Abstract
Autophagy is an intracellular degradation system that is present in most eukaryotes. In the process of autophagy, double membrane vesicles called autophagosomes sequester a wide variety of cellular constituents and deliver them to lytic organelles: lysosomes in mammals and vacuoles in yeast and plants. Although autophagy used to be considered a non-selective process in its target sequestration into autophagosomes, recent studies have revealed that autophagosomes can also selectively sequester certain proteins and organelles that have become unnecessary or harmful for the cell. We recently discovered that the endoplasmic reticulum (ER) is degraded by autophagy in a selective manner in the budding yeast Saccharomyces cerevisiae, and identified “receptor proteins” that play pivotal roles in this “ER-phagy” pathway. Moreover, several ER-phagy receptors in mammalian cells have also been reported. This report provides an overview of our current knowledge on ER-phagy and discuss their mechanisms, physiological roles, and possible links to human diseases.
Keywords: autophagy, endoplasmic reticulum, ER-phagy
Introduction
Autophagy is defined as a collective term for cellular processes that mediate degradation of the cell’s own components in lysosomes or vacuoles. There are three types of autophagy, which differ in their membrane dynamics: macroautophagy,1,2) microautophagy,3) and autophagy mediated by direct target translocation across the lysosomal membrane, such as chaperon-mediated autophagy,4) DNautophagy,5) and RNautophagy.6) In microautophagy, lysosomal/vacuolar membranes are invaginated and pinched off to form intralumenal vesicles containing cytoplasmic components, which are degraded by lysosomal/vacuolar enzymes.3) In macroautophagy, a membrane cisterna called the isolation membrane (or phagophore) forms in the cytoplasm, and it expands while curving, becomes spherical, and eventually closes to complete a double membrane vesicle called the autophagosome (Fig. 1).1,2) The outer membrane of the autophagosome then fuses with the lysosomal/vacuolar membrane, and the inner autophagosomal membrane and sequestered material are degraded within the lysosome/vacuole. Among these different pathways for autophagy, macroautophagy has been studied most extensively, and thus its mechanism and physiological significance are relatively well understood.7–11) This review outlines our current understanding of selective degradation of the endoplasmic reticulum (ER) and nucleus by macroautophagy (hereafter simply referred to as autophagy).
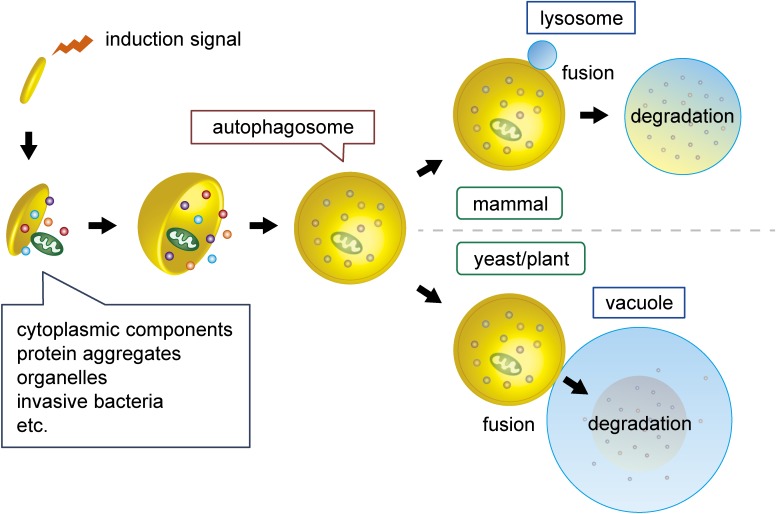
Figure 1.
(Color online) The process of autophagy. When autophagy is induced, small flattened vesicles called isolation membranes or phagophores appear in the cytoplasm and expand to form double membrane vesicles called autophagosomes, which enclose various cellular material in a selective or non-selective manner. The outer autophagosomal membrane fuses with the lysosome in mammalian cells or the vacuole in yeast and plant cells to allow degradation of the inner membrane and sequestered material by hydrolases within the lysosome/vacuole. Degradation products are exported from the lysosomal/vacuolar lumen to the cytoplasm and recycled in various ways.
