ABSTRACT
RNA-binding proteins (RBPs) are key regulators of posttranscriptional processes such as RNA maturation, localization, turnover and translation. Despite their dysregulation in various diseases including cancer, the landscape of RBP expression in human cancer has not been well elucidated. Here, we built a comprehensive expression landscape of 1504 RBPs across 16 human cancer types, which revealed that RBPs are predominantly upregulated in tumours and this phenomenon is affected by the tumour immune subtypes and microenvironment. Across different cancer types, 109 RBPs are consistently upregulated while 41 RBPs are consistently downregulated. These up-regulated and down-regulated RBPs show distinct molecular characteristics and prognostic effects, whereas their dysregulation is mediated by distinct cis/trans-regulatory mechanisms. Finally, we validated one candidate PABPC1L that might promote colon tumorigenesis by regulating mRNA splicing. In summary, we built a comprehensive expression landscape of RBPs across different cancer types and identified consistently dysregulated RBPs which could be novel targets for developing broad-spectrum anticancer agents.
KEYWORDS: Expression landscape, RNA binding protein, cancer
Introduction
RNA-binding proteins (RBPs) are a group of conserved proteins in eukaryotes, which play essential roles in co-transcriptional and posttranscriptional gene regulation, including RNA maturation, RNA turnover, RNA localization and translation [1]. RBPs can interact with RNAs to form protein-RNA complexes, such as ribosomes that serve as the basic translational machinery, and small nuclear ribonucleoproteins (snRNPs) which are core component of pre-mRNA splicing machinery. RBPs can also regulate splicing, translation by controlling accessibility and activity of these basic machineries, for instance, the splicing enhancer/repressor and translation regulators [2,3]. Based on RNA-binding domain prediction and manually selection of RBPs from literature, a census of 1542 human RBPs has been established [4]. However, only a small proportion of them have been functionally characterized.
Since RBPs have diverse functions in posttranscriptional gene regulation, they are critical to many biological processes, such as cell differentiation, proliferation and cell fate transition. For example, RBP MBNL1 and MBNL2 control splicing of the 18th exon of transcription factor FOXP1 to switch pluripotency and reprogramming of embryonic stem cells (ESCs), while RBP NUDT21 directs alternative polyadenylation of thousands of transcripts to control cell fate transition of ESCs [5–7]. Dysregulation of RBPs could cause severe human diseases, including cancer [8]. For instance, SF3B1 is mutated in around 10-15% of chronic lymphocytic leukaemia (CLL) patients and is associated with poor survival rate [9,10]. In addition, mutations within four RBP genes U2AF, ZRSR2, SRSF2 and SF3B1 were very frequent (~70%) in a cohort of myelodysplasia (MDS) patients [11]. Besides mutation, expressional dysregulation of genes encoding core splicing and translational machinery component is required for MYC-mediated lymphomagenesis in mice mode [12,13]. Besides, it has been reported that RBP NELFE promote cancer progression via selectively regulating MYC-associated genes [14].
Despite extensive studies on selected RBPs, the general role of RBPs in cancer development is still unclear. Recently, somatic mutation landscape of splicing factors across 33 human cancer types, and mutation landscape of RBPs across 26 cancer types have been established [15,16]. However, the expression landscape of RBPs across different human cancer types is still under debate. In normal human tissues, expression of RBPs is significantly higher than that of transcription factors (TFs), other protein-coding genes (PCGs), miRNAs and lincRNAs [4,17]. By comparing tumour samples from The Cancer Genome Atlas (TCGA) to corresponding normal tissue from human body map, Kechavarzi showed that RBPs are overrepresented in top quantile upregulated genes [17]. However, a recent study found that RBPs are predominantly downregulated in tumours comparing to the adjacent normal tissue across 15 human cancer types only using TCGA data [18]. In contrast, another study revealed that RBPs tend to be upregulated in hepatocellular carcinoma comparing with normal liver tissues using the same TCGA data and one additional dataset [14]. Therefore, there is a pressing need to clarify the true expression landscape of RBPs across different human cancer types, explore the associations between expressional dysregulation and somatic mutation, understand the underlying regulatory mechanisms and investigate their roles in cancer development.
Here we built a comprehensive expression landscape of 1504 RBPs in ~6700 clinical samples across 16 human cancer types. We showed that RBPs are preferentially upregulated in tumours compared to their adjacent normal and fractions of upregulated RBPs were correlated with proliferation and wound healing signatures and impacted by the tumour microenvironment, such as stromal and leukocyte fractions. Across 16 cancer types, we identified 109 consistently upregulated RBPs (cuRBPs) and 41 consistently downregulated RBPs (cdRBPs). The cuRBPs are enriched in RNA modification, transcription termination (3ʹ end processing) and tRNA functional categories. Furthermore, we found that upregulation of cuRBPs is largely associated with amplification of copy number, whereas downregulation of many cdRBPs is likely a result of epigenetic silencing mediated by DNA methylation. Besides, we constructed TF/miRNA-cuRBP/cdRBP regulatory networks, in which we identified several hub TFs and miRNAs. Finally, we validated one candidate, PABPC1L might promote cancer progression in colon adenocarcinoma (COAD) by regulating mRNA splicing. Our study provides a comprehensive resource for studying RBPs across different human cancers and the consistently dysregulated RBPs we identified might be potentially helpful for developing broad-spectrum anti-cancer agents.
Results
Predominantly upregulation of RBPs in TCGA tumour samples is associated with tumour microenvironment
To dissect the dysregulation of RBPs in human cancers, we analysed mRNA expression of 1504 RBPs in ~6700 clinical samples from TCGA across 16 cancer types (Table 1). The other 17 cancer types were excluded due to insufficient normal samples (n < 10). For each cancer type, dysregulated RBPs were identified by comparing the mRNA expression in tumours to adjacent normal tissues at BH-adjusted p < 0.001, two-sided test and average RSEM value >1 in either normal or tumour samples [14]. Interestingly, we found more upregulated RBPs than downregulated RBPs in 14 out of the 16 cancer types, while this trend was not observed for TFs and other PCGs (Fig. 1(a)). Furthermore, we analysed the global expression changes of the entire set of RBPs and observed a positive fold change in almost all 16 cancer types, except KICH and THCA (Supplementary Fig. S1A). Next we analysed the expression of RBPs from 10 functional categories, including RNA splicing, translation, transcription termination (RNA 3ʹ end processing), RNA localization & transport, RNA surveillance & degradation, RNA modification, ribosome, tRNA, mitochondrial and enzymes (including helicase, nuclease, ATPase and ligase), as well as unclassified RBPs. Interestingly, RBPs are predominantly upregulated in BRCA, COAD, LIHC, LUAD and LUSC for almost all categories. In prostate cancer (PRAD), the most pervasively upregulated categories are a translation, transcription termination, ribosome, tRNA and mitochondrial. In head and neck squamous cell carcinoma (HNSC), the upregulated RBPs are enriched in transcription termination (Fig. 1(b)). These results suggest that tumour samples from different cancer types have distinct posttranscriptional dysregulation signatures.
Table 1.
The number of samples used in this study.
| RNA sequencing data |
DNA methylation data |
Copy Number data |
||||
|---|---|---|---|---|---|---|
| TCGA abbreviation |
Cancer types | Normal | Tumour | Normal | Tumour | Tumour |
| BLCA | Bladder Urothelial Carcinoma | 19 | 408 | 17 | 410 | 404 |
| BRCA | Breast invasive carcinoma | 113 | 1094 | 84 | 786 | 1088 |
| COAD | Colon adenocarcinoma | 41 | 284 | 19 | 284 | 274 |
| ESCA | Oesophageal carcinoma | 11 | 184 | - | - | 184 |
| HNSC | Head and Neck squamous cell carcinoma | 44 | 520 | 20 | 521 | 514 |
| KICH | Kidney Chromophobe | 25 | 65 | - | - | 65 |
| KIRC | Kidney renal clear cell carcinoma | 72 | 533 | 24 | 320 | 517 |
| KIRP | Kidney renal papillary cell carcinoma | 32 | 290 | 23 | 274 | 284 |
| LIHC | Liver hepatocellular carcinoma | 50 | 371 | 41 | 373 | 366 |
| LUAD | Lung adenocarcinoma | 59 | 515 | 21 | 459 | 496 |
| LUSC | Lung squamous cell carcinoma | 51 | 502 | - | - | 484 |
| PRAD | Prostate adenocarcinoma | 52 | 496 | 35 | 496 | 492 |
| READ | Rectum adenocarcinoma | 10 | 93 | - | - | 93 |
| STAD | Stomach adenocarcinoma | 35 | 415 | - | - | 413 |
| THCA | Thyroid carcinoma | 59 | 504 | 50 | 511 | 506 |
| UCEC | Uterine Corpus Endometrial Carcinoma | 24 | 176 | 24 | 173 | 177 |
Figure 1.
