ABSTRACT
MicroRNAs (miRNAs) have emerged as key regulators orchestrating a wide range of inflammatory and fibrotic diseases. However, the role of miRNAs in degenerative shoulder joint disorders is poorly understood. The aim of this explorative case‐control study was to identify pathology‐related, circulating miRNAs in patients with chronic rotator cuff tendinopathy and degenerative rotator cuff tears (RCT). In 2017, 15 patients were prospectively enrolled and assigned to three groups based on the diagnosed pathology: (i) no shoulder pathology, (ii) chronic rotator cuff tendinopathy, and (iii) degenerative RCTs. In total, 14 patients were included. Venous blood samples (“liquid biopsies”) were collected from each patient and serum levels of 187 miRNAs were determined. Subsequently, the change in expression of nine candidate miRNAs was verified in tendon biopsy samples, collected from patients who underwent arthroscopic shoulder surgery between 2015 and 2018. Overall, we identified several miRNAs to be progressively deregulated in sera from patients with either chronic rotator cuff tendinopathy or degenerative RCTs. Importantly, for the several of these miRNAs candidates repression was also evident in tendon biopsies harvested from patients who were treated for a supraspinatus tendon tear. As similar expression profiles were determined for tendon samples, the newly identified systemic miRNA signature has potential as novel diagnostic or prognostic biomarkers for degenerative rotator cuff pathologies. © 2019 The Authors. Journal of Orthopaedic Research ® published by Wiley Periodicals, Inc. on behalf of Orthopaedic Research Society. Inc. J Orthop Res 38:202–211, 2020
Keywords: biomarker, circulating microRNA, rotator cuff, tendinopathy, tendon degeneration
Traumatic injuries and degenerative changes of the rotator cuff are one of the major causes of shoulder pain and dysfunction causing persistent pain and restricted mobility. Due to an increasingly ageing population, shoulder pain in general represents a major future socio‐economic burden, with currently more than 4.5 million visits to physicians annually and associated costs of US$ 5 billion in the United States alone. A rotator cuff tear (RCT) is either traumatic or degenerative with a strong age‐dependent prevalence.1, 2 Tendinopathy as part of the degenerative tendon continuum is linked to both intrinsic and extrinsic patient‐specific factors. While the majority of RCTs have traditionally been mainly related to wear‐related processes in consequence of anatomic variations or repetitive overuse, recent studies have proposed a multifactorial pathogenesis of chronic RCTs.3 Based on clinical and experimental data, microdisruption of tendon fibers is followed by inflammatory (e.g., overexpression of cytokines) and degenerative (e.g., thinning and disorientation of collagen fibers) changes of the normal tendon structure followed by a neo‐angiogenic response and degradation of the extracellular matrix.4, 5 Since tendinopathy is a chronic, long lasting condition, a prognostic statement about either a favorable spontaneous course toward self‐healing or a potential progression toward tendon failure is always difficult.
Generally, early diagnosis, prognosis, and risk assessment based on analyses of minimally invasive sample materials (“liquid biopsies”) are preferred.6 Recently, several studies have demonstrated the feasibility of employing circulating miRNA signatures as diagnostic tools.7, 8 MicroRNAs (miRNAs) are small, non‐coding RNA species and play an important regulatory role in almost any biological process. They regulate gene expression post‐transcriptionally, that is, they are able to control the amount of coding messenger RNA and thus influence the development and function of certain tissues. With regard to tendon disease, specific miRNAs identified in human tissue specimens were found to influence various stages of tendinopathy, for example, miR‐28 mediates oxidative stress‐induced tenocyte apoptosis or miR‐421‐5p regulates local expression of metalloproteases (MMP) 2 and 9 and drives angiogenesis by enhancing vascular endothelial growth factor (VEGF) production.9 Further, Millar et al.10 found a correlation of miR‐29a expression and the development of tendinopathy. However, to our knowledge, no published data exist characterizing a correlation between systemically expressed miRNA species and degenerative rotator cuff pathologies. Therefore, the aim of this exploratory study was to identify candidate miRNA profiles from peripheral blood samples of patients with chronic tendinopathy and of patients with degenerative RCTs. These data will contribute to determine the relevance of circulating miRNAs as a potential novel diagnostic tool for degenerative rotator cuff disorders.
MATERIAL AND METHODS
An application was submitted to the local ethics committee to conduct the study and was approved prior to commencement (201609_EK06 and EK‐2014‐1809). Informed consent was obtained from every enrolled patient. This explorative case‐control study (level of evidence: III) included two independent in vitro analyses. First, the expression profile of circulating miRNAs was assessed. In 2017, a total of 15 patients were prospectively recruited at the outpatient clinic of the Department of Orthopedics (St. Vincent Hospital, Vienna, Austria). Second, the expression level of specific miRNAs in ruptured human supraspinatus (SSP; n = 4) or intact subscapularis (SSC) tendons from patients who had suffered a complete SSP tendon rupture (n = 7) were obtained during arthroscopic rotator cuff repair between 2015 and 2018 at (i) the Department of Orthopedics and Traumatology (Paracelsus Medical University, Salzburg, Austria), (ii) Center for Musculoskeletal Surgery (Charité Universitaetsmedizin, Berlin, Germany) and (iii) German Shoulder Centre (ATOS Clinic, Munich, Germany). The arthroscopic procedure itself was indicated due to a preexisting pathology and not by the study protocol.
