Abstract
Genetic risk factors frequently affect multiple common human diseases, providing insight into shared pathophysiological pathways and opportunities for therapeutic development. However, systematic identification of genetic profiles of disease risk is limited by the availability of both comprehensive clinical data on population-scale cohorts and the lack of suitable statistical methodology that can handle the scale of and differential power inherent in multi-phenotype data. Here, we develop a disease-agnostic approach to cluster genetic risk profiles for 3,025 genome-wide independent loci across 19,155 disease classification codes from 320,644 participants in the UK Biobank, representing a large and heterogeneous population. We identify 339 distinct disease association profiles and use multiple approaches to link clusters to underlying biological pathways. We show how clusters can decompose the variance and covariance in risk for disease, thereby identifying underlying biological processes and their impact. We demonstrate the use of clusters in defining disease relationships and their potential in informing therapeutic strategies.
Genome-wide association studies (GWAS) of risk for common diseases have revealed widespread pleiotropy, such that individual genetic loci are often associated with multiple disorders 1–4 and many pairs of traits show substantial genome-wide correlation in effects5,6. However, while overlap in genetic risk, such as is seen among the immune-mediated diseases (IMDs) 6–9, implies sharing of aetiological mechanism, clinical practice is largely organised by the tissues or organs affected, leading to potential inefficiency in treatment and challenging drug development 10. Nevertheless, patterns of pleiotropy are complex. For example, within IMDs, some variants, such as rs34536443 in TYK2, are consistent in effect direction across all associated disorders 11, while others, such as rs1800693 in TNFRSF1A, confer risk in some and protection in others 12,13. Moreover, genetic risk scores, which sum effects over all associated variants, are typically highly precise for the corresponding disorder 9, indicating that the specific constellation of genetic risk factors for a disorder are typically not shared.
These observations suggest that systematic characterisation of patterns of pleiotropy can lead to better definition of pathways of risk that affect common human diseases 14–16 and pave the way towards improved clinical care and effective therapeutic development 10,16–18. To date, however, it has not been possible to integrate and interrogate information from the full range of clinical phenotypes that are required to achieve this, as GWAS have focused on a relatively small number of traits and diseases and have often studied patients with only the most clear-cut diagnoses and uniform clinical manifestations. The availability of population-based cohorts with genome-wide variation data, such as the UK Biobank (UKB) 9,19,20, provides a unique opportunity to take a disease-agnostic perspective to investigate cross-trait genetic associations. The UKB has collected genetic and routine healthcare data from over 500,000 participants, including 19,155 diagnostic terms from hospitalization episode statistics (HES), recorded using the tree of International Classification of Diseases, Tenth Revision (ICD-10) codes. This ontology is not intended to reflect biological processes, though nevertheless captures many important relationships between related disorders, subtypes and complications.
Previously, we developed a Bayesian approach for mapping genetic risk across disease classification codes within a hierarchical ontology, referred to as TreeWAS 9, which uses the ontology to shape prior belief about the profile of pleiotropy. The method allows shared signal across related codes (for example subtypes of a disease) to be combined effectively, but also allows for distinct patterns of risk (or absence of risk) in other parts of the ontology. The approach measures the evidence that a variant has any effect on any disease classification code, quantified by the tree Bayes Factor, or BFtree, and enables posterior decoding to identify affected nodes within the ontology. Here, we have applied the TreeWAS method to 654,546 SNPs genotyped in the UKB using the ICD-10 HES data, identifying 3,025 independent loci with strong evidence for association. We then developed and applied a novel clustering method to identify 339 distinct profiles of risk across the ontology and used gene ontology enrichment, overlap with the GWAS Catalog 21, and cluster-specific genetic risk scores to identify associated biological processes and intermediate traits. We show how a cluster-based approach can partition genetic variance and covariance within and among traits as well as generating therapeutic hypotheses.
Results
Genome-wide associations in UKB routine healthcare data
To identify variants that are associated with clinical terms recorded within the ICD-10 HES data, we first ran TreeWAS genome-wide across the 320,644 UKB individuals identified as having British Isles ancestry, correcting for age, sex, genotyping array and the first seven principle components from the genome-wide array data. To enable subsequent comparison between variants, we simplified genetic effects into null, risk and protection for each code, integrating over a prior on effect size. This results in strong correlation of BFtree with the original implementation (Pearson ρ = 0.99; Extended Data Figure 1). Of the 654,546 SNPs, we observed associations for 1.78%; and with 7.35% of the ontology terms showing evidence of an association with at least one tested variant (posterior probability (PP) ≥ 0.99; threshold used throughout) (Fig. 1A). Genome-wide, the strongest evidence of association was observed within the major histocompatibility complex (MHC), with the SNP rs532965 being the most significant (log10 BFtree (lBFtree) = 522.85). This SNP tags the class II alleles HLA-DQA1*03:01 (r = 0.95) and HLA-DRB1*04:01 (r = 0.75) and is observed, in line with previous findings 22, to be associated with 82 ICD-10 codes, including terms related to rheumatoid arthritis, type 1 diabetes and several other IMDs (Fig. 1B). Outside the extended MHC, we identified 3,025 independent lead SNPs with a MAF of at least 1%, with a false positive rate (FPR) of 1% (Extended Data Figure 2), and where any pair of SNPs within the same locus and not in linkage-disequilibrium (LD) had independent phenotype associations (see Supplementary Note). Results are available at www.treewas.org.