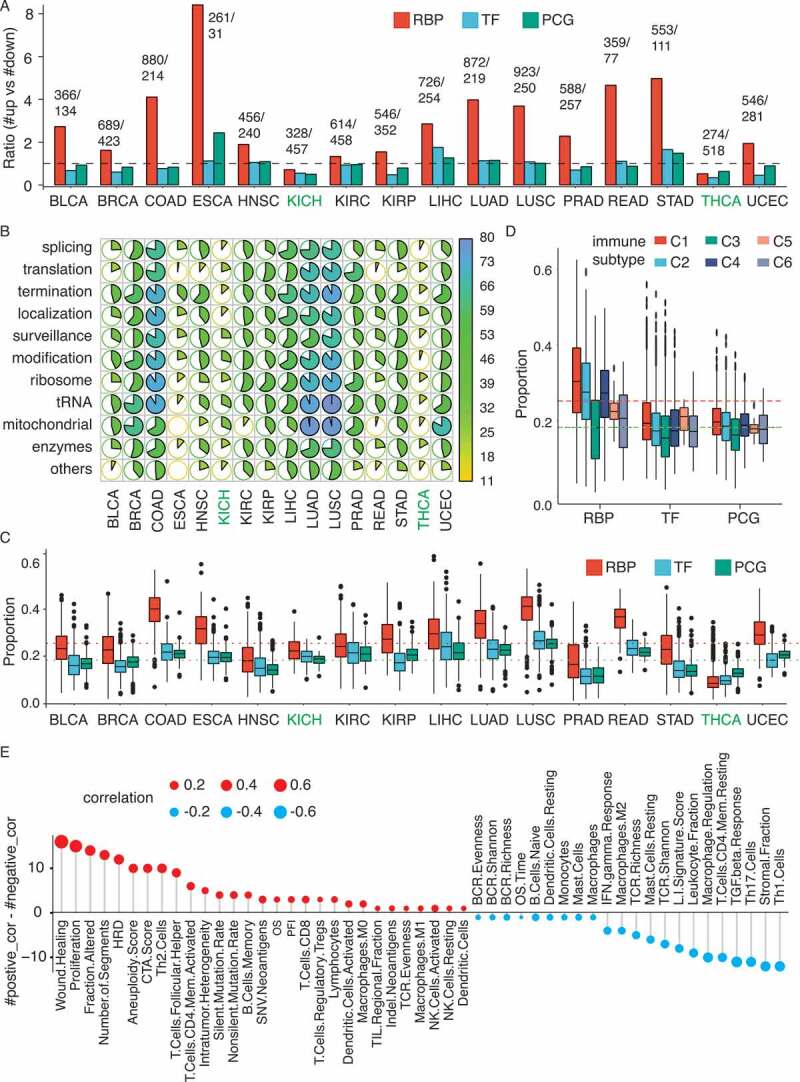
Expression landscape RBPs across 16 human cancer types. (A) The ratio of the number of upregulated genes to that of downregulated ones when comparing tumour with normal samples across 16 cancer types. (B) Fraction of RBPs in 11 functional categories that is significantly upregulated in tumour across 16 types. Colour key and pie chart illustrating the percentage. (C) Boxplot illustrating the fraction of upregulated RBPs, TFs and PCGs across 16 types. (D) Boxplot illustrating fraction of upregulated RBPs, TFs and PCGs across six immune subtype samples. (E) Distribution of Pearson correlation between upregulated RBPs and xx signatures. Y-axis indicating the difference in cohort numbers between positively correlated (r > 0.2) and negatively correlated (r < −0.2). Dot size and colour representing average correlation across 16 types.
Contradictory to our results and previous studies [14,17], Wang et al. showed that RBP expression is predominantly downregulated in tumour [18]. As they used the Voom function in the Limma package to identify differentially expressed genes, while we applied the t-test for the normalized expression data from TCGA, this might be the cause of discrepancy. To investigate this, we compared the expression fold change (tumour vs normal) of significant dysregulated RBPs in their study with our results. It turns out that the expression fold changes in these two are extremely similar (r = 0.978, p = 0, Supplementary Fig. S1B), suggesting a high reproducibility between our study and their study. However, without an arbitrary expression fold change cut-off, we got much more significant dysregulated RBPs (BH adjusted p < 0.001). By employing the same cut-off (fold change >2) as Wang et al. for the selection of dysregulated genes between tumour and normal, cancer types such as BLCA, BRCA, KICH, PRAD and THCA, indeed have more downregulated RBPs than upregulated ones (Supplementary Table S1, marked in red). Glioblastoma (GBM), which has much more upregulated RBPs than downregulated ones in Wang et al., was excluded from our study as there were only five normal samples. Additionally, Wang et al. performed a combined analysis of colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ), resulting in the comparison between 609 tumour and 51 normal samples. We analysed them separately: 281 tumour compared to 41 normal samples in COAD and 93 tumour compared to 10 normal samples in READ. This might also be a cause of the differences between their results and ours. Nevertheless, the histogram of log2 (fold change) of all dysregulated RBPs at BH-adjusted p < 0.001 shows a clear skewed trend towards RBP upregulation, while many of them have fold changes slightly lower than 2 (Supplementary Fig. S1C). To tackle with this problem in terms of setting an arbitrary fold change cut-off, an alternative approach was used in our study to measure RBP dysregulations. Rather than comparing the entire tumour group to normal group, we measured the RBP expression in each tumour sample versus the whole normal group (See Method, significantly upregulated: tumour expression higher than 95% quantile of normal expression; significantly downregulated: tumour expression lower than 5% quantile of normal expression). Next, we calculated the proportion of upregulated RBPs for each tumour sample and showed that the proportions of upregulation in RBPs are much higher than that of PCGs and TFs except in THCA (Fig. 1(c)). In contrast, the proportions of downregulated RBPs are quite close to that of TFs and PCGs (Supplementary Fig. S1D). The above results demonstrated that RBPs are predominantly upregulated in tumour, especially for those RBPs with well-known posttranscriptional functions such as splicing, translation, transcription termination, in BRCA, COAD, LIHC, LUSC and LUAD.
Both our analysis and Wang et al. showed that THCA and KICH do not exhibit any preferential upregulation of RBPs, which could be due to the intrinsic characteristics of these two cancer types. Based on the recently established immune landscape across 33 TCGA cancer types, which classified tumour samples into six immune subtypes including C1: Wound Healing, C2: IFN-g Dominant, C3: Inflammatory, C4: Lymphocyte Depleted, C5: Immunologically Quiet and C6: TGF-b Dominant according to a series of gene expression signatures [19], tumour samples from THCA and KICH are enriched for subtype C3. We found that the proportion of upregulated RBPs is much higher in C1, C2 and C4 samples comparing with the rest, and is the lowest in C3 samples (Fig. 1(d)). This is coherent with our results, as most BRCA, COAD and LUSC samples are C1 and C2 subtypes, while the majority of THCA and KICH samples are C3 subtype. Since the six immune subtypes were defined based on 60 signatures, next we directly correlated the upregulated RBP proportions to each signature in 16 cancer types, respectively. Among them, 53 signatures show significant correlation (|r| > 0.2, p < 0.001) in at least one cancer type, while the most consistent and strongest correlations across different cancers are from wound healing, proliferation and SCNA related signatures (e.g. Aneuploidy score, number of fragments) (Fig. 1(e)). Furthermore, the proportion of upregulated RBPs is inversely correlated with stromal and leukocyte fraction, across different cancers, suggesting that the upregulation of RBPs in pure tumour cells might be even more evident than we observed (Fig. 1(e)). These were consistent with a recent finding that aneuploidy is positively correlated with the expression of proliferation genes, while negatively correlated with the expression of immune signalling genes [20].
Identification of consistently dysregulated RBPs across 16 human cancer types
As our results suggested that different cancer types have distinct posttranscriptional dysregulation signatures (Fig. 1(b)) and dysregulated RBPs, we hypothesized that there might be some consistently dysregulated RBPs across 16 cancer types analysed. Thus, we sought to find consistently up and down-regulated ones by examining two features for each RBP: 1) directionality in terms of difference of number of cancer types (BH-adjusted p-value < 0.001) between up regulated and down regulated (#up regulated cancer types – #down regulated cancer types); 2) log2 fold change tumour/normal. The definition of directionality was adapted from a previous study [21]. Only the RBPs showing consistent up/down-regulated expression across multiple cancer types could get high absolute value, whereas those RBPs only dysregulated in certain cancer types or upregulated in some cancers but downregulated in other cancer types will get small values. Based on this we identified 109 cuRBPs and 41 cdRBPs (Fig. 2(a), Material and Methods).
Figure 2.

Consistently dysregulated RBPs across 16 human cancer types. (A) directionality and amplitude of the expression change in tumour compared with normal for each RBP. The X axis represents the directionality that is defined by the number of cancer types RBP showing upregulated expression minus that showing downregulated expression. The Y-axis is the average of the fold change across 16 cancer types. cuRBPs and cdRBPs were within the area with green and blue colour respectively. SUR and significantly dysregulated RBPs defined in the previous study were marked with red and blue colours. RBPs pointed by yellow colour arrows are RBPs showing bidirectional expression changes across cancer, while RBPs pointed by red arrows are examples showing consistently dysregulation but not included in significantly dysregulated RBPs. (B) and (C) Heatmap illustrating expression fold change of 41 cdRBPs and 109 cuRBPs, respectively. The colour key illustrating the log2 fold change of gene expression between tumour and normal. RBPs mentioned in the main text were marked in red.