Study 1 ‐ Patient Population
The patient groups to be examined was composed as follows: (i) patients without any shoulder complaints (healthy control, CTRL), (ii) patients with chronic rotator cuff tendinopathy (chronic tendinopathy, CT) and (iii) patients with degenerative RCTs (degenerative rotator cuff tear [degRCT]). Five patients meeting the inclusion and exclusion criteria were consecutively assigned to each group. In general, males and females aged between 50 and 70 years were included. The specific diagnosis was made according to the anamnesis and clinical as well as radiological assessment, which was performed by the principal investigator on the day of initial presentation. Specifically, patients with chronic tendinopathy complained of extensive shoulder pain, especially while moving the affected arm. Consequently, clinical assessment revealed a weak shoulder due to associated pain. Radiographic criteria were defined independently for each group. In group 1, bilateral ultrasound assessment was performed to rule out any rotator cuff lesion in patients without any shoulder complaints. Magnetic resonance imaging (MRI) of the affected shoulder was used to evaluate the rotator cuff in groups 2 and 3. A chronic tendinopathy (group 2) was defined as an inhomogeneous rotator cuff tendon with increased intrasubstance signal intensity on oblique coronal proton density (PD) and fat‐suppressed T2 images (Fig. 1B).11 In group 3, a full‐thickness tear of the posterosuperior rotator cuff was defined as a radiological inclusion criterion (Fig. 1A). Tendon retraction was evaluated on coronal T2 images and classified according to the Patte grading system,12 whereby retraction not further than to the humeral apex (grade 2) was accepted. Muscle degeneration was graded according to Goutallier et al. on sagittal T1‐weighted images.13 Patients showing signs of progressed degeneration (stages 3 and 4) were not included. Analysis was performed by the principal investigator as well as a highly specialized musculoskeletal radiologist who was blinded to the study protocol, working in consensus. Exclusion criteria for all groups are shown in Table 1.
Figure 1.
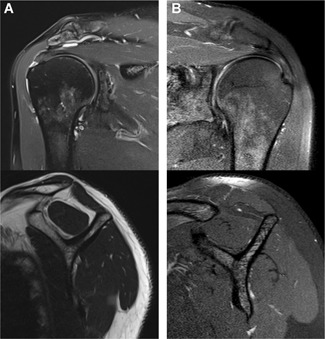
Magnetic resonance imaging of the affected shoulder. The oblique coronal imaging is shown on the top row and the sagittal images on the bottom row, respectively. (A) A full‐thickness rotator cuff tear with slight tendon retraction and mild muscle degeneration. (B) A chronic tendinopathy without any tearing of the supraspinatus tendon and muscle degeneration.
Table 1.
Exclusion Criteria for all Groups
| Patient age below 50 years or above 70 years |
| Clinical symptoms for over 12 months |
| Type 1 and 2 diabetes mellitus |
| Thyroid dysfunction |
| Rheumatic disease |
| Positive smoking anamnesis |
| Traumatic rotator cuff tear |
| Corticosteroid intake within 3 weeks |
| Previous surgery on both shoulders |
| Regular overhead sports activity |
| Overhead occupation |
| Shoulder arthrosis |
| Immunosuppression |
| Tumor disease |
| Pregnancy |
Liquid Biopsy Collection and miRNA Profiling
Fasting venous blood was collected from each patient. A total of two serum tubes with approximately 2 ml of venous blood each were drawn and subsequently coagulated at room temperature for 10–30 min. Samples were then centrifuged at 15,000 rpm for 10 min and subsequently, sera were immediately stored at −80°C until further use.
Total RNA Extraction
Total RNA was extracted from sera using the miRNeasy Mini Kit (Qiagen, Hilden, Germany). Samples were thawed on ice and centrifuged at 12.000 rpm for 5 min to remove any remaining cellular debris. For each sample, 200 µl of serum was mixed with 1,000 µl Qiazol and 1 µl of a mix of three synthetic spike‐in controls (Exiqon, Vedbaek, Denmark). After 10 min incubation at RT, 200 µl chloroform was added to the lysates followed by centrifugation at 12,000 rpm for 15 min at +4°C. Precisely 650 µl of the upper aqueous phase was mixed with 7 µl glycogen (50 mg/ml) to enhance precipitation. Samples were then transferred to a miRNeasy mini‐column, and RNA was precipitated with 750 µl ethanol followed by automated washing with RPE and RWT buffer in a QiaCube liquid handling robot. Finally, total RNA was eluted in 30 µl nuclease‐free water and stored at −80°C until further analysis.
Reverse‐Transcription and Quantitative Polymerase Chain Reaction (RT‐qPCR)
RT‐qPCR based miRNA screening was performed by TAmiRNA (Vienna, Austria). Starting from total RNA samples, complementary deoxyribonucleic acid (cDNA) was synthesized using the Universal cDNA Synthesis Kit II (Exiqon). Reaction conditions were set according to recommendations by the manufacturer. In total, 2 µl of total RNA was used per 10 µl RT reaction. To monitor RT efficiency and presence of impurities with inhibitory activity, a synthetic RNA spike‐in (cel‐miR‐39‐3p) was added to the RT reaction. PCR amplification was performed in a 384‐well plate format using custom Pick&Mix plates (Qiagen) in a Roche LC480 II instrument (Roche, Vienna, Austria) and EXiLENT SYBR Green Master Mix (Exiqon) with the following settings: 95°C for 10 min, 45 cycles of 95°C for 10 s and 60°C for 60 s, followed by melting curve analysis. To calculate the cycle of quantification values (Cq‐values), the second derivative method was used. Cq‐values were normalized to the spike‐in controls to account for analytical variability due to RT (cel‐miR‐39) and RNA extraction (UniSp4) using the following equation: Cq = Cq (miRNA) − ΔCq (cel‐miR‐39‐3p) – ΔCq (Cq UniSp4 – Cq cel‐miR‐39‐3p).
Hemolysis was assessed in all samples using the ratio of miR‐23a‐3p versus miR‐451a and applying a cut‐off of > 7 to the ratio for calling a sample hemolytic.
