ABSTRACT
Entamoeba histolytica (Eh) is a protozoan parasite of humans that colonizes the outer colonic mucus layer. Under conditions not fully understood, Eh breaches innate host defenses and invades the intestinal mucosa-causing amebic colitis and liver abscess. In asymptomatic infection, Eh interacts with and feeds on resident microbiota that forms biofilms on the outer colonic mucus layer. Despite the close association between Eh and commensal microbiota, we still lack basic knowledge on whether microbiota and/or their metabolites influence Eh virulence traits critical in disease pathogenesis. In the pathogenesis of intestinal amebiasis, Eh overcomes the protective mucus layer using a combination of mucinase/glycosidase and potent mucus secretagogue activity. In this addendum, we discuss the interconnected role of a healthy mucus barrier and the role commensal microbiota play in shaping innate host defense against Eh-induced pro-inflammatory and secretory responses critical in disease pathogenesis.
KEYWORDS: Entamoeba histolytica, mucus, Muc2 mucin, bacteria, innate immunity, pro-inflammatory cytokines, secretion, mucosal epithelium, goblet cell
Introduction
The intestinal epithelium constitutes a barrier between the luminal content and the lamina propria. This barrier is multilayered and regulates the flow of cells, gases, nutrients and other molecules between both compartments.1 In the colon, the epithelial barrier is composed of three layers: a single layer of intestinal epithelial cells (IEC) on top of the lamina propria, the mucus layer in the middle and the microbiota and secreted products at the luminal side.2 The colonic mucus layer can be divided into two compartments: right above the apical side of epithelial cells there is a condensed inner mucus layer which is devoid of bacteria,3 while the superficial loose outer mucus layer is colonized by commensal microorganisms4 (Figure 1a).
Figure 1.
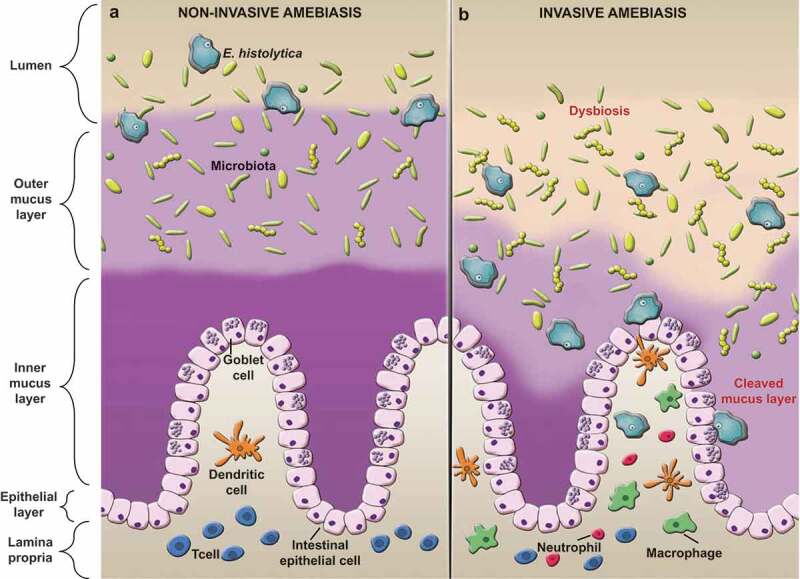
Entamoeba histolytica disrupts the mucus layer and cause invasive disease. (a) When Eh infects a healthy individual with an intact intestinal barrier, mucus and bacteria keep Eh in the outer mucus layer and away from the surface epithelium. Here, Eh feeds on bacteria, cell detritus and sugars from mucus establishing asymptomatic infections. (b) Under unknown conditions, Eh breaches innate host defenses by cleaving mucin glycans and MUC2 N terminal proteins with parasite glycosidase and cysteine proteases. Eh-induced mucus degradation compromises the epithelial barrier and alters microbial composition. Eh then contact epithelial cells via parasite Gal/GalNAc lectin and EhCP-A5 to evoke mucus and water secretion and elicits acute pro-inflammatory responses in disease pathogenesis.