Figure 1. Genome-wide evidence for association to the UK Biobank hospital episode statistics (HES) phenotype data set.
(A) Manhattan plot depicting evidence of association (log10 BFtree) across the HES data set. SNPs labelled with gene names exemplify notable associations to common human diseases (see text). (B) Posterior decoding of genetic effect direction and strength of evidence for the rs532965 SNP in the MHC class II region. The ICD-10 classification is depicted as a radial tree where the first orbit represents the 22 ICD-10 Chapters, followed by an orbit representing blocks of categories, and then by two consecutive orbits representing ICD-10 categories including the observed annotation codes. To simplify the representation of the posterior decoding of the ICD-10 codes (left tree) we only colour ICD-10 codes with a posterior probability of association above 0.99 (right tree). Posterior decoding for the SNPs rs4420638 (C), rs10455872 (D) and rs505922 (E) in the APOE, LPA and ABO genes, respectively.
To assess the power of the UKB data for recovering previously described genetic associations we measured association at 25,640 SNPs present in the GWAS Catalog 21 in the UKB cohort. We found evidence for association (lBFtree ≥ 0) with 54.2% and strong evidence for association (lBFtree ≥ 5) for 10.2% (Fig. 2A), though the fraction varies among experimental factor ontology (EFO) groupings and was observed higher for SNPs annotated for cardiovascular diseases (21.48%) and lower for SNPs annotated for biological processes (3.54%). For each group we identified the node with the strongest evidence of association, thus providing a data-driven mapping between terms (Fig. 2A). These results imply that the ICD-10 codes within UKB capture a substantial fraction of variants known to impact human phenotypes, though we note that variants affecting rarer disorders or quantitative traits with no strong disease risk association will be under-represented. In addition, we assessed the evidence of association of the 3,025 independent SNPs and the 25,640 GWAS Catalog SNPs in the self-reported phenotypes from the verbal questionnaires and found correlated evidence of association (Pearson ρ = 0.56 and 0.87, respectively; Extended Data Figure 3)
Figure 2. ICD-10 ontology within UKB HES data captures a substantial fraction of variants known to impact human disease phenotypes in the GWAS Catalog.
(A) Measure of association at GWAS Catalogue SNPs. GWAS Catalog SNPs were grouped into 16 experimental factor ontology (EFO) categories based on the individual SNP annotation found in the GWAS Catalog. For each category we identified the ICD-10 code with the highest evidence of association by taking the product of the posterior of each SNP in the category for all ICD-10 codes. (B) Relationship between the evidence of association of a SNP and the number of phenotypes associated with the SNP (PP ≥ 0.99).
The ability to capture disease-wide measurement enables discovery of the full clinical impact of common variants. For example, the rs4420638 minor allele, which tags the APOE*ε4 haplotype, is the strongest genetic determinant for Alzheimer’s disease 23, and is also associated with cardiovascular diseases 24 and lipid levels 25. We found the variant to confer risk for 53 ICD-10 terms in six clades within the ontology, including those with parent nodes G30-G32 “Other degenerative diseases of the nervous system”; Chapter IX “Diseases of the circulatory system”; E78 “Disorders of lipoprotein metabolism and other lipidaemias”; and Z95 “Presence of cardiac and vascular implants and grafts” (Fig. 1C). Unexpectedly, the same allele also shows evidence (PP = 0.76) for protection against one clade whose parent node is K70-K77 “Diseases of the liver”, demonstrating that implementing our approach across the HES data set can potentially reveal previously unrecognised disease associations for even well-studied pleiotropic risk variants, though we note that this specific result has relatively low evidence (logistic regression OR = 0.93, P = 0.0067) and has yet to be validated in a different cohort.
Cross-trait association patterns also reveal distinctions between genes thought to affect similar biological pathways. For example, for rs2289252 in the F11 blood clotting factor locus, that is associated with venous thromboembolism 26, we observed a restricted set of diseases associations, only including I26.9 “Pulmonary embolism without mention of acute cor pulmonale”; I80.2 “Phlebitis and thrombophlebitis of other deep vessels of lower extremities”; Z86.7 “Personal history of diseases of the circulatory system”; and Z92.1 “Personal history of long-term (current) use of anticoagulants”. However, whilst rs6025 (Arg534Gln, MAF = 3%), known as the Leiden mutation 27 in the F5 blood clotting factor gene, has also been reported to affect venous thromboembolism 28,29, we observed a much more diverse range of additional associations for this SNP. These include other vascular traits, such as I26-I28 “Pulmonary heart disease and diseases of pulmonary circulation”; infections (e.g. J18.9 “Pneumonia, unspecified”); and drug allergies (e.g. Z88.8 “Personal history of allergy to other drugs, medicaments and biological substances”). Therefore, despite both SNPs influencing blood coagulation, their only partially overlapping disease association profiles suggest some disparity in the biological mechanisms they impact and motivates a quantitative assessment of pleiotropy and the similarities and differences between variant effects.