Study 2 ‐ Patient Population and Tendon Biopsy Collection
Both human SSP tendon (n = 4) and SSC tendon (n = 7) samples were obtained from 11 patients with a full‐thickness tear of the posterosuperior rotator cuff who underwent arthroscopic rotator cuff repair between 2015 and 2018. Furthermore, SSC tendon samples were harvested from 8 patients without any lesion of the rotator cuff, henceforth referred to as controls.
The indication for surgical treatment was given independently from the study protocol. Arthroscopic treatment was performed in a standard manner with the patient placed in a beach‐chair position. A sample of the ruptured SSP tendon (app. 2 × 2 × 2 mm) was obtained arthroscopically from the medium tendon portion after debridement of degenerative tissue.
Collection of human SSC tendons has previously been reported.14 Briefly, tendon samples with a size of 2 × 2 × 2 mm were harvested from the upper portion of the intact SSC tendon, 10 mm lateral to the articular surface of the glenoid with the arm in neutral position.
All tendon samples were directly placed in a test tube filled with RNAlater (Qiagen, Germany) and stored at −20°C for subsequent analysis, which was performed at the Institute of Tendon and Bone Regeneration (Paracelsus Medical University, Salzburg, Austria).
Total RNA Isolation From Tendon Tissue
Total RNA was isolated from tendon tissues using TRI® Reagent (Sigma‐Aldrich, Vienna, Austria) according to the manufacturer's protocol with slight modifications. Tendon tissue was homogenized using TRI Reagent® on ice using an Ultra‐Turrax (IKA, Staufen, Germany). Two additional chloroform extraction steps were performed. Total RNA was precipitated for 30 min at −20°C with an equal volume of ice‐cold isopropanol followed by centrifugation for 30 min at 13.000 rpm at +4°C. RNA pellets were washed in 75% EtOH, air dried and resuspended in RNase‐free water supplemented with 20 units Ribolock RNase inhibitor (Thermo Fisher Scientific, Vienna, Austria) and stored at −80°C. RNA yield and purity were determined using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific) and RNA integrity was verified by the Experion Automated Electrophoresis System (Bio‐Rad, Munich, Germany). An RNA with an RNA quality indicator (RQI) > 7.5 was defined as intact RNA.
qPCR Analysis of Tissue Samples
RT and RT‐qPCR for total RNA extracted from tissue samples were essentially performed as described further above. Sample reactions were carried out in a 96‐well format using a CFX96 Touch™ Real‐Time PCR Detection System (Bio‐Rad). All samples were run in technical duplicates and a minimum of two independent experiments were performed. Cq‐values were analyzed using qBasePlus (v. 2.4; Biogazelle NV, Zwijnaarde, Belgium) and normalized relative quantities were calculated by normalizing the data to the expression of two previously validated endogenous controls (5SrRNA, u6 snRNA).15
Statistics
Descriptive statistics including means, standard deviations, minimum and maximum values of continuous variables were calculated using SPSS 21.0 (IBM, Armonk, NY) or Prism 5.04 (GraphPad Software Inc., San Diego, CA). Differences between the three group means (Study 1) were calculated using the one‐way analysis of variance. Exploratory data analysis was performed using ClustVis.16 Principal component analysis (PCA) and hierarchical clustering analysis were performed using the default settings, that is, singular value decomposition with imputation, Pearson's correlation as the distance metric, and average distance for clustering. Differential expression analysis was performed based on spike‐in normalized ΔΔCq‐values under the assumption of the normal distribution, which was assessed visually, using two‐sided t tests. p‐Values were adjusted for multiple testing based on the method of Benjamini–Hochberg to compute false‐discovery rates.17 Differences in expression in tissue biopsy samples were analyzed using the Mann–Whitney test. An α value below 0.05 indicated the statistical significance and all variables were considered two‐tailed.
RESULTS
Study 1 ‐ Patient Characteristics (Liquid Biopsies)
In total, 15 consecutive patients were included of whom one patient was retrospectively excluded due to insufficient sample quality resulting in complete inhibition of enzyme activity observed in two independent replicate analyses (Supplementary Fig. S1).
Therefore, group 1 (healthy controls) consisted of five patients with a mean age of 58.1 ± 6.0 years (range, from 51 to 64 years). Group 2 (chronic tendinopathy) consisted of five patients with a mean age of 57.0 ± 5.9 years (range, from 51 to 66 years) and group 3 (degRCT) of four patients with a mean age of 60.1 ± 8.4 years (range, from 51 to 69 years). Additional patient demographics are summarized in Table 2, demonstrating no inter‐group significant difference.
Table 2.
Patient Demographics
| Variables | Group 1 (CTRL) | Group 2 (CT) | Group 3 (degRCT) | p‐Value |
|---|---|---|---|---|
| Patient age (years, mean ± SD) | 58.1 ± 6.0 | 57.0 ± 5.9 | 60.1 ± 8.4 | 0.787 |
| Body mass index (kg/m2, mean ± SD) | 30.7 ± 2.4 | 25.7 ± 4.0 | 27.9 ± 4.1 | 0.801 |
| Male gender (%) | 40 | 60 | 50 | 0.350 |
CTRL, healthy controls; CT, chronic tendinopathy; degRCT, degenerative rotator cuff tear.
Study 1 ‐ Circulating miRNA Expression Profiles
Levels of 187 endogenous miRNAs and five quality controls were assessed by RT‐qPCR in all 14 samples. Of these, 160 were detected (Cq < 40) in all 14 samples, and 179 were detected in more than 10 samples (Supplementary Table S1).