Microbiota refers to the community of microorganisms gathered together in a determined ecological niche. The human digestive tract harbors numerous, diverse and dynamic population of microorganisms, mostly bacteria, but also important numbers of protozoa, fungi and viruses. These microorganisms have been adapted to live on/in the mucus surface and in the intestinal lumen.5 Intestinal microbiota is essential for correct nutrition, metabolism and immune function.6,7 E. histolytica (Eh) is a human protozoa parasite and the causative agent of amebiasis presented as amebic colitis and amebic liver abscess. Of every 10 individual that are infected one will develop symptoms of the disease.8 In symptomatic cases, the intensity of the disease varies from mild diarrhea to more intense manifestation characterized by anorexia, asthenia, abdominal pain, alterations in intestinal transit and mucoid diarrhea. If left untreated, intestinal obstruction, fever and vomit appear producing dehydration. When Eh overcomes the intestinal mucosa it causes necrosis extended to the submucosa and muscularis, producing the typical amebic “flask-shaped ulcer.” Untreated mucosal ulceration could progress to peritonitis, sepsis and death.8,9 Another species of Entamoeba morphologically indistinguishable from Eh called E. dispar also colonizes the colon. While nonpathogenic, recent studies suggest it could play a role in causing intestinal lesions. The only successful way to differentiate the species is by molecular methods such as PCR and antigen detection tests by ELISA.10
Eh have two stages in its life cycle: the trophozoite or vegetative form that colonizes the gut, and the cyst. In the trophozoite form, Eh inhabits the colon, where it interacts with resident microbiota and the outer MUC2 mucus layer (Figure 1a). It is known that Eh requires commensal bacteria presence for its pathogenicity.11 Eh feeds on microbiota or on bacteria-induced mucin glycans. To counteract this effect, goblet cells increase mucus release, keeping Eh away from the epithelial surface.12,13 Increase mucus secretion could potentially promote differential bacterial colonization,14 supporting the concept that interaction with microbiota-ameba-mucus could be critical in onset and progression of disease.
Mucus-bacteria interactions
The host-microbe interaction starts in the outer mucus layer. MUC2 O-glycosylated side-chains are rich in N-acetylglucosamine, galactose, N-acetylgalactosamine and fucose.15 These saccharides residues are keys for commensals and pathogenic organisms binding through their lectins. Binding to the outer mucus layer expels pathogens and opportunistic organisms. Indigenous commensal bacteria harness this bond to have an ecological niche protected by mucus. Thus mucus plays a protective role in maintaining a healthy and stable microbial community, since commensals and pathogens are in constant competition for binding sites to the mucus layer.16 Recent studies have showed that the O-glycan complex is highly conserved between individuals, suggesting a role in the selection of commensal microbiota providing binding sites to certain sugars.17-18 This specificity suggests that intestinal mucin influences the composition of the microbiota community. Bacteria degrade mucus and use mucin glycans as an energy source, simultaneously, the released monosaccharaides are turn into SCFA and used as an energy source for intestinal epithelial cells.19 Butyrate and propionate induce MUC2 transcription via AP1 binding and histones acetylation and methylation on the MUC2 promoter.20 Mucin is an important carbohydrate source, mainly in the distal colon where the abundance of fermentable carbohydrate substrates is low.21 Therefore, bacterial stimulation of mucus secretion, as well as mucin degradation is part of the normal process of mucus turnover and contributes to maintain a functional protective barrier to potential hazards.
Mucus layer, a niche for E. histolytica
The first barrier Eh must overcome to invade the intestinal mucosa is the MUC2 mucin layers. Eh gains access to the intestinal cells by proteolysis of the mucin layer that covers the intestinal epithelium. This process is fundamental for the parasite to exert its pathogenic potential on host cells. Eh strongly binds to mucin via the Gal/GalNAc-lectin, which has high affinity for galactose and N-acetyl-D-galactosamine glycans present on the O-linked sugar side chains of the MUC2 molecule.22 This interaction is essential for Eh colonization.