Structure of genetic pleiotropy in the UKB hospital data
To characterise the structure of genetic pleiotropy in the UKB data we determined the relationship between the evidence of association for the 3,025 lead SNPs and the number of ICD-10 codes associated with it. We find that 96.9% of associated SNPs affect more than one diagnostic term, with the top three most pleiotropic variants being well-studied variants near LPA 30,31 (Fig. 1D), CDKN2B 32 and APOE 25,33 (Fig. 1C) (rs10455872 with 61 codes; rs10757274 with 59 codes; and rs4420638 with 53 codes respectively). Overall, we observed a positive correlation between the evidence of association and the number of affected diagnostic terms (ρ = 0.14, P < 10-16, Fig. 2B). However, we also observed variants with very strong evidence of association (lBFtree > 20) that affect only a small number of phenotypes (2.5% affect only one or two codes). For example, rs2981575 and rs4784227 (both lBFtree > 90) localise (on different chromosomes) near FGFR2 and TOX3, respectively, and are associated with nearly identical nodes (14 and 17, respectively) in the ICD-10 ontology, all related to breast cancer (including C50 “Malignant neoplasm of breast” and its child nodes) and procedures such as Z90.1 “Acquired absence of breast”. These SNPs have a similar association profile, displaying a strong evidence of association with a high precision in the phenotypes affected, which likely reflects a strong similarity in the biological pathways they influence. Overall, we found that 82.5% of SNPs were associated with at least 2 of the 24 disease coding chapters of ICD-10 (I-XXII), providing evidence that most genetic variants affecting risk to a diagnostic term will often also affect risk to other terms distant in the ontology.
Decoding cross-trait associations through SNP clustering
Across independently associated variants we observed several repeated patterns of risk and protection, suggestive of distinct genes modulating similar underlying biological processes. To test this hypothesis, we calculated, for every pair of variants, a Bayes factor, BFidentical, comparing a model in which they share the same profile, to a model in which they are independent, thus considering differential uncertainty of individual variant-code associations and their ontological relatedness. We then used hierarchical clustering to define relationships among variants. We chose a threshold of lBFidentical > -5 to group variants into separate clusters, consistent with the threshold chosen for single variant significance (that is, no pair of variants shows greater evidence for having distinct profiles than this threshold) (Fig 3A; Extended Data Figure 4). For each cluster identified we computed a joint posterior decoding to identify associated diagnostic terms.
Figure 3. Genetic risk profiles across common diseases in the HES data set.
(A) Schematic of the study design from genome-wide TreeWAS analysis to hierarchical genetic-risk SNP profile clustering and enrichment analyses. A hierarchical tree was constructed using the pairwise distances between the 3,025 lead SNPs. SNP clusters were determined by cutting the tree at a threshold (see methods). For each cluster a joint genetic risk profile was inferred. (B) Relationship between the number of SNPs and the number of associated ICD-10 codes for the 339 identified clusters. (C) Evidence for enrichment of Biological Processes Gene Ontology terms in SNP sets assigned to each cluster. For each cluster SNP set we calculate enrichment statistics for all GO terms and record the minimal P-value observed across all terms. We then, for each cluster, calculate an empirical P-value which is the proportion of times the minimal GO term P-value is smaller than those observed by randomly generating SNP sets from background of the same size (see Methods).
For the 3,025 independent variants observed, we identified 339 distinct clusters with sizes ranging from 1-37 SNPs, with a median of 76 nodes affected, but ranging from one to 755 (Fig. 3B). Overall, 50% of SNPs occurred in the largest 82 clusters of 13 or more SNPs each and 16 clusters were of a single SNP. For example, the low frequency rs11591147 SNP (Arg46His; MAF ≃ 2%) in the PCSK9 locus, which is correlated with reduced low-density lipoprotein cholesterol levels and coronary artery disease (CAD) risk 34, lies in a cluster of 16 variants (Cluster 34), many of which are near previously-identified CAD risk loci associated with LDL (Fig. 4). The diagnostic code with the greatest number of distinct clusters showing association is I25.8 “Other forms of chronic ischaemic heart disease” (48 clusters), which likely reflects power within UKB (with an I25.8 prevalence of 2.3%). We emphasize that the biological impact of variants in the same cluster are not likely to be identical, rather their clinical consequences are similar across the UKB hospital data.
Figure 4. Posterior decoding for cluster 34 and a selection of individuals variants assigned to this cluster.
For each profile ICD-10 codes with PP ≥ 0.99 are shown. Individual SNP profiles for six out of the 16 variants assigned to Cluster 34 are shown (figures for all variants can be accessed at www.treewas.org.
