ABSTRACT
ATP is generated in mitochondria of eukaryotic cells by oxidative phosphorylation (OXPHOS). The OXPHOS complex, which is crucial for cellular metabolism, comprises of both nuclear and mitochondrially encoded subunits. Also, the occurrence of several pathologies because of mutations in the mitochondrial translation apparatus indicates the importance of mitochondrial translation and its regulation. The mitochondrial translation apparatus is similar to its prokaryotic counterpart due to a common origin of evolution. However, mitochondrial translation has diverged from prokaryotic translation in many ways by reductive evolution. In this review, we focus on mammalian mitochondrial translation initiation, a highly regulated step of translation, and present a comparison with prokaryotic translation.
KEYWORDS: Mammalian mitochondria, ribosome, mitoribosome, protein synthesis, translation initiation, mitochondrial disease
Introduction
Mitochondria are sub-cellular organelles that perform a number of functions crucial for cellular homoeostasis. According to the endosymbiont theory, mitochondria are descendants of α-proteobacteria that were endocytosed by pre-eukaryotic cells [1] and retained, since they produce ATP. A vast majority of the mitochondrial genes except those encoding mitochondrial tRNAs, rRNAs and 13 subunits of the electron transport chain moved to the nuclear genome over time [2]. Mitochondria have retained a translational apparatus to synthesize members of the electron transport chain encoded from its own genome. Broadly, the mitochondrial translational system is similar to its eubacterial counterpart [3]. However, mitochondrial translation is unique in the structure and composition of ribosomes, presence of leaderless mRNAs, codon assignments and in having a smaller number of tRNAs [3]. The process of translation initiation is a highly regulated step of protein synthesis, and perturbations of mitochondrial translation initiation have been associated with human diseases. In this article, we survey the peculiarities of mammalian mitochondrial translation initiation based on the features of the mitochondrial ribosome (mitoribosome), initiator tRNA (i-tRNA) and initiation factors (IFs). While we have occasionally discussed yeast mitochondrial translation to better understand the evolution of mammalian mitochondrial translation, this review is primarily focussed on the mammalian mitochondrial system (for yeast mitochondrial translation reviews, see [4–6]). In this review, we compare and contrast prokaryotic and mammalian mitochondrial translation initiation, to understand how mitochondrial translation has diverged from its evolutionary ancestors and adapted to its new environment. This is followed by a discussion of the various pathologies associated with the dysfunction of mammalian mitochondrial translation initiation. Finally, we propose methods of studying mitochondrial translation in vivo.
The mitochondrial ribosome
The bovine mitoribosome is a 55S molecule composed of a 28S small subunit (SSU) and a 39S large subunit (LSU). The mitoribosome is more protein-rich than eukaryotic (cytosolic) ribosomes with an RNA: protein ratio of 1:2 as opposed to 2:1 [3]. The reversal in ratio is not due to loss of individual nucleotides, but deletion of large regions, such as the anti-Shine-Dalgarno (aSD) region from rRNAs. Consistent with the loss of aSD region from the SSU rRNA, the mammalian mitochondrial mRNAs also lack the Shine-Dalgarno (SD) sequence. However, regions such as helix 45 and the sarcin/ricin loop (SRL) of the LSU are conserved. Some, but not all of the excised regions have been replaced by ‘new’ ribosomal proteins that have no known homologs outside of mitoribosomes. Besides, the proteins have extra sequences compared to their bacterial counterparts [7,8]. This has led to a rather ‘porous’ and low density structure of mitoribosome.
The SSU is composed of 12S rRNA and 29 proteins and the LSU is made up of 16S rRNA and 48 proteins [2]. About half the mitoribosomal proteins have homologs in prokaryotes while the rest are unique to mitochondria. A 5S rRNA is not encoded by the mitochondrial genome [9] and it was thought to be imported into mitochondria through the mitochondrial intermembrane enzyme polynucleotide phosphorylase [10]. Interestingly, recent cryoelectron microscopy (cryoEM) studies have shown that the mitochondrial tRNAVal (mt-tRNAVal) is present in the human mitochondrial LSU and it plays an integral structural role [11] (Fig. 1). The mt-tRNAVal interacts with mL46, mL40, mL48, uL18m, mL38, bL27m in the mt-LSU (Fig. 1A) while in the prokaryotic LSU, the 5S rRNA interacts with bL25, uL5, uL18 and bL27 (Fig. 1B). The L18 and L27 homologues are conserved as interacting partners of 5S rRNA/mt-tRNAVal across bacteria and mammalian mitochondria. Interestingly, while yeast mitoribosomes have also lost the 5S rRNA along with 5S RNA-binding proteins L18 and L25 during the course of evolution, there is currently no evidence that the 5S rRNA has been replaced by a tRNA or a structural mimic of the 5S rRNA [9]. It has been proposed that RNA expansion clusters, the extensions of uL5m, uL16m, bL27m, bL31m and bL33m, and mitochondria-specific proteins such as mL38, mL40, and mL46 occupy the ribosomal region that is occupied by the 5S rRNA in prokaryotes.
Figure 1.

Structures of (A) human mitochondrial (PDB ID: 3J9M) and (B) E. coli ribosomes (PDB ID: 6O9J) bound to tRNAVal and 5S rRNA, respectively.
The Ramakrishnan group solved the structure of the human mitoribosome to a resolution of 3.5 Å using single particle cryoEM [11]. Their studies revealed that the SSU lacks homologs of the ribosomal proteins uS8, uS13, uS19, and bS20; while uS4 has been replaced by a mitochondria-specific counterpart. The yeast mitoribosomal SSU has a nearly complete complement of proteins (with the exception of bS20) when compared with the bacterial homologs [12]. Strikingly, due to the absence of uS4 and a shortening of the C-terminal domain of uS3m (which is partially compensated for by uS5m) in the human mitoribosome, the mRNA tunnel is distinct from that of prokaryotic ribosomes. This has led to a shift and an expansion of the channel. The mRNA exit region in bacterial ribosomes contains the aSD sequence, which is crucial for translation initiation in bacteria. As stated earlier, both the SD (in mRNA) and aSD (in 12S rRNA) sequence features are absent from the mitochondrial translation machinery. The mRNA exit tunnel, through which the mitochondrial mRNA emerges during translation, is composed of mS37, bS21m and bS1m. The structural conservation of bS1m with bS1, coupled with their similar positions on the respective ribosomes and the presence of a large electropositive patch in the direction of the mRNA indicates that bS1m may be involved in RNA binding. The mRNA channel is lined with positively charged conserved amino acids contributed by an extension of uS5m. Nenad Ban’s group has proposed that the small subunit protein mS39 present at the mRNA entrance may initially tether mitochondrial mRNAs [13]. Subsequently, the uS5m extension with its positively charged residues may guide the mRNA through the channel and then the codon-anticodon interaction may stabilize mRNA binding. Additionally, the N-terminus of mL45 is shown to be important for mitochondrial translation, in recruiting the translocation machinery after translation initiation.