In a previous study, Kechavarzi et al. identified a group of RBPs (SUR RBPs, n = 33) showing strong upregulation in cancers by comparing RBP expression in tumour from TCGA to corresponding normal tissue from human body map [17]. The log2 fold change (tumour/normal) of these RBPs are required be above 9 in their study, whereas no RBPs showing such dramatic overexpression when comparing tumour to their adjacent normal in the present study. Moreover, seven of them even showed significant downregulated expression across different cancer types (Fig. 2(a)). These results strongly suggested to include adjacent normal samples to study RBP expression changes in tumour. In addition, a recent study identified 36 significantly dysregulated RBPs by requiring they are significantly dysregulated in at least 10 cancer types using the TCGA data [18]. Indeed, many of them were also included in our cuRBPs and cdRBPs list. However, their study missed many RBPs showing similar consistently dysregulation and some of their candidates are bidirectional dysregulated in terms of upregulated in some cancers but downregulated in other cancers. Besides the expression, we also overlapped cuRBPs/cdRBPs with RBPs and splicing factors that identified as tumour driver based on somatic mutations [15,16]. It turns out that they are poorly overlapped (Supplementary Fig. S2A), suggesting that dysregulation of expression and mutation might contribute to cancer development independently, consistent with the view that alternative mechanisms of gene regulation could function distinctly in tumorigenesis [22].
Among 109 cuRBPs, IGF2BP1, MEX3A, DNMT3B, PABPC1L, IGF2BP3, EXO1, TERT, EZH2 and RDM1 showed the most dramatic overexpression pattern (Fig. 2(b), marked in red). DNMT3B and EZH2 are well-known epigenetic regulators and both have strong oncogenic functions, while the promoter of TERT is recurrently mutated in many cancers [23–28]. IGF2BP1 and IGF2BP3 are two oncofetal proteins which have been reported to promote adhesion, migration and invasiveness of tumour cells by mediating mRNA stability and translation [29,30]. However, the functional roles of PABPC1L and MEX3A in tumorigenesis remain unclear. For the 41 cdRBPs, CPEB1, NOVA1, RBPMS2, PPARGC1A, RBM20, RBM24 and AFF3 exhibited drastic downregulation pattern in many cancer types (Fig. 2(c), marked in red). The CPEB family proteins has been implicated in translation control of cancer cells, while NOVA1 has been found to be downregulated in the gastric cancer microenvironment [31–33]. Taken together, our study identified a group of RBPs that were consistently up-/down-regulated in the majority of cancer types we analysed in the present study. Even though many of them overlap with previously identified dysregulated RBPs in cancers or have well-known oncogenic/tumour suppressive functions, there are still dozens of them, whose function in tumorigenesis is not very clear. These largely expanded the current scope of universally dysregulated RBPs across different human cancer types.
cuRBPs and cdRBPs show distinct molecular characteristics and prognostic effects
As cuRBPs and cdRBPs are consistently dysregulated across human cancers, we sought to explore their potential roles in cancer development by investigating a series of characteristics, including molecular function, oncogenic/tumour suppressive potential, association with prognosis, connectivity between each other and capacity to form protein complexes. Interestingly, among 10 functional categories, cuRBPs are significantly enriched in RNA modification, transcription termination and tRNA categories. In contrast, cdRBPs are only slightly overrepresented in unclassified RBPs outside of the 10 functional categories (Fig. 3(a)). One interesting example is TLR3, which is not involved in common posttranscriptional regulations, but can sensor double-stranded RNA and trigger type I interferon (IFN), as well as induce apoptosis in human cancer cells [34].
Figure 3.

Distinct characteristics of cuRBPs and cdRBPs in molecular function, prognostic effect and connectivity with other proteins. (A) Enrichment of cuRBPs and cdRBPs across 10 functional categories. Circular size indicating their proportion in each category. (B) Distribution of the prognostic favourable, unfavourable and ambiguous RBPs. (C) PPI network of cuRBPs. Three most extensively connected modules are shown in red (snRBPs), blue (ribosome proteins) and green (ribosome biogenesis related) colours, while other proteins are shown in grey colour.
Based on the TUSON explorer, 300 tumour suppressor genes (TSGs) and 250 oncogenes (OGs) were identified in a previous study [35]. Among 1478 RBPs (RSEM value >1), 42 of them are potential TSGs and 22 of them are potential OGs, the proportion of TSG is higher than the background (all PCGs) with statistical significance (Hypergeometric test, p < 0.0004, one-sided). However, there is a slightly overrepresentation of tumour suppressors among 44 cdRBPs (Hypergeometric test, p < 0.1, one-sided). As the TUSON explorer is based on the mutation pattern of genes across thousands of tumour sample, these results are consistent with the comparisons between cuRBPs/cdRBPs and RBPs as tumour driver (Supplementary Fig. S2A).
The recently established human pathology atlas provides the association between overall survival rate and expression of PCGs, in which 3755 favourable and 4476 unfavourable prognostic genes were identified [21]. Interestingly, comparing to TFs and PCGs, a higher proportion of RBPs are prognostically unfavourable (chi-squared test, p < 1.1e-18), in which increased expression is associated with poor overall survival rate. A more extreme trend is observed for cuRBPs, in which more than ~65% of cuRBPs are prognostically unfavourable while less than 5% are prognostically favourable. On the contrary, cdRBPs have a higher proportion of favourable prognostic genes than unfavourable ones (Fig. 3(b)).
As the consistently dysregulated RBPs might come from some common protein complex or functional modules, we checked the protein connectivity within cuRBPs and cdRBPs, while consistently upregulated TFs (cuTFs) and consistently downregulated (cdTFs) were used as a background as RBPs are a key regulator of posttranscriptio while TFs are key regulators of transcription. Based on protein–protein interactions (PPIs, requiring experimental evidence, see Materials and Methods) extracted from the STRING database [36], we found that the number of PPIs within cuRBPs was much higher than those in the other three groups (Supplementary Fig. S2B). By constructing a PPI network, we observed three most extensively interacted modules: 1) small nuclear ribonucleoproteins (snRNPs), 2) ribosomal proteins, and 3) ribosome biogenesis-related proteins (Fig. 3(c)). Although there are 363 RBPs in the RNA splicing functional category based on our classification, majority of them are splicing factors and only 12 of them are components of snRNPs, while we found that 8 of them are cuRBPs. Another interesting observation is that in module 2 there are six mitochondria ribosomal proteins (6 out of 75), but only two are ribosomal proteins (2 out of 93). The PPI only shows the interaction within each group, what is their potential to interact with other outside group proteins remains unclear. To address this, we scanned through each RBP for its potential to form protein complexes based on the protein complex annotation from CORUM [37]. We found that approximately half of the cuRBPs have the potential to form protein complexes, while more than 80% of the cdRBPs could not form any annotated protein complex (Supplementary Fig. S2C). Taken together, we showed that RBPs involved in RNA modification, transcription termination and tRNA related functions, as well as a component of snRNPs were consistently upregulated in tumours and they are extensively interacted with each other. Their overexpression might promote cancer progression as they were associated with poor prognosis.
cis- and trans-regulatory mechanisms underlying the dysregulation of cuRBPs and cdRBPs across cancers
cis- and trans-regulatory mechanisms underlying the dysregulation of cuRBPs and cdRBPs across cancers
Gene expression can be modulated by cis- and trans-regulatory mechanisms, such as copy number alteration (cis-), the abundance of transcription factors and miRNAs (trans-), and DNA methylation at the promoter region (cis-) [38–40]. To explore their relationship with dysregulation of RBP expression in cancer, we first analysed somatic copy number alteration (SCNA) by comparing the tumour to normal samples. As expected, cuRBPs have significant amplification of copy numbers (Mann-Whitney test, p < 4.9e-77, one-sided, 16 cancer types together), while cdRBPs have significant loss of copy numbers (Mann-Whitney test, p < 6.7e-22, one-sided, 16 cancer types together). This trend is consistent across almost all analysed cancer types except KICH and THCA (Fig. 4(a)). To investigate whether the SCNA do result in the dysregulation of RBPs in cancer, we analysed the correlation between SCNA and mRNA expression for each PCG. Interestingly, we observed a much stronger positive correlation for the cuRBPs (median of r = 0.42, p < 2.1e-160, Mann-Whitney test) compared to the PCGs background (median of r = 0.2) (Fig. 4(b)). This suggests that the overexpression of cuRBPs in tumours is largely associated with amplification of copy numbers, which might be critical to drive cancer progression.
Figure 4.