Hemolysis can be a strong confounder of circulating miRNA data.18 Therefore, we applied the established ratio of miR‐23a and miR‐451a to confirm the absence of hemolysis in all samples19 (Supplementary Fig. S2). In order to investigate the ability of chronic tendinopathy and degenerative RCT to induce circulating miRNA variability above the “noise background,” we performed PCA using levels of the 50 most variant miRNAs based on the coefficient of variation (Fig. 2A). Indeed, we observed clustering of samples on principal component 2 (PC2, 12.8% miRNA variance) according to the severity of tendinopathy, suggesting that the associated pathophysiologic cellular and molecular changes can result in distinct miRNA profiles. Next, PCA was performed using only the top 24 miRNAs (sorted by t‐test derived p‐value) showing a difference in expression between degenerative RCTs and healthy controls (Fig. 2B).
Figure 2.
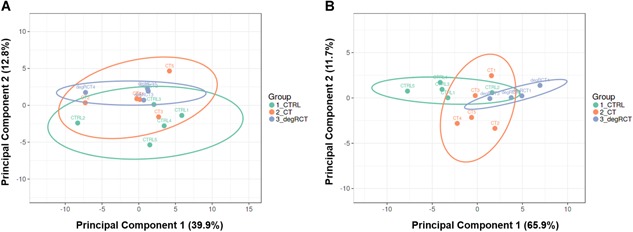
Principal component analysis (PCA). PCA was performed to assess clustering of patients based on their serum microRNA (miRNA) profiles reflected by principal components 1 and 2. (A) Clustering of samples based on the 50 most variant miRNAs in the data set based on the coefficient of variation (CV%). (B) Clustering of samples based on the 24 miRNAs differentially regulated miRNAs (p < 0.05) based on the comparison between degenerative rotator cuff tear (degRCT) and healthy controls (CTRL). [Color figure can be viewed at wileyonlinelibrary.com]
Principal component 1 (PC1, 68.4% miRNA variance) allowed good discrimination between patients with degenerative RCTs and healthy controls, while chronic tendinopathy group resided between healthy controls and patients with degenerative RCTs. This finding implies the existence of a disease‐related circulating miRNA signature. In order to identify the relevant miRNA species, we performed differential expression analysis comparing (i) degenerative RCTs and healthy controls, (ii) chronic tendinopathy and healthy controls, and (iii) degenerative RCTs and chronic tendinopathy (Supplementary Fig. S3) by applying an adjusted p‐value of 0.05 and fold change (FC) > 1.5 as cut‐off.
As shown in the VENN diagram (Fig. 3), six miRNAs (hsa‐miR‐19b‐3p, hsa‐miR‐192‐5p, hsa‐miR‐25‐3p, hsa‐miR‐19a‐3p, hsa‐miR‐18b‐5p, hsa‐miR‐93‐5p) were exclusively repressed in degRCT versus CTRL or CT (Fig. 4 and Supplementary Figs. S4 and S5). In addition, we identified six miRNAs (hsa‐miR‐30a‐5p, hsa‐miR‐324‐3p, hsa‐miR‐210‐3p, hsa‐miR‐140‐3p, hsa‐miR‐425‐5p, hsa‐miR‐222‐3p) that showed differential expression for both, degRCT and CT serum samples when compared with the CTRL samples (Fig. 5 and Supplementary Figs. S4 and S5).
Figure 3.
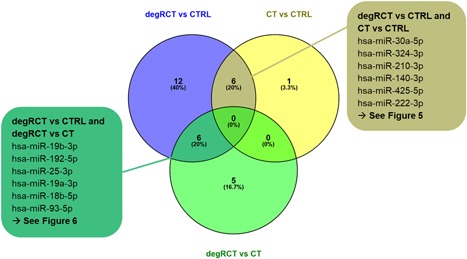
VENN overlap of microRNA (miRNA) lists derived from differential expression analysis. Six miRNAs were regulated in degenerative rotator cuff tears (degRCT) versus healthy controls (CTRL) and degRCT versus chronic tendinopathy (CT) (left, green box). Six miRNAs were regulated in degRCT versus CTRL and CT versus CTRL (right, brown box). [Color figure can be viewed at wileyonlinelibrary.com]
Figure 4.
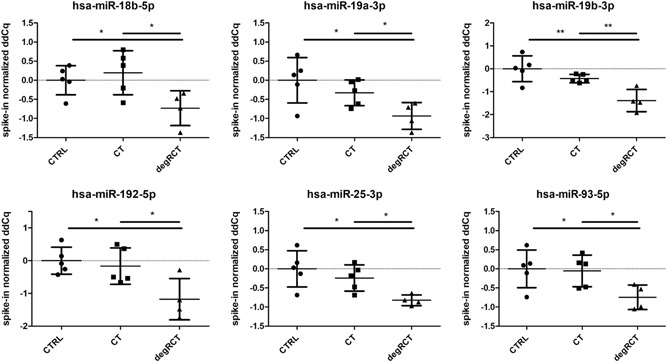
Scatterplots depicting the levels of six miRNAs that were found to be only downregulated in degenerative rotator cuff tear samples (degRCT), but not in healthy controls (CTRL) or chronic tendinopathy samples (CT).
Figure 5.

Serum levels of microRNAs (miRNAs) commonly repressed in chronic tendinopathy (CT) as well as degenerative rotator cuff tear samples (degRCT) when compared with healthy controls (CTRL).
As a proof of concept, species of the miR‐29 family (hsa‐miR‐29a‐3p and hsa‐miR‐29c‐3p), which have been previously shown to be regulated in rotator cuff pathology, were analyzed. As expected, both were only significantly repressed in degRCT samples but not CT samples, although a similar trend was seen (Fig. 6).
Figure 6.
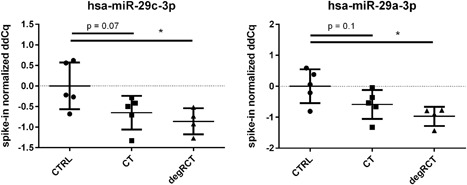
Comparison of serum expression levels of has‐miR‐29c and has‐miR‐29a between the three groups. Significant repression was found between healthy controls (CTRL) and patients with degenerative rotator cuff tears (degRCT), while the difference between CTRL and patients with chronic tendinopathy was not significant.