Each cluster represents a potentially distinct biological mechanism or pathway conferring risk for common diseases, with distinct patterns of potential comorbidity. To investigate the potential for identifying pathways, we assessed enrichment of variants within each cluster among SNPs reported previously in the GWAS Catalog (at the level of EFO terms) and to gene ontology (GO) terms for biological processes. We find 113 (33.3%) clusters that show overlap with EFO terms (permutation P < 0.05) and, 66 clusters with evidence for enrichment in GO terms (permutation P < 0.05; Fig. 3C). For example, the previously-mentioned Cluster 34 is associated with 36 ICD-10 codes (Fig. 4A), including metabolic traits, e.g., E78.0 “Pure hypercholesterolaemia”, diseases of the circulatory system, e.g., I20.9 “Angina pectoris, unspecified”, and complications, such as T82.8 “Other complications of cardiac and vascular prosthetic devices, implants and grafts”. SNPs in this cluster are enriched for GWAS Catalog SNPs reported for 29 EFO terms (Supplementary Table 1), including circulatory system diseases, e.g, atherosclerosis, and metabolic measurements, such as HDL and LDL cholesterol measurements. GO terms enriched in the cluster include “lipoprotein metabolic process” and “very-low-density lipoprotein particle receptor binding” (Supplementary Table 2).
A cluster-based approach reveals the different pathways that contribute to any single clinical endpoint. To illustrate this, we considered the single most common code within the UKB HES data, I10 “Essential (primary) hypertension” (for which there are 24.37% of individuals with at least one record of this code). We observed 27 distinct clusters (with a median number of SNPs of six) with strong association to the code, each affecting between one and 259 ICD-10 codes. Among these clusters, one affects hypertension only; eight are associated with type 2 diabetes (code E11); eight are associated with hypercholesterolaemia (code E78); 17 with angina (code I20), myocardial infarction (codes I21 and I22) or ischaemic heart disease (codes I24 and I25); four are associated with chronic kidney disease (code N18); two are associated with disorders of the gallbladder and bile duct (code K80); and three associate with obesity (code E66) (Fig. 5). Importantly, this heterogeneity in risk profile among clusters is obscured by genome-wide measures of genetic correlation between traits.
Figure 5. Heterogeneity in genetic risk profiles associated with hypertension.
27 risk profiles for clusters associated with the ICD-10 term I10 “Essential (primary) hypertension” (PP ≥ 0.99). Colour labels indicate terms mentioned in the text.
To quantify the relationship between clusters in terms of the phenotypes they affect we estimated (taking into account uncertainty) two measures of association; the Jaccard index (JI) and a metric analogous to the |D’| statistic measure of LD 35 (Extended Data Figure 5 and Supplementary Note). Combined, these metrics can identify whether clusters affect subsets of disorders, disjoint sets, similar profiles or independent profiles. We find that only 0.138% of all pairs have a subset relationship (|D’| ≥ 0.99 and JI ≥ 0.99), while 12.2% have similar profiles (0.5 ≤ |D’| < 0.99), 35.2% are disjoint (JI = 0.0) and 7.11% are effectively independent (|D’| < 0.1); the remaining 45.4% being weakly correlated (0.1 ≤ |D’| < 0.5). These results imply that biological pathways identified through clusters of variants typically impact partially overlapping sets of diseases, with complex and diverse patterns of genetic covariance, typified by low phenotypic disequilibrium.
Identifying focal phenotypes
Clusters may associate with multiple phenotypes either because the pathway affects risk for a specific disease that, in turn, creates risk for a series of clinical complications and comorbidities, or because disruption of the pathway may lead to different diseases in different individuals. Inferring causal structures from multiple categorical variables with genetic instruments and the potential for hidden (or latent) factors remains an open problem 36. We therefore adopted a simpler approach to characterise the relationship among clinical terms within a cluster, aiming to identify ‘focal phenotypes’ whose variance in risk is most (causally) explained by the cluster-specific latent factor (see Methods and Supplementary Note). To achieve this, we note that in a simple model in which there is a latent factor (that is directly influenced by genetics) and two downstream phenotypes, one of which has a much stronger correlation to the latent factor, then the relative impact of genetics on the two observed phenotypes is a measure of relative correlation to the latent factor. We therefore estimated genetic effects for each variant and associated observed clinical term within a cluster and used these to construct cluster-specific genetic risk scores (GRSs) for each phenotype. We then estimated the effect size for these GRSs on all other associated phenotypes (Extended Data Figure 6). Phenotypes within a cluster are ranked by the median effect size of the cross-trait GRS comparisons.