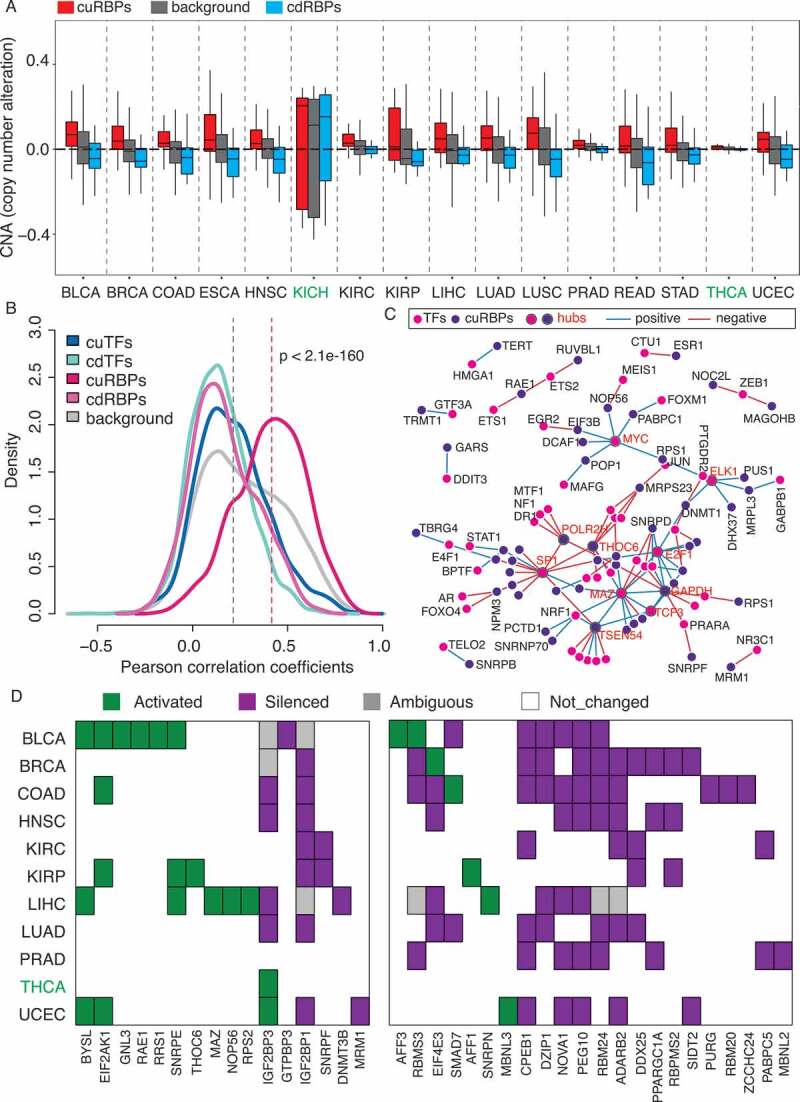
Cis- and trans-regulatory mechanisms underlying dysregulation of cuRBPs and cdRBPs across cancers. (A) The SCNA distribution of the cuRBPs, cdRBPs and background (all PCGs) across 16 cancer types in TCGA. The black horizontal-dashed line indicates the median of the SCNA of all PCGs across 16 types. (B) Density distribution of Pearson correlation coefficient between SCNA and gene expression for different gene groups. The p value represents the significance between cuRBPs and background. (C) regulatory network between TFs and their potential target cuRBPs (see Material and Methods). Hub TFs and cuRBPs were marked with grey boundary. (D) Epigenetically activated, silenced, ambiguous and no changed cuRBPs and cdRBPs genes across 11 types. Left panel are the 16 cuRBPs and right panel are 22 cdRBPs.
Besides changes in copy numbers, variation in the abundance of TFs such as MYC, and miRNAs could also result in dysregulated expression of RBPs [13,41]. To identify potential TFs and miRNAs which might be involved in the dysregulation of cuRBPs and cdRBPs across different cancers, we utilized the gene sets (C3 sub-collection MIR: microRNA targets and C3 sub-collection TFT: transcription factor targets) from the GSEA molecular signature database (MSigDB), in which targets of TFs/miRNAs were predicted by searching the sequence motif either at the promoter region or within 3ʹ UTR, respectively, [42]. In total, 500 TF and 221 miRNA gene sets were obtained from the MSigDB. For each TF/miRNA-RBP pair, we calculated the correlation in RNA expression across different samples between TF and its putative RBP targets predicted by binding motif at promoter, as well as miRNAs and its RBPs targets predict by motif within 3ʹ UTRs in each cancer type, respectively. To find consistent TF/miRNA-RBP targeting relationships across different cancers, we required the correlation should be consistent in more than eight cancer types. Finally, 113 TF-cuRBP pairs (51 TFs, 50 cuRBPs), 84 TF-cdRBP pairs (48 TFs, 20 cdRBPs) and 16 miRNA-cdRBP pairs (13 miRNAs, 5 cdRBPs) passed these criteria, while no miRNA-cuRBP pairs managed to fulfil this requirement (Supplementary Table S2).
For TFs that potentially target cuRBPs, 21 of them are positive correlated (r > 0.2) and 32 are negative correlated (r < −0.2), while 10 TFs are positive and 38 TFs are negative correlated with cdRBPs (Supplementary Fig. S2D). By constructing a putative TF-cuRBP regulatory network, we found that most of TFs only target one to two cuRBPs, while SP1, E2F1, MAZ, MYC, TCF3 and ELK1 target multiple RBPs (n 5) which might be hub TFs. On the other hand, several cuRBPs, including GAPDH, TSEN54, THOC6 and POLR2H, are targeted by multiple TFs (n 5), which could be hub cuRBPs (Fig. 4(c)). Interestingly, SP1 is positively correlated with three and negatively correlated with 10 cuRBPs, which is consistent with the previous view that SP1 is able to both enhance or repress promoter activity [43]. Among 16 miRNA-cdRBP pairs, 13 miRNAs were consistently negative correlated with 5 putative RBP targets, including CPEB2, CPEB3, CPEB4, RBMS3 and ZCCHC24 (Supplementary Fig. S2E). Therein, CPEB2, CPEB3 and CPEB4 might be potentially co-regulated by MIR25, while CPEB4 and RMBS3 could be co-regulated by MIR19A. Surprisingly, eight miRNAs out of 13, including MIR30B/D, MIR141, MIR191, MIR200A/B/C and MIR429, are all potentially regulating ZCCHC24. Unlike CPEB and RBMS3, functional relevance of ZCCHC24 in tumorigenesis is unknown.
DNA methylation has been shown to be an important epigenetic mechanisms for the regulation of gene expression [39]. Based on TCGA data, we were able to analyse DNA methylation in 4607 tumours and 358 normal samples for around 0.3 million CpG sites across 11 cancer types (for the other five cancer types data were not available). Adapted from the method as described in previous study [44], we classified CpG sites into three groups: epigenetically activated group (DNA hypermethylated (beta value >0.3) in more than 90% of samples, while became hypomethylated (beta value <0.1) in at least 10% tumour samples), epigenetically silenced group (DNA hypomethylated (beta value < 0.1) in more than 90% of samples, while became hypermethylated in at least 10% of tumour samples) and non-significantly changed group. Genes are defined as epigenetically activated or silenced if their promoter region only contains either epigenetically activated or silenced CpG sites, respectively. If the promoter region contains both epigenetically activated and silenced CpG sites, it will be considered as ambiguous. In total, 9 out of 109 (8%) cuRBPs were epigenetically activated, while 22 out of 44 (50%) cdRBPs were epigenetically silenced in at least one cancer types (Fig. 4(d)). We noted that even though IGF2BP1 is predicted to be potentially epigenetically silenced based on DNA methylation at the promoter, its mRNA expression level still significantly increased in tumour comparing with normal. This might be due to the gain of additional copies (Supplementary Table S3).
All these results suggest that dysregulation of cuRBPs/cdRBPs could be due to either cis- and trans-regulatory mechanisms across cancers. In cis, the upregulation of cuRBPs is correlated with amplification of copy number, while downregulated expression of many cdRBPs could be due to epigenetic silencing mediated by DNA methylation. In trans-, the following TFs, SP1, E2F1, MAZ, MYC, TCF3 and ELK1 might function as hub regulators of cuRBPs. Besides, five cdRBPs were potential regulated by miRNAs, including ZCCHC24 which might be regulated by multiple miRNAs and function in tumorigenesis is totally unexplored.
Characterization of potential novel cancer-related RBPs in colon adenocarcinoma (COAD)
As TCGA-COAD is one of the cancers showing strongest upregulation of RBPs, we sought to confirm this pattern with additional expression datasets. Indeed, transcriptome analysis of a public dataset (GSE104836), and in-house microarray (Clariom) of 10 normal colon tissue versus 10 COAD tumours, reveals that both of them show similar predominant upregulation of RBPs as TCGA-COAD (Fig. 5(a)). As the mRNA abundance might not truly reflect the gene expression, we next analysed the protein expression in COAD and normal colon tissues from a previous study [45]. Again, we found that protein expression of RBPs is significantly upregulated in tumours and this trend is even more evident for cuRBPs (Fig. 5(b)).
Figure 5.