Study 2 ‐ Study Group and miRNA in Human Tendon Samples
In order to correlate the systemic expression of the candidate miRNAs identified for CT and degRCT samples with local, tissue‐resident miRNA expression, SSP biopsy samples and SSC samples harvested from patients who were treated for a complete SSP tear14 were collected. SSP samples were retrieved from three males and one female with a mean age of 62.4 ± 10.1 years (range, from 50 to 70 years) were collected and analyzed by pPCR. The average body mass index (BMI) was 22.5 ± 1.4 (range, from 20.8 to 24.2). SSC samples were collected from seven patients (five males and two females) with a mean age at the time of surgery of 64.8 ± 7.9 years (range, from 55 to 76 years) and a mean BMI of 28.0 ± 5.1 (range, from 21 to 36).
The control tendon samples were harvested from intact SSC tendons of eight patients (eight males), who underwent surgery due to a post‐traumatic shoulder instability or a SLAP lesion without any lesion of the rotator cuff tendons. Based on the etiology of the injury and the observation that the SSC tendons of these patients were macroscopically intact, these samples were considered as suitable control samples. The mean patient age at time of surgery was 29.8 ± 8.1 years (range, from 18 to 40 years) and the mean BMI was 24.9 ± 4.6 (range, from 19 to 34).
In total, nine miRNA candidate targets, which were found to be repressed in CT and degRCT sera, were evaluated for their expression. As shown in Figure 7, for the majority of the samples the candidate miRNAs were indeed significantly repressed, or showed a clear trend when compared with the control samples. For hsa‐miR‐29a‐3p, hsa‐miR‐29c‐3p, hsa‐miR‐30a‐5p, hsa‐miR‐140‐3p, and hsa‐miR‐192‐5p, expression levels were significantly lower in comparison to the healthy control (p < 0.05). The results for, hsa‐miR‐210‐3p, hsa‐miR‐324‐3p, and hsa‐miR‐425‐5p also indicated repression of the miRNA species by trend (p = 0.0759). Merely hsa‐miR‐222‐3p did not show a significant difference between control and test samples (p = 0.4828). Taken together, these data demonstrate that the majority of the candidate miRNAs show a reduced expression profile both systemically and locally.
Figure 7.
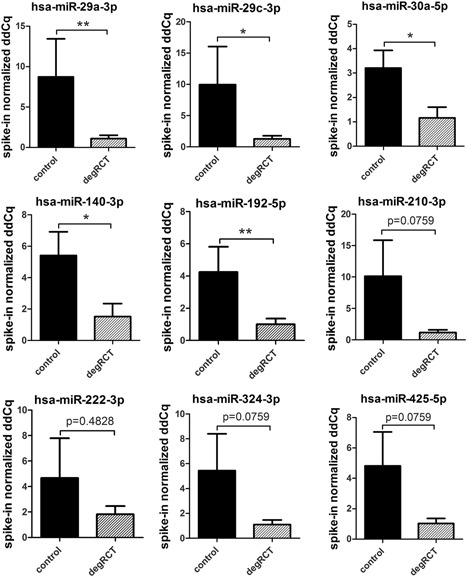
Change in microRNAs (miRNAs) expression in tissue specimens. The majority of the tested miRNAs were significantly repressed in biopsy samples harvested from patients undergoing rotator cuff surgery (n = 11; black gray bars) when compared with control samples (n = 8; black bars) (*p < 0.05; **p < 0.01; Mann–Whitney test).
DISCUSSION
MiRNAs have emerged as key regulators orchestrating a wide range of inflammatory and fibrotic diseases.20, 21, 22 However, the role of miRNAs for the etiology of degenerative shoulder joint disorders is poorly understood. Dubin et al. recently reported on tissue‐resident miRNAs to be important for the maintenance of tendon homeostasis.23 Several miRNAs were found to impact on tenocyte function and tendon‐related gene expression playing an important role during normal tendon development as well as the pathophysiology of tendon tissue degeneration. Therefore, the purpose of the study was to identify differentially expressed, circulating miRNA profiles in liquid biopsies (serum) retrieved from patients with a rotator cuff pathology when compared with healthy controls. As the variability of serum miRNA levels is caused by both controllable and uncontrollable biological factors such as diurnal variation, exercise, nutrition, and sex, as well as pathophysiologic processes due to disease,6 we applied stringent exclusion criteria to minimize confounding factors (Table 1).