Across all 339 clusters we find that 257 (75.8%) have at least one phenotype with a median GRS of greater than one (which we refer to as a focal phenotype; Fig. 6A and Supplementary Table 3). To illustrate the approach, Fig. 6B shows Cluster 34, which contains the previously-mentioned PCSK9 variant. We find that code E78.2, “Mixed hyperlipidaemia”, consistently has the largest effect size (median relative effect size of 1.72, greater than one in 72% of comparisons). In some cases the causal biological process is clear. For example, Cluster 110, which includes the Factor 5 variant rs6025, has focal phenotypes D68.2 “Hereditary deficiency of other clotting factors” and D68.5 “Primary Thrombophilia” (Fig. 6C), while Cluster 184 has the focal phenotype E55.9 “Vitamin D deficiency, unspecified” (Fig. 6D). For other clusters, the driver phenotypes identified are indirect. For example, Cluster 328 is associated with ICD-10 codes within the C43 and C44 branches “Malignant melanoma of skin” and “Other malignant neoplasms of skin”, respectively, it is enriched for GWAS Catalog SNPs for EFO term melanoma, and it is enriched in GO terms melanin_biosynthetic_process, pigmentation, and UV-damage_excision_repair. However, the focal phenotype identified is W01.6 “Industrial and construction area” with parent code W01 “Fall on same level from slipping, tripping and stumbling”, which may potentially be a proxy for unprotected exposure to sunlight among construction workers (Fig. 6E). However, for 24.2% of the clusters, including 2 out of the 27 hypertension-associated clusters, there is no focal phenotype, indicating that, at least among clinical codes, there are likely distinct manifestations of disruption of the pathway that are observed in different individuals (e.g., Cluster 52; Fig. 6F).
Figure 6. Identification of focal phenotypes within clusters.
(A) Relationship between the median cross-trait GRS effect-size for the driver phenotype in each cluster and the fraction of cross-trait GRS effects that are above one. (B)-(F) Individual cross-trait GRS effect size heatmaps for five of the 339 clusters, cluster 34, 110, 184, 328 and 52, respectively. In each heatmap the ICD-10 codes are sorted by the sum of their cross-trait GRS effect-sizes, with the putative focal phenotype of the left-hand side of the heatmap.
Discussion
The genetic dissection of complex disease has been revolutionised by large-scale biobanks, which link detailed biological measurement, including genomics, to longitudinal data on disease, treatment and response 20,37. However, the statistical analysis of such high dimensional data is still very much in its infancy. Here, we have extended the TreeWAS methodology 9 to the problem of finding groups of variants that have similar impact across diseases. Such clustering has multiple potential benefits. First, by identifying a group of variants, rather than a single one, commonalities among loci, for example in terms of the nature of nearby genes or overlap with genetic studies of intermediate phenotypes, can be used to generate hypotheses about the biological processes modulated. Second, for the same reason, the approach at least partially addresses the challenge of pleiotropy in searching for causal relationships between phenotypes, because it identifies a biologically homogenous set of genetic instruments for applications such as Mendelian randomisation 38. An approach similar to the focal phenotype analysis could potentially be used to search for causal relationships between quantitative trait measurements and disease clusters. Finally, the approach can provide a much more precise definition of the impact of disrupting specific targets, by borrowing information across both phenotypes (through the use of the hierarchical phenotype structure) and loci.
There are multiple potential applications of the relationships characterised in this study. In addition to the well-established use of genetic association data to provide a natural mimic of perturbation of specific targets, thus helping to prioritise candidates for therapeutic development 18, the partitioning of genetic risk into a limited set of pathways or axes has implications for individual patient risk prediction and potentially diagnosis, prognosis and treatment 16. For example, two individuals may have identical genetic risk for hypertension, though differ substantially in terms of risk for potential comorbidities such as diabetes, kidney disease, heart disease and substance abuse. Indeed, we identified a cluster that appears to affect hypertension but no other disorders. However, further work is required to develop and test the use of such partitioned risk, and interpretability requires a much stronger understanding of the biological basis for each axis of risk.
Finally, we acknowledge that the approach described here has several limitations that need to be addressed in future research. Some are technical, including over-estimation of evidence resulting from non-genetic associations between traits and an ad hoc approach for analysing variants in LD. Some arise because of reliance on a single ontology for diseases, which almost certainly fails to capture many of the subtle relationships between disorders and their consequences and introduces biases as a result. More generally, in the search for causal biological explanations for disease risk, onset and progression, additional sources of information, such as molecular and quantitative traits and the longitudinal aspect of multiple data sources, should be utilised. Statistical frameworks for the analysis of multiple trait sexist, including model-comparison 39–41, Mendelian randomisation 42,43 and longitudinal analysis 44. However, typically, these do not scale to the size and complexity of biobank-style data. The high throughput analysis of complex, heterogeneous and multi-modal biomedical data, integrating data on molecular pathways, cellular processes, cell types, tissues, organs and physiology, remains a major obstacle to our understanding of complex disease.
Online Methods
UK Biobank data
The UK Biobank is a prospective cohort of over 500,000 men and women aged 40 to 69 years when recruited in 2006–2010. Participants have provided medical history through an interview and completion of a questionnaire; biological samples for genotyping; and informed consent to long-term medical follow-up through linkage of national health registries, including the hospital episode statistics and cancer registry. The UK Biobank has obtained ethical approval covering this study from the National Research Ethics Committee (REC reference 11/NW/0382).
We use the phenotypic data set available in the UK Biobank participants derived from linkage with the hospital episode statistics registry (data fields 41142 and 41078; accessed on 25-07-2017). This data set includes 2,779,598 records with 7,719,358 diagnoses, and 395,978 participants contained at least one record. Clinical diagnoses in this data set are described with the ICD-10 list compiled by the World Health Organization which follows a hierarchical structure. The ICD-10 contains a total of 19,155 clinical terms, 16,310 of which are terms where diagnoses can be made. Each hospitalisation episode in the data set has a primary diagnosis associated with the event, and an event maybe annotated with one or more secondary diagnoses. Disease outcomes for each individual, as a binary trait, were generated for the combined primary and secondary diagnosis annotations. Individuals were considered unaffected for any given diagnostic term unless the diagnosis was reported in a hospitalization event.