The P site of the bacterial 30S ribosomal subunit includes parts of the 3ʹ major domain, central domain and h44 regions of the 16S rRNA, and the C-terminal tails of ribosomal proteins uS13 and uS9 [14–16]. These elements are crucial for the specificity of the i-tRNA binding. Thus, some components of the human mitoribosome, such as uS12m, uS9m, bS1m and the rRNAs could be important for translation initiation. X-ray crystallography of prokaryotic ribosomes indicates that IF1 contacts the ribosomal protein uS12 and the helices 18 and 44 of 16S rRNA in the ribosomal A site [17]. IF3 interacts with bS1, uS2, uS3, uS7, uS11, uS13, bS18, uS19, bS21 and particularly strongly with uS12 [18–20]. The ribosomal protein, uS12 serves to modulate translocation of mRNA-tRNA complex through the ribosome [21] and it is one of the strongest interacting partners of IF3. IF3mt cross-links to mitochondrial homologs of the bacterial ribosomal proteins uS5, uS9, uS10, and bS18 and to the unique mitochondrial ribosomal proteins mS29, mL42, mS36 and mS39 [22]. Surprisingly, no cross-links were obtained between IF3mt and uS12m.
A well-studied protein involved in i-tRNA discrimination, in the ribosomal P site, is uS9. According to the crystal structure of Thermus thermophilus ribosomes, the C-terminal tail of the uS9 protein contacts the i-tRNA at positions 33 and 34 [16]. The C-terminal tail sequence (SKR) of uS9 is highly conserved amongst bacteria and its deletion leads to a modest decrease in cellular growth at 37°C, but causes significant increase in cold sensitivity and a decrease in i-tRNA binding to the 30S ribosome [23]. Additionally, in vivo reporter studies from our lab have established that the absence of the C-terminal SKR sequence leads to an increase in initiation with anticodon stem mutants of i-tRNAs [24]. In many species of Mycoplasma, the SKR sequence of uS9, is found to be represented by TKR, which correlates with a change of the conserved R131 to P, F or Y in their IF3 [25]. Multiple sequence alignments of uS9m from mitochondria and its prokaryotic homologue have revealed the presence of KKR instead of SKR at the C-terminal tail (Fig. 2). A detailed exploration of the role of the C-terminal tail of mitochondrial uS9m would probably serve to reveal a great deal about the mechanism of fidelity of translation initiation.
Figure 2.

Multiple sequence alignment of uS9m C-terminal tail (in the red box) from mammalian mitochondria and uS9 from E. coli. Accession numbers: E. coli uS9- BAE77273.1; R. norvegicus uS9m- AAI58705.1, H. sapiens uS9m- NP_872578.1, B. taurus uS9m- XP_005212500.1, S. scrofa uS9m- NP_001231482.1, P. vampyrus uS9m- XP_011368436.1.
Mitochondrial initiation factors
Three initiation factors are present in all prokaryotes: IF1, IF2 and IF3. In the absence of initiation factors, initiation complex (IC) can be formed with an elongator tRNA in the P site, but the presence of all the factors distinctly favours the accommodation of i-tRNA (fMet-tRNAfMet) at the P site [26]. Unlike the eubacterial system, mammalian mitochondria have only two initiation factors: IF2mt and IF3mt.
Mitochondrial initiation factor 2
A mitochondrial initiation factor equivalent to the E. coli IF2 (EcoIF2) was characterized and studied from bovine mitochondria [27] and yeast [28]. The bovine IF2mt was first identified as a factor that promoted binding of yeast i-tRNA to mitoribosomes, where the efficiency of binding was tremendously enhanced by GTP, but not GDP [27]. Bovine IF2mt bears 39% sequence identity with EcoIF2 and can stimulate binding of formylated i-tRNAs from bacteria and yeast, more efficiently than their non-formylated forms.
IF2mt is a single subunit protein which is compatible to function with the mitochondrial, chloroplast and prokaryotic ribosomes [29]. On the contrary, EcoIF2 is not functional on mitoribosomes [27]. IF2mt also consists of a conserved 37 amino acid insertion between its domains V and VI (Fig. 3A, red box; Fig. 3B, pink region). IF1 homologs exist in the organellar translational system of chloroplasts but not in mitochondria. This phenomenon has been explained by Gaur et al. (2008) using an E. coli system wherein the 37 amino acid protrusion in IF2mt discharges the functions of IF1. Thus, IF2mt has a bifunctional role in mammalian mitochondria [30]. Subsequently, cryoEM studies indicated that the 37 amino acid domain interacts with the same region on the ribosome where IF1 is known to bind [31]. Thus, this helical insertion domain blocks the A site of the ribosome, much like the bacterial IF1. IF2mt consists of domains III to VI, which are homologous to the corresponding domains in EcoIF2. Of these, the domain VI binds to i-tRNA [32]. Recent cryoEM studies have shown that H678 of this domain interacts with the formyl group of i-tRNA while F632 interacts with the amino acid methionine [13]. Surprisingly, although the loss of IF1 from mitochondria appears to be universal, the insertion in mammalian IF2mt that serves the functions of IF1, is not seen in yeast [33]. This indicates that the loss of IF1 from mitochondria may have occurred prior to the gain of the insertion in mammalian IF2mt.
Figure 3.

Initiation factor 2. (A) Multiple sequence alignment of IF2 from E. coli, T. thermophilus and human mitochondria. Structures of (B) Human IF2mt (PDB ID: 6GAZ) (C) T. thermophilus IF2 (PDB ID: 3J4J). The region in pink in (B) and the red box in (A) denote the extension of human IF2mt that discharges the functions of bacterial IF1. Accession numbers: Human IF2mt- NP_001307933.1, T. thermophilus IF2- CAA88038.1, E. coli IF2- WP_000133051.1.
Mitochondrial initiation factor 3
There is a rather low sequence homology between the bacterial IF3 and IF3mt (21–26%) and the sequences of IF3mt from various eukaryotes are not very conserved [34]. However, IF3mt has a central region with homology to bacterial IF3. Interestingly, IF3mt possesses N- and C-terminal extensions (Next and Cext, respectively), which are absent from eubacterial IF3 (Fig. 4). The closest prokaryotic relatives of IF3mt are those from the Mycoplasma species [35]. However, a greater homology of human IF3mt is observed with many of the chloroplast IF3s than with the prokaryotic IF3s. Previously, the NTD of IF3mt had been modelled on the structure of the Bacillus stearothermophilus IF3NTD. Further, the structure of mouse IF3mt CTD (without the Cext) was solved by NMR [36]. The N- and C- terminal extensions were predicted to be disordered. More recently, cryoEM structures of IF3mt complexed with SSU of the mammalian mitochondria were obtained at 3.3–3.5 Å resolution [37]. The structures indicate that the NTD is composed of an α-helix and four β-stranded sheets, which are packed against two α-helices (Fig. 4B). The two globular domains are joined by a flexible linker region.
Figure 4.

Initiation factor 3. (A) Multiple sequence alignment of IF3 from E. coli, T. thermophilus and human mitochondria. Structures of (B) Human IF3mt (PDB ID: 6NEQ) (C) T. thermophilus IF3 (PDB ID: 5LMN). The colours in the structures represent the domains shown by arrows in (A). Accession numbers: Human IF3mt- NP_001159735.1, T. thermophilus IF3- WP_014629997.1, E. coli IF3- KIG24527.1.