Basic mechanism of target recognition in selective autophagy
Since early electron microscopy studies showed that autophagosomes formed in conditions of nutrient starvation contain random portions of the cytoplasm, autophagic degradation was considered a non-selective process for many years.12–14) However, recent studies have revealed that an increasing number of cellular components, including proteins and organelles, are selectively degraded by autophagy and revealed how cells “mark” these targets for autophagic degradation.15–17) Proteins called “autophagy receptors” play pivotal roles in target recognition during selective autophagy. Autophagy receptors recognize specific targets, and in the budding yeast Saccharomyces cerevisiae, these proteins interact with the adaptor protein Autophagy-related 11 (Atg11), which recruits the “core” Atg proteins that mediate autophagosome formation (Fig. 2).18) This role for autophagy receptors still remains unclear in mammalian cells (also see below). In both yeast and mammals, autophagy receptors contain an amino acid sequence motif called the Atg8-family interacting motif (AIM) or LC3 interacting region (LIR).16,19) Using this motif, autophagy receptors bind to Atg8-family proteins (LC3s and GABARAPs in mammals), which are anchored to growing isolation membranes via conjugation to phosphatidylethanolamine,20–22) and thereby links targets to the membrane, leading to their efficient sequestration into the autophagosome (Fig. 2). To date, proteins, including ubiquitinated proteins and ferritin, and organelles including ribosomes, mitochondria, peroxisomes, the ER, and nucleus, as well as receptors for these targets have been discovered, and the physiological impact of these selective autophagy pathways have been described.15–17)
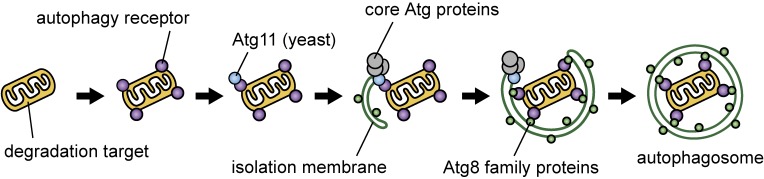
Figure 2.
(Color online) The mechanism of target recognition, initiation of autophagosome formation, and target sequestration into the autophagosome. Cellular components targeted to selective autophagy are first recognized by “autophagy receptors”. These receptors interact with the adaptor proteins Atg11 in yeast or probably FIP200 in mammals, which recruits the core Atg proteins, initiating autophagosome formation on the degradation target. The receptors also bind to Atg8 family proteins (Atg8 in yeast and LC3s and GABARAPs in mammals) which are anchored to forming autophagosomal membranes via conjugation to phosphatidylethanolamine, leading to the efficient sequestration of the degradation target into the autophagosome.
Discovery of ER-phagy in yeast
In an early study, electron microscopy of S. cerevisiae in starvation conditions revealed that most autophagosomes contain fragments of the ER.23) This finding raised the possibility that the ER is efficiently degraded by autophagy. Our analyses using yeast cells lacking Atg11 and those expressing an Atg8 mutant deficient in AIM binding, which are defective in receptor-mediated selective autophagy, convinced us that this autophagic degradation of the ER is mediated by a yet-unknown autophagy receptor.24) Given that previously described autophagy receptors all interacted with Atg8, we aimed to identify it among Atg8-binding proteins. Immunoprecipitates of Atg8 prepared from yeast cells treated with rapamycin, which inhibits Tor kinase complex 1 (TORC1) and thereby induces the starvation response, including autophagic degradation of the ER,25) were analyzed by mass spectrometry. Among the proteins identified, we focused on two proteins of unknown function and named them Atg39 and Atg40. It soon turned out that both of these proteins are important for autophagy of the ER in starvation conditions. We showed that Atg39 and Atg40 interact with Atg8 and Atg11 and also identified AIMs in these proteins and an Atg11-binding motif in Atg39 (Fig. 3). Mutant proteins defective in these protein–protein interactions could not cause ER degradation. These results led us to conclude that these proteins function as receptors for autophagic degradation of the ER, i.e., ER-phagy.24)
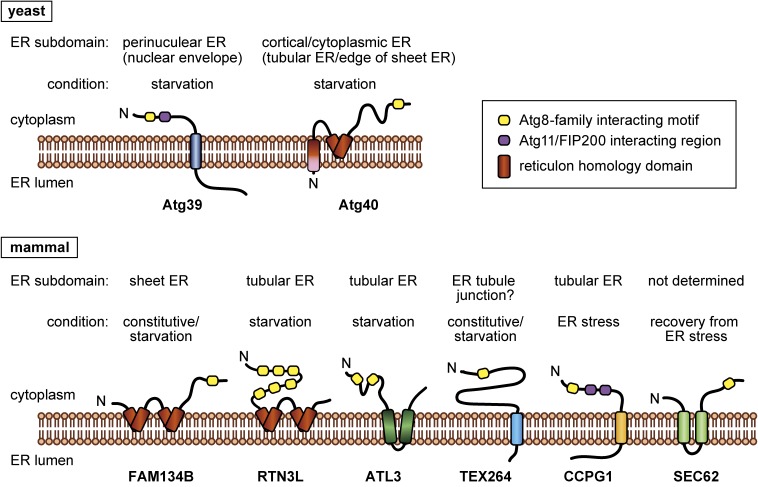
Figure 3.