The UK Biobank genetic data used for this study includes 488,377 individuals, 320,644 of whom were determined to be of British Isles ancestry (Extended Data Figure 7) and included in the analysis. Of the total cohort, 49,949 individuals were genotyped on the Affymetrix UK BiLEVE Axiom array as part of a pilot study described elsewhere 45, and the remaining 438,414 individuals were genotyped on the Affymetrix UK Biobank Axiom array. Quality control of SNP data and whole-genome SNP imputation was performed by the UK Biobank analysis team 20. We analysed a total of 654,546 genotyped SNPs. For the GWAS Catalog SNPs not present in the genotype calls we extracted the imputed genotype calls from the whole-genome imputation files and transformed the probabilistic genotypes into allele counts of the minor allele by taking the genotype with the maximum posterior probability.
TreeWAS analysis
The previously-described 9 TreeWAS methodology was applied to the UK Biobank data with two extensions. First, for a given SNP we infer genetic effects as null, risk or protection for each code in the ICD-10, by integrating over a prior on effect size. And second, we allow for the inclusion of covariates to control for population structure, sex and age (details available in the Supplementary Note).
For a SNP, TreeWAS calculates the evidence of an association with at least one code in the tree as a Bayes factor, BFtree. Allele-frequency-specific permutations were carried out to assess the distribution of variant BFtree under the null hypothesis of no association, with randomisation of observed genotypes carried out at the level of the entire cohort and within recruitment centres, to control for geographical variation in clinical coding practice, environmental exposure and fine-scale population structure not captured by broad principal components, while maintaining the observed phenotypic correlation.
We also analysed 25,641 variants reported within the GWAS Catalog (v. 1.0.1, e87, released 2017-03-13) that had been directly genotyped or imputed into the UK Biobank data. Variants with significant association were grouped into sets of independent signals (Supplementary Note). A posterior probability of at least 0.99 was used for level of significance.
Genetic risk profile clustering
To identify clusters of independent variants within similar profiles of risk and protection across diseases, we calculated, for each pair, a Bayes factor, BFidentical, comparing the hypothesis of identical profiles, to the hypothesis of independent profiles. We then used hierarchical clustering with complete linkage to identify clusters, with a threshold equal to that used for identifying variants with non-zero effects (Supplementary Note). For each cluster, we used permutation analyses to estimate the significance of enrichment in GWAS Catalog EFO terms and Gene Ontology annotations for nearby genes (Supplementary Note). Posterior decoding of associated variants and clusters was carried out as described previously. For clusters, we assume that all variants have the same profile of risk and protection.
Focal phenotype analysis
To identify focal phenotypes for each cluster of size N variants we identified the M associated (PP ≥ 0.99) ‘selectable’ ICD-10 codes. For each code we estimated the additive genetic effects, βmn, using a multivariate logistic regression framework:
where C is the set of covariates (PCs, sex, genotyping chip and age at baseline; effects measured relative to the risk allele), xc is the value of the cth covariate, xn is the genotype of the nth variant and ym is the probability of observing code m. For each code m and individual i, we then constructed a GRS with the inferred genetic effects:
where xni is the genotype of individual i on SNP n. This resulted in the construction of M GRSs, each with a set of genetic effects inferred on code m. Then we quantified the effect of GRSm on code w (for all w in M), βGRSmw, using a logistic regression framework with the same set of covariates:
Additional details of the focal phenotype approach are given in the Supplementary Note.
Extended Data
Extended Data Fig. 1. Comparison of estimated log10(BFtree) in the two implementations of TreeWAS for 25,000 SNPs in the hospital episode statistics data set.
Pearson correlation between the two analysis is noted in text.
Extended Data Fig. 2. Derivation of an allele frequency-specific log10(BFtree) significance threshold to maintain a false positive rate below 1%.
The threshold for each allele frequency bin was set to be at least log10(BFtree) = 5.
Extended Data Fig. 3. Concordance of TreeWAS analysis results in the two sources of phenotype data from the UK Biobank, self-reported (SR) data-field 20002 and hospitalisation in-patient records (HES) data-fields 41142 and 41078.
We observed high concordance of the observed evidence of association (log10(BFtree)) for 3,025 independent SNPs and 25,640 GWAS catalog SNPs, with Pearson’s correlation of 0.87 and 0.56, respectively.
Extended Data Fig. 4. Hierarchical clustering of 3,025 SNP risk profiles across the ICD-10 classification tree in the UK Biobank HES data set.
Y-axis is the distance between pairs. Blue line is at height value 0 and red line at height value -5.
Extended Data Fig. 5. Estimates of relationship between the genetic risk profiles for 339 clusters.
For all pairwise comparisons we computed the |D'| statistic and the Jaccard index (see Section Disease ontology analyses in the Supplementary Note).