Human IF3mt has been expressed in E. coli and the purified protein (mass 27 kDa) has properties similar to those of eubacterial IF3 [34]. According to the in vitro experiments performed by Koc and Spremulli, IF3mt acts as an anti-association factor (and a dissociation factor, discussed later) for the two mitochondrial ribosomal subunits. It allows the formation of the IC on mitoribosomes when IF2mt, fMet-tRNAMet and poly (A, U, G) or transcripts of a mitochondrial gene are present, but not on 70S ribosomes in the presence of EcoIF2 and E. coli IF1 (EcoIF1). However, upon inclusion of IF2mt in place of EcoIF2, IF3mt becomes active on 70S ribosomes. EcoIF2 has two more domains (domains I and II) at its N-terminus, which are not found in IF2mt [38]. It is unclear if these two domains may obstruct the binding site of IF3mt. IF3mt can allow binding of bacterial mRNAs to 55S ribosome even though these differ from the mitochondrial mRNAs in possessing 5ʹ untranslated regions. The Spremulli group has also subsequently demonstrated that the addition of even a few nucleotides to the 5ʹ end of the AUG reduces the efficiency of translation initiation in the in vitro mitochondrial system [39].
The roles of the isolated NTD and CTD of IF3mt have been studied in vitro [40]. Most of the interactions of IF3mt with the SSU are mediated by its CTD. The CTD alone binds to the mitoribosome with two- to threefold lower affinity than the full length IF3mt. The IF3mt NTD binds the mitoribosome with an affinity that is about ten-fold lower than that of the full length protein. Interestingly, recent cryoEM studies show that the NTD interacts with the mitoribosomal protein uS11m, and h23 of the 12S rRNA [37]. In the presence of the LSU, while both the NTD and CTD of IF3mt are displaced from SSU, the CTD is displaced more readily. The binding site of IF3mt CTD with the SSU is in a region where the SSU has contacts with the LSU. The NTD alone (with or without the linker region) is incapable of dissociating the 55S ribosome. However, the CTD alone has weak dissociation activity. Interestingly, CTD possessing the linker region becomes active in dissociating the 55S mitoribosome. Thus, the linker plays an important role in the dissociation activity of the CTD. In bacteria, a highly conserved residue Y75 in the linker region plays an important role in the fidelity function of IF3 and in interaction with C701 of 16S rRNA [41]. In IF3mt, the residues R140 and R144 that are conserved amongst mammals allow the linker to bind to the SSU platform and may have evolved to compensate for the loss of Y75 [37]. Only the C-terminal domain in the presence of the linker region is able to reduce fMet-tRNAMet binding to the SSU or the 30S ribosomal subunit in the absence of mRNA. The action of the NTD in this regard, even in the presence of the linker region, is very weak. Both the NTD and CTD play a significant role in promoting complex formation between fMet-tRNAMet and IF2mt. The NTD is more effective than the CTD in this process and the formylation of fMet-tRNAMet is essential.
The N- and C- termini of IF3mt have 30 amino acids long extensions compared to the bacterial IF3. These extensions are not essential for promoting IC formation on mitochondrial 55S ribosomes. However, the Cext was found to be essential for the dissociation of fMet-tRNAMet bound to the SSU in the absence of mRNA [42]. The currently accepted evolutionary role of the Cext is to facilitate an ordered pathway of mRNA binding prior to fMet-tRNAMet binding during initiation. Full length IF3mt interacts weakly with the LSU. However, when either the Next or the Cext are deleted, the factor has a higher affinity for the LSU [43]. Thus, another role for the extensions is to reduce the affinity of IF3mt for the protein-rich LSU that could lead to regulation of joining of the two subunits during formation of IC. A truncated derivative of IF3mt missing the extensions crosslinks to the same mitoribosomal proteins as the full length IF3mt except that no cross-links were detected to mS36. This indicates that the terminal extensions of IF3mt do not contribute significantly to its site of interaction on the mitoribosome. The uS10m, which is near the head region of the SSU, interacts with the NTD of IF3mt. The CTD of IF3mt does not interact with the proteins (such as uS11m and uS15m) of the platform region. Thus, it is possible that the binding of IF3mt to the mitoribosome differs from the binding of bacterial IF3 to the 30S subunit.
Our group has recently utilized an E. coli strain, in conjunction with plasmid-borne IF3mt and its truncated derivatives [44] to characterize IF3mt in vivo. Using the CAT reporter system and by analysing polysome profiles, we have shown that IF3mt allows ‘promiscuous’ translation initiation from the non-AUG codons such as AUA, AUU and ACG with the wild type i-tRNA but not with i-tRNAs that lack the universally conserved 3GC base pairs in their anticodon stems. Interestingly, IF3mt devoid of its Next, and Cext, curtailed initiation from the non-AUG codons. This observation suggests that the N- and C-terminal extensions of IF3mt may have evolved to relax the fidelity of translation initiation in order to accommodate classically non-canonical initiation codons such as AUU and AUA in mitochondria. Our findings also indicated that the NTD adds to the fidelity function of IF3mt for initiation codon and i-tRNA (through its anticodon stem) selections, which are reminiscent of the fidelity functions of the NTD of EcoIF3 [45]. This finding is supported by structural studies which indicate that the Next may interact with A424 (E. coli A790) of h24 of 12S rRNA and thus the NTD may play a role in i-tRNA binding in the mitoribosomal P site, through its extension [37].
Mitochondrial mRNA and mitochondrial initiator tRNA
The protein-coding genes of the mammalian mitochondrial genome are transcribed, post-transcriptionally modified and translated within mitochondria [2]. The ORFs are separated by very few nucleotides. In fact, in the two instances of ND4 and ND4L; and in ATPase 6 and ATPase 8, the genes overlap. Mammalian mitochondrial mRNAs are distinct from bacterial mRNAs in their usage of non-AUG initiation codons such as AUU (for NADH dehydrogenase subunit 2 or ND2 mRNA) and AUA (for ND1, ND3 and ND5 mRNAs) and for the presence of the very short or non-existent leader sequences. Although. yeast mitochondrial mRNAs also lack SD sequences, they retain long 5ʹ untranslated regions (UTR). Thus, it is likely that during the course of evolution, the loss of SD sequences happened prior to the shortening of the 5ʹ UTR [46]. To compensate for the absence of SD sequences, yeast mitochondrial mRNAs rely on translational activators, which align the initiation codon of mRNAs with the mitoribosomal P site [42]. Yeast translational activators connect 5ʹ UTRs of transcripts with the mitoribosome and the inner mitochondrial membrane to position mitoribosomes containing the specific mRNAs near the mitochondrial membrane to allow insertion of the nascent polypeptides into the membrane [46]. Translational activators are specific for individual yeast mitochondrial RNAs, for instance Pet309 is a translational activator for the COX1 mRNA, while Pet111 is an activator for the COX2 mRNA.
Studies have also shown the efficient usage of the GUG codon in mammalian mitochondria under diseased conditions [47]. Accurate recognition of the AUA codon as an initiation codon is facilitated by methylation of cytosine 34 in the mitochondrial tRNAMet by NSUN3 [48]. According to in vitro studies from the Spremulli group, the 5ʹ phosphate group of leaderless mRNAs is not required for their recruitment to the ribosome [39].