Our data confirmed our hypothesis and 6 out of 187 circulating miRNAs (3%) were found to be significantly downregulated only in patients with degenerative RCTs when compared with both healthy controls and patients with chronic rotator cuff tendinopathy: miR‐18b, miR‐19a, miR‐19b, miR‐25, miR‐93 and miR‐192 (Fig. 4). It is well known, that increased pro‐inflammatory cytokine expression profiles negatively affect shoulder pain and tendon integrity.24, 25 miR‐25 was reported to be an important regulator of inflammatory responses by inhibiting the expression of several cytokines such as tumor necrosis factor α (TNF‐α) and also high mobility group (HMG) protein (e.g., HMGB1).26 Thus, downregulation of miR‐25 in rotator cuff tendinopathy potentially leads to an increased expression of inflammatory cytokines and HMGB1. Importantly, HMGB1 expression has been shown to be increased in samples harvested from injured rotator cuff tendons.27
miR‐19 regulates the expression of modulators of the JAK‐STAT signaling pathway and a decrease in miR‐19 enhances inflammation‐related cytokines release (e.g. IL‐6 or MMP‐3) and thus, promotes local inflammation.28 Generally, degenerative RCTs are associated with hypocellularity, higher rates of tenocyte apoptosis and premature cellular senescence.29 Along these lines, the expression of miR‐93 has been shown to positively correlate with cell survival and proliferation.30 Furthermore, miR‐93 has been shown to limit inflammation by controlling the expression of the pro‐inflammatory interleukin 1 (IL‐1) receptor‐associated kinase 4 (IRAK4) in cerebral ischemia–reperfusion (CIR) injuries.31 Therefore, the repression of miR‐93 further potentiates an inflammatory response, thereby potentially contributing to chronic tendinopathy. The expression of miR‐18 was described to negatively correlate with age and thus, repression was correlated with the overexpression of pro‐fibrotic genes.32 Fibrotic responses are central to the development of chronic tendinopathy, affecting the tendon‐bone junction but also affecting repair processes after tendon microruptures, ultimately leading to biomechanically inferior tendon tissue.33, 34 Several studies have also reported a pro‐fibrotic role for miR‐192 in TGF‐β‐dependent renal fibrosis which was observed in rodent models of kidney disease.35
Interestingly, we also identified several miRNAs to be significantly deregulated in both chronic tendinopathy and degenerative RCTs (Fig. 5)—miR‐30a‐5p, miR‐140‐3p, miR‐210‐3p, miR‐222‐3p, miR‐324‐3p, miR‐425‐5p. Importantly, for those candidate miRNAs a progressive decline in the expression levels was evident depending on the severity of tendon degeneration, suggesting that those miRNAs potentially contribute to the pathogenesis and/or progression of degenerative rotator cuff disorders. Therefore, our data indicate that these miRNAs have potential as prognostic miRNA profiles for chronic tendinopathy. However, additional studies including a larger cohort number is required to further substantiate this hypothesis. Nevertheless, analysis of tendon biopsies harvested from patients who underwent arthroscopic SSP repair confirmed that these miRNAs were repressed when compared with control samples (Fig. 7). Up to now, mainly the miR‐29 family has been reported to mediate degenerative tendon disorders of the shoulder joint.5, 10, 23, 36 Our results confirm these findings, as we observed repression of miR‐29a and miR‐29c on the tissue and systemic level. Interestingly, both miR‐29a and miR‐29c showed progressive repression depending on the severity of the tendon pathology (Fig. 6). A recent investigation found the miR‐29 family to directly regulate collagen expression in rotator cuff tendon tissue samples.10 By controlling interleukin 33 syntheses, miR‐29a positively regulated collagen 3 (Col‐3) expression in tenocytes and miR‐29a downregulation was correlated with the overexpression of Col‐3, resulting in an unbalanced ratio of collagen type 1 and 3, which is a hallmark of tendon degeneration. Further, in an equine therapeutic model Watts et al. demonstrated a significant reduction of Col‐3 synthesis by overexpression of miR‐29a, illustrating the important role of miRNAs in fine‐tuning ECM composition in tendon tissue.37
MMPs are considered to be major regulators of ECM degradation and overexpression is linked to the pathogenesis of tendinopathy.38 According to Cao and colleagues, miR‐324 inhibits the expression of MMPs, such as MMP‐2 and MMP‐9.39 Although miR‐324 was only significantly repressed in the serum samples and only by trend in the tissue biopsies, repression of miR‐324 might promote tendon disorganization thereby contributing to rotator cuff tendinopathy.
miR‐30a and miR‐140‐3p were also significantly repressed in RCT sera and tissue biopsies. miR‐30a has been shown to promote cancer cell apoptosis and significantly inhibit cell proliferation.40 Further, its repression has been correlated with hepatic fibrosis and hence an extensive and aberrant ECM deposition.41 miR‐140‐3p negatively regulates nuclear factor‐κB (NF‐κB) inflammatory signaling by limiting the expression of nuclear receptor co‐activator 1 (NCOA1) and nuclear receptor‐interacting protein 1 (NRIP1), both of which are NF‐κB co‐activators.42 As canonical NF‐κB signaling has been shown to play a role in both chronic and acute tendon injuries,43, 44 it is tempting to speculate that repression of miR‐140‐3p might promote tendon inflammation and degeneration. Finally, miR‐425 repression also has been demonstrated to correlates with upregulation of inflammatory cytokines (e.g. IL‐2).45
Taken together, the majority of the miRNAs identified have been shown to impact upon pro‐inflammatory and pro‐fibrotic responses, both hallmarks of tendinopathy.
Our study has some limitations that need to be taken into consideration. First, the sample size is small. However, despite the low number of patients, we were able to identify a circulating miRNA signature correlating with degenerative rotator cuff pathologies for the first time and demonstrate similarities between local and systemic expression profiles. Further, it is important to note that individual genes are usually not controlled by a single miRNA and the ramifications of the identified miRNA species on tendon homeostasis and degeneration need to be further evaluated. Nevertheless, our data indicate the presence of an RTC‐related miRNA profile, warranting further investigations to harness circulating miRNAs as a non‐invasive diagnostic or prognostic biomarker for rotator cuff tendinopathies.
CONCLUSION
With this study, we identified a circulating miRNA signature correlating with degenerative rotator cuff pathologies. Repression of these miRNAs may influence different molecular biological processes in the development and progression of degenerative rotator cuff disorders, further enhancing our understanding of tendinopathy. Further, our data will contribute to establishing miRNA signatures as potential novel diagnostic and prognostic biomarkers for degenerative rotator cuff pathologies.
AUTHORS' CONTRIBUTION
All listed authors have contributed substantially to this work (FP, PH, MH, and AT. conceived the study; FP, RG, HT, CL, NW, AW, MW, and MH. designed the experiments; P.H. and J.F. recruited patients and collected blood samples; RG, HT, CL, NW, AW, and M.W. conducted the experiments; FP, RG, HAT, AW, MW, PM, and MH. analyzed the data; all authors contributed to manuscript preparation and critically revised the manuscript. All authors approved the final manuscript.) and have approved the submission.