Extended Data Fig. 6. Schematic illustration of the model that is used to motivate the focal phenotype analysis.
We hypothesize that a set of variants, G, that influences risk for a common set of disease phenotypes, Z, can be acting through a single underlying biological process, X. Typically, we are unlikely to have direct measurement of this variable, though of those disease codes that are mediated by this latent variable, some are likely to be closer to it than others, where closer means a larger absolute value for the regression coefficient of the latent variable on the observed outcome (See Supplementary Note).
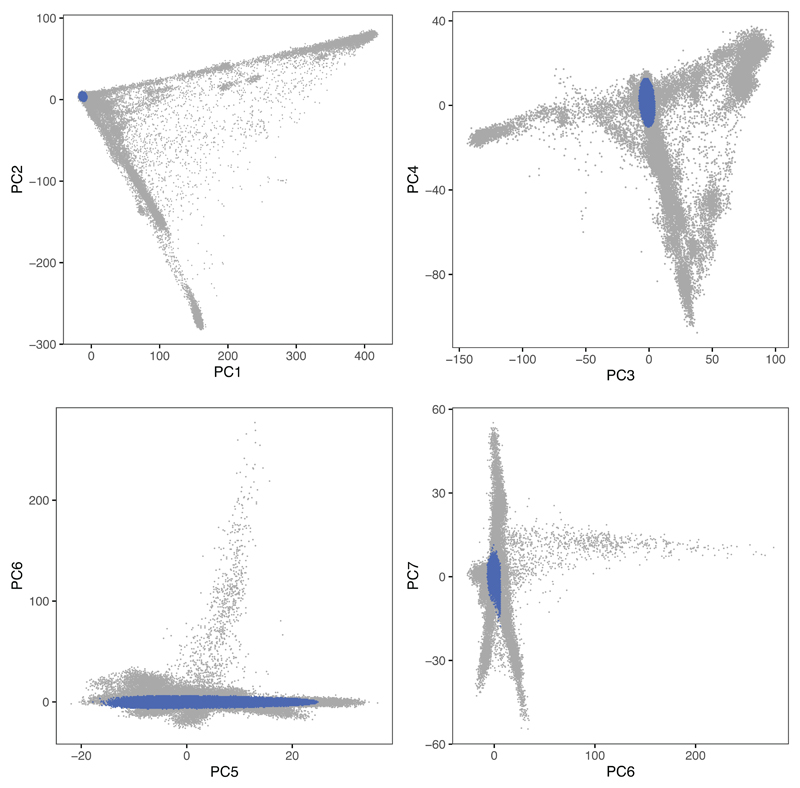
Extended Data Fig. 7. Principal component analysis of genome-wide genotype data in the UK Biobank cohort.
Each plot corresponds to a projection into two dimensions of the principal component analysis. Individuals in blue were determined to be of recent and genome-wide British Isles ancestry.
Supplementary Material
Acknowledgements
This research has been conducted using the UK Biobank Resource (application number 10625). This work uses data provided by patients and collected by the NHS as part of their care and support. Computation used the BMRC facility, a joint development between the Wellcome Centre for Human Genetics and the Big Data Institute supported by the NIHR Oxford BRC. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Funding: This research has been conducted with the support of the Wellcome Trust (100956/Z/13/Z and 090532/Z/09/Z to G.M. and 100308/Z/12/Z to L.F.), the Danish National Research Foundation (L.F.), Takeda to L.F., the Medical Research Council (MC_UU_12010/3 to L.F.), the Oak Foundation (OCAY-15-520 to L.F.), the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre (BRC) to L.F., the Wellcome Trust/Royal Society (204290/Z/16/Z to C.A.D.) and the Li Ka Shing Foundation (to G.M.).
Footnotes
Data availability: The data shown in this paper are available at www.treewas.org. Code for TreeWAS analysis is available at https://github.com/mcveanlab/TreeWASDir.
Competing interests: G.M. is a cofounder of, holder of shares in, and consultant to Genomics PLC, and is a partner in Peptide Groove LLP. The other authors declare no competing financial interests.
Author contributions: A.C. and G.M. performed the analyses with contributions from C.A.D. and L.F. A.C., L.F. and G.M. conceived the study. A.C., C.A.D., L.F. and G.M. wrote the manuscript. P.K.A. design and created the website www.treewas.org and prepared manuscript figures.