Although, mitochondria of lower eukaryotes such as yeast possess distinct initiator and elongator tRNAMet, mammalian mitochondria are characterized by the presence of a single tRNAMet [35] for its functions in initiation and elongation. The mammalian mitochondrial i-tRNA harbours a combination of features present in eukaryotic cytoplasmic and prokaryotic i-tRNAs and also elongator tRNAs [35]. (i) Mitochondrial tRNAMet has three consecutive GC base pairs in the anticodon stem, which is found virtually in all i-tRNAs. (ii) Quite like cytoplasmic i-tRNAs, mammalian mitochondrial tRNAMet has an A:U pair at the beginning of the acceptor helix. This serves as an optimal pair to allow interaction with both IF2mt and EF-Tumt, since many mitochondrial elongator tRNAs also have an A:U pair at the end of the acceptor stem. (iii) Prokaryotic i-tRNAs have a purine 11: pyrimidine 24 pair, which mammalian mitochondrial tRNAMet also possesses. (iv) Mammalian mitochondrial tRNAMet partially resembles prokaryotic tRNAs due to the presence of U54 and pyrimidine 60, except that the 54th position is un-methylated. In general, mammalian mitochondrial tRNAMet has very few modifications compared to the prokaryotic and eukaryotic (cytosolic) i-tRNAs. These modifications in the mitochondrial tRNAMet include Ψ at positions 27 and 50 and f5C in the anticodon. To facilitate initiation with the AUA codon, the C of the CAU anticodon of mitochondrial i-tRNA is modified to 5-formylcytidine (f5C) by the successive actions of the RNA methyltransferase NSUN3 and the dioxygenase ABH1 [48–50]. Recently, our group has generated and characterized mutants of E. coli tRNAfMet, which sustained E. coli for its requirement of both the initiator and elongator tRNAMet. The i-tRNA mutant that was most adept at both the initiation and elongation functions was also the one that was more similar to mitochondrial tRNAMet. Thus, the bacterial elongator and initiator tRNAMet may have originated from a single dual function tRNA [51].
Events in mitochondrial translation initiation
Although the SSU can interact with mRNAs in a sequence independent manner, the mRNA needs to be at least 350 nucleotides long for suitable binding [27]. It has also been shown by in vitro studies that an mRNA may bind to the SSU in the absence of any initiation factors [52]. Mitochondria do not have polysomes and only a single SSU binds to a single mRNA at a time [27].
The initial model for mitochondrial translation postulated by the Spremulli lab proposed that binding of fMet-tRNAMet by IF2mt in the absence of mRNA is destabilized by IF3mt, leading to the formation of a non-productive complex [42]. In the productive pathway, the SSU may bind the mRNA first but its position on the mRNA may be uncertain due to the absence of an SD-aSD interaction. IF3mt might alter the position of the mRNA such that the AUG (or an alternate initiation codon) is positioned at the P site (Fig. 5). Subsequently, IF2mt might promote fMet-tRNAMet binding to the P site. This is followed by LSU binding and dissociation of the initiation factors to form the 55S IC. In the absence of the Cext, IF3mt is not able to dissociate a non-productive complex. Therefore, the Cext may have evolved to create an ordered translation initiation pathway. Further, in the absence of extensions, IF3mt has an enhanced affinity for the LSU and it may allow its premature docking [43]. Therefore, another reason for the occurrence of the terminal extensions could be to reduce the affinity of IF3mt to the LSU to prevent the formation of incorrect ICs. Subsequent studies by the Spremulli group allowed them to propose that IF3mt is a dissociation factor and not just an anti-association factor for the ribosomal subunits [36]. While studying the preference for the leaderless mRNAs in mitochondrial translation [39], the Spremulli group proposed that after dissociation of the 55S complexes by IF3mt, an mRNA enters the ribosome and the ribosome pauses when the first 17 nucleotides enter to inspect the codon at the 5ʹ end of the mRNA, even if it is not an AUG codon. During this step, IF2mt allows fMet-tRNAMet binding at the P site. In case the initiation codon or the tRNA at the P site are incorrect, the mRNA moves through the ribosome and the monosome eventually dissociates. If the initiation codon and the tRNA are canonical, IF2mt hydrolyzes GTP, initiation factors are released and the LSU associates to form the 55S IC.
Figure 5.
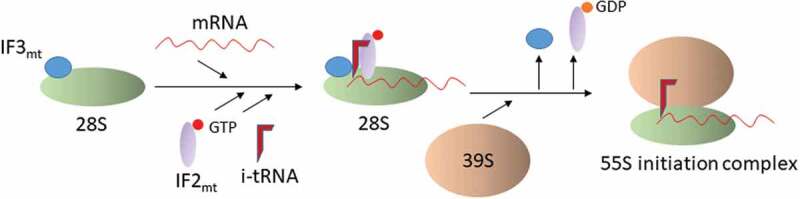
Mitochondrial translation initiation pathway.
Work from our lab has shown that both IF3mt and EcoIF3 are capable of examining the highly conserved feature of the 3GC base pairs in the anticodon stem of the i-tRNA [44]. However, when both the Next and Cext [IF3mtΔ(NextCext)] are absent, there is an improvement in the 3GC base pair-mediated fidelity of i-tRNA selection (i. e. binding of i-tRNA becomes more dependent on the presence of the 3GC base pairs). Based on the in vitro data, the absence of the extensions may facilitate i-tRNA accommodation at the P-site even in the absence of mRNA [42]. However, the, i-tRNA binding would be stabilized only after mRNA binding [42] in the presence of IF3mt. Since non-AUG initiation codons are present in the mitochondrial system, such an ejection of i-tRNA would slow down mitochondrial protein synthesis. Thus, the extensions of IF3mt may have evolved to somewhat lessen anticodon stem-based discrimination, to allow initiation with non-canonical initiation codons such as AUU and AUA.
Recent in vivo studies [45] and structural studies [53] have suggested that prokaryotic IF3 may remain bound to the 70S IC due to a recently discovered binding site on the 50S ribosome. These findings are crucial in explaining the occurrence of the 70S mode of initiation for translation of leaderless mRNAs. The Spremullli group has seen that IF3mt can bind to the 55S ribosome, for the purpose of ribosome dissociation. Further IF3mt only permits initiation with leaderless mRNAs [39]. Given that the mammalian mitochondrial system utilizes leaderless mRNAs, we propose that it is entirely possible that the mammalian mitochondrial system may also be capable of initiating with monosomes under certain circumstances. It would be interesting to investigate if 55S initiation does indeed occur in mammalian mitochondria and what factors might influence such non-canonical modes of initiation.
In vivo studies of IF2mt have been crucial in deciphering the order of the departure of prokaryotic initiation factors from the IC [30]. Earlier studies in prokaryotes have indicated that the affinity of IF2 for the 30S subunit increases while IF1 is present, whereas the release of IF1 from the ribosomal subunit leads to ejection of IF2 [54]. Since IF2mt is a protein that is capable of complementing E. coli for the functions of eubacterial IF1 and IF2, it may be proposed that in the bacterial system also, IF1 and IF2 may depart from the 70S complex simultaneously.
Diseases of mitochondrial translation initiation
Mitochondrial pathologies, which can be caused by either nuclear or mitochondrial DNA mutations, affect 1 in 5000 people [55,56]. These diseases cause OXPHOS dysfunction that can lead to a variety of disorders, which may be multi-systemic or tissue-specific with varying degrees of severity. Defects in mitochondrial translation are responsible for a large number of these pathologies. Mutations in elongation factors have been known to cause encephalopathy with liver or heart involvement [57] while mutations in the mitochondrial termination factor C12orf65 can cause Leigh Syndrome [58]. Mutations in mitoribosomal proteins such as bS16m may cause neonatal lactic acidosis [59] while an uL3m mutation causes cardiomyopathy [60]. Mitochondrial DNA rearrangements as seen in Kearns Sayre Syndrome can lead to mitochondrial tRNA or rRNA defects [55]. A large number of mitochondrial DNA mutations affect tRNA genes such as tRNALys (MERRF) and tRNALeu (MELAS). Additionally, mutations of factors involved in mitochondrial mRNA maturation, and mitochondrial tRNA processing and aminoacylation have also been implicated in a wide variety of mitochondrial pathologies [56]. In this review, we will focus on diseases of mitochondrial translation initiation (Table 1).