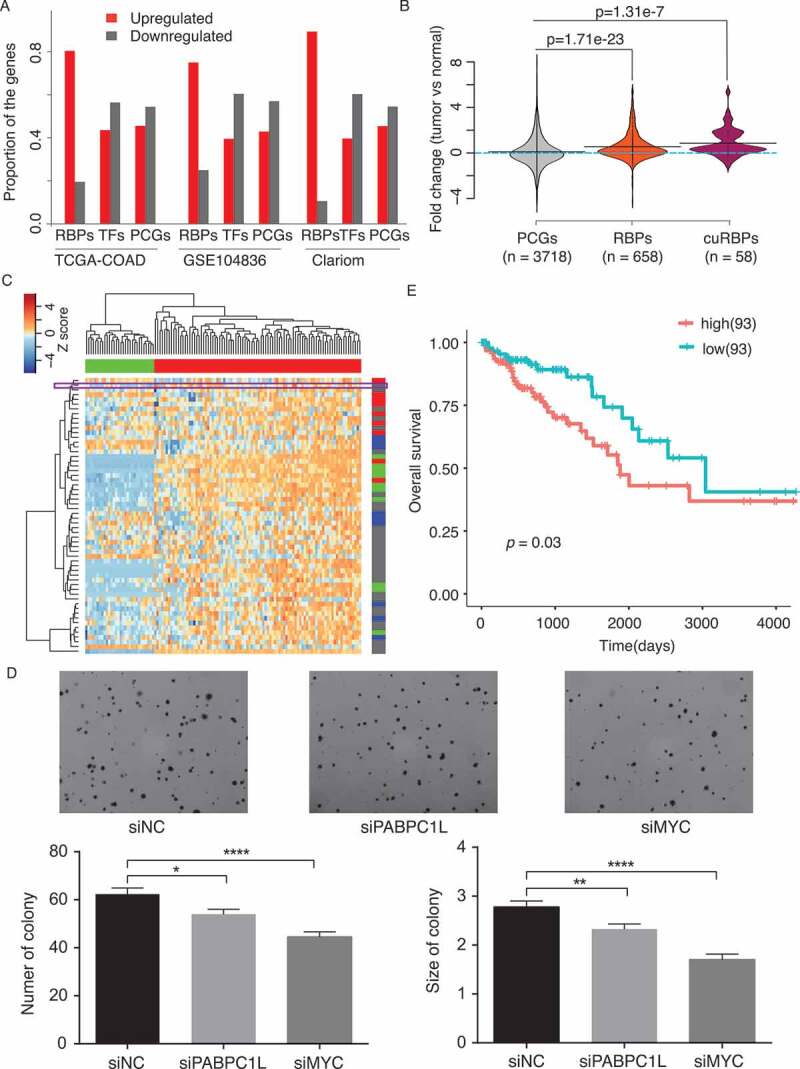
Characterization of potential novel cancer-related RBPs in COAD. (A) Proportion of dysregulated genes (tumour versus normal) in three independent datasets. TCGA-COAD: COAD dataset from TCGA project; GSE104836: public dataset containing the RNA sequencing data of 10 normal and 10 tumour COAD samples. Clariom: in-house microarray data for 10 COAD and 10 normal colon samples. (B) Distribution of the protein expression fold change between COAD tumour and normal colon samples. (C) Heatmap illustrating the protein expression of cuRBPs in normal and tumour samples. The horizontal bar: green stands for 30 normal samples and red for 91 tumour samples. The right vertical bar: red stands for the proteins related to splicing machinery, green for the proteins involved in ribosome biogenesis and the blue for the proteins related to translation machinery. The colour and definition in the vertical bar is the same as Fig. 3B. PABPC1L is highlighted in the purple box. (D) Soft agar assay of DLD cells upon PABPC1L, MYC knockdown. Error bar: SEM. t-test. *: p < 0.05, **: p < 0.01; ****: p < 0.0001. (E) Kaplan-Meier survival analysis of TCGA-COAD datasets based on segmentation values of PABPC1L expression.
Hierarchical clustering of samples based on the protein expression of cuRBPs could distinctively separate tumour and normal, while proteins from the same functional module shared similar expression pattern and clustered together, such as the snRNPs and proteins related to ribosome biogenesis (Fig. 5(c)). Interestingly, we found several RBPs (RAE1, ALYREF, PPIH and PABPC1L) which has similar protein expression profile across 120 samples to the RBPs in snRNPs, while XPO5, XPOT and OAS3, were clustered together with the ribosome biogenesis related RBPs based on expression pattern (Fig. 5(c)). Among them, PABPC1L and RAE1 are frequently (~10%) amplified in COAD tumour samples (Supplementary Fig. S3A). While RAE1 has been revealed to function in tumour immunity, chromosome segregation and RNA transport [46–48], functions of PABPC1L is not very clear. Moreover, among the many cancer types in which PABPC1L is overexpressed or amplified, large intestine ranked as the most significant (Overexpressed in more than 32% and amplified in more than 12% of tumour samples, see Table 2). As its expression is correlated with RBPs in snRNPs across 120 samples, such as SNRPD1, SNRPE and SNRPF, we hypothesized that it should have a global effect on pre-mRNA splicing. To check this, we compared the index of aberrant splicing between samples with high expression of PABPC1L (PABPC1L_high: 77 samples) and low expression of PABPC1L (PABPC1L_low: 77 samples) from TCGA-COAD. Indeed, 2502 significant aberrant events were more spliced in PBBPC1L_high group, while only 84 events were more spliced in PABPC1L_low group, suggesting that suggesting that high expression of PABPC1L might potentially induce aberrant mRNA splicing in TCGA-COAD (Supplementary Fig. S3B). Furthermore, we performed q-PCR in the COAD cell line DLD1 to validate candidates which showed aberrant splicing in PABPC1L_high group comparing with PABPC1L_low group. Among the eight candidates we selected, we were able to detect aberrant-spliced transcripts for six of them. Even though 5 out of 6 showed decreased expression for both normal and aberrant spliced isoforms, the ratios between aberrant spliced and normal transcripts were decreased for three candidates upon PABPC1L knockdown, while none of the candidates showed increased ratios (Supplementary Fig. S3C and S3D).
Table 2.
Somatic point mutation, RNA expression and somatic copy number alteration of PABPC1L across different cancer types based on the data obtained from COSMIC database (https://cancer.sanger.ac.uk/cosmic).
| Points mutations |
RNA expression |
Copy number |
||||
|---|---|---|---|---|---|---|
| Mutated (%) |
Total tested samples |
Over-expressed (%) | Total tested samples |
Gain Copy (%) |
Total tested samples |
|
| Large intestine | 2.8 | 2320 | 32.8 | 610 | 12.4 | 773 |
| Stomach | 1.3 | 861 | 24.9 | 285 | 8.8 | 501 |
| Oesophagus | 0.3 | 1513 | 13.6 | 125 | 2.4 | 546 |
| Cervix | 0.9 | 329 | 13.4 | 307 | 1.3 | 313 |
| Ovary | 0.1 | 895 | 11.3 | 266 | 1.5 | 729 |
| Skin | 1.3 | 1288 | 9.7 | 473 | 0.8 | 650 |
| Liver | 0.7 | 2163 | 8.3 | 373 | 0.4 | 692 |
| Breast | 0.5 | 2522 | 7.9 | 1104 | 1.7 | 1544 |
| Urinary tract | 0.6 | 697 | 7.8 | 408 | 2.4 | 419 |
| Kidney | 0.2 | 2177 | 6.7 | 600 | 0.2 | 1027 |
| Lung | 1.1 | 2468 | 6.2 | 1019 | 1.1 | 1185 |
| Pancreases | 0.1 | 1815 | 6.2 | 179 | 0.1 | 929 |
| Prostate | 0.2 | 2051 | 5.6 | 498 | 0.1 | 1037 |
Therefore, to further investigate the oncogenic function of PABPC1L, we performed soft-agar assay to assess its impact on cell proliferation in DLD1 cells. siRNA-mediated knockdown of the PABPC1L transcript resulted in a ~ 50% reduction in PABPC1L mRNA expression (Supplementary Fig. S3E) and a concomitant, significant reduction in cell proliferation relative to the control (Fig. 5(d)). Moreover, higher expression of PABPC1L is correlated with poor overall survival rate in both our analysis and the result from human pathology atlas (http://www.proteinatlas.org/ENSG00000101104_PABPC1L/pathology/tissue/colorectal+cancer), suggesting that its dysregulation does contribute to cancer progression (Fig. 5(e)).
Discussion
In this study, with transcriptome analysis of RBP transcript expression in ~6700 samples, we built a comprehensive mRNA expression landscape of RBPs across 16 human cancer types. Although the landscape has been characterized previously in several studies, their results are not quite consistent and even contradictory with each other. The first study compared mRNA expression of ~800 RBPs in tumour samples from TCGA to corresponding normal tissue from human body map across nine cancer types. They found RBPs are overrepresented in top quantile of upregulated genes across different cancers [17]. Another study analysed mRNA expression of ~1500 RBPs in TCGA-LIHC and one public dataset and revealed that RBPs are preferentially upregulated in tumour [14]. In addition, a recently study, which characterizes the expression of ~1500 RBPs across 15 human cancer types in TCGA, claimed that RBPs are predominantly downregulated in tumour comparing to normal [18].