References
- 1.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pickrell JK, et al. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet. 2016;48:709–717. doi: 10.1038/ng.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malik R, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50:524–537. doi: 10.1038/s41588-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warren HR, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet. 2017;49:403–415. doi: 10.1038/ng.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cross-Disorder Group of the Psychiatric Genomics Consortium et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellinghaus D, et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet. 2016;48:510–518. doi: 10.1038/ng.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14:661–673. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 8.Inshaw JRJ, Cutler AJ, Burren OS, Stefana MI, Todd JA. Approaches and advances in the genetic causes of autoimmune disease and their implications. Nat Immunol. 2018;19:674–684. doi: 10.1038/s41590-018-0129-8. [DOI] [PubMed] [Google Scholar]
- 9.Cortes A, et al. Bayesian analysis of genetic association across tree-structured routine healthcare data in the UK Biobank. Nat Genet. 2017;49:1311–1318. doi: 10.1038/ng.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oprea TI, et al. Unexplored therapeutic opportunities in the human genome. Nat Rev Drug Discov. 2018;17:317–332. doi: 10.1038/nrd.2018.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dendrou CA, et al. Resolving TYK2 locus genotype-to-phenotype differences in autoimmunity. Sci Transl Med. 2016;8:363ra149. doi: 10.1126/scitranslmed.aag1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.International Multiple Sclerosis Genetics Consortium (IMSGC) et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.International Genetics of Ankylosing Spondylitis Consortium (IGAS) et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet. 2013;45:730–738. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Timpson NJ, Greenwood CMT, Soranzo N, Lawson DJ, Richards JB. Genetic architecture: the shape of the genetic contribution to human traits and disease. Nat Rev Genet. 2018;19:110–124. doi: 10.1038/nrg.2017.101. [DOI] [PubMed] [Google Scholar]
- 15.Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 2013;14:483–495. doi: 10.1038/nrg3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Udler MS, et al. Type 2 diabetes genetic loci informed by multi-trait associations point to disease mechanisms and subtypes: A soft clustering analysis. PLoS Med. 2018;15:e1002654. doi: 10.1371/journal.pmed.1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanseau P, et al. Use of genome-wide association studies for drug repositioning. Nat Biotechnol. 2012;30:317–320. doi: 10.1038/nbt.2151. [DOI] [PubMed] [Google Scholar]
- 18.Nelson MR, et al. The support of human genetic evidence for approved drug indications. Nature Genetics. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 19.Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacArthur J, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raychaudhuri S, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–296. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.CARDIoGRAMplusC4D Consortium et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willer CJ, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, et al. Genetic variants associated with deep vein thrombosis: the F11 locus. J Thromb Haemost. 2009;7:1802–1808. doi: 10.1111/j.1538-7836.2009.03544.x. [DOI] [PubMed] [Google Scholar]
- 27.Bertina RM, et al. Mutation in blood coagulation factor V associated with resistance to activated protein C. Nature. 1994;369:64–67. doi: 10.1038/369064a0. [DOI] [PubMed] [Google Scholar]
- 28.Klarin D, et al. Genetic analysis of venous vhromboembolism in UK Biobank identifies the ZFPM2 locus and implicates obesity as a causal risk factor. Circ Cardiovasc Genet. 2017;10:e001643. doi: 10.1161/CIRCGENETICS.116.001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerhardt A, et al. Prothrombin and factor V mutations in women with a history of thrombosis during pregnancy and the puerperium. N Engl J Med. 2000;342:374–380. doi: 10.1056/NEJM200002103420602. [DOI] [PubMed] [Google Scholar]
- 30.Clarke R, et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 31.Thanassoulis G, et al. Genetic associations with valvular calcification and aortic stenosis. N Engl J Med. 2013;368:503–512. doi: 10.1056/NEJMoa1109034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McPherson R, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao W, et al. Identification of new susceptibility loci for type 2 diabetes and shared etiological pathways with coronary heart disease. Nat Genet. 2017;49:1450–1457. doi: 10.1038/ng.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abifadel M, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 35.Lewontin RC. The Interaction of Selection and Linkage. I. General Considerations; Heterotic Models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frot B, Jostins L, McVean G. Graphical Model Selection for Gaussian Conditional Random Fields in the Presence of Latent Variables. Journal of the American Statistical Association. 2018:1–12. doi: 10.1080/01621459.2018.1434531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evangelou E, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50:1412–1425. doi: 10.1038/s41588-018-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23:R89–98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trochet H, et al. Bayesian meta-analysis across genome-wide association studies of diverse phenotypes. Genet Epidemiol. 2019;43:532–547. doi: 10.1002/gepi.22202. [DOI] [PubMed] [Google Scholar]
- 40.Giambartolomei C, et al. A Bayesian framework for multiple trait colocalization from summary association statistics. Bioinformatics. 2018;34:2538–2545. doi: 10.1093/bioinformatics/bty147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephens M. A unified framework for association analysis with multiple related phenotypes. PLoS One. 2013;8:e65245. doi: 10.1371/journal.pone.0065245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richardson TG, Harrison S, Hemani G, Davey Smith G. An atlas of polygenic risk score associations to highlight putative causal relationships across the human phenome. Elife. 2019;8 doi: 10.7554/eLife.43657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frot B, Jostins L, McVean G. Graphical Model Selection for Gaussian Conditional Random Fields in the Presence of Latent Variables. Journal of the American Statistical Association. 2019;114:723–734. doi: 10.1080/01621459.2018.1434531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding L, et al. Modeling of multivariate longitudinal phenotypes in family genetic studies with Bayesian multiplicity adjustment. BMC Proc. 2014;8:S69. doi: 10.1186/1753-6561-8-S1-S69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wain LV, et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir Med. 2015;3:769–781. doi: 10.1016/S2213-2600(15)00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.