In disease pathogenesis, Eh cleaves the polysaccharides present in the outer structure of MUC2 utilizing various glycosidases, such as N-acetylgalactosamidase, N-acetyl-glucosaminidase,23 β-galactosidase and β-N-acetyl-hexosaminidase.24 Simultaneously, Eh proteases target the peptides in the poor glycosylated regions of the MUC2 C-terminus. Eh produces several enzymes with proteolytic activity, among them: cysteine proteases, serine proteases, metalloproteases and aspartic proteases.25 Cysteine proteases have an important role in invasion and digestion of phagocytosed material, as they degrade different components of the extracellular matrix (fibronectin, laminin and collagen) among others.26 Cysteine proteases are present in Eh-secreted components and have a crucial role in mucin degradation.12 In addition to degrading MUC2 mucin, the parasite also evokes mucus hypersecretion by goblet cells. Eh is a potent mucus secretagogue that leads to goblet cells cavitation and mucus depletion. This is accomplished by cysteine protease-5 (EhCP-A5) arginine-glycine-aspartate peptide (RGD) binding to αvβ3 integrin on goblet cells.13 We have recently showed that in response to Eh, mucus secretion from goblet cells was modulated by a classical exocytosis manner mediated by the SNARE vesicle-associated membrane protein 8 (VAMP8) present in mucin granules,27 advancing significantly our understanding on mucus secretion basally and in response to Eh. Recently, an interesting mechanism for host cell destruction by Eh has been described28 called trogocytosis. This is characterized by Eh nibbling and ingesting pieces of epithelial cells consequently causing cell death.
Microbiota and ameba interactions
In the colon, Eh interacts with multiple components that can modulate the course of tissue invasion and destruction. Contact between Eh and resident microbiota constitutes the beginning of the first host-parasite interaction that could potentially initiate disease. Eh in the outer mucus layer29 shares a niche rich in a diverse community of microbiota, directly interacting with them and benefiting from their presence. Colonic microbiota breaks down complex carbohydrates into glycans19 that can serve as a nutrient source for Eh, at the same time, Eh can also feed on the resident microbiota. Infection of Eh in germ-free animals failed to induce disease, but Eh pathogenicity was re-established after inoculation with bacteria.11 Similarly, in vitro cultures of Eh in axenic conditions decreased Eh virulence that was restored after inoculation and incubation with live bacteria.30 More recently, it was shown that bacteria from the Enterobacteriacea family stimulated Eh gene response associated with high oxidative stress survival that was not observed when Eh was co-cultured with a probiotic strain.31 The exact mechanism whereby the presence of enteric bacteria is required for Eh to express virulence-associated genes remains elusive, but it demonstrates the complex dynamism that exists between Eh and commensal organisms, and could potentially provide insights on explaining why only about 10% of infected individuals with Eh develop intestinal amebiasis.
E. histolytica disturbs microbiota and promotes translocation
The recent growth in molecular and bioinformatics techniques has made possible significant advances in the identification, classification and characterization of organisms that make up the microbiota. By using these techniques and under the premise that certain bacteria have been associated to different pathologies, several studies have been carried out to propose that specific bacteria could be relevant in the development of amebiasis. Recent studies from endemic areas have shown that Eh infection alters resident bacterial composition (Figure 1b). A study in Northern India linked Eh-positive patients with a dysbiotic state, characterized by a decrease in Bacteroides, Clostridium coccoides, C. leptum, Lactobacillus, Campylobacter and Eubacterium with a corresponding increase in Bifidobacterium species.14 Another study in the Cameroon showed that individuals positive for Eh infection presented with an augmented number of bacterial species (alpha diversity) and a decrease in inter-individual variation (beta diversity). This study also linked Eh presence with an increase in Clostridiales and Ruminococcaceae and a corresponding decrease in Prevotella copri and Fusobacteria.32 A longitudinal study performed in children from a rural zone in Bangladesh associated parasite-induced diarrhea with an expansion of Prevotella copri,33 a bacteria that has been associated with intestinal inflammation. A different study done in patients with amebic liver abscess (ALA) could not relate specific bacteria to ALA incidence; however, most of the ALA patients were co-infected with different bacteria, and particularly presented with a high abundance of Klebsiella.34 The fact that Eh induces the production of antimicrobial peptides but are resistant to their cytopathic effect35 could explain the alteration in microbial composition observed in Eh infections. All these association studies came to the same conclusion: it is not known if the observed dysbiosis was a cause or an effect of Eh infection. Recently, a metagenomics study on the vitro association between Eh and enteric bacteria showed that Eh preferentially phagocytose Lactobacillus ruminus, Faecalibacterium prausnitzii, Bifidobacterium longum and B. ruminantium. These findings suggest that Eh preferentially phagocytose commensal bacteria that are part of the healthy microbiota that have important beneficial effects in the host.36 We have previously shown that Eh alters tight junction proteins and affect epithelial barrier permeability37 and promoted the translocation of intestinal bacteria into mucosal surfaces as well as dissemination to other organs.38 Numerous factors, such as the glycobiome (the microbiota carbohydrate breakdown), the dynamics of opportunistic pathogens under a dysbiotic state and the host immune responses against Eh infection, and their effects on microbial composition, among others, need to be considered to clarify this complex relationship.