Here, our study provides much more evidences supporting that RBPs are predominant upregulated in tumour across different cancers by using two different comparison approaches, additional public RNA-seq dataset beside TCGA, in-house microarray data and protein expression data. This unbalanced upregulation pattern is most evident in BRCA, COAD, LIHC, LUSC and LUAD for those RBPs genes with known functions in common posttranscriptional regulations, such as splicing, transcription termination, translation, RNA modification. Moreover, our results reveal that fractions of upregulated RBPs across different samples were impacted by their tumour immune subtypes and microenvironment, and tumour samples with high upregulated RBP factions are associated with high proliferation rate and a low fraction of stromal and leukocyte (Fig. 1(e)). These findings could even explain the heterogeneities in RBPs upregulation amplitude across different cancers. For instance, two outliers, THCA and KICH, which do not show preferential upregulation of RBPs could be due to majority samples from them are C3: inflammatory immune subtype, which is characterized with low or moderate proliferation and low overall SCNA.
Despite the observation that dysregulated RBPs are quite heterogeneous across different cancers, we still identified more than 100 consistently dysregulated RBPs in terms of cuRBPs and cdRBPs. Many of them have been extensively studied, while the remaining few have unclear functions in tumorigenesis. These RBPs are poorly overlapped with previous identified dysregulated RBPs based on both mutation and expression, which greatly expanded the current scope of RBPs that are commonly dysregulated across different cancer types. Interestingly, we found that cuRBPs are significantly overrepresented in RNA modification, transcription termination and tRNA related functional categories. Indeed, widespread 3ʹ UTR shortening actives oncogenes in cancer cells, and intronic polyadenylation induced truncation of tumour suppressor genes in leukaemia has been revealed previously [49,50]. Both of them could be mediated by upregulation of RBPs involved in transcription termination, however why the upregulated activity of transcription termination specifically affects those oncogenes and tumour suppressor genes needs to be explored in the future. Besides, METTL3, a RBP belongs to N6-adenosine-methytransferase has been showed promoting translation in human cancer cells [51]. However, compared to splicing and translation, studies on these three functional categories are much less investigated. Thus, our results not only provide insights to understand posttranscriptional dysregulations in tumorigenesis, but also implies some novel RBPs with oncogenic and tumour suppressive potential.
Furthermore, we revealed that overexpression of cuRBPs is correlated with amplification of copy numbers, while the downregulation of cdRBPs could be due to DNA-methylation-mediated epigenetic silencing. These suggest that dysregulation of cuRBPs and cdRBPs are not just passenger effect, but may have driver function in tumorigenesis. Additionally, we constructed TF/miRNA-RBP regulatory networks for cuRBPs/cdRBPs and found that several TFs might function as common hub regulators of cuRBPs across cancers. Among them, SP1, E2F1, MYC, TCF3 have been extensively investigated in thousands of studies for their critical roles in tumorigenesis, while MAZ and ELK1 were much less studied, especially, their roles as hub TFs controlling overexpression of cuRBPs across cancers.
Finally, we found several RBP candidates which might contribute to tumorigenesis in COAD, including PABPC1L and RAE1, whose expression patterns are quite similar to the RBPs in snRNPs (Fig. 4(c)). As the copy numbers of PABPC1L and RAE1 are frequently amplified in COAD, we speculate that PABPC1L and RAE1 might regulate the expression of RBPs in snRNPs. Both these two belong to RNA transport pathway, while PABPC1L is also involved in mRNA decay. Further studies should be performed to determine how they regulate snRNPs. Besides, we showed that the expression of PABPC1L is associated with mRNA splicing, cell proliferation and overall survival rate in COAD, suggesting that PABPC1L might promote cancer progression via regulating snRNPs expression. Therefore, dysregulation of PABPC1L might have similar functional consequences as other regulators of snRNPs biogenesis, such as PRMT5, and it could be a potentially valuable target for drug discovery to inhibit cancer progression as well.
Materials and methods
Reagents
Reagents are as follows: siGENOME SMARTpool siRNA reagents (Dharmacon) for negative control (NC) (siNC), PABPC1L (siPABPC1L) and MYC (siMYC); DharmaFECT 1 transfection reagent (Dharmacon); Trizol reagent (ThermoFisher), Dulbecco’s modified Eagle medium (DMEM) (ThermoFisher), OptiMem reduced serum media (ThermoFisher), foetal bovine serum (FBS) (ThermoFisher), Trypsin-EDTA (ThermoFisher).
Primers
hs-EZH1-L_F_qP: CCCAACTGTTATGCCAAAGGT
hs-EZH1-L_R_qP: ACTAAGATTGAGAGGGGCCT
hs-CDK10-L/S_F: GGCCTCCAGTATCTGCACA
hs-CDK10-L_R: CGAAATCCGCTGTCTTCACA
hs-CDK10-s_R: TTCAGGGGCTCGGTACAAAT
hs-XAF1-L_F: GGTTCCTGGTCCTGTGTCC
hs-XAF1-L_R: CACATCGTACACCCAACCTG
hs-XAF1-s_F: TCTCTGCCAACTTCACCCTC
hs-XAF1-s_R: CACATCGTACACCCAACCTG
hs-SMARCD3-L_F: CATCAGgtgaggtggccc
hs-SMARCD3-L_R: aagtccagcccttcgtgtc
hs-SMARCD3-s_F: CATGTCATCAGCGTGGACC
hs-SMARCD3-s_R: GTGGATAGGAGGAAGCTGCT
hs-NOXA1-L_F: TTGGGCAACTCAGgtggg
hs-NOXA1-L_R: GTCCTCACCTGGGGCTAG
hs-NOXA1-s_F: ACTCAGTTACCTAGCCCCAG
hs-NOXA1-s_R: ACATCGGGCTCTTCACACAG
hs-ACCS-L_F: CACTCTGAGGTCTGGGGATC
hs-ACCS-L_R: GGTTCTGCCTGACTCCCA
hs-ACCS-s_F: CACCCCTTACTATGGCGCTA
hs-ACCS-s_R: TGACCTTCACACCCTCAGAG
hs-CD46-L_F: ATTCAGTGTCGACTTCTTCCAC
hs-CD46-L_R: ACAGCAATGACCCAAACATCC
hs-CD46-s_F: TCCAAAGTGTCTTAAAGTGTCGA
hs-CD46-s_R: CGGGACAACACAAATTACTGC
hs-METTL3-L_F: tggggcccaattcaataggt
hs-METTL3-L_R: TGACACCAACctgctcacc
hs-METTL3-s_F: CACTGCTTGGTTGGTGTCAA
hs-METTL3-s_R: CGAGTGCCAGGAGATAGTCT
hs-CPNE7-s_F: CTTCACCGTGGCCATTGAC
hs-CPNE7-s_R: ATAGTCCTGGCAGATCTCGC
hs-CPNE7-L_F: TCCACTTCACCACAAAACGT
hs-CPNE7-L_R: ATAGTCCTGGCAGATCTCGC
hs-PABPC1L_F: GCCAGCCTATTCGCATCATG
hs-PABPC1L_R: CACGCCACCTTGCAAGAG
hs-GAPDH_F: AGCCACATCGCTCAGACAC
hs-GAPDH_R: GCCCAATACGACCAAATCC
Soft agar assay
A 0.6% agarose base was prepared in six-well dishes. At 24 h post-transfection, DLD-1 cells were trypsinized, re-suspended and counted. The cells (15 x 103) were mixed with complete growth medium and agarose to a final agarose concentration of 0.3%, which was added above the base. The cells were grown at 37°C in a humidified atmosphere with 5% CO2 and were fed with complete growth medium every 2 days. After 8 days, the colonies were imaged under 4× magnification and quantified using ImageJ v.1.51k.
Omics data and annotation resource
The RNA sequencing and the somatic copy number alteration (SCNA) as well as the DNA methylation data from TCGA project were downloaded and processed by TCGA-Assembler2 [52]. In total, the normalized mRNA expression (RSEM value, provided by The Genomic Data Commons) of 20,530 genes in ~6700 clinical samples across 16 cancer types was obtained (Supplemental Table S1). The processed SCNA results contain the somatic copy number alteration of around ~16,000 genes in each cancer in average. To exclude the impact of different platforms on the DNA methylation data, only the data from Infinium HumanMethylation450K BeadChip (Illumina) were used for the analysis, which includes the normalized beta value for more than ~50 million CpG site across the genome.