Table 1.
Human diseases due to defects in mitochondrial translation initiation.
| Gene | Protein/RNA | Clinical phenotypes | Reference |
|---|---|---|---|
| MTIF2 | Mitochondrial initiation factor 2 | Poor prognosis in inorganic arsenic induced lung cell malignancy | [61] |
| MTIF3 | Mitochondrial initiation factor 3 | Sporadic Parkinson’s disease, obesity, autoantibodies identified in Type I diabetes patients | [62–66] |
| MTFMT | mitochondrial methionyl-tRNA formyltransferase | Leigh syndrome and combined OXPHOS deficiency | [67] |
| NSUN3 | Sun RNA Methyltransferase 3 | Combined OXPHOS deficiency in skeletal muscle, combined developmental disability, microcephaly, failure to thrive, recurrent increased lactate levels in plasma, muscular weakness, proximal accentuated, external ophthalmoplegia and convergence nystagmus | [76] |
| MT-TM | tRNAMet | Maternally inherited hypertension, mitochondrial myopathy, LHON, intellectual disability, hypotonia, seizure, muscle weakness, lactic acidosis, hearing loss, MELAS | [50,68–75] |
| MT-COII | Mitochondrially encoded Cytochrome C Oxidase II | Mitochondrial encephalomyopathy | [77] |
| MT-ND1 | Mitochondrially encoded NADH:Ubiquinone Oxidoreductase Core Subunit 1 | Bilateral striatal necrosis and MELAS | [78] |
A recent study has shown that upregulation of IF2mt may be indicative of poor prognosis in inorganic arsenic induced lung cell malignancy [61]. IF3mt has been implicated in the pathogenesis of various disorders like Parkinson’s disease (PD), obesity and diabetes. A synonymous polymorphism (D266D, resulting from C798T nucleotide change) has been associated with sporadic PD [62]. IF3mt mutations have also been implicated in obesity, where an SNP in IF3mt (rs4771122) in Mexican children was associated with increased body mass index (BMI) [63]. Interestingly, the same SNP was associated with long-term weight loss after bariatric surgery [64]. Each copy of the minor G allele of IF3mt (rs1885988) was associated with greater weight loss following lifestyle intervention of the Diabetes Prevention Program[65]. Additionally, autoantibodies to IF3mt were identified in type I diabetes patients [66], implicating IF3mt in diabetes. Disruptions in modifications of the i-tRNA have been implicated in various disease phenotypes. Mutations in MTFMT (encoding mitochondrial methionyl-tRNA formyltransferase, which formylates tRNAMet) have been isolated in patients with Leigh syndrome and combined OXPHOS deficiency [67]. Patient fibroblasts had very low levels of fMet-tRNAMet, which were rescued by overexpression of wild type MTFMT. There are numerous documented cases of mitochondrial disease caused by mitochondrial tRNAMet mutations [68–75], all of which are characterized by either mitochondrial myopathy or hypertension. In addition, pathogenic mutations in the anticodon stem loop region of mitochondrial tRNAMet have been isolated, which led to impaired NSUN3 binding causing hypomodification of f5C34 [50]. Similarly, a compound heterozygous mutation in NSUN3 led to severe mitochondrial disease characterized by numerous clinical presentations including combined OXPHOS deficiency in skeletal muscle, microcephaly and failure to thrive [76]. Mutations of the AUG initiation codon of the gene encoding subunit II of cytochrome c oxidase and the AUA initiation codon of the ND1 gene to ACG and ACA, respectively, were also shown to be pathogenic [77,78].
Available methods of studying mitochondrial translation in vivo
In vitro mitochondrial translation studies have been limited due to technical difficulties in reconstituting the entire translational machinery [79], and in vivo studies have not been feasible due to inadequate methodologies to manipulate mitochondria [3]. Mitochondria are incapable of propagation and survival outside eukaryotic cells. Therefore, a robust in vivo system is crucial to study mitochondrial translation. The resemblances between bacterial and mitochondrial protein synthesis have allowed us to establish that IF2mt can complement E. coli for the essential roles of EcoIF1 and EcoIF2 [30]. It was shown that the 37 amino acid protrusion of IF2mt was functionally equivalent to EcoIF1 and this in vivo data was further validated by structural studies [31]. Thus, E. coli was found to be an able ‘substitute in vivo system’ to characterize proteins involved in mitochondrial translation. Such a system can be exploited to a greater extent by using reporter genes to functionally characterize a mitochondrial protein. Our lab has recently developed an E. coli strain (infCΔ55- lacking 55 amino acids from the N-terminus of IF3), with compromised IF3 activity [45]. This strain, in conjunction with the chloramphenicol acetyltransferase reporter system has proved to be a useful heterologous system for characterization of IF3mt [44]. Our group was also able to use a system of tRNA gene deletions in E. coli to investigate how initiator and elongator tRNAs may have evolved from a single bifunctional tRNAMet [51].
The ideal in vivo method of studying mitochondrial translation would be to carry out reporter-based experiments in human cell lines or whole organisms. Reporter systems are imperative to study initiation codon usage, i-tRNA selection, mitoribosome stalling and the roles of the various domains of mitochondrial translation factors in health and disease. Unfortunately, both DNA and RNA import are difficult in mammalian mitochondria [3]. To recapitulate, mammalian mitochondria only utilize mRNAs transcribed from their own genomes for translation. Unlike yeast and kinetoplastid protozoa, mammalian mitochondria are self-sufficient for all the tRNAs required during translation and they do not need to import tRNAs from the cytosol. Similarly, the mammalian mitochondrial genome also encodes its own rRNAs (12S and 16S) and mitochondrially encoded tRNAVal is found in the position of bacterial 5S rRNA [11,80]. Mitochondrial RNase P, which is an endonuclease that plays a role in the maturation of tRNAs, was earlier thought to have a nuclear-encoded RNA and multiple protein components like its bacterial and cytosolic counterparts [81]. However, it was subsequently shown that human mitochondrial RNase P is a protein enzyme without a mitochondrially imported RNA component [82]. Therefore, it is important to note that there seems to be no requirement for RNA import in mammalian mitochondria and the broader impact of RNA import is still to be shown. Recent studies have also shown that mammalian mitochondrial genomes can be modified by transcription activator-like effector nucleases (TALENs), Zinc Finger Nucleases and CRISPR Cas9 [83–87]. We hope that these techniques will be harnessed in the near future to insert reporter genes into mammalian mitochondrial genomes.
Although reporter studies in mammalian mitochondria appear difficult, Koehler’s group has shown that the fusion of the H1 RNA (earlier thought to be a part of RNase P [82]) import signal to a test RNA facilitated its import into mitochondria [88]. Subsequently, studies from the Tarassov and Adhya groups have shown that the yeast and Leishmania model systems function for RNA import. Kinetoplastid protozoa such as the Leishmania species import all the required tRNAs from the cytosol into mitochondria as their mitochondrial genomes do not encode any tRNAs. Work from the Adhya group has shown that tRNA import is mediated by a large multiprotein complex (RNA import complex or RIC) and it is dependent on a tRNA motif (similar to the D arm of tRNATyr) which is present in a majority of Leishmania tRNAs [89,90]. Mitochondrial translation can potentially be studied in the Leishmania system, with the help of reporter mRNAs tagged with the D arm of tRNATyr [91–95]. The Tarassov lab has shown that short synthetic RNAs (referred to as FD-RNA) comprising two domains of the tRNALys were individually sufficient to deliver RNAs into mitochondria. This technology could be adopted to send reporter mRNAs into yeast mitochondria in order to characterize mitochondrial translation in vivo. Such systems would help us study initiation codon preference, codon usage and the influence of a 5ʹ UTR on mitochondrial translation in vivo. Microprojectile bombardment has been successfully utilized to deliver DNA sequences to yeast mitochondria, and these DNA sequences are subsequently integrated into the mitochondrial genome by homologous recombination [96]. Such systems have been successfully utilized to carry out reporter assays for translation initiation in yeast [97].