Microbiota role for an appropriate immune response against E. histolytica
As mentioned above, several studies have shown the importance of commensal microbiota in Eh virulence. We have recently demonstrated that a dysbiotic state renders the host hyper responsive for increased pro-inflammatory cytokine and chemokine production with hyper secretory responses toward Eh38 (Figure 2a,b). These finding are of great relevance, as individuals with dysbiosis (either by disease, antibiotics or due to a poor diet) that are infected with Eh are at high risk to develop severe intestinal amebiasis associated with acute inflammation compared to individuals with a healthy microbiota. Within the clinical setting, this is pertinent, since amebiasis is endemic in developing countries and in sectors of the population (children, elderly or those immunosuppressed) in which they usually administer significant amounts of antibiotics, even without the need for a medical prescription. In this same study, we also demonstrated using germ-free mice that in the absence of commensal microbiota, the normal host immune responses toward Eh infection are severely impaired (Figure 2c). These results could be due to a combination of factors such as: (1) naivety of the germ-free mice immune system,39 caused by the absence of microbiota, and (2) the biochemical nature of Muc2 is different in germ-free mice. These mice posses a thinner and penetrable Muc2 layer,40 and their O-glycan monomers are shorter than wildtype,41 thus modifying Eh-mucus interactions. With the absence of bacteria in the lumen from which to feed, Eh could be forced to approach mucosal epithelial cells in search for alternative energy sources, explaining why Eh are in close contact with the epithelium in infected germ-free mice (Figure 2c). Of extreme relevance was that Eh-induced inflammation provoked dysregulation of the transcription factor for secretory goblet cells Math1, through a mechanism that was microbiota dependent. These results lead us to hypothesize that Eh could manipulate innate host defense by modulating goblet cell differentiation to decrease mucus production, which in turn, enabling Eh to reach the intestinal epithelium. To date, there is no concrete information on the effect of Math1 on inflammatory-related infectious processes. Certainly, more research must be done to understand the role of this secretory cell transcription factor in the maintenance of homeostasis. Interestingly, this phenomenon also promoted bacterial translocation in the proximal colon and the ileum. Our results showed that Eh-induced inflammation in the proximal colon promoted bacterial translocation and disrupted goblet cell lineage. Further investigation will be needed to better understand the connection between commensal microbiota and the immune response to this parasite. Intestinal bacteria can affect the outcome of parasitic helminths infections (reviewed in42) as well as those associated with some parasitic protozoa (reviewed in43). Studies done in germ free mice have showed that microbiota is important for protozoa to mount a characteristic immune response with Giardia duodenalis,44 Leishmania amazonensis45 and Schistosoma mansoni infection.46 Interestingly this is not the case for Trypanosoma cruzi infection,47 where germ-free mice presented with a worse disease outcome. Parasitic infections in germ-free mice are in its infancy and more studies will be needed to delineate the role play by intestinal bacteria in shaping innate and systemic innate host defenses and the role specific bacterium play in educating the immune system.
Figure 2.