Besides the data from the TCGA project, a public dataset which contains 10 normal and 10 COAD tumour samples (GSE67526) were obtained from GEO. The protein expression data of 30 normal and 90 COAD tumour samples were obtained from a previous study [45]. The 1542 RBPs were defined from RBP census [4], while only 1502 of them have available expression data in the TCGA datasets. The functional categories of RBPs were manually generated by combining relevant GO terms and KEGG pathways, while top 10 categories with the largest number of RBPs were selected. Among them, there are 363 RBPs in splicing, 289 RBPs in translation, 122 RBPs in transcription termination (3ʹ end processing), 205 RBPs in RNA localization & transport, 119 RBPs in RNA surveillance & degradation, 157 RBPs in RNA modification, 374 RBPs in ribosome, 158 in tRNA, 158 in mitochondrial, 262 in enzymes (helicase & nuclease & ATPase & ligase). These 10 categories are not exclusive as some RBPs might have multiple functions. It needs to be noted that spliceosome is integrated into splicing categories, while ribosome is separated from translation. This is due to 98% of RBPs from GO terms spliceosome is also included in GO terms RNA splicing, whereas only half of RBPs were overlapped between ribosome and translation categories. One thousand two hundred and ninety TFs were derived from DBD human transcription factor database [53]. The oncogenes and tumour suppressor genes were defined based on the TUSON explorer score as described in a previous study [35]. In brief, 18,679 genes were ranked by q value, the top 300 with smallest TUSON_q_value_TSG and top 250 with smallest TUSON_q_value_OG were selected as oncogenes and tumour suppressors, respectively. Six thousand one hundred and fourteen prognostic favourable genes and 6834 unfavourable genes were obtained from the human pathology atlas [21]. Therein, 2357 genes that could be observed in both prognostic favourable and unfavourable lists were defined as ambiguous. Three thousand seven hundred and fifty-six prognostic favourable genes and 4476 unfavourable genes were defined by removing those ambiguous ones. The genetic alteration of seven candidate RBPs were obtained from cBioPortal [54].
Transcriptome data analysis
As TCGA datasets we studied contain at least 10 normal and many more tumour samples (usually hundreds of samples) in each cancer cohort (Supplemental Table S1), we performed the t-test to evaluate expression difference between tumour and normal samples in each cancer cohort and used Benjamin-Hochberg [15] approach to adjust the p-value similar to previous study (Dang et al. 2017). The genes with BH-adjusted p-value smaller than 0.001 were considered as dysregulated genes. For each cancer type, the ratio between upregulated RBPs and downregulated RBPs was compared to that of TF and background that contains all the 20,531 PCGs. The significance of the difference between RBPs and TFs, RBPs and PCGs were estimated by the hypergeometric test.
We also used an alternative approach to estimate dysregulation of gene expression in tumour. Simply, for each gene in each cohort, instead of comparing the entire tumour samples to normal samples directly, we compared its expression in each tumour sample to the entire normal samples to check whether it’s significantly dysregulated in this sample. If its expression in this tumour sample is above 95% quantile of normal expression, it will be defined as upregulated, while lower than 5% quantile will be defined as downregulated. With these approaches, we could access fraction of upregulated RBPs/TFs/PCGs in each tumour samples. In each cohort, we further correlated the fractions of upregulated RBPs with 60 signatures from immune landscape paper [19].
The consistently up/down-regulated RBPs and TFs were defined based on two metrics: the directionality and amplitude. For each gene, the directionality was the difference between the number of cancer types, in which the gene is upregulated and the number of cancer types, in which the gene is downregulated. The amplitude was the average of the expression fold change across 16 cancer types. The RBPs with directionality larger than 8 and amplitude larger than log2(1.5) was defined as cuRBPs, while those with directionality smaller than −8 and amplitude smaller than – log2(1.5) were defined as cdRBPs. In total, 109 cuRBPs and 41 cdRBPs were obtained. Similarly, 45 cuTFs and 137 cdTFs were defined by the same criteria. All of the heatmaps of gene expression in normal and tumour samples were generated by R function heatmap.2 from ‘gplots’ package.
Microarray preparation
RNA quality was assessed by using the Agilent Model 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). 150nanogram of total RNA was processed for use on the microarray by using the Affymetrix WT plus kit according to the manufacturer’s recommended protocols. The resulting biotinylated cRNA was fragmented and then hybridized to the Clariom D array (Applied Biosystems). The arrays were washed, stained, and scanned using the Affymetrix Model 450 Fluidics Station and Affymetrix Model 3000 7G scanner using the manufacturer’s recommended protocols by the Microarray Facility. Expression values were generated by using Expression Console software (Affymetrix). Each sample and hybridization underwent a quality control evaluation.
SCNA analysis
Based on the SCNA data from TCGA, copy number alterations of cuRBPs and cdRBPs were compared to the corresponding background of all the PCGs with Mann-Whitney-Wilcoxon test for each cancer type. In addition, the Pearson correlation coefficient between the gene expression (log-transformed normalized RSEM value) and SCNA were calculated for each gene in a group. The significance of difference of the correlation coefficients between the gene sets (cuRBPs/cdRBPs/cuTFs/cdTFs) and background (PCGs) were estimate by Mann-Whitney-Wilcoxon test.
Tf/miRNA analysis
To identify potential TF/miRNA regulators of RBPs, 500 gene sets of TFs, 221 gene set of miRNAs were obtained from molecular signature database (MSigDB, version 6), in which targets of TFs/miRNAs were predicted based on the motif at the promoters/3ʹ UTRs. To determine whether dysregulation of a TF/miRNA in tumour is really functional to its target cuRBPs or cdRBPs, we calculated the correlation in expression between this TF/miRNA and each of its target cuRBP and cdRBP in each cancer type separately. The TF-cuRBP and TF-cdRBP pairs with correlation larger than 0.2 in more than eight cancer types were defined as positive regulation, while correlation smaller than −0.2 were defined as negative regulation. For the miRNA-cuRBP and miRNA-cdRBP pairs, the correlation smaller than −0.2 in more than eight cancer types were required.
DNA methylation analysis
To investigate the link between DNA methylation and transcription of a gene, only the CpG sites at its promoter region were selected. Based on the GENCODE (Version 27 liftover to hg19) annotation and definition from the DNA methylation database [55], 5ʹ UTR and 1700bp (TSS200 and TSS1500) upstream region of transcription start site [21] of a gene were defined as a promoter. By comparing normalized DNA methylation values (β) between tumour and normal samples, the epigenetically silenced and activated CpG sites were determined with a similar approach as described in the TCGA project. The epigenetically silenced CpG site must fit the following three criteria: 1), more than 90% of normal samples are un-methylated (β < 0.1); 2) the difference of DNA methylation average between tumour and normal is larger than 0.2; 3) The difference between tumour and normal should be significant (BH-adjusted p < 0.05). The epigenetically activated CpG sites were identified by a similar approach but the change is in the opposite way: 1), at least 90% of normal samples is methylated (β > 0.3), 2) the difference is smaller than −0.2; 3) BH-adjusted p < 0.05. Those genes, whose promoter only contains epigenetically activated/silenced CpG site were defined as epigenetically activated/silenced. If the promoter of a gene contains both the epigenetically silenced and activated CpG sites, it was defined as ambiguous.
Protein–protein interactions and protein complexes
The putative protein–protein interactions (PPIs) within each groups, including the cuRBPs, cdRBPs, cuTFs and cdTFs were identified based on the STRING database (version 10.5) [36]. The medium confidence with interaction score >0.4 was set as cut-off for an existing interaction between two proteins and only the interactions with experimental evidence were used. To evaluate the capacity of each protein to form protein complexes, the complex annotation was downloaded from the CORUM database [37]. To compare the connectivity within each group, the ratio of the number of nodes (proteins) and edges (PPIs) was assessed and the significance between different groups was estimated by the hypergeometric test.
Funding Statement
Y.T. was supported by a Singapore National Research Foundation Fellowship, a National University of Singapore President’s Assistant Professorship and the RNA Biology Center at CSI Singapore, NUS, from funding by the Singapore Ministry of Education’s Tier 3 grant number [MOE2014-T3-1-006].
Acknowledgments
We thank all members in Tay lab and Henry lab for critical comments and suggestions. We thank Parul Saxena for microarray service and National Supercomputing Centre Singapore (NSCC) for computing service.
Author contribution
Y.T and H.Y conceived and led the project and B.Z performed the data analysis and wrote the manuscript. K.R.B and C.Y.L did the soft-agar assay experiment and K.Z.H performed microarray experiment. J.L provided splicing results and S.Z and K.R.B helped to modify the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability
A public GEO datasets GSE67526 (https://www.ncbi.nlm.nih.gov/geo/) was used in this study. The Clariom microarray was deposited to GEO under accession number (GSE115261).