In addition, there are a number of untapped resources to potentially target nucleic acids to mammalian mitochondria. For instance, triphenylphosphine-coated nanoparticles have been used to target cancer drugs to the mitochondria of breast cancer cells [98]. This technology could be engineered to target reporter mRNAs to human cells.
In the absence of reporter genes, another method of studying mitochondrial translation initiation is by means of gene deletions and knockdowns. Currently, our group is in the process of obtaining and analysing mitochondrial initiation factor knockdowns and knockouts. Such a system can also be used to study the functions of individual domains of translational factors as well as their disease-causing variants.
Concluding remarks
Considerable progress has been made in the last decade to decipher the cryptic process of mitochondrial translation initiation: from superior structural analyses of the human mitoribosome to clearer biochemical characterizations of mitochondrial i-tRNA and initiation factors, and studies of naturally occurring pathogenic mutations. However, the need of the hour is to develop suitable RNA or DNA import mechanisms for efficient reporter studies to comprehensively characterize translation in mammalian mitochondria.
Funding Statement
The work in the laboratory of UV is supported by research grants from the Department of Biotechnology (DBT), Ministry of Science and Technology; Science and Engineering Research Board (SERB); Department of Science and Technology (DST), Ministry of Science and Technology; and the J.C. Bose Fellowship of DST (to U.V.). The authors acknowledge the DBT-IISc partnership programme, University Grants Commission, New Delhi for the Centre of Advanced Studies, and the DST-FIST level II infrastructure supports.
Acknowledgments
We thank our laboratory colleagues for their suggestions on the manuscript.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
References
- [1].Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 2012;4:a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. [DOI] [PubMed] [Google Scholar]
- [3].Lightowlers RN, Rozanska A, Chrzanowska-Lightowlers ZM. Mitochondrial protein synthesis: figuring the fundamentals, complexities and complications, of mammalian mitochondrial translation. FEBS Lett. 2014;588:2496–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Towpik J. Regulation of mitochondrial translation in yeast. Cell Mol Biol Lett. 2005;10:571–594. [PubMed] [Google Scholar]
- [5].Kuzmenko A, Atkinson GC, Levitskii S, et al. Mitochondrial translation initiation machinery: conservation and diversification. Biochimie. 2014;100:132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Derbikova KS, Levitsky SA, Chicherin IV, et al. Activation of yeast mitochondrial translation: who is in charge? Biochemistry (Mosc). 2018;83:87–97. [DOI] [PubMed] [Google Scholar]
- [7].Cavdar Koc E, Burkhart W, Blackburn K, et al. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J Biol Chem. 2001;276:19363–19374. [DOI] [PubMed] [Google Scholar]
- [8].Koc EC, Burkhart W, Blackburn K, et al. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J Biol Chem. 2001;276:43958–43969. [DOI] [PubMed] [Google Scholar]
- [9].Amunts A, Brown A, Bai XC, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang G, Chen HW, Oktay Y, et al. PNPASE regulates RNA import into mitochondria. Cell. 2010;142:456–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Amunts A, Brown A, Toots J, et al. Ribosome. The structure of the human mitochondrial ribosome. Science. 2015;348:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Desai N, Brown A, Amunts A, et al. The structure of the yeast mitochondrial ribosome. Science. 2017;355:528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kummer E, Leibundgut M, Rackham O, et al. Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. Nature. 2018;560:263–267. [DOI] [PubMed] [Google Scholar]
- [14].Steitz TA. A structural understanding of the dynamic ribosome machine. Nat Rev Mol Cell Biol. 2008;9:242–253. [DOI] [PubMed] [Google Scholar]
- [15].Moore PB, Steitz TA. The involvement of RNA in ribosome function. Nature. 2002;418:229–235. [DOI] [PubMed] [Google Scholar]
- [16].Selmer M, Dunham CM, Murphy FVT, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. [DOI] [PubMed] [Google Scholar]
- [17].Carter AP, Clemons WM Jr., Brodersen DE, et al. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. [DOI] [PubMed] [Google Scholar]
- [18].MacKeen LA, Kahan L, Wahba AJ, et al. Photochemical cross-linking of initiation factor-3 to Escherichia coli 30 S ribosomal subunits. J Biol Chem. 1980;255:10526–10531. [PubMed] [Google Scholar]
- [19].Cooperman BS, Expert-Bezancon A, Kahan L, et al. IF-3 crosslinking to Escherichia coli ribosomal 30 S subunits by three different light-dependent procedures: identification of 30 S proteins crosslinked to IF-3–utilization of a new two-stage crosslinking reagent, p-nitrobenzylmaleimide. Arch Biochem Biophys. 1981;208:554–562. [DOI] [PubMed] [Google Scholar]
- [20].Boileau G, Butler P, Hershey JW, et al. Direct cross-links between initiation factors 1, 2, and 3 and ribosomal proteins promoted by 2-iminothiolane. Biochemistry. 1983;22:3162–3170. [DOI] [PubMed] [Google Scholar]
- [21].Cukras AR, Southworth DR, Brunelle JL, et al. Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA: tRNAcomplex. Mol Cell. 2003;12:321–328. [DOI] [PubMed] [Google Scholar]
- [22].Haque ME, Koc H, Cimen H, et al. Contacts between mammalian mitochondrial translational initiation factor 3 and ribosomal proteins in the small subunit. Biochim Biophys Acta. 2011;1814:1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hoang L, Fredrick K, Noller HF. Creating ribosomes with an all-RNA 30S subunit P site. Proc Natl Acad Sci U S A. 2004;101:12439–12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Arora S, Bhamidimarri SP, Bhattacharyya M, et al. Distinctive contributions of the ribosomal P-site elements m2G966, m5C967 and the C-terminal tail of the S9 protein in the fidelity of initiation of translation in Escherichia coli. Nucleic Acids Res. 2013;41:4963–4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ayyub SA, Dobriyal D, Shah RA, et al. Coevolution of the translational machinery optimizes initiation with unusual initiator tRNAs and initiation codons in mycoplasmas. RNA Biol. 2018;15:70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hartz D, McPheeters DS, Gold L. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev. 1989;3:1899–1912. [DOI] [PubMed] [Google Scholar]
- [27].Liao HX, Spremulli LL. Identification and initial characterization of translational initiation factor 2 from bovine mitochondria. J Biol Chem. 1990;265:13618–13622. [PubMed] [Google Scholar]
- [28].Vambutas A, Ackerman SH, Tzagoloff A. Mitochondrial translational-initiation and elongation factors in Saccharomyces cerevisiae. Eur J Biochem. 1991;201:643–652. [DOI] [PubMed] [Google Scholar]
- [29].Liao HX, Spremulli LL. Initiation of protein synthesis in animal mitochondria. Purification and characterization of translational initiation factor 2. J Biol Chem. 1991;266:20714–20719. [PubMed] [Google Scholar]
