D, Beynon RJ, Moots RJ, et al. Caught in a trap? proteomic analysis of neutrophil extracellular traps in rheumatoid arthritis and systemic lupus erythematosus. Front Immunol. (2019) 10:423. 10.3389/fimmu.2019.00423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petretto A, Bruschi M, Pratesi F, Croia C, Candiano G, Ghiggeri G, et al. Neutrophil extracellular traps (NET) induced by different stimuli: a comparative proteomic analysis. PLoS ONE. (2019) 14:e0218946. 10.1371/journal.pone.0218946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fessler MB, Malcolm KC, Duncan MW, Worthen GS. A genomic and proteomic analysis of activation of the human neutrophil by lipopolysaccharide and its mediation by p38 mitogen-activated protein kinase. J Biol Chem. (2002) 277:31291–302. 10.1074/jbc.M200755200 [DOI] [PubMed] [Google Scholar]
- 33.Aquino EN, Neves AC, Santos KC, Uribe CE, Souza PE, Correa JR, et al. Proteomic analysis of neutrophil priming by PAF. Protein Pept Lett. (2016) 23:142–51. 10.2174/0929866523666151202210604 [DOI] [PubMed] [Google Scholar]
- 34.Tomazella GG, da Silva I, Laure HJ, Rosa JC, Chammas R, Wiker HG, et al. Proteomic analysis of total cellular proteins of human neutrophils. Proteome Sci. (2009) 7:32. 10.1186/1477-5956-7-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lippolis JD, Reinhardt TA. Proteomic survey of bovine neutrophils. Vet Immunol Immunopathol. (2005) 103:53–65. 10.1016/j.vetimm.2004.08.019 [DOI] [PubMed] [Google Scholar]
- 36.Piubelli C, Galvani M, Hamdan M, Domenici E, Righetti PG. Proteome analysis of rat polymorphonuclear leukocytes: a two-dimensional electrophoresis/mass spectrometry approach. Electrophoresis. (2002) 23:298–310. [DOI] [PubMed] [Google Scholar]
- 37.Bright LA, Dittmar W, Nanduri B, McCarthy FM, Mujahid N, Costa LR, et al. Modeling the pasture-associated severe equine asthma bronchoalveolar lavage fluid proteome identifies molecular events mediating neutrophilic airway inflammation. Vet Med. (2019) 10:43–63. 10.2147/VMRR.S194427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khandpur R, Carmona-Rivera C, Vivekanandan-Giri A, Gizinski A, Yalavarthi S, Knight JS, et al. NETs are a source of citrullinated autoantigens and stimulate inflammatory responses in rheumatoid arthritis. Sci Transl Med. (2013) 5:178ra140. 10.1126/scitranslmed.3005580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zschaler J, Schlorke D, Arnhold J. Differences in innate immune response between man and mouse. Crit Rev Immunol. (2014) 34:433–54. 10.1615/CritRevImmunol.2014011600 [DOI] [PubMed] [Google Scholar]
- 40.Horohov DW. The equine immune responses to infectious and allergic disease: a model for humans? Mol Immunol. (2015) 66:89–96. 10.1016/j.molimm.2014.09.020 [DOI] [PubMed] [Google Scholar]
- 41.Rickards KJ, Page CP, Lees P, Cunningham FM. Phosphodiesterase activity in neutrophils from horses with chronic obstructive pulmonary disease. Vet Immunol Immunopathol. (2000) 76:319–30. 10.1016/S0165-2427(00)00220-8 [DOI] [PubMed] [Google Scholar]
- 42.Deeg CA, Hauck SM, Amann B, Pompetzki D, Altmann F, Raith A, et al. Equine recurrent uveitis–a spontaneous horse model of uveitis. Ophthalmic Res. (2008) 40:151–3. 10.1159/000119867 [DOI] [PubMed] [Google Scholar]
- 43.Herteman N, Vargas A, Lavoie JP. Characterization of circulating low-density neutrophils intrinsic properties in healthy and asthmatic horses. Sci Rep. (2017) 7:7743. 10.1038/s41598-017-08089-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vargas A, Boivin R, Cano P, Murcia Y, Bazin I, Lavoie JP. Neutrophil extracellular traps are downregulated by glucocorticosteroids in lungs in an equine model of asthma. Respir Res. (2017) 18:207. 10.1186/s12931-017-0689-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheats MK. A comparative review of equine SIRS, sepsis, and neutrophils. Front Vet Sci. (2019) 6:69. 10.3389/fvets.2019.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Busch M, Wefelmeyer KL, Walscheid K, Rothaus K, Bauer D, Deeg CA, et al. Identification of ocular autoantigens associated with juvenile idiopathic arthritis-associated uveitis. Front Immunol. (2019) 10:1793. 10.3389/fimmu.2019.01793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scudamore CL, Pemberton A, Watson ED, Miller HR. Neutrophil chemotaxis in the horse is not mediated by a complex of equine neutrophil elastase and equine alpha-1-proteinase inhibitor. Br Vet J. (1993) 149:331–8. 10.1016/S0007-1935(05)80074-0 [DOI] [PubMed] [Google Scholar]
- 48.Brazil TJ, Dixon PM, Haslett C, Murray J, McGorum BC. Constitutive apoptosis in equine peripheral blood neutrophils in vitro. Vet J. (2014) 202:536–42. 10.1016/j.tvjl.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferguson GJ, Milne L, Kulkarni S, Sasaki T, Walker S, Andrews S, et al. PI(3)Kgamma has an important context-dependent role in neutrophil chemokinesis. Nat Cell Biol. (2007) 9:86–91. 10.1038/ncb1517 [DOI] [PubMed] [Google Scholar]