Supplementary material
supplemental data for this article can be acessed here
References
- [1].Glisovic T, Bachorik JL, Yong J, et al. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582(14):1977–1986. PubMed PMID: 18342629; PubMed Central PMCID: PMCPMC2858862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fu XD, Ares M Jr.. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15(10):689–701. PubMed PMID: 25112293; PubMed Central PMCID: PMCPMC4440546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Moore MJ, Proudfoot NJ.. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136(4):688–700. PubMed PMID: 19239889. [DOI] [PubMed] [Google Scholar]
- [4].Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15(12):829–845. PubMed PMID: 25365966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Brumbaugh J, Di Stefano B, Wang X, et al. Nudt21 controls cell fate by connecting alternative polyadenylation to chromatin signaling. Cell. 2018;172(1–2):106–20 e21. PubMed PMID: 29249356; PubMed Central PMCID: PMCPMC5766360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gabut M, Samavarchi-Tehrani P, Wang X, et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147(1):132–146. PubMed PMID: 21924763. [DOI] [PubMed] [Google Scholar]
- [7].Han H, Irimia M, Ross PJ, et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498(7453):241–245. PubMed PMID: 23739326; PubMed Central PMCID: PMCPMC3933998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yeo GW. Systems biology of RNA binding proteins. New York: Springer; 2014. p. x, 468. [Google Scholar]
- [9].Quesada V, Conde L, Villamor N, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2011;44(1):47–52. PubMed PMID: 22158541. [DOI] [PubMed] [Google Scholar]
- [10].Wang L, Lawrence MS, Wan Y, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365(26):2497–2506. PubMed PMID: 22150006; PubMed Central PMCID: PMCPMC3685413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–69. PubMed PMID: 21909114. [DOI] [PubMed] [Google Scholar]
- [12].Barna M, Pusic A, Zollo O, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456(7224):971–975. PubMed PMID: 19011615; PubMed Central PMCID: PMCPMC2880952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Koh CM, Bezzi M, Low DH, et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015;523(7558):96–100. PubMed PMID: 25970242. [DOI] [PubMed] [Google Scholar]
- [14].Dang H, Takai A, Forgues M, et al. Oncogenic activation of the RNA binding protein NELFE and MYC Signaling in hepatocellular carcinoma. Cancer Cell. 2017;32(1):101–14 e8. PubMed PMID: 28697339; PubMed Central PMCID: PMCPMC5539779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Neelamraju Y, Gonzalez-Perez A, Bhat-Nakshatri P, et al. Mutational landscape of RNA-binding proteins in human cancers. RNA Biol. 2018;15(1):115–129. PubMed PMID: 29023197; PubMed Central PMCID: PMCPMC5786023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Seiler M, Peng S, Agrawal AA, et al. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 2018;23(1):282–96 e4. PubMed PMID: 29617667; PubMed Central PMCID: PMCPMC5933844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kechavarzi B, Janga SC. Dissecting the expression landscape of RNA-binding proteins in human cancers. Genome Biol. 2014;15(1):R14. PubMed PMID: 24410894; PubMed Central PMCID: PMCPMC4053825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang ZL, Li B, Luo YX, et al. Comprehensive genomic characterization of RNA-binding proteins across human cancers. Cell Rep. 2018;22(1):286–298. PubMed PMID: 29298429. [DOI] [PubMed] [Google Scholar]
- [19].Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity. 2018;48(4):812–30e14. PubMed PMID: 29628290; PubMed Central PMCID: PMCPMC5982584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Taylor AM, Shih J, Ha G, et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell. 2018;33(4):676–89 e3. Epub 2018/04/07. PubMed PMID: 29622463; PubMed Central PMCID: PMCPMC6028190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352). PubMed PMID: 28818916. DOI: 10.1126/science.aan2507 [DOI] [PubMed] [Google Scholar]
- [22].Santarius T, Shipley J, Brewer D, et al. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer. 2010;10(1):59–64. PubMed PMID: 20029424. [DOI] [PubMed] [Google Scholar]
- [23].Beaulieu N, Morin S, Chute IC, et al. An essential role for DNA methyltransferase DNMT3B in cancer cell survival. J Biol Chem. 2002;277(31):28176–28181. PubMed PMID: 12015329. [DOI] [PubMed] [Google Scholar]
- [24].Huang FW, Hodis E, Xu MJ, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957–959. PubMed PMID: 23348506; PubMed Central PMCID: PMCPMC4423787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rhee I, Bachman KE, Park BH, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416(6880):552–556. PubMed PMID: 11932749. [DOI] [PubMed] [Google Scholar]
- [26].Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647(1–2):21–29. PubMed PMID: 18723033. [DOI] [PubMed] [Google Scholar]
- [27].Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–629. PubMed PMID: 12374981. [DOI] [PubMed] [Google Scholar]
- [28].Vinagre J, Almeida A, Populo H, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4:2185. PubMed PMID: 23887589. [DOI] [PubMed] [Google Scholar]
- [29].Bell JL, Wachter K, Muhleck B, et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013;70(15):2657–2675. PubMed PMID: 23069990; PubMed Central PMCID: PMCPMC3708292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lederer M, Bley N, Schleifer C, et al. The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin Cancer Biol. 2014;29:3–12. PubMed PMID: 25068994. [DOI] [PubMed] [Google Scholar]
- [31].D’Ambrogio A, Nagaoka K, Richter JD. Translational control of cell growth and malignancy by the CPEBs. Nat Rev Cancer. 2013;13(4):283–290. PubMed PMID: 23446545. [DOI] [PubMed] [Google Scholar]
- [32].Fernandez-Miranda G, Mendez R. The CPEB-family of proteins, translational control in senescence and cancer. Ageing Res Rev. 2012;11(4):460–472. PubMed PMID: 22542725. [DOI] [PubMed] [Google Scholar]
- [33].Yoon SO, Kim EK, Lee M, et al. NOVA1 inhibition by miR-146b-5p in the remnant tissue microenvironment defines occult residual disease after gastric cancer removal. Oncotarget. 2016;7(3):2475–2495. PubMed PMID: 26673617; PubMed Central PMCID: PMCPMC4823049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Salaun B, Coste I, Rissoan MC, et al. TLR3 can directly trigger apoptosis in human cancer cells. J Immunol. 2006;176(8):4894–4901. PubMed PMID: 16585585 [DOI] [PubMed] [Google Scholar]
- [35].Davoli T, Xu AW, Mengwasser KE, et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155(4):948–962. PubMed PMID: 24183448; PubMed Central PMCID: PMCPMC3891052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D8. PubMed PMID: 27924014; PubMed Central PMCID: PMCPMC5210637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ruepp A, Waegele B, Lechner M, et al. CORUM: the comprehensive resource of mammalian protein complexes–2009. Nucleic Acids Res. 2010;38(Database issue):D497–501. PubMed PMID: 19884131; PubMed Central PMCID: PMCPMC2808912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319(5871):1785–1786. PubMed PMID: 18369135. [DOI] [PubMed] [Google Scholar]
- [39].Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. PubMed PMID: 22641018. [DOI] [PubMed] [Google Scholar]
- [40].Stranger BE, Forrest MS, Dunning M, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315(5813):848–853. PubMed PMID: 17289997; PubMed Central PMCID: PMCPMC2665772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ciafre SA, Galardi S. microRNAs and RNA-binding proteins: a complex network of interactions and reciprocal regulations in cancer. RNA Biol. 2013;10(6):935–942. PubMed PMID: 23696003; PubMed Central PMCID: PMCPMC4111733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. PubMed PMID: 16199517; PubMed Central PMCID: PMCPMC1239896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Li L, He S, Sun JM, et al. Gene regulation by Sp1 and Sp3. Biochem Cell Biol. 2004;82(4):460–471. PubMed PMID: 15284899. [DOI] [PubMed] [Google Scholar]
- [44].Cancer Genome Atlas Research N . Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159(3):676–690. PubMed PMID: 25417114; PubMed Central PMCID: PMCPMC4243044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang B, Wang J, Wang X, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513(7518):382–387. PubMed PMID: 25043054; PubMed Central PMCID: PMCPMC4249766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Pritchard CE, Fornerod M, Kasper LH, et al. RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. Epub 1999/04/20 J Cell Biol. 1999;145(2):237–254. PubMed PMID: 10209021; PubMed Central PMCID: PMCPMC2133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Diefenbach A, Jensen ER, Jamieson AM, et al. Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature. 2001;413(6852):165–171. Epub 2001/ 09/15. PubMed PMID: 11557981; PubMed Central PMCID: PMCPMC3900321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Babu JR, Jeganathan KB, Baker DJ, et al. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;160(3):341–353. Epub 2003/01/29. PubMed PMID: 12551952; PubMed Central PMCID: PMCPMC2172680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lee SH, Singh I, Tisdale S, et al. Widespread intronic polyadenylation inactivates tumour suppressor genes in leukaemia. Nature. 2018;561(7721):127–131. 10.1038/s41586-018-0465-8. PubMed PMID: 30150773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Mayr C, Bartel DP. Widespread shortening of 3ʹUTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138(4):673–684. PubMed PMID: 19703394; PubMed Central PMCID: PMCPMC2819821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lin S, Choe J, Du P, et al. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62(3):335–345. PubMed PMID: 27117702; PubMed Central PMCID: PMCPMC4860043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wei L, Jin Z, Yang S, et al. TCGA-assembler 2: software pipeline for retrieval and processing of TCGA/CPTAC data. Bioinformatics;2017. PubMed PMID: 29272348. DOI: 10.1093/bioinformatics/btx812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wilson D, Charoensawan V, Kummerfeld SK, et al. DBD–taxonomically broad transcription factor predictions: new content and functionality. Nucleic Acids Res. 2008;36(Database issue):D88–92. PubMed PMID: 18073188; PubMed Central PMCID: PMCPMC2238844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. PubMed PMID: 23550210; PubMed Central PMCID: PMCPMC4160307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Huang WY, Hsu SD, Huang HY, et al. MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 2015;43(Database issue):D856–61. PubMed PMID: 25398901; PubMed Central PMCID: PMCPMC4383953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A public GEO datasets GSE67526 (https://www.ncbi.nlm.nih.gov/geo/) was used in this study. The Clariom microarray was deposited to GEO under accession number (GSE115261).