Supporting information
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
ACKNOWLEDGMENT
This work was supported by the German Society for Arthroscopy and Joint Surgery (AGA) (Project award Nr. 69).
Conflicts of interest: F.P. received a funding by the German Society for Arthroscopy and Joint Surgery (AGA); M.H. is co‐founder and employee of TAmiRNA GmbH; P.H. and P.M. are consultants for Arthrex Inc., not related to the subject of the study; and A.T. is Head of Preclinical Development of Celericon Therapeutics GmbH, which activities are not related to the subject of the study.
References
REFERENCES
- 1. Minagawa H, Yamamoto N, Abe H, et al. 2013. Prevalence of symptomatic and asymptomatic rotator cuff tears in the general population: from mass‐screening in one village. J Orthop 10:8–12. 10.1016/j.jor.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lähteenmäki HE, Virolainen P, Hiltunen A, et al. 2006. Results of early operative treatment of rotator cuff tears with acute symptoms. J Shoulder Elbow Surg 15:148–153. 10.1016/j.jse.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 3. Fu SC, Rolf C, Cheuk YC, et al. 2010. Deciphering the pathogenesis of tendinopathy: a three‐stages process. Sports Med Arthrosc Rehabil Ther Technol 2:30 10.1186/1758-2555-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abate M, Silbernagel KG, Siljeholm C, et al. 2009. Pathogenesis of tendinopathies: inflammation or degeneration? Arthritis Res Ther 11:235 10.1186/ar2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thankam FG, Boosani CS, Dilisio MF, et al. 2018. MicroRNAs associated with inflammation in shoulder tendinopathy and glenohumeral arthritis. Mol Cell Biochem 437:81–97. 10.1007/s11010-017-3097-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hackl M, Heilmeier U, Weilner S, et al. 2016. Circulating microRNAs as novel biomarkers for bone diseases—complex signatures for multifactorial diseases? Mol Cell Endocrinol 432:83–95. 10.1016/j.mce.2015.10.015 [DOI] [PubMed] [Google Scholar]
- 7. Kocijan R, Muschitz C, Geiger E, et al. 2016. Circulating microRNA signatures in patients with idiopathic and postmenopausal osteoporosis and fragility fractures. J Clin Endocrinol Metab 101:4125–4134. 10.1210/jc.2016-2365. [DOI] [PubMed] [Google Scholar]
- 8. Qian L, Yu S, Chen Z, et al. 2018. The emerging role of circRNAs and their clinical significance in human cancers. Biochim Biophys Acta Rev Cancer 1870:247–260. 10.1016/j.bbcan.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 9. Nakasa T, Nagata Y, Yamasaki K, et al. 2011. A mini‐review: microRNA in arthritis. Physiol Genomics 43:566–570. 10.1152/physiolgenomics.00142.2010 [DOI] [PubMed] [Google Scholar]
- 10. Millar NL, Gilchrist DS, Akbar M, et al. 2015. MicroRNA29a regulates IL‐33‐mediated tissue remodelling in tendon disease. Nat Commun 6:6774 10.1038/ncomms7774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kjellin I, Ho CP, Cervilla V, et al. 1991. Alterations in the supraspinatus tendon at MR imaging: correlation with histopathologic findings in cadavers. Radiology 181:837–841. 10.1148/radiology.181.3.1947107 [DOI] [PubMed] [Google Scholar]
- 12. Patte D. 1990. Classification of rotator cuff lesions. Clin Orthop Relat Res 254:81–86. [PubMed] [Google Scholar]
- 13. Goutallier D, Postel JM, Bernageau J, et al. 1994. Fatty muscle degeneration in cuff ruptures. Pre‐ and postoperative evaluation by CT scan. Clin Orthop Relat Res 304:78–83. [PubMed] [Google Scholar]
- 14. Plachel F, Moroder P, Gehwolf R, et al. 2019. Risk factors for rotator cuff disease: an experimental study on intact human subscapularis tendons. J Orthop Res 10.1002/jor.24385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vandesompele J, De Preter K, Pattyn F, et al. 2002. Accurate normalization of real‐time quantitative RT‐PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:research0034.1. Research003410.1186/gb‐2002‐3‐7‐research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Metsalu T, Vilo J. 2015. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res 43:W566–W570. 10.1093/nar/gkv468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300. [Google Scholar]
- 18. Kirschner MB, Kao SC, Edelman JJ, et al. 2011. Haemolysis during sample preparation alters microRNA content of plasma. PLoS One 6. e2414510.1371/journal.pone.0024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blondal T, Jensby Nielsen S, Baker A, et al. 2013. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 59:S1–S6. 10.1016/j.ymeth.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 20. Raj Christian SD, Thirugnanasambantham K, Islam MIH, et al. 2019. Identification of expressed miRNAs in human rheumatoid arthritis using computational approach ‐ Discovery of a new miR‐7167 from human. MicroRNA 8:147–154. 10.2174/2211536608666181204111438. [DOI] [PubMed] [Google Scholar]
- 21. Churov AV, Oleinik EK, Knip M. 2015. MicroRNAs in rheumatoid arthritis: altered expression and diagnostic potential. Autoimmun Rev 14:1029–1037. 10.1016/j.autrev.2015.07.005 [DOI] [PubMed] [Google Scholar]