(Color online) ER-phagy receptors. A schematic illustration of the structures of ER-phagy receptors in yeast and mammals, their localization to ER subdomains, and conditions in which they act as ER-phagy receptors.
In yeast, the ER consists of three distinct subdomains: the cortical (cell peripheral) ER, cytoplasmic ER, and perinuclear ER.26) Unexpectedly, we found that Atg39 and Atg40 differ in their localization to these ER subdomains. Atg39 was specifically observed in the perinuclear ER, whereas Atg40 mainly localized to the cortical and cytoplasmic ER. Consistent with this characteristic localization, further analyses revealed that Atg39 and Atg40 are responsible for degradation of the perinuclear ER and the cortical/cytoplasmic ER, respectively (Fig. 4). Thus, yeast cells are equipped with two autophagy receptors for degradation of different ER subdomains.24)
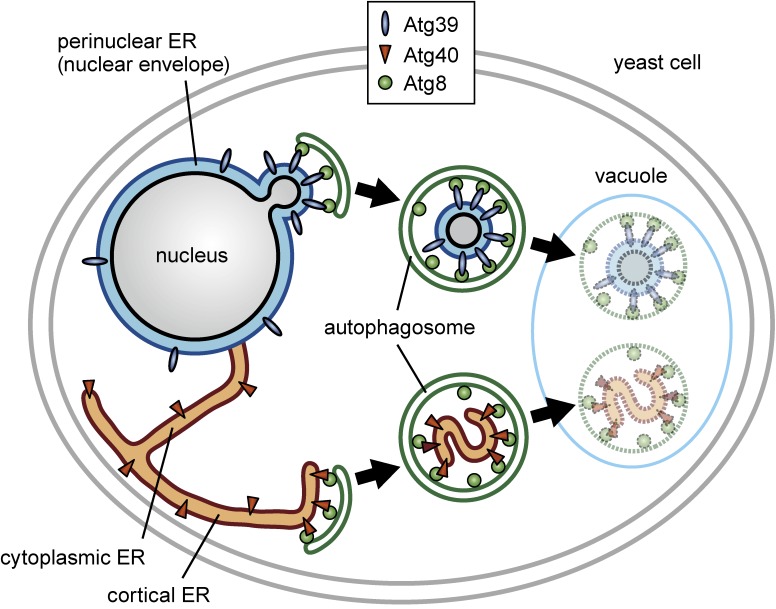
Figure 4.
(Color online) Two ER-phagy receptors mediate degradation of distinct ER subdomains in yeast. Yeast cells induce expression of Atg39 and Atg40 in response to nitrogen starvation (TORC1 inactivation). Atg39 and Atg40 localize to the perinuclear ER and cortical/cytoplasmic ER, respectively, and in association with Atg11 and Atg8, trigger autophagic degradation of the corresponding ER subdomains. While tubular/sheet fragments of the cortical/cytoplasmic ER are loaded into autophagosomes in Atg40-mediated ER-phagy, double membrane vesicles budded from the nucleus are sequestered into autophagosomes in Atg39-mediated ER-phagy. Because these double membrane vesicles encapsulate intranuclear proteins, Atg39 should also be regarded as a receptor for “nucleophagy”.
Electron microscopy showed that the perinuclear ER is sequestered into the autophagosome as double membrane vesicles of ∼200 nm in diameter depending on Atg39, whereas fragments of tubules or sheets of the cortical/cytoplasmic ER are loaded into the autophagosome by Atg40 (the diameter of autophagosomes in these conditions is ∼500 nm). In addition, immunoelectron microscopy detected an ER/nuclear envelope lumenal protein between the two membranes of double membrane vesicles derived from the perinuclear ER, and an intranuclear protein inside these vesicles. Moreover, given the fact that the perinuclear ER in yeast is poorly developed and, therefore, almost equivalent to the nuclear envelope, parts of the nucleus, rather than the perinuclear ER, should be considered as the target of Atg39-mediated selective autophagy.24) Thus, currently we also regard Atg39 as a receptor for selective autophagy of (parts of) the nucleus, i.e., “nucleophagy”. How is the nuclear envelope budded and pinched off to generate double membrane vesicles to be degraded by nucleophagy? How does the cortical/cytoplasmic ER cause fragmentation to be sequestered into the autophagosome during ER-phagy? These intriguing questions still remain to be addressed.