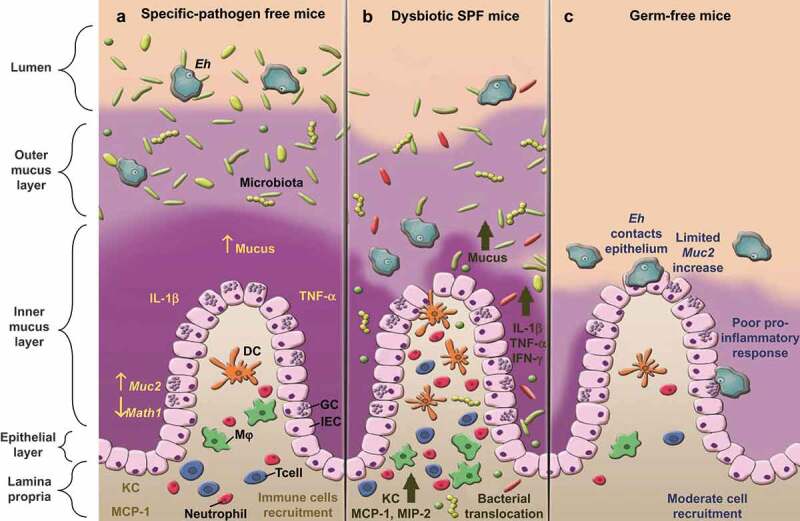
Host response to E. histolytica is enhanced when intestinal barriers are disrupted. (a) In specific-pathogen free mice (SPF), with a functional intestinal barrier, the presence of Eh evokes an inflammatory response characterized with increase mucus secretion, pro-inflammatory cytokines IL-1β and TNF-α and chemokines KC and MCP-1 with corresponding immune cells recruitment. There is also a reduction of Math1 transcription factor and an increase in Muc2 expression. (b) Under antibiotic-induced dysbiosis, Eh evokes robust mucus secretion, increased pro-inflammatory and chemokines that promoted bacterial translocation. (c) In germ-free mice, Math1 and Muc2 transcript, as well as pro-inflammatory cytokine expression are unaltered. Due to a thinner and biochemically altered mucus layer, Eh readily contact and disrupt the intestinal epithelium. GC: goblet cell, IEC: intestinal epithelial cell, Mφ: macrophage, DC: dendritic cells.
Concluding remarks and future directions
Undoubtedly, there is a complex Eh-mucus-microbiota relationship. Being able to glimpse and accurately understand the dynamism and the way in which these entities are related is of utmost importance to advance our understanding on the pathogenesis of intestinal amebiasis. Our recent work reveals an interconnection among these three entities, demonstrating that Eh in a dysbiotic environment exacerbates not only the immune response but also mucus and water secretion. We also reported that Eh decreased Math1 transcription which was required for goblet cell differentiation.48 These findings require further investigation to unravel the mechanism by which Eh targets secretory lineage maturation. Our study is the first to characterize the host response to Eh in germ-free mice. An important finding was that despite the closeness of Eh to epithelial cells, the typical pro-inflammatory and secretory response to Eh was absent in these mice. These findings reinforce the notion that microbiota plays a critical role in educating and shaping innate host defenses at mucosal surfaces. The importance of intestinal microbiota in maintaining homeostasis is evident and new important roles are continually attributed to it. New and modern molecular techniques will allow us to interrogate its composition, ecology and metabolism in future studies. Being able to understand the relationship and interaction that this has with a parasite of global importance would be highly relevant to treat and manage, not only amebiasis, but other parasitic problems in developing countries. A recent study49 aimed to identify human protozoa using metagenomics found that co-infection of different protozoa species, mostly commensals, is fairly common. Interestingly, this study was not able to identify a single positive sample for Eh, either because the individuals were reported as healthy, and/or because of the challenge in extracting genetic material from Eh cyst. This study reinforces the importance of recognizing commensal protozoa as part of our microbiota.50 This is critical for understanding the dynamics and interactions between different communities that make up the intestinal microbiota. Our studies are just beginning to understand the complex interconnected relationship between Eh, mucus and bacteria; however, an explanation of the mechanisms underlying this phenomenon requires further investigation.
Funding Statement
This work was supported by the Canadian Institutes of Health Research [KC; MOP-34045] and the Mexican Council of Science and Technology [ALC; CONACyT-314101].
References
- 1.Camilleri M, Madsen K, Spiller R, Meerveld BGVAN.. Intestinal barrier function in health and gastrointestinal disease. Neurogastroenterol Motil. 2012;24:503–512. doi: 10.1111/j.1365-2982.2012.01921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9(11):799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 3.Johansson MEV, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105(39):15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johansson MEV, Larsson JMH, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. PNAS. 2011;108(1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7(7):688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Reddy DN. Role of the normal gut microbiota. World J Gastroenterol. 2015;21(29):8787. doi: 10.3748/wjg.v21.i10.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y-J, Li S, Gan R-Y, Zhou T, Xu D-P, Li H-B. Impacts of gut bacteria on human health and diseases. Int J Mol Sci. 2015;16:7493–7519. doi: 10.3390/ijms16047493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mortimer L, Chadee K. The immunopathogenesis of Entamoeba histolytica. Exp Parasitol. 2010;126(3):366–380. doi: 10.1016/j.exppara.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Ralston KS, Petri WA. Tissue destruction and invasion by Entamoeba histolytica. Trends Parasitol. 2011;27(6):254–263. doi: 10.1016/j.pt.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliveira FMS, Neumann E, Gomes MA, Caliari MV. Entamoeba dispar: could it be pathogenic. Trop Parasitol. 2015;5(1):9–14. doi: 10.4103/2229-5070.149887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phillips B, Wolfe PA, Rees CW, Gordon HA, Wright WH, Reyniers JA. Comparative results of the intracecal inoculation of germfree, monocontaminated, and conventional guinea pigs with Entamoeba histolytica. Am J Trop Med Hyg. 1955;4(4):675–692. doi: 10.4269/ajtmh.1955.4.675. [DOI] [PubMed] [Google Scholar]
- 12.Moncada D, Keller K, Chadee K. Entamoeba histolytica -secreted products degrade colonic mucin oligosaccharides. Infect Immun. 2005;73(6):3790–3793. doi: 10.1128/IAI.73.6.3790-3793.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornick S, Moreau F, Chadee K. Entamoeba histolytica cysteine proteinase 5 evokes mucin exocytosis from colonic goblet cells via αvβ3 integrin. PLoS Pathog. 2016;12(4):1–24. doi: 10.1371/journal.ppat.1005579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verma AK, Verma R, Ahuja V, Paul J. Real-time analysis of gut flora in Entamoeba histolytica infected patients of Northern India. BMC Microbiol. 2012;12:183. doi: 10.1186/1471-2180-12-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birchenough GMH, Johansson MEV, Gustafsson JK, Bergström JH, Hansson GC. New developments in goblet cell mucus secretion and function. Mucosal Immunol. 2015;8(4):712–719. doi: 10.1038/mi.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pelaseyed T, Bergstrom J, Gustafsson JK, Ermund A, Birchenough GMH, Schütte A, van der Post S, Svensson F, Rodríguez-Piñeiro AM, Nyström EEL, et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. 2014;260(1):8–20. doi: 10.1111/imr.2014.260.issue-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juge N. Microbial adhesins to gastrointestinal mucus. Trends Microbiol. 2012;20(1):30–39. doi: 10.1016/j.tim.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez-Piñeiro AM, Johansson MEV. The colonic mucus protection depends on the microbiota. Gut Microbes. 2015;6(5):326–330. doi: 10.1080/19490976.2015.1086057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sicard J-F, Le Bihan G, Vogeleer P, Jacques M, Harel J. Interactions of intestinal bacteria with components of the intestinal mucus. Front Cell Infect Microbiol. 2017;7:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burger-van Paassen N, Vincent A, Puiman PJ, Van Der Sluis M, Bouma J, Boehm G, Van Goudoever JB, Van Seuningen I, Renes IB. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem J. 2009;420(2):211–219. doi: 10.1042/BJ20082397. [DOI] [PubMed] [Google Scholar]
- 21.Marcobal A, Southwick AM, Earle KA, Sonnenburg JL. A refined palate: bacterial consumption of host glycans in the gut. Glycobiology. 2013;23(9):1038–1046. doi: 10.1093/glycob/cwt040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chadee K, Petri WA, Innes DJ, Ravdin JI. Rat and human colonic mucins bind to and inhibit adherence lectin of Entamoeba histolytica. J Clin Invest. 1987;80(5):1245–1254. doi: 10.1172/JCI113199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frederick JR, Petri WAJ. Roles for the galactose-/N -acetylgalactosamine-binding lectin of Entamoeba in parasite virulence and differentiation. Glycobiology. 2005;15(12):53–59. doi: 10.1093/glycob/cwj007. [DOI] [PubMed] [Google Scholar]
- 24.Thibeaux R, Weber C, Hon C-C, Dillies, M-A, Avé, P, Coppée, J-Y, Labruyère, E, Guillén, N, Petri, WA. Identification of the virulence landscape essential for Entamoeba histolytica invasion of the human colon. PLoS Pathog. 2013;9(12):e1003824. doi: 10.1371/journal.ppat.1003824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tillack M, Biller L, Irmer H, Freitas M, Gomes MA, Tannich E, Bruchhaus I. The Entamoeba histolytica genome: primary structure and expression of proteolytic enzymes. BMC Genomics. 2007;8:170. doi: 10.1186/1471-2164-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serrano-Luna J, Piña-Vázquez C, Reyes-López M, Ortiz-Estrada G, de la Garza M. Proteases from Entamoeba spp. and pathogenic free-living amoebae as virulence factors. J Trop Med. 2013;2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cornick S, Moreau F, Gaisano HY, Chadee K. Entamoeba histolytica-induced mucin exocytosis is mediated by VAMP8 and is critical in mucosal innate host defense. MBio. 2017;8(5):1–14. doi: 10.1128/mBio.01323-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ralston KS, Solga MD, Mackey-Lawrence NM, Bhattacharya A, Petri WA. Trogocytosis by Entamoeba histolytica contributes to cell killing and tissue invasion. Nature. 2014;508:526–530. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lidell ME, Moncada DM, Chadee K, Hansson GC. Entamoeba histolytica cysteine proteases cleave the MUC2 mucin in its C-terminal domain and dissolve the protective colonic mucus gel. PNAS. 2006;103(24):9298–9303. doi: 10.1073/pnas.0600623103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wittner M, Rosenbaum R. Role of bacteria in modifying virulence of Entamoeba histolytica. Am J Trop Med Hyg. 1970;19(5):755–761. doi: 10.4269/ajtmh.1970.19.755. [DOI] [PubMed] [Google Scholar]
- 31.Varet H, Shaulov Y, Sismeiro O, Trebicz M, Legendre R. Enteric bacteria boost defences against oxidative stress in Entamoeba histolytica. Sci Rep. 2018;8(9042):1–12. doi: 10.1038/s41598-018-27086-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morton ER, Lynch J, Froment A, Lafosse, S, Heyer, E, Przeworski, M, Blekhman, R, Ségurel, L. Variation in rural african gut microbiota is strongly correlated with colonization by Entamoeba and subsistence. PLoS Genet. 2015;11(11):1–28. doi: 10.1371/journal.pgen.1005658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilchrist CA, Petri SE, Schneider BN, Reichman DJ, Jiang N, Begum S, Watanabe K, Jansen CS, Elliott KP, Burgess SL, et al. Role of the gut microbiota of children in diarrhea due to the protozoan parasite Entamoeba histolytica. JID. 2016;213:1579–1585. doi: 10.1093/infdis/jiv772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reyna-Fabián ME, Zermeño V, Ximénez C, Valadez A, Romero MF, Ximénez C, Argueta J, Moran P, Diaz D, Reyna-Fabián ME, et al. Analysis of the bacterial diversity in liver abscess : differences between pyogenic and amebic abscesses. Am J Trop Med Hyg. 2016;94(1):147–155. doi: 10.4269/ajtmh.15-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cobo ER, He C, Hirata K, Hwang G, Tran U, Eckmann L, Gallo RL, Reed SL. Entamoeba histolytica induces intestinal cathelicidins but is resistant to cathelicidin-mediated killing. Infect Immun. 2012;80(1):143–149. doi: 10.1128/IAI.06224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iyer LR, Verma AK, Paul J, Bhattacharya A. Phagocytosis of gut bacteria by Entamoeba histolytica. Front Cell Infect Microbiol. 2019;9:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kissoon-Singh V, Moreau F, Trusevych E, Chadee K. Entamoeba histolytica exacerbates epithelial tight junction permeability and proinflammatory responses in Muc2−/− mice. Am J Pathol. 2013;182(3):852–865. doi: 10.1016/j.ajpath.2012.11.035. [DOI] [PubMed] [Google Scholar]