- [30].Gaur R, Grasso D, Datta PP, et al. A single mammalian mitochondrial translation initiation factor functionally replaces two bacterial factors. Mol Cell. 2008;29:180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yassin AS, Haque ME, Datta PP, et al. Insertion domain within mammalian mitochondrial translation initiation factor 2 serves the role of eubacterial initiation factor 1. Proc Natl Acad Sci U S A. 2011;108:3918–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Spencer AC, Spremulli LL. Interaction of mitochondrial initiation factor 2 with mitochondrial fMet-tRNA. Nucleic Acids Res. 2004;32:5464–5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Atkinson GC, Kuzmenko A, Kamenski P, et al. Evolutionary and genetic analyses of mitochondrial translation initiation factors identify the missing mitochondrial IF3 in S. cerevisiae. Nucleic Acids Res. 2012;40:6122–6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Koc EC, Spremulli LL. Identification of mammalian mitochondrial translational initiation factor 3 and examination of its role in initiation complex formation with natural mRNAs. J Biol Chem. 2002;277:35541–35549. [DOI] [PubMed] [Google Scholar]
- [35].Spremulli LL, Coursey A, Navratil T, et al. Initiation and elongation factors in mammalian mitochondrial protein biosynthesis. Prog Nucleic Acid Res Mol Biol. 2004;77:211–261. [DOI] [PubMed] [Google Scholar]
- [36].Christian BE, Spremulli LL. Evidence for an active role of IF3mt in the initiation of translation in mammalian mitochondria. Biochemistry. 2009;48:3269–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Koripella RK, Sharma MR, Haque ME, et al. Structure of human mitochondrial translation initiation factor 3 bound to the small ribosomal subunit. iScience. 2019;12:76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ma J, Spremulli LL. Expression, purification, and mechanistic studies of bovine mitochondrial translational initiation factor 2. J Biol Chem. 1996;271:5805–5811. [DOI] [PubMed] [Google Scholar]
- [39].Christian BE, Spremulli LL. Preferential selection of the 5ʹ-terminal start codon on leaderless mRNAs by mammalian mitochondrial ribosomes. J Biol Chem. 2010;285:28379–28386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Haque ME, Spremulli LL. Roles of the N- and C-terminal domains of mammalian mitochondrial initiation factor 3 in protein biosynthesis. J Mol Biol. 2008;384:929–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hussain T, Llacer JL, Wimberly BT, et al. Large-scale movements of IF3 and tRNA during bacterial translation initiation. Cell. 2016;167:133–44 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bhargava K, Spremulli LL. Role of the N- and C-terminal extensions on the activity of mammalian mitochondrial translational initiation factor 3. Nucleic Acids Res. 2005;33:7011–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Haque ME, Grasso D, Spremulli LL. The interaction of mammalian mitochondrial translational initiation factor 3 with ribosomes: evolution of terminal extensions in IF3mt. Nucleic Acids Res. 2008;36:589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ayyub SA, Aswathy SL, Dobriyal D, et al. Fidelity of translation in the presence of mammalian mitochondrial initiation factor 3. Mitochondrion. 2017;39:1–8. [DOI] [PubMed] [Google Scholar]
- [45].Ayyub SA, Dobriyal D, Varshney U. Contributions of the N- and C-terminal domains of initiation factor 3 to its functions in the fidelity of initiation and antiassociation of the ribosomal subunits. J Bacteriol. 2017;199(11):e00051–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Herrmann JM, Woellhaf MW, Bonnefoy N. Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochim Biophys Acta. 2013;1833:286–294. [DOI] [PubMed] [Google Scholar]
- [47].Dubot A, Godinot C, Dumur V, et al. GUG is an efficient initiation codon to translate the human mitochondrial ATP6 gene. Biochem Biophys Res Commun. 2004;313:687–693. [DOI] [PubMed] [Google Scholar]
- [48].Haag S, Sloan KE, Ranjan N, et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 2016;35:2104–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Takemoto C, Spremulli LL, Benkowski LA, et al. Unconventional decoding of the AUA codon as methionine by mitochondrial tRNAMet with the anticodon f5CAU as revealed with a mitochondrial in vitro translation system. Nucleic Acids Res. 2009;37:1616–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Nakano S, Suzuki T, Kawarada L, et al. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNA(Met). Nat Chem Biol. 2016;12:546–551. [DOI] [PubMed] [Google Scholar]
- [51].Govindan A, Ayyub SA, Varshney U. Sustenance of Escherichia coli on a single tRNAMet. Nucleic Acids Res. 2018;46:11566–11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Liao HX, Spremulli LL. Interaction of bovine mitochondrial ribosomes with messenger RNA. J Biol Chem. 1989;264:7518–7522. [PubMed] [Google Scholar]
- [53].Goyal A, Belardinelli R, Rodnina MV. Non-canonical binding site for bacterial initiation factor 3 on the large ribosomal subunit. Cell Rep. 2017;20:3113–3122. [DOI] [PubMed] [Google Scholar]
- [54].Celano B, Pawlik RT, Gualerzi CO. Interaction of Escherichia coli translation-initiation factor IF-1 with ribosomes. Eur J Biochem. 1988;178:351–355. [DOI] [PubMed] [Google Scholar]
- [55].Boczonadi V, Horvath R. Mitochondria: impaired mitochondrial translation in human disease. Int J Biochem Cell Biol. 2014;48:77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Mai N, Chrzanowska-Lightowlers ZM, Lightowlers RN. The process of mammalian mitochondrial protein synthesis. Cell Tissue Res. 2017;367:5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Valente L, Tiranti V, Marsano RM, et al. Infantile encephalopathy and defective mitochondrial DNA translation in patients with mutations of mitochondrial elongation factors EFG1 and EFTu. Am J Hum Genet. 2007;80:44–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Antonicka H, Ostergaard E, Sasarman F, et al. Mutations in C12orf65 in patients with encephalomyopathy and a mitochondrial translation defect. Am J Hum Genet. 2010;87:115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Miller C, Saada A, Shaul N, et al. Defective mitochondrial translation caused by a ribosomal protein (MRPS16) mutation. Ann Neurol. 2004;56:734–738. [DOI] [PubMed] [Google Scholar]
- [60].Galmiche L, Serre V, Beinat M, et al. Exome sequencing identifies MRPL3 mutation in mitochondrial cardiomyopathy. Hum Mutat. 2011;32:1225–1231. [DOI] [PubMed] [Google Scholar]
- [61].Zhang L, Huang Y, Ling J, et al. Screening of key genes and prediction of therapeutic agents in Arsenic-induced lung carcinoma. Cancer Biomark. 2019;25:351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Anvret A, Ran C, Westerlund M, et al. Possible involvement of a mitochondrial translation initiation factor 3 variant causing decreased mRNA levels in Parkinson’s disease. Parkinson’s Dis. 2010;2010:491751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Abadi A, Peralta-Romero J, Suarez F, et al. Assessing the effects of 35 European-derived BMI-associated SNPs in Mexican children. Obesity. 2016;24:1989–1995. [DOI] [PubMed] [Google Scholar]