Identification of specific receptors allowed us to examine the physiological significance of ER-phagy and nucleophagy. We found that when ATG39 knockout cells were exposed to prolonged nitrogen starvation, these cells exhibited abnormally extended nuclear morphology and died earlier than wild-type cells, demonstrating that nucleophagy is critical for cellular homeostasis in starvation conditions.24) On the other hand, the cortical ER was more densely reticulated in ATG40 knockout cells starved of nitrogen, but no decrease in cell viability was observed in these cells. A recent study reported that Atg40-mediated ER-phagy is important for the clearance of the Z variant of human alpha-1 antitrypsin, which is an aggregation-prone protein and accumulates in the ER when it is overexpressed in yeast.27) In addition, the unfolded protein response was shown to be enhanced in ATG40 knockout cells in both the absence and presence of ER stress. These results suggested that ER-phagy is involved in the maintenance of ER homeostasis by eliminating abnormal proteins in yeast. Nevertheless, it is still unclear what components in the ER and the nucleus should be degraded by ER-phagy and nucleophagy in nitrogen starvation (TORC1 inactivation), which most strongly induces these pathways. Degradation of these organelles may supply certain molecules as degradation products, which are required to maintain cellular functions in starvation conditions. It is also possible that some toxic materials accumulate within these organelles in these conditions, which should be gotten rid of by ER-phagy and nucleophagy.
Whereas receptors for ER-phagy have also been identified in mammals (see the next section), a nucleophagy receptor other than Atg39 has not been reported yet. However, there are some observations that likely represent nucleophagy in other organisms including mammals,28–30) which may involve receptor proteins like Atg39.
ER-phagy receptors in mammals
1. FAM134B — house-keeping ER-phagy.
At the same as our identification of Atg39 and Atg40 in yeast, Ivan Dikic’s group reported the mammalian ER-phagy receptor FAM134B (family with sequence similarity 134 member B), which was identified as an LC3-binding protein by yeast two-hybrid screening.31) Unlike yeast, in which ER-phagy is induced by nutrient starvation, ER-phagy, as well as the other selective autophagy pathways, constitutively occurs in mammalian cells to maintain cellular homeostasis. FAM134B knockout causes ER swelling, and these cells are sensitive to different stresses including nutrient starvation and ER stress.31) Remarkably, FAM134B was known as a gene responsible for human hereditary sensory and autonomic neuropathy type II (HSAN II).32) The mutations that cause this disease impaired ER-phagy, and FAM134B knockout mice exhibited phenotypes similar to sensory neuronal disease.31) These results demonstrated that not only yeast but mammals also have evolved ER-phagy, and that impairment of ER-phagy can cause neuropathy in humans.
There is no amino acid sequence similarity between Atg40 and FAM134B. However, the domain architectures of these proteins are similar to each other, with reticulon-family domains in the N-terminal regions and the AIM/LIR in the C-terminal regions (Fig. 3). Therefore, FAM134B is likely to be a functional counterpart of Atg40 in mammals. The reticulon-family domains adopt wedge-like transmembrane structures, which are inserted into membranes but do not penetrate them, and thereby generating membrane curvature.33) Reticulons and reticulon-family proteins use these domains to shape ER tubules and the edge of ER sheets. Recombinant FAM134B was shown to cause fragmentation of liposomes (artificial membrane vesicles) in vitro.31) Recent work also suggested that FAM134B generates membrane curvature and senses curvature with its reticulon-like domain, leading to its enrichment in curved regions.34) It is possible that FAM134B and Atg40 are involved in bending and fragmentation of the ER during ER-phagy by exerting their reticulon-like functions for efficient loading of ER fragments into autophagosomes.
2. RTN3L and ATL3 — starvation-induced ER-phagy.
Dikic’s group also identified RTN3 (reticulon 3) as another ER-phagy receptor.35) RTN3 is a reticulon-family protein and known to be involved in the regulation of ER morphology. Its longest splicing variant (RTN3L), which possesses six LIR motifs in the cytoplasmic N-terminal region (Fig. 3), functions in ER-phagy in nutrient-deprived conditions. Remarkably, whereas FAM134B resides in ER sheets,31) RTN3L preferentially localizes to ER tubules and triggers autophagic degradation of these ER regions in response to starvation.35) Thus, yeast Atg40 is structurally closer to FAM134B, but similar to RTN3L in that it mediates degradation of the tubular (cortical/cytoplasmic) ER (Fig. 3). