Abstract
Say-Meyer syndrome is a rare and clinically heterogeneous syndrome characterized by trigonocephaly, short stature, developmental delay and hypotelorism. Nine patients with this syndrome have been reported thus far although no causative gene has yet been identified. Here, we report two siblings with clinical phenotypes of Say-Meyer syndrome with moderate to severe intellectual disability and autism spectrum disorder. Cytogenetics and array-based comparative genomic hybridization did not reveal any chromosome abnormalities or copy number alterations. Exome sequencing of the patients revealed a novel X-linked recessive splice acceptor site variant c.145-2A>G in intron 5 of HUWE1 gene in both affected siblings. RT-PCR and sequencing revealed the use of an alternate splice acceptor site downstream, which led to deletion of six nucleotides resulting loss of two amino acids p.(Cys49-Glu50del) in HUWE1 protein. Deletion of these two amino acids, which are located in a highly conserved region, is predicted to be deleterious and quite likely to affect the function of HUWE1 protein. This is the first report of a potential candidate gene mutation for Say-Meyer syndrome, which was initially described four decades ago.
Keywords: Craniosynostosis, developmental delay, metopic suture, calvarial sutures, cranial sutures
Introduction
Say-Meyer syndrome (OMIM: 314320) was first described almost four decades ago in a family of three males in two successive generations with premature closure of metopic sutures and/or other sutures, short stature and developmental delay (Say and Meyer, 1981). Additional clinical phenotypes observed in those patients included hypotelorism, small anterior fontanel, closed posterior fontanel, prominent metopic ridge, narrow forehead, epicanthal folds, wide nasal bridge, high arched palate and low set ears. Synostosis of metopic, sagittal and lambdoid sutures was reported in one of the three patients and metopic and lambdoid suture synostosis was reported in another patient. Apart from these, clinodactyly of fifth finger along with seizures were reported in one patient. Moderate intellectual disability was reported in the older patient and the other two patients were too young to evaluate intellectual disability (6 months and 3 years) (Say and Meyer, 1981). X-linked recessive inheritance was apparent in the family; however, the authors concluded that autosomal dominant inheritance with low expressivity in females should not be ignored (Say and Meyer, 1981). Subsequently, six more Say-Meyer syndrome patients were reported with several additional phenotypes (Azimi et al., 2003; Karthik et al., 2013; Martinon Sanchez et al., 1985; Salinas-Torres, 2015).
One of the clinical features in the patients with Say-Meyer syndrome is trigonocephaly that arises due to premature fusion of the metopic suture and/or other cranial sutures. Metopic suture generally closes between 18 months to three years of life. In the case of patients with trigonocephaly, the metopic suture closes before birth or shortly after birth. Exogeneous factors as well as genetic alterations can cause premature fusion of sutures (Johnson and Wilkie, 2011). Craniosynostosis/trigonocephaly is implicated in several syndromes including C syndrome (also known as Opitz trigonocephaly syndrome caused by mutations in CD96 (Kaname et al., 2007)), Saethre-Chotzen syndrome (TWIST1) (Howard et al., 1997), Bohring-Opitz syndrome (ASXL1) (Bohring et al., 1999; Hoischen et al., 2011), Trigonocephaly 1 (FGFR1) (Frydman et al., 1984; Kress et al., 2000), Pfeiffer syndrome type 2 (FGFR2) (Priolo et al., 2000), Muenke coronal craniosynostosis (FGFR3) (Bellus et al., 1996), trigonocephaly 2 (FREM1) (Vissers et al., 2011), Greig cephalopolysyndactyly syndrome (GLI3) (Elson et al., 2002) and Jacobsen syndrome (Jacobsen et al., 1973).
Overlapping phenotypes across distinct syndromes lead to diagnostic dilemmas and underscore the need for better diagnostic modalities. New patients with additional phenotypes can further complicate the diagnosis, especially in rare disorders. Precise identification of molecular cause is critical for clinical management, prenatal diagnosis and carrier counseling as well as to understand the molecular mechanism of the disorder. Next generation sequencing has revolutionized molecular medicine to diagnose and characterize various syndromes based on the genetic mutations found in certain genes that are potential cause for the disorders observed in patients. These methods overcome the challenges associated with diagnosis and characterization of several disorders.
Say-Meyer syndrome is one such disorder with heterogeneous clinical presentation for which diagnosis is usually confusing and the molecular cause is unknown. In this study, we performed whole exome sequencing in an Indian family with a clinical phenotype resembling Say-Meyer syndrome and identified a potentially pathogenic novel splice site mutation, c.145-2A>G, in intron 5 of HUWE1 gene. The identified splice site mutation, which was shared by both patients, led to disruption of a normal splice site and use of an alternate cryptic splice site in exon 6 of HUWE1 gene as confirmed by RT-PCR analysis and sequencing. This resulted in deletion of two amino acids at position 49 and 50 located in the N-terminus of HUWE1 protein. HUWE1 gene anomalies have been reported in patients with moderate to severe intellectual disability with or without craniosynostosis (Moortgat et al., 2017). Our study reports the first genetic variant and a potential molecular cause for the Say-Meyer syndrome.
Patient Data
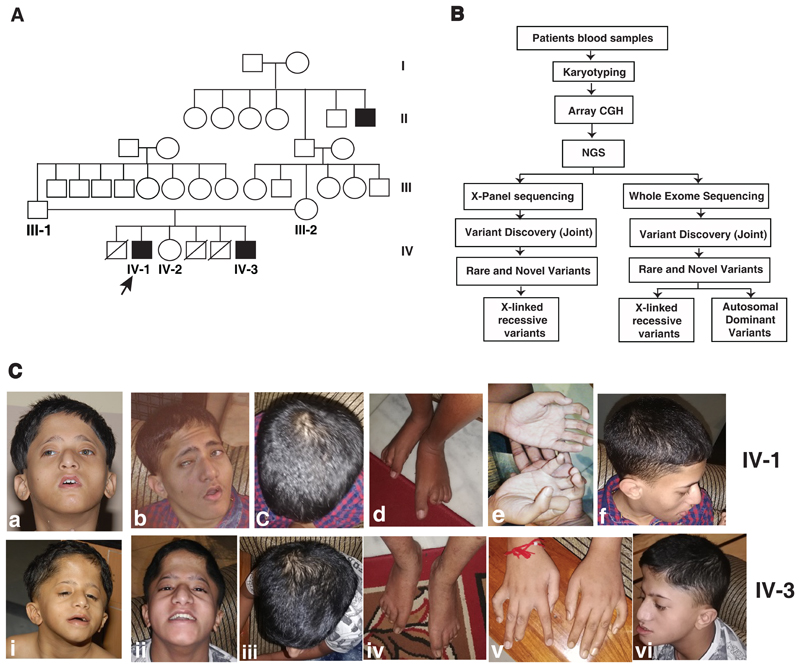
We recruited a non-consanguineous Indian family with two male siblings affected with moderate to severe intellectual disability, trigonocephaly, developmental delay, short stature and hypotelorism. At the time of their first presentation in the clinic, the father was 43 years old, mother was 40 years old and the patients were 11 and 5 years old, respectively. During the second review of the patients, the patients were 20 years and 14 years old, respectively. The family comprised of two affected males and an unaffected female child. In addition, there was a history of three neonatal deaths, all being male children in the family (Fig. 1A). The parents and unaffected sister were healthy and phenotypically normal. One of the brothers of maternal grandfather was affected with intellectual disability with no information on other clinical features (Fig. 1A).
Fig. 1. Pedigree, workflow and clinical photographs of the patients.
A) Pedigree showing affected and unaffected members in the family. Whole exome, X-panel and Sanger sequencing were carried out in proband (IV-1), affected brother (IV-3) and mother (III-2). The identified mutation in HUWE1 gene was tested in father (III-1) and unaffected sister (IV-2) using Sanger sequencing. B) Illustration of the workflow to identify potential causal variants using various methods. C) Clinical photographs of patients IV-1 and IV-3: Upper panel showing clinical phenotypes of IV-1 and lower panel showing clinical phenotypes of IV-3. IV-1:a and IV-3:i are the photographs of patients when they were 11 and 5 years old and the rest of the photographs are when they were 21 and 14 years old.
The motor and language milestones were severely delayed in both siblings and they have not attained speech yet. Both had highly distinctive and almost identical facial and other phenotypic characteristics. Both patients consistently exhibited trigonocephaly, bifrontal narrowing and flat occiput. Facial dysmorphic features were observed in both the patients with elongated triangular face with malar hypoplasia, hypotelorism, ptosis, blepharophimosis, strabismus, low set ears, wide nasal bridge, small pointed nose, narrow mouth, micrognathia, tented upper lip and high arched palate. Other notable features were short neck, camptodactyly, elongated thumb, clinodactyly of fifth finger, pectus excavatum, hypospadias and pes planus. Behaviorally, both affected individuals were diagnosed with autism spectrum disorder. Detailed phenotypic characteristics of both the patients are described in Table 1 and two panels of photographs of the patients depicting the clinical phenotypes are provided in Fig. 1C.
Table 1. Clinical phenotypes of patients.
| Patient 1 (IV-1) | Patient 2 (IV-3) | |
|---|---|---|
| Age | 11 years | 5 years |
| Intellectual disability | Moderate to Severe | Moderate to Severe |
| Age at independent walking | 8 years | 2 years |
| First words | 4 years | 4 years |
| Speaking in sentences | Not attained | Not attained |
| Seizures | No | Febrile seizures |
| Head circumference | 49.5 cm | 48.5 cm |
| Microcephaly | Yes, 2.9 SD below mean | Yes, 2.2 SD below mean |
| Stature | Short | Short |
| External genitalia | Hypospadias | Hypospadias |
| Face | Elongated, triangular, with frontal bossing and coarse facies | Elongated, triangular, with frontal bossing |
| Eyes | Closely set, nystagmus + | Closely set, nystagmus |
| Mouth | Small | Small |
| Ears | Low set | Low set |
| Skull | Marked trigonocephaly | Marked trigonocephaly |
| Occiput | Flat | Flat |
| Chest | Pectus excavatum | Pectus excavatum |
| Fingers | Clinodactyly of fifth finger and camptodactyly | Clinodactyly of fifth finger and camptodactyly |
| Thumb | Elongated | Elongated |
| Pes planus | Yes | Yes |
| Hallux | Elongated (noticeable at 14 years) | Elongated (noticeable at 20 years) |
| Behaviour | Features of autism spectrum disorder | Features of autism spectrum disorder and hyperactivity |
Ethics statement
Informed written consent was obtained from the parents of the patients for genetic research and for the publication of clinical photographs of the patients. All procedures in this study were performed in accordance with the rules and guidelines of the institutional ethics committee at the National Institute of Mental Health and Neurosciences (NIMHANS), Bangalore, India. The patients were assessed by experienced clinicians who followed international classification of diseases (ICD-10) for diagnosis.
Methods
Blood samples were collected from the proband, affected brother, unaffected sister and unaffected parents after obtaining written informed consent from the parents. Cytogenetic analysis, array-based comparative genomic hybridization, whole exome sequencing, X-chromosome panel sequencing and data analysis were carried out as described previously (Muthusamy et al., 2017). Briefly, genomic DNA was isolated from both of the affected siblings and processed for cytogenetic analysis. Chromosomes were stained using G-banding (Seabright, 1971). In order to identify copy number variations in the genomic DNA of patients with reference to control samples, array comparative genomic hybridization was carried out using Agilent human whole genome 8 x 44K and 8 x 60K CGH microarrays (Agilent, USA).
Whole exome and X-chromosome panel target enrichment was carried out using Agilent SureselectXT Human all exon V5 kit and SureselectXT custom 6-11.9 Mb (Agilent, USA), respectively. Paired-end sequencing was performed on HiSeq 2500 (Illumina, USA) to obtain 2 x 75 bp reads for whole exome and MiSeq (Illumina, USA) to obtain 2 x 100 bp reads for X-chromosome panel (X-panel) sequencing. The raw sequencing reads were aligned to the human reference genome hg19 (GRCh37) using BWA-mem (version 0.7.10) (Li and Durbin, 2010). Picard toolkit (version 1.126) was used to remove duplicate PCR reads. Post alignment quality control measures such as indel realignment and base quality score recalibration were performed using GATK (version 3) (DePristo et al., 2011). Genomic variants were called using GATK-HaplotypeCaller and gVCF files were generated. Further, joined variant calling was performed with these gVCF files using GenotypeGVCFs walker. Variants with a minimum base quality score of 20 were retained and annotated using Annovar (Wang et al., 2010). Common variants were removed from 1000 genome project (Genomes Project et al., 2012), Exome Aggregation Consortium (ExAC) database (http://exac.broadinstitute.org/) or NHLBI-EVS Exome Sequencing Project (http://evs.gs.washington.edu/EVS/) with the minor allele frequency filtering cutoff of greater than 0.01. SIFT (Kumar et al., 2009), Polyphen (Adzhubei et al., 2010) and MutationTaster (Schwarz et al., 2010) were used to predict the functional effect of the mutation in protein function. American College of Medical Genetics and Genomics (ACMG) criteria (Richards et al., 2015) were followed for considering the variants.
These variants were further filtered by carrying out segregation analysis across the patients and their mother’s sequencing data. Say-Meyer syndrome has previously been suggested to follow an X-linked recessive or an autosomal dominant mode of inheritance (Say and Meyer, 1981). Thus, we analyzed the variants to account for both modes of inheritance (Fig. 1B). To identify variants that support an X-linked recessive inheritance, we started with variants on the X-chromosome and filtered for protein altering variants that were heterozygous in mother and hemizygous in both the affected individuals. We also included variants occurring at splice sites. We then filtered for variants associated with XLID genes and further analyzed them. To account for an autosomal dominant mode of inheritance, we filtered for protein altering variants and splice site variants that were heterozygous and common between the affected individuals and the mother. Further, the identified splice site mutation in HUWE1 gene was tested in patients, unaffected sister and parents using genomic PCR and Sanger sequencing using the forward and reverse primers F:5’-CGGAGTTCTCCATCTAAAGACC-3’ and R:5’- AGTTGCTCTCTTTCTGGCCTATC-3’, respectively.
Transcript analysis
In order to study the X-inactivation pattern in carrier females and the effect of the splice site mutation, we performed transcript analysis using reverse transcription PCR. Fresh blood samples were collected from patients, unaffected sister and unaffected parents in PAXgene blood RNA tubes and total RNA was extracted using QIAamp RNA blood kits using standard protocols (Qiagen GmbH, Hilden, Germany). The obtained mRNA quality was checked using Nanodrop. cDNA synthesis was carried out using Invitrogen Super Script III One-Step RT-PCR System (Invitrogen, #12574-026) according to the manufacturer’s instructions. The synthesized cDNA was further amplified using the forward primer 5’-GGACTAAACTGAAGAAGACACC-3’ and the reverse primer 5’- GGGAAAAGCTGTACTCAATG-3’. Next, the reaction products were electrophoresed on 1% agarose gel and were extracted using the Gel Purification kit (Favorgen, # FAGCK001). The eluted products were sequenced using Sanger sequencing.
Results
Clinical phenotypes observed in both patients are summarized in Table 1. We set out to determine the genetic basis for the phenotypes observed in these patients. We first performed chromosome analysis using G-banding technique on both patients and found normal 46, XY karyotype. Further, array comparative genomic hybridization was carried out on patient and control samples but did not observe any significant large structural variants or duplications. Next, we carried out whole exome and a targeted X-chromosome panel sequencing on two affected siblings and their mother (Fig. 1A). We obtained an average of 40 million and 13 million paired-end reads from whole exome and the targeted X-chromosome panel sequencing, respectively. Of these reads, ~95% were high quality and mapped to the reference genome (hg19). We obtained 45x and 218x mean depth of coverage for the whole exome and targeted X-chromosome panel sequencing data, respectively. On average, 89% and 92% of the aligned bases provided at least 15x coverage for the whole exome and targeted X-chromosome panel alignment, respectively. Joint variant calling was carried out in all three samples resulted in a total of 6,98,148 and 2,00,158 variants from whole exome sequencing and targeted X-chromosome panel sequencing data, respectively. We removed common variants with minor allele frequency >0.01 in 1000 genome project (Genomes Project et al., 2012), EVS and ExAC (Lek et al., 2016) and obtained 1,13,176 and 1,779 variants from whole exome and targeted X-chromosome panel sequencing, respectively. The entire analysis workflow is depicted in Fig. 1B.
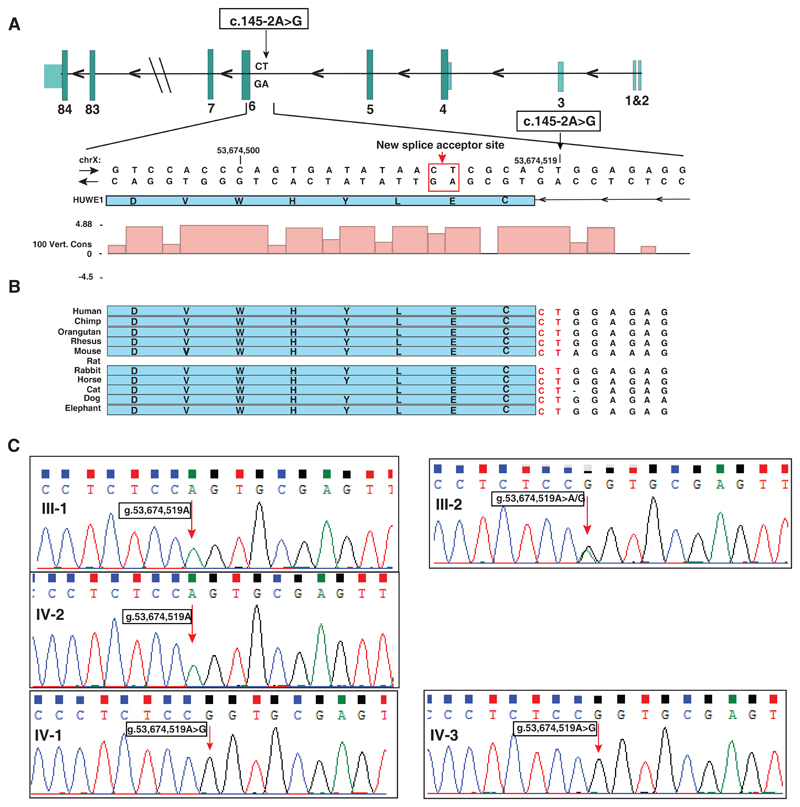
Analysis of variants associated with an X-linked recessive mode of inheritance identified a novel variant g.53674519T>C (genome build: hg19) on the X-chromosome. This is a splice acceptor site variant (c.145-2A>G) in the fifth intron of HUWE1 gene (RefSeq accession: NM_031407.6) (Table 2; Fig. 2A). HUWE1 gene codes for HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase located at Xp11.22. Conservation analysis across 100 vertebrate species using PhyloP algorithm revealed higher conservation of the splice site variant (PhyloP score: 4.88) (Fig. 2B). This variant was not found in 1000 genome project (Genomes Project et al., 2012), EVS and ExAC (Lek et al., 2016). MutationTaster (Schwarz et al., 2014) predicted this variant to be ‘disease causing’ with probability score of 1 (Table 2).
Table 2. Potential causative mutation.
| Gene symbol | Nucleotide change | Protein change | Nucleotide accession | Location | Effect | Inheritance pattern |
|---|---|---|---|---|---|---|
| HUWE1 | c.145-2A>G | p.(Cys49-Glu50del) | NM_031407.6 | Intron 5 | Disruption of the normal splice site led to use of a downstream splice acceptor site resulting in deletion of two amino acids in HUWE1 protein. | X-linked recessive |
Fig. 2. HUWE1 splice site mutation.
A) Schematic representation of exon-intron architecture of HUWE1 gene and depiction of the splice site mutation c.145-2A>G in chromosome X along with a zoomed in view of the mutated residue to depict the conservation of splice site across species. The pink color bars represent conservation across 100 vertebrates using PhyloP algorithm shown in UCSC browser. The normal splice acceptor site is shown with black arrow and alternate splice acceptor site due to the mutation is shown with red arrow and a red rectangle. B) Depiction of conservation of exon-intron junction sequences across species as shown in UCSC conservation track. Intronic sequences shown in red are the conserved splice acceptor of intron 5 of HUWE1. C) Genomic DNA sequencing of a region around HUWE1 splice site mutation g.53674519T>C in father (III-1), mother (III-2), unaffected sister (IV-2) and the patients (IV-1 and IV-3) showing X-linked recessive inheritance. The mutated nucleotide is indicated by a red arrow.
Further, Sanger sequencing of genomic DNA obtained from patients, unaffected sister and unaffected parents confirmed that the HUWE1 variant was consistent with an X-linked recessive mode of inheritance (Fig. 2C). Indeed, HUWE1 variant was hemizygous in both the patients, heterozygous in the mother and absent in both father and unaffected sister. Thus, the unaffected sister is not a carrier for this mutation. Based on the mutation data available in ExAC (Lek et al., 2016), HUWE1 gene is categorized to be in a class of genes that are intolerant to loss-of-function mutations (pLI score= 1.0) which implies that complete loss of function of HUWE1 may lead to lethality. Of note, HUWE1 is reported as a developmental gene and loss of it leads to embryonic lethality in mice (Zhao et al., 2008).
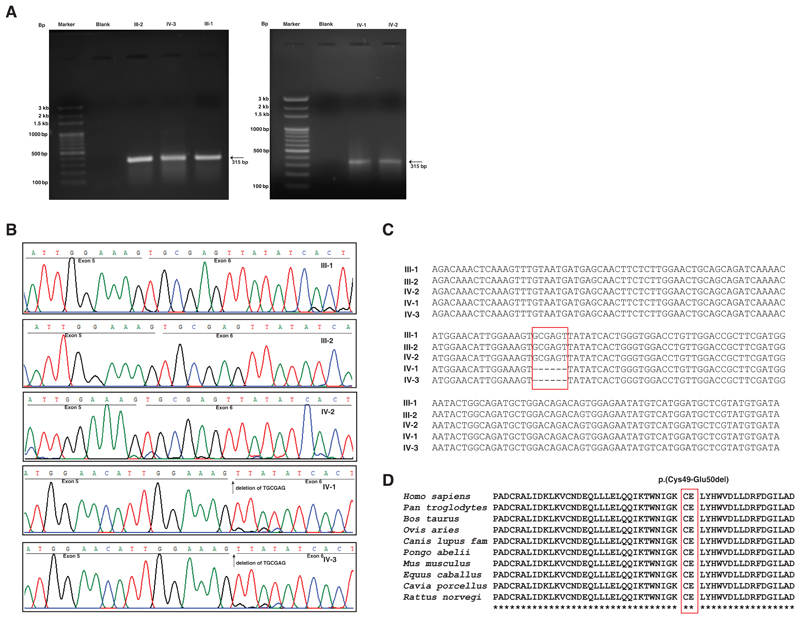
In order to study the effect of the identified mutation in splicing and X-chromosome inactivation in carrier females, we collected fresh blood samples and isolated total RNA from the patients, unaffected sister and parents’ samples and performed RT-PCR. An agarose gel run on the RT-PCR products resulted in a single band in all samples (Fig. 3A). Further, Sanger sequencing of the RT-PCR products revealed the use of a downstream cryptic splice acceptor site in exon 6 in both patients that led to a deletion of 6 nucleotides in exon 6. No such deletion was observed in the father, mother or unaffected sister (Fig. 3B). Alignment of the transcript sequences obtained by Sanger sequencing from unaffected parents, unaffected sister and patients showed the deletion of six nucleotides only in the patient samples (Fig. 3C). The alternate splice site in the exonic region caused a deletion of six nucleotides leading to an in-frame deletion of two amino acids in the N-terminus of HUWE1 p.(Cys49-Glu50del). Conservation analysis of this deletion mutation across species showed high conservation around the mutation p.(Cys49-Glu50del) (Fig 3D). PROVEAN predicted the effect of deletion of these two amino acids as deleterious (Score: -19.148; Default threshold: -2.5 and below scores considered deleterious). This variant has been submitted to ClinVar (Accession: SCV000882449).
Figure 3. Transcript analysis of patients (IV1 and IV-3), unaffected sister (IV-2) and unaffected parents (III-1 and III-2) assessed using reverse transcription-PCR and Sanger sequencing.
A) Agarose gel images of reverse transcription-PCR product across brother of proband (IV-3), unaffected parents (III-1 and III-2), proband (IV-1) and unaffected sister (IV-2) as compared to the ladders M1 and M2. A single band at ~315 bp was observed in all the three samples which is the targeted amplicon size. B) Sanger sequencing of patients (IV-1 and IV-3), unaffected sister (IV-2), unaffected mother (III-2) and father (III-1). The underlined sequences depict the regions corresponding to exon 5 and exon 6 (black). Deletion of the 6 nucleotides is observed in both the patients but not in the unaffected parents or the sister. The sequences of carrier mother and unaffected sister are same as that of reference sequence indicating that the mother had X-inactivation of the mutant X-chromosome. C) Depiction of deletion of six nucleotides in patients using multiple sequence alignment of transcript sequences obtained by Sanger sequencing of cDNA from parents, unaffected sister and the two patients. The red rectangle indicates the 6 nucleotides that are deleted in patients. D) Conservation analysis of HUWE1 protein sequence surrounding the two amino acids that are deleted p.(Cys49-Glu50del) (marked in red).
Sanger sequencing of obligate carrier mother and unaffected sister showed expression the wild type allele in consistent with their normal phenotypes observed. Screening for variants that follow autosomal dominant mode of inheritance did not yield any other significant mutations that could explain the phenotypes observed in the patients.
Discussion
In this study, we report the phenotype and genotype characteristics of two patients with clinical phenotypic features consistent with Say-Meyer syndrome. Table 3 summarizes the clinical phenotypic correlation observed between these two patients and nine patients reported with Say-Meyer syndrome (Salinas-Torres, 2015). The patients consistently exhibited trigonocephaly, short stature, developmental delay and hypotelorism, the characteristic of Say-Meyer syndrome. We carried out exome sequencing and filtered the variants to look for both X-linked recessive and autosomal dominant inheritance with low penetrance as suggested by Say and Meyer (Say and Meyer, 1981). We did not identify any significant autosomal dominant variants but identified a novel X-linked recessive splice acceptor site mutation in HUWE1 gene as a potential cause for the phenotypes observed in the patients. Transcript analysis and sequencing of the transcripts around HUWE1 mutation revealed disruption of normal splice site and acquisition of an alternative downstream cryptic splice acceptor signal downstream. The resulting alternate splicing even led to deletion of six nucleotides in exon 6 resulting in an in-frame deletion of two amino acids p.(Cys49-Glu50del) in HUWE1 protein. As predicted by ExAC intolerance score and consistent with previous knockout studies, the HUWE1 splice site mutation observed in the patients did not cause complete loss of HUWE1 gene but rather resulted in a deletion of two amino acids that could result in the phenotypes observed in the patients. Alternate splicing due to splice site mutation has been reported in a patient with intellectual disability, developmental delay and hypotelorism which caused skipping of exon 8 in HUWE1 gene (Moortgat et al., 2018). PROVEAN predicted the deletion of two amino acids as deleterious and conservation analysis revealed that the deleted amino acids are indeed located in a highly conserved region across multiple species (Fig. 3D).
Table 3. Comparison of phenotypes observed in patients with Say-Meyer syndrome.
| Patients from this study (IV-1 and IV-3) (two male siblings) | Say-Meyer syndrome review Salinas-Torres, 2015 (nine patients – 8 males and 1 females) |
|
|---|---|---|
| Craniofacial manifestations | ||
| Trigonocephaly | ++ | 7/7 |
| Microcephaly | ++ | 2/6 |
| Narrow bi-frontal diameter | ++ | 8/8 |
| Triangular face | ++ | ? |
| Prominent metopic suture / metopic ridge | ++ | 8/8 |
| Bitemporal narrowing | - | 6/6 |
| Small anterior fontanel | - | 3/4 |
| Closed posterior fontanel (at birth) | - | 2/2 |
| Flat occiput | ++ | ? |
| Strabismus | - | 5/7 |
| Deep set eyes | - | 6/7 |
| Epicanthal folds | - | 6/6 |
| Hypotelorism | ++ | 9/9 |
| Arched eye-brows | + | 5/5 |
| Wide nasal bridge | - | 6/6 |
| Small pointed nose | - | 7/7 |
| Anteverted nostrils | - | 7/7 |
| Malar flattening | ++ | 5/5 |
| Long philtrum | ++ | 6/6 |
| Narrow mouth | + | 5/6 |
| Thin lips | - | 5/5 |
| Tented upper lip | ++ | ? |
| High palate | ++ | 3/5 |
| Micrognathia | - | 6/6 |
| Blepharophimosis | ++ | ? |
| Low-set ears | ++ | 5/6 |
| Frontal bossing | ++ | ? |
| Retroverted ears | ++ | ? |
| Nystagmus | ++ | ? |
| Microstomia | ++ | ? |
| Skeletal manifestations | ||
| Clinodactyly | ++ | 1/6 |
| Pectus excavatum | + | ? |
| Pes planus | ++ | ? |
| Short neck | + | ? |
| Neurological manifestations | ||
| Intellectual disability | ++ | 1/9 |
| Developmental delay | ++ | 8/8 |
| Short stature | ++ | 9/9 |
| Seizures | + | 4/7 |
| EEG anomalies | Not done | 2/6 |
| CNS anomalies | Not done | 4/4 |
| Autism spectrum disorder | ++ | ? |
| Other findings | ||
| Hypospadias | ++ | |
| Hand pattern anomalies | + | 2/3 |
| Inguinal hernia | - | 2/5 |
| Cardiovascular defects | - | 3/5 |
| Renal anomalies | - | 1/3 |
| Hearing loss | - | 2/3 |
HUWE1 is a ubiquitously expressed pleiotropic E3 ubiquitin ligase, which is involved in several cellular processes including neuronal development and proliferation (D'Arca et al., 2010; Zhao et al., 2009; Zhao et al., 2008) and synaptogenesis (Sieburth et al., 2005). The association of HUWE1 in intellectual disability has been previously demonstrated. X-linked intellectual disability, Turner type (OMIM: 300706) is reported with HUWE1 mutations with the patients exhibiting mild to severe intellectual disability, microcephaly, hyper/hypotelorism, obesity, coarse facial appearance and holoprosencephaly in male infants (Turner et al., 1994). In addition, mutations in HUWE1 have been reported in patients with heart diseases (Zaidi et al., 2013), autism spectrum disorders (Nava et al., 2012), Juberg-Marsidi and Brooks syndromes (Friez et al., 2016) and several non-syndromic X-linked intellectual disability cases (Froyen et al., 2012; Froyen et al., 2008; Moortgat et al., 2018). Submicroscopic duplications resulting into increased copy number of HUWE1 has been reported in patients with mild to moderate intellectual disability (Froyen et al., 2012; Froyen et al., 2008; Orivoli et al., 2016; Santos-Reboucas et al., 2015). Recently, 21 moderate to profound intellectually disabled patients including two female patients with craniosynostosis and skewed X-inactivation of normal X-chromosome have been reported with mutations in HUWE1 (Moortgat et al., 2017). Females presenting with HUWE1 alterations were asymptomatic and a few female patients showed random or skewed X-inactivation (Froyen et al., 2012; Moortgat et al., 2018; Santos-Reboucas et al., 2015). The non-syndromic XLID patients reported with HUWE1 mutations exhibited moderate to profound ID, delayed or absent speech, delayed motor milestones, short stature and facial dysmorphisms including broad nasal tip, deep set eyes, epicanthic folds, short palpebral fissures, short philtrum and many other heterogeneous clinical presentations. There were very few clinical phenotypes that overlapped with the HUWE1 associated syndromic XLID patients. The patients reported in this study showed a significant match with the clinical phenotypes reported in Say-Meyer syndrome (Table 3). Although nine patients were described with Say-Meyer syndrome, genetic analysis was not carried out in those patients and thus, no mutations were reported. Our study reports the first potential genetic cause for Say-Meyer syndrome. Association of HUWE1 mutations with Say-Meyer syndrome warrants validation in additional patients with Say-Meyer syndrome.
XLID causing HUWE1 mutations had led to alterations in the maintenance of genome stability and oxidative stress which in turn results in defects in neurodevelopment (Bosshard et al., 2017). Around 32 mutations reported to date in HUWE1 gene are associated with intellectual disability. Among them, one splice site mutation and deletion of 20 amino acids have been reported (Moortgat et al., 2017) while the rest are predicted deleterious missense mutations. Several duplications and microduplications involving HUWE1 gene leading to gain of function have also been reported. However, complete HUWE1 deletions or truncating variants in HUWE1 gene have not been reported in males thus far. Further, based on mutations catalogued in ExAC, missense mutations in HUWE1 are rare and truncating mutations are not reported thus far. HUWE1 is a dosage sensitive gene and complete loss or gain of function are presumably not tolerated.
The predominant phenotype in Say-Meyer syndrome is trigonocephaly resulting from craniosynostosis which is premature closure of one or more cranial sutures of skull that can occur in isolation or as part of a syndrome. Genetic and/or environmental factors play crucial role in the manifestation of craniosynostosis. More than 57 genes have been known to be associated with craniosynostosis and HUWE1 is rarely known to be associated with craniosynostosis (Miller et al., 2017). A total of five patients have been reported thus far with HUWE1 mutation and craniosynostosis. The first study that reported the HUWE1 association in craniosynostosis was on a female patient with synostosis of all sutures and learning difficulties. She was identified with a de novo missense mutation p.Arg110Trp (heterozygous) in HUWE1 (Taylor et al., 2015). Microcephaly was diagnosed in utero and synostosis of all sutures was observed. The patient was observed with tall skull with several palpable soft spots. Marked transverse occipital constriction was also noticed. She was dysmorphic with exorbitism, slightly upslanting palpebral fissures and arched eyebrows. She had high arched palate and thin upper lip. Decreased attention and concentration with significant distraction were noted. She had severe language disability. This patient expressed only the mutant allele which was mutated de novo in the DNA inherited paternally and the maternally derived wild-type X-chromosome was inactivated. In order to validate this genotype-phenotype correlation, the authors sequenced 280 patients with craniosynostosis and identified the same mutation in a boy presenting with metopic craniosynostosis, moderate-severe learning disability and many other dysmorphic features (Taylor et al., 2015).
Followed by this study, a de novo missense mutation p.Arg110Trp in HUWE1 was reported in a male with clinical manifestations of craniosynostosis involving premature fusion of metopic sutures with moderate to severe intellectual disability. In addition, flat midface, facial asymmetry, downslanting palpebral fissures, low set ears, retracted pre-maxilla, right choanal stenosis, micrognathia, abnormal teeth arrangements, pectus excavatum, scoliosis, long palms, slight digital shortening, mild 4/5 syndactyly of toes and Chiari malformation were observed in the patient (Miller et al., 2017).
Further, two female patients with skewed inactivation of wild-type X chromosome were reported with HUWE1 mutation in the same site (p.Arg110Gln) with the clinical characteristics of mild ID (1/2), global developmental delay (2/2), delayed or absent speech (2/2), craniosynostosis (2/2), microcephaly (1/2), hypertelorism (1/2), small nose (2/2), broad nasal tip (1/2), thin upper lip (2/2), long face (1/2), high forehead (2/2) and strabismus (2/2). Intellectual disability was not evaluated in one of the patient as she was too young to evaluate (age= 1.5 years) (Moortgat et al., 2018). Craniosynostosis of metopic sutures was consistently observed in all the patients and mild to severe intellectual disability has been reported in these patients. Consistently, mutation of Arg110 was observed in all these patients and skewed inactivation of wild type X-chromosome was observed in female patients. The mutation identified in our study is located in the region 49-50 which is close to the Arg110 mutation. The N-terminus spanning 49-110 could be probably associated with synostosis of sutures and/or cognition. Additional reports are warranted to support this observation. Since skewed inactivation of wild type X-chromosome was observed in all the female patients, we tested for the mutant allele expression using RT-PCR and found that the mother and the sister expressed the wild type X-chromosome rather than the mutant X-chromosome which is consistent with the normal phenotypes observed in the two females tested in this family.
Since the association of HUWE1 with XLID has been demonstrated previously, the effect of the splice defect may lead to a loss of function and this variant could be a potential candidate mutation causing intellectual disability and/or craniosynostosis in the patients. Further animal model studies and additional familial studies reporting HUWE1 mutations will help to confirm the exact role of HUWE1 in Say-Meyer syndrome.
Conclusions
Symptoms of Say-Meyer syndrome overlap with several other syndromes such as C or Opitz’s syndrome and Bohring-Opitz syndrome where trigonocephaly is the major phenotype. The patients described in this study had distinctive phenotypes to distinguish from several of these disorders with other overlapping phenotypes. Accurate finding of the molecular cause is essential for appropriate clinical management, prenatal diagnosis, family counseling and follow up of patients. Our study expanded the spectrum of clinical phenotypes known associated with HUWE1 mutation. The mutation identified in our study will provide a molecular tool that will aid in the clinical diagnosis. However, further studies investigating the effects of the mutation in HUWE1 would shed light on the role of this gene in craniofacial manifestations and neurodevelopmental disorders.
Acknowledgements
We thank the family for participating in this study and for providing samples and consent. We thank the Department of Biotechnology (DBT), Government of India for research support to the Institute of Bioinformatics (IOB). We thank Infosys Foundation for research support to IOB.
Funding Sources
This work was supported by the Wellcome Trust/DBT India Alliance Margdarshi Fellowship [grant number IA/M/15/1/502023] awarded to Akhilesh Pandey. A part of this work was funded by DBT through two grants on X-linked intellectual disability (BT/PR10345/Med/30/79/2007) and (BT/PR18182/BIC/101/937/2016). Dr. Babylakshmi Muthusamy is a recipient of DBT-BioCARe Women Scientist award, Department of Biotechnology, Government of India. Aravind K. Bandari is a recipient of Senior Research Fellowship from CSIR, Government of India.
Footnotes
Disclosure and Conflict of interest
SS holds Roche shares. The other authors declare no conflicts of interest.
Accession Numbers
CRediT author statement
Babylakshmi Muthusamy: Methodology, Software, Validation, Analysis, Investigation, Data Curation, Writing – Original Draft, Writing – Review & Editing, Visualization, Project Administration and Funding Acquisition. Thong T. Nguyen: Methodology, Software, Validation, Formal Analysis, Writing – Review & Editing. Aravind K. Bandari: Investigation. Salah Basheer: Writing – Review & Editing. Lakshmi Dhevi N. Selvan: Investigation, Writing – Review & Editing. Jesna Manoj: Methodology.Deepshikha Chandel – Investigation. Srimonta Gayan – Investigation, review. Somasekar Seshagiri: Methodology, Review & Editing and Supervision. Satish Chandra Girimaji: Conceptualization, Review & Editing and Supervision. Akhilesh Pandey: Conceptualization, Methodology, Resources, Review & Editing, Supervision, Project Administration and Funding Acquisition.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nature methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimi C, Kennedy SJ, Chitayat D, Chakraborty P, Clarke JT, Forrest C, Teebi AS. Clinical and genetic aspects of trigonocephaly: a study of 25 cases. American journal of medical genetics. Part A. 2003;117A(2):127–135. doi: 10.1002/ajmg.a.10021. [DOI] [PubMed] [Google Scholar]
- Bellus GA, Gaudenz K, Zackai EH, Clarke LA, Szabo J, Francomano CA, Muenke M. Identical mutations in three different fibroblast growth factor receptor genes in autosomal dominant craniosynostosis syndromes. Nature genetics. 1996;14(2):174–176. doi: 10.1038/ng1096-174. [DOI] [PubMed] [Google Scholar]
- Bohring A, Silengo M, Lerone M, Superneau DW, Spaich C, Braddock SR, Poss A, Opitz JM. Severe end of Opitz trigonocephaly (C) syndrome or new syndrome? American journal of medical genetics. 1999;85(5):438–446. doi: 10.1002/(sici)1096-8628(19990827)85:5<438::aid-ajmg2>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Bosshard M, Aprigliano R, Gattiker C, Palibrk V, Markkanen E, Hoff Backe P, Pellegrino S, Raymond FL, Froyen G, Altmeyer M, Bjoras M, et al. Impaired oxidative stress response characterizes HUWE1-promoted X-linked intellectual disability. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-15380-y. 15050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arca D, Zhao X, Xu W, Ramirez-Martinez NC, Iavarone A, Lasorella A. Huwe1 ubiquitin ligase is essential to synchronize neuronal and glial differentiation in the developing cerebellum. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(13):5875–5880. doi: 10.1073/pnas.0912874107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson E, Perveen R, Donnai D, Wall S, Black GC. De novo GLI3 mutation in acrocallosal syndrome: broadening the phenotypic spectrum of GLI3 defects and overlap with murine models. Journal of medical genetics. 2002;39(11):804–806. doi: 10.1136/jmg.39.11.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friez MJ, Brooks SS, Stevenson RE, Field M, Basehore MJ, Ades LC, Sebold C, McGee S, Saxon S, Skinner C, Craig ME, et al. HUWE1 mutations in Juberg-Marsidi and Brooks syndromes: the results of an X-chromosome exome sequencing study. BMJ Open. 2016;6(4):e009537. doi: 10.1136/bmjopen-2015-009537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froyen G, Belet S, Martinez F, Santos-Reboucas CB, Declercq M, Verbeeck J, Donckers L, Berland S, Mayo S, Rosello M, Pimentel MM, et al. Copy-number gains of HUWE1 due to replication- and recombination-based rearrangements. American journal of human genetics. 2012;91(2):252–264. doi: 10.1016/j.ajhg.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froyen G, Corbett M, Vandewalle J, Jarvela I, Lawrence O, Meldrum C, Bauters M, Govaerts K, Vandeleur L, Van Esch H, Chelly J, et al. Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. American journal of human genetics. 2008;82(2):432–443. doi: 10.1016/j.ajhg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman M, Kauschansky A, Elian E. Trigonocephaly: a new familial syndrome. American journal of medical genetics. 1984;18(1):55–59. doi: 10.1002/ajmg.1320180109. [DOI] [PubMed] [Google Scholar]
- Genomes Project, C. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoischen A, van Bon BW, Rodriguez-Santiago B, Gilissen C, Vissers LE, de Vries P, Janssen I, van Lier B, Hastings R, Smithson SF, Newbury-Ecob R, et al. De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nature genetics. 2011;43(8):729–731. doi: 10.1038/ng.868. [DOI] [PubMed] [Google Scholar]
- Howard TD, Paznekas WA, Green ED, Chiang LC, Ma N, Ortiz de Luna RI, Garcia Delgado C, Gonzalez-Ramos M, Kline AD, Jabs EW. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nature genetics. 1997;15(1):36–41. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- Jacobsen P, Hauge M, Henningsen K, Hobolth N, Mikkelsen M, Philip J. An (11;21) translocation in four generations with chromosome 11 abnormalities in the offspring. A clinical, cytogenetical, and gene marker study. Hum Hered. 1973;23(6):568–585. doi: 10.1159/000152624. [DOI] [PubMed] [Google Scholar]