- 38.Leon-Coria A, Kumar M, Moreau F, Kris C. Defining cooperative roles for colonic microbiota and Muc2 mucin in mediating innate host defense against Entamoeba histolytica. PLoS Pathog. 2018;14(11):1–22. doi: 10.1371/journal.ppat.1007466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahern PP, Faith JJ, Gordon JI. Mining the human gut microbiota for effector strains that shape the immune system. Immunity. 2014;40(6):819–823. doi: 10.1016/j.immuni.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johansson MEV, Jakobsson HE, Holmén-Larsson J, Schütte A, Ermund A, Rodríguez-Piñeiro A, Arike L, Wising C, Svensson F, Bäckhed F, et al. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe. 2015;18(5):582–592. doi: 10.1016/j.chom.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arike L, Holmén-Larsson J, Hansson GC. Intestinal Muc2 mucin O-glycosylation is affected by microbiota and regulated by differential expression of glycosyltranferases. Glycobiology. 2017;27:318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaiss MM, Harris NL. Interactions between the intestinal microbiome and helminth parasites. Parasite Immunol. 2016;38(1):5–11. doi: 10.1111/pim.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burgess SL, Gilchrist CA, Lynn TC, Petri WA. Parasitic protozoa and interactions with the host intestinal microbiota. Infect Immun. 2017;85(8):1–12. doi: 10.1128/IAI.00101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torres MF, Uetanabaro AP, Costa AF, Farias, LM, Penna, FJ, Alves, CA, Bambirra, EA, Nicoli, JR, Vieira, EC. Influence of bacteria from the duodenal microbiota of patients with symptomatic giardiasis on the pathogenicity of Giardia duodenalis in gnotoxenic mice. J Med Microbiol. 2000;49(2000):209–215. doi: 10.1099/0022-1317-49-3-209. [DOI] [PubMed] [Google Scholar]
- 45.Vieira E, Nicoli J, Moraes-Santos T, Silva, ME, Costa, CA, Mayrink, W, Bambirra, EA. Cutaneous leishmaniasis in germfree, gnotobiotic, and conventional mice. Rev Inst Med Trop Sao Paulo. 1987;29(6):385–387. doi: 10.1590/S0036-46651987000600009. [DOI] [PubMed] [Google Scholar]
- 46.Amiri P, Locksley RM, Parslow TG, Sadickt, M, Rector, E, Ritter, D, McKerrow, JH. Tumour necrosis factor α restores granulomas and induces parasite egg-laying in schistosome-infected SCID mice. Nature. 1992;356:604–607. doi: 10.1038/356604a0. [DOI] [PubMed] [Google Scholar]
- 47.Furarah A, Crocco-Afonso L, Silva M, Bambirra EA, Vieira EC, Nicoli JR. Immune responses of germfree mice to experimental infection with Trypanosoma cruzi. Braz J Med Biol Res. 1991;24(12):1991. [PubMed] [Google Scholar]
- 48.van Es JH, de Geest N, van de Born M, Clevers H, Hassan BA. Intestinal stem cells lacking the Math1 tumour suppressor are refractory to notch inhibitors. Nat Commun. 2010;1(2):1–5. doi: 10.1038/ncomms1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lokmer A, Cian A, Froment A, Gantois, N, Viscogliosi, E, Chabé, M, Ségurel, L. Use of shotgun metagenomics for the identification of protozoa in the gut microbiota of healthy individuals from worldwide populations with various industrialization levels. PLoS One. 2019;14(2):e0211139. doi: 10.1371/journal.pone.0211139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chabé M, Lokmer A, Ségurel L. Gut protozoa : friends or foes of the human gut microbiota ? Trends Parasitol. 2017;33(12):925–934. doi: 10.1016/j.pt.2017.08.005. [DOI] [PubMed] [Google Scholar]