- [64].Rasmussen-Torvik LJ, Baldridge AS, Pacheco JA, et al. rs4771122 predicts multiple measures of long-term weight loss after bariatric surgery. Obes Surg. 2015;25:2225–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Papandonatos GD, Pan Q, Pajewski NM, et al. Genetic predisposition to weight loss and regain with lifestyle intervention: analyses from the diabetes prevention program and the look AHEAD randomized controlled trials. Diabetes. 2015;64:4312–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Bian X, Wasserfall C, Wallstrom G, et al. Tracking the antibody immunome in type 1 diabetes using protein arrays. J Proteome Res. 2017;16:195–203. [DOI] [PubMed] [Google Scholar]
- [67].Tucker EJ, Hershman SG, Kohrer C, et al. Mutations in MTFMT underlie a human disorder of formylation causing impaired mitochondrial translation. Cell Metab. 2011;14:428–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Peverelli L, Gold CA, Naini AB, et al. Mitochondrial myopathy with dystrophic features due to a novel mutation in the MTTM gene. Muscle Nerve. 2014;50:292–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Vissing J, Salamon MB, Arlien-Soborg P, et al. A new mitochondrial tRNA(Met) gene mutation in a patient with dystrophic muscle and exercise intolerance. Neurology. 1998;50:1875–1878. [DOI] [PubMed] [Google Scholar]
- [70].Zhu HY, Wang SW, Liu L, et al. Genetic variants in mitochondrial tRNA genes are associated with essential hypertension in a Chinese Han population. Clin Chim Acta. 2009;410:64–69. [DOI] [PubMed] [Google Scholar]
- [71].Bortot B, Barbi E, Biffi S, et al. Two novel cosegregating mutations in tRNAMet and COX III, in a patient with exercise intolerance and autoimmune polyendocrinopathy. Mitochondrion. 2009;9:123–129. [DOI] [PubMed] [Google Scholar]
- [72].Qu J, Li R, Zhou X, et al. The novel A4435G mutation in the mitochondrial tRNAMet may modulate the phenotypic expression of the LHON-associated ND4 G11778A mutation. Invest Ophthalmol Vis Sci. 2006;47:475–483. [DOI] [PubMed] [Google Scholar]
- [73].Tang S, Wang J, Zhang VW, et al. Transition to next generation analysis of the whole mitochondrial genome: a summary of molecular defects. Hum Mutat. 2013;34:882–893. [DOI] [PubMed] [Google Scholar]
- [74].Sternberg D, Chatzoglou E, Laforet P, et al. Mitochondrial DNA transfer RNA gene sequence variations in patients with mitochondrial disorders. Brain. 2001;124:984–994. [DOI] [PubMed] [Google Scholar]
- [75].Liu Y, Li Y, Zhu C, et al. Mitochondrial biogenesis dysfunction and metabolic dysfunction from a novel mitochondrial tRNA(Met) 4467 C>A mutation in a Han Chinese family with maternally inherited hypertension. Sci Rep. 2017;7:3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Van Haute L, Dietmann S, Kremer L, et al. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat Commun. 2016;7:12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Clark KM, Taylor RW, Johnson MA, et al. An mtDNA mutation in the initiation codon of the cytochrome C oxidase subunit II gene results in lower levels of the protein and a mitochondrial encephalomyopathy. Am J Hum Genet. 1999;64:1330–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Campos Y, Martin MA, Rubio JC, et al. Bilateral striatal necrosis and MELAS associated with a new T3308C mutation in the mitochondrial ND1 gene. Biochem Biophys Res Commun. 1997;238:323–325. [DOI] [PubMed] [Google Scholar]
- [79].Dekker PJ, Papadopoulou B, Grivell LA. In-vitro translation of mitochondrial mRNAs by yeast mitochondrial ribosomes is hampered by the lack of start-codon recognition. Curr Genet. 1993;23:22–27. [DOI] [PubMed] [Google Scholar]
- [80].Rorbach J, Gao F, Powell CA, et al. Human mitochondrial ribosomes can switch their structural RNA composition. Proc Natl Acad Sci U S A. 2016;113:12198–12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Walker SC, Engelke DR. A protein-only RNase P in human mitochondria. Cell. 2008;135:412–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Holzmann J, Frank P, Loffler E, et al. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. [DOI] [PubMed] [Google Scholar]
- [83].Bian WP, Chen YL, Luo JJ, et al. Knock-In strategy for editing human and zebrafish mitochondrial DNA using mito-CRISPR/Cas9 system. ACS Synth Biol. 2019;8:621–632. [DOI] [PubMed] [Google Scholar]
- [84].Gammage PA, Gaude E, Van Haute L, et al. Near-complete elimination of mutant mtDNA by iterative or dynamic dose-controlled treatment with mtZFNs. Nucleic Acids Res. 2016;44:7804–7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Gammage PA, Viscomi C, Simard ML, et al. Genome editing in mitochondria corrects a pathogenic mtDNA mutation in vivo. Nat Med. 2018;24:1691–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Bacman SR, Williams SL, Pinto M, et al. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat Med. 2013;19:1111–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Loutre R, Heckel AM, Smirnova A, et al. Can mitochondrial DNA be CRISPRized: pro and contra. IUBMB Life. 2018;70:1233–1239. [DOI] [PubMed] [Google Scholar]
- [88].Wang G, Shimada E, Zhang J, et al. Correcting human mitochondrial mutations with targeted RNA import. Proc Natl Acad Sci U S A. 2012;109:4840–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Adhya S, Mahato B, Jash S, et al. Mitochondrial gene therapy: the tortuous path from bench to bedside. Mitochondrion. 2011;11:839–844. [DOI] [PubMed] [Google Scholar]
- [90].Mukherjee S, Basu S, Home P, et al. Necessary and sufficient factors for the import of transfer RNA into the kinetoplast mitochondrion. EMBO Rep. 2007;8:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Martin RP, Schneller JM, Stahl AJ, et al. Study of yeast mitochondrial tRNAs by two-dimensional polyacrylamide gel electrophoresis: characterization of isoaccepting species and search for imported cytoplasmic tRNAs. Nucleic Acids Res. 1977;4:3497–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Kamenski P, Kolesnikova O, Jubenot V, et al. Evidence for an adaptation mechanism of mitochondrial translation via tRNA import from the cytosol. Mol Cell. 2007;26:625–637. [DOI] [PubMed] [Google Scholar]
- [93].Entelis N, Brandina I, Kamenski P, et al. A glycolytic enzyme, enolase, is recruited as a cofactor of tRNA targeting toward mitochondria in Saccharomyces cerevisiae. Genes Dev. 2006;20:1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Tarassov I, Entelis N, Martin RP. Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA synthetases. EMBO J. 1995;14:3461–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Tarassov I, Entelis N, Martin RP. An intact protein translocating machinery is required for mitochondrial import of a yeast cytoplasmic tRNA. J Mol Biol. 1995;245:315–323. [DOI] [PubMed] [Google Scholar]
- [96].Bonnefoy N, Fox TD. Directed alteration of Saccharomyces cerevisiae mitochondrial DNA by biolistic transformation and homologous recombination. Methods Mol Biol. 2007;372:153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Bonnefoy N, Fox TD. In vivo analysis of mutated initiation codons in the mitochondrial COX2 gene of Saccharomyces cerevisiae fused to the reporter gene ARG8m reveals lack of downstream reinitiation. MGG. 2000;262:1036–1046. [DOI] [PubMed] [Google Scholar]
- [98].Mallick A, Kuman MM, Ghosh A, et al. Cerberus nanoparticles: cotargeting of mitochondrial DNA and mitochondrial topoisomerase i in breast cancer cells. ACS Appl Nano Mater. 2018;1:2195–2205. [Google Scholar]