- Johnson D, Wilkie AO. Craniosynostosis. European journal of human genetics : EJHG. 2011;19(4):369–376. doi: 10.1038/ejhg.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaname T, Yanagi K, Chinen Y, Makita Y, Okamoto N, Maehara H, Owan I, Kanaya F, Kubota Y, Oike Y, Yamamoto T, et al. Mutations in CD96, a member of the immunoglobulin superfamily, cause a form of the C (Opitz trigonocephaly) syndrome. American journal of human genetics. 2007;81(4):835–841. doi: 10.1086/522014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthik TS, Prasad NR, Rani PR, Maheshwari R, Reddy PA, Chakradhar BV, Menon B. A rare case of short stature: Say Meyer syndrome. Indian J Endocrinol Metab. 2013;17(Suppl 1):S130–131. doi: 10.4103/2230-8210.119531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress W, Petersen B, Collmann H, Grimm T. An unusual FGFR1 mutation (fibroblast growth factor receptor 1 mutation) in a girl with non-syndromic trigonocephaly. Cytogenet Cell Genet. 2000;91(1-4):138–140. doi: 10.1159/000056834. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature protocols. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon Sanchez ML, Garcia Rodriguez C, Martinon Sanchez JM, Martinon Sanchez F. [Say-Meyer syndrome. Report of a new case] An Esp Pediatr. 1985;23(5):375–377. [PubMed] [Google Scholar]
- Miller KA, Twigg SR, McGowan SJ, Phipps JM, Fenwick AL, Johnson D, Wall SA, Noons P, Rees KE, Tidey EA, Craft J, et al. Diagnostic value of exome and whole genome sequencing in craniosynostosis. Journal of medical genetics. 2017;54(4):260–268. doi: 10.1136/jmedgenet-2016-104215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moortgat S, Berland S, Aukrust I, Maystadt I, Baker L, Benoit V, Caro-Llopis A, Cooper NS, Debray FG, Faivre L, Gardeitchik T, et al. HUWE1 variants cause dominant X-linked intellectual disability: a clinical study of 21 patients. European journal of human genetics : EJHG. 2017 doi: 10.1038/s41431-017-0038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moortgat S, Berland S, Aukrust I, Maystadt I, Baker L, Benoit V, Caro-Llopis A, Cooper NS, Debray FG, Faivre L, Gardeitchik T, et al. HUWE1 variants cause dominant X-linked intellectual disability: a clinical study of 21 patients. European journal of human genetics : EJHG. 2018;26(1):64–74. doi: 10.1038/s41431-017-0038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy B, Selvan LDN, Nguyen TT, Manoj J, Stawiski EW, Jaiswal BS, Wang W, Raja R, Ramprasad VL, Gupta R, Murugan S, et al. Next-Generation Sequencing Reveals Novel Mutations in X-linked Intellectual Disability. OMICS. 2017;21(5):295–303. doi: 10.1089/omi.2017.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava C, Lamari F, Heron D, Mignot C, Rastetter A, Keren B, Cohen D, Faudet A, Bouteiller D, Gilleron M, Jacquette A, et al. Analysis of the chromosome X exome in patients with autism spectrum disorders identified novel candidate genes, including TMLHE. Transl Psychiatry. 2012;2:e179. doi: 10.1038/tp.2012.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orivoli S, Pavlidis E, Cantalupo G, Pezzella M, Zara F, Garavelli L, Pisani F, Piccolo B. Xp11.22 Microduplications Including HUWE1: Case Report and Literature Review. Neuropediatrics. 2016;47(1):51–56. doi: 10.1055/s-0035-1566233. [DOI] [PubMed] [Google Scholar]
- Priolo M, Lerone M, Baffico M, Baldi M, Ravazzolo R, Cama A, Capra V, Silengo M. Pfeiffer syndrome type 2 associated with a single amino acid deletion in the FGFR2 gene. Clinical genetics. 2000;58(1):81–83. doi: 10.1034/j.1399-0004.2000.580116.x. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine : official journal of the American College of Medical Genetics. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas-Torres VM. Say-Meyer syndrome: additional manifestations in a new patient and phenotypic assessment. Childs Nerv Syst. 2015;31(7):1181–1187. doi: 10.1007/s00381-015-2704-8. [DOI] [PubMed] [Google Scholar]
- Santos-Reboucas CB, de Almeida LG, Belet S, Dos Santos SR, Ribeiro MG, da Silva AF, Medina-Acosta E, Dos Santos JM, Goncalves AP, Bahia PR, Pimentel MM, et al. Novel microduplications at Xp11.22 including HUWE1: clinical and molecular insights into these genomic rearrangements associated with intellectual disability. Journal of human genetics. 2015;60(4):207–211. doi: 10.1038/jhg.2015.1. [DOI] [PubMed] [Google Scholar]
- Say B, Meyer J. Familial trigonocephaly associated with short stature and developmental delay. Am J Dis Child. 1981;135(8):711–712. doi: 10.1001/archpedi.1981.02130320025008. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nature methods. 2014;11(4):361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nature methods. 2010;7(8):575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- Seabright M. A rapid banding technique for human chromosomes. Lancet. 1971;2(7731):971–972. doi: 10.1016/s0140-6736(71)90287-x. [DOI] [PubMed] [Google Scholar]
- Sieburth D, Ch'ng Q, Dybbs M, Tavazoie M, Kennedy S, Wang D, Dupuy D, Rual JF, Hill DE, Vidal M, Ruvkun G, et al. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436(7050):510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- Taylor JC, Martin HC, Lise S, Broxholme J, Cazier JB, Rimmer A, Kanapin A, Lunter G, Fiddy S, Allan C, Aricescu AR, et al. Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nature genetics. 2015;47(7):717–726. doi: 10.1038/ng.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner G, Gedeon A, Mulley J. X-linked mental retardation with heterozygous expression and macrocephaly: pericentromeric gene localization. American journal of medical genetics. 1994;51(4):575–580. doi: 10.1002/ajmg.1320510456. [DOI] [PubMed] [Google Scholar]
- Vissers LE, Cox TC, Maga AM, Short KM, Wiradjaja F, Janssen IM, Jehee F, Bertola D, Liu J, Yagnik G, Sekiguchi K, et al. Heterozygous mutations of FREM1 are associated with an increased risk of isolated metopic craniosynostosis in humans and mice. PLoS genetics. 2011;7(9):e1002278. doi: 10.1371/journal.pgen.1002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, Carriero NJ, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498(7453):220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, D DA, Lim WK, Brahmachary M, Carro MS, Ludwig T, Cardo CC, Guillemot F, Aldape K, Califano A, Iavarone A, et al. The N-Myc-DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev Cell. 2009;17(2):210–221. doi: 10.1016/j.devcel.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Heng JI, Guardavaccaro D, Jiang R, Pagano M, Guillemot F, Iavarone A, Lasorella A. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol. 2008;10(6):643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]