- 22. Lino Cardenas CL, Henaoui IS, Courcot E, et al. 2013. miR‐199a‐5p Is upregulated during fibrogenic response to tissue injury and mediates TGFbeta‐induced lung fibroblast activation by targeting caveolin‐1. PLoS Genet 9. e100329110.1371/journal.pgen.1003291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dubin JA, Greenberg DR, Iglinski‐Benjamin KC, et al. 2018. Effect of micro‐RNA on tenocytes and tendon‐related gene expression: a systematic review. J Orthop Res 36:2823–2829. 10.1002/jor.24064. [DOI] [PubMed] [Google Scholar]
- 24. Shih CA, Wu KC, Shao CJ, et al. 2018. Synovial fluid biomarkers: association with chronic rotator cuff tear severity and pain. J Shoulder Elbow Surg 27:545–552. 10.1016/j.jse.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 25. Millar NL, Hueber AJ, Reilly JH, et al. 2010. Inflammation is present in early human tendinopathy. Am J Sports Med 38:2085–2091. 10.1177/0363546510372613. [DOI] [PubMed] [Google Scholar]
- 26. Zhu C, Chen T, Liu B. 2018. Inhibitory effects of miR‐25 targeting HMGB1 on macrophage secretion of inflammatory cytokines in sepsis. Oncol Lett 16:5027–5033. 10.3892/ol.2018.9308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thankam FG, Roesch ZK, Dilisio MF, et al. 2018. Association of inflammatory responses and ECM disorganization with HMGB1 upregulation and NLRP3 inflammasome activation in the injured rotator cuff tendon. Sci Rep 8:8918 10.1038/s41598-018-27250-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Z, Cai J, Cao X. 2016. MiR‐19 suppresses fibroblast‐like synoviocytes cytokine release by targeting toll like receptor 2 in rheumatoid arthritis. Am J Transl Res 8:5512–5518. [PMC free article] [PubMed] [Google Scholar]
- 29. Lundgreen K, Lian OB, Scott A, et al. 2014. Rotator cuff tear degeneration and cell apoptosis in smokers versus nonsmokers. Arthroscopy 30:936–941. 10.1016/j.arthro.2014.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choi JY, Shin HJ, Bae IH. 2018. miR‐93‐5p suppresses cellular senescence by directly targeting Bcl‐w and p21. Biochem Biophys Res Commun 505:1134–1140. 10.1016/j.bbrc.2018.10.010 [DOI] [PubMed] [Google Scholar]
- 31. Tian F, Yuan C, Hu L, et al. 2017. MicroRNA‐93 inhibits inflammatory responses and cell apoptosis after cerebral ischemia reperfusion by targeting interleukin‐1 receptor‐associated kinase 4. Exp Ther Med 14:2903–2910. 10.3892/etm.2017.4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seeger T, Boon RA. 2016. MicroRNAs in cardiovascular ageing. J Physiol 594:2085–2094. 10.1113/jp270557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gigliotti D, Xu MC, Davidson MJ, et al. 2017. Fibrosis, low vascularity, and fewer slow fibers after rotator‐cuff injury. Muscle Nerve 55:715–726. 10.1002/mus.25388 [DOI] [PubMed] [Google Scholar]
- 34. Tillander B, Franzén L, Norlin R. 2002. Fibronectin, MMP‐1 and histologic changes in rotator cuff disease. J Orthop Res 20:1358–1364. 10.1016/s0736-0266(02)00057-8 [DOI] [PubMed] [Google Scholar]
- 35. Chung ACK, Lan HY. 2015. MicroRNAs in renal fibrosis. Front Physiol 6:50 10.3389/fphys.2015.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thankam FG, Boosani CS, Dilisio MF, et al. 2016. MicroRNAs associated with shoulder tendon matrisome disorganization in glenohumeral arthritis. PLoS One 11. e016807710.1371/journal.pone.0168077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watts AE, Millar NL, Platt J, et al. 2017. MicroRNA29a treatment improves early tendon injury. Mol Ther 25:2415–2426. 10.1016/j.ymthe.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Del Buono A, Oliva F, Osti L, et al. 2019. Metalloproteases and tendinopathy. Muscles Ligaments Tendons J 03:51–57. 10.11138/mltj/2013.3.1.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cao L, Xie B, Yang X, et al. 2015. MiR‐324‐5p suppresses hepatocellular carcinoma cell invasion by counteracting ECM degradation through post‐transcriptionally downregulating ETS1 and SP1. PLoS One 10. e013307410.1371/journal.pone.0133074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jiang L, Zhang H, Tang J. 2018. MiR‐30a: a novel biomarker and potential therapeutic target for cancer. J Oncol 2018:1–9. 10.1155/2018/5167829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen J, Yu Y, Li S, et al. 2017. MicroRNA‐30a ameliorates hepatic fibrosis by inhibiting Beclin1‐mediated autophagy. J Cell Mol Med 21:3679–3692. 10.1111/jcmm.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Takata A, Otsuka M, Kojima K, et al. 2011. MicroRNA‐22 and microRNA‐140 suppress NF‐κB activity by regulating the expression of NF‐κB coactivators. Biochem Biophys Res Commun 411:826–831. 10.1016/j.bbrc.2011.07.048. [DOI] [PubMed] [Google Scholar]
- 43. Best KT, Lee FK, Knapp E, et al. 2019. Deletion of NFKB1 enhances canonical NF‐κB signaling and increases macrophage and myofibroblast content during tendon healing. Sci Rep 9:10926 10.1038/s41598-019-47461-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Abraham AC, Shah SA, Golman M, et al. 2019. Targeting the NF‐κB signaling pathway in chronic tendon disease. Sci Transl Med 11 10.1126/scitranslmed.aav4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nakagawa R, Muroyama R, Saeki C, et al. 2017. miR‐425 regulates inflammatory cytokine production in CD4(+) T cells via N‐Ras upregulation in primary biliary cholangitis. J Hepatol 66:1223–1230. 10.1016/j.jhep.2017.02.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.