- 50.Feng S, Zhou L, Zhang Y, Lu S, Long M. Mechanochemical modeling of neutrophil migration based on four signaling layers, integrin dynamics, and substrate stiffness. Biomech Model Mechanobiol. (2018) 17:1611–30. 10.1007/s10237-018-1047-2 [DOI] [PubMed] [Google Scholar]
- 51.Schmidt M, Finley D. Regulation of proteasome activity in health and disease. Biochim Biophys Acta. (2014) 1843:13–25. 10.1016/j.bbamcr.2013.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Cimermancic P, Yu C, Schweitzer A, Chopra N, Engel JL, et al. Molecular details underlying dynamic structures and regulation of the human 26S proteasome. Mol Cell Proteomics. (2017) 16:840–54. 10.1074/mcp.M116.065326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pashevin DO, Nagibin VS, Tumanovska LV, Moibenko AA, Dosenko VE. Proteasome inhibition diminishes the formation of neutrophil extracellular traps and prevents the death of cardiomyocytes in coculture with activated neutrophils during anoxia-reoxygenation. Pathobiology. (2015) 82:290–8. 10.1159/000440982 [DOI] [PubMed] [Google Scholar]
- 54.Wang J, Maldonado MA. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cell Mol Immunol. (2006) 3:255–61. [PubMed] [Google Scholar]
- 55.Sjakste T, Paramonova N, Osina K, Dokane K, Sokolovska J, Sjakste N. Genetic variations in the PSMA3, PSMA6 and PSMC6 genes are associated with type 1 diabetes in Latvians and with expression level of number of UPS-related and T1DM-susceptible genes in HapMap individuals. Mol Genet Genomics. (2016) 291:891–903. 10.1007/s00438-015-1153-0 [DOI] [PubMed] [Google Scholar]
- 56.Shi CX, Kortum KM, Zhu YX, Bruins LA, Jedlowski P, Votruba PG, et al. CRISPR genome-wide screening identifies dependence on the proteasome subunit PSMC6 for bortezomib sensitivity in multiple myeloma. Mol Cancer Ther. (2017) 16:2862–70. 10.1158/1535-7163.MCT-17-0130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dempsey PW, Doyle SE, He JQ, Cheng G. The signaling adaptors and pathways activated by TNF superfamily. Cytokine Growth Factor Rev. (2003) 14:193–209. 10.1016/S1359-6101(03)00021-2 [DOI] [PubMed] [Google Scholar]
- 58.Futosi K, Fodor S, Mocsai A. Neutrophil cell surface receptors and their intracellular signal transduction pathways. Int Immunopharmacol. (2013) 17:638–50. 10.1016/j.intimp.2013.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spagnuolo PJ, Fain M, Bass SN. Dissociation of neutrophil aggregation, adhesiveness, and Fc receptor activity. Am J Hematol. (1987) 26:221–8. 10.1002/ajh.2830260303 [DOI] [PubMed] [Google Scholar]
- 60.Jablonska J, Granot Z. Neutrophil, quo vadis? J Leukoc Biol. (2017) 102:685–8. 10.1189/jlb.3MR0117-015R [DOI] [PubMed] [Google Scholar]
- 61.Garley M, Jablonska E. Heterogeneity among neutrophils. Arch Immunol Ther Exp. (2018) 66:21–30. 10.1007/s00005-017-0476-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Battaglia M, Petrelli A, Vecchio F. Neutrophils and type 1 diabetes: current knowledge and suggested future directions. Curr Opin Endocrinol Diabetes Obes. (2019) 26:201–6. 10.1097/MED.0000000000000485 [DOI] [PubMed] [Google Scholar]
- 63.Rungelrath V, Kobayashi SD, DeLeo FR. Neutrophils in innate immunity and systems biology-level approaches: an update. Wiley Interdiscip Rev Syst Biol Med. (2019) 12:e1458 10.1002/wsbm.1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karlsson A, Nixon JB, McPhail LC. Phorbol myristate acetate induces neutrophil NADPH-oxidase activity by two separate signal transduction pathways: dependent or independent of phosphatidylinositol 3-kinase. J Leukoc Biol. (2000) 67:396–404. 10.1002/jlb.67.3.396 [DOI] [PubMed] [Google Scholar]
- 65.Koga H, Kasprick A, Lopez R, Auli M, Pont M, Godessart N, et al. Therapeutic effect of a novel phosphatidylinositol-3-kinase delta inhibitor in experimental epidermolysis bullosa acquisita. Front Immunol. (2018) 9:1558. 10.3389/fimmu.2018.01558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fingerhut L, Ohnesorge B, von Borstel M, Schumski A, Strutzberg-Minder K, Morgelin M, et al. Neutrophil extracellular traps in the pathogenesis of equine recurrent uveitis (ERU). Cells. (2019) 8:E1528. 10.3390/cells8121528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neubert E, Senger-Sander SN, Manzke VS, Busse J, Polo E, Scheidmann SEF, et al. Serum and serum albumin inhibit in vitro formation of neutrophil extracellular traps (NETs). Front Immunol. (2019) 10:12. 10.3389/fimmu.2019.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takami M, Terry V, Petruzzelli L. Signaling pathways involved in IL-8-dependent activation of adhesion through Mac-1. J Immunol. (2002) 168:4559–66. 10.4049/jimmunol.168.9.4559 [DOI] [PubMed] [Google Scholar]
- 69.Naegelen I, Beaume N, Plancon S, Schenten V, Tschirhart EJ, Brechard S. Regulation of neutrophil degranulation and cytokine secretion: a novel model approach based on linear fitting. J Immunol Res. (2015) 2015:817038. 10.1155/2015/817038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. (2019) 47:D442–50. 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (70) partner repository with the dataset identifier PXD013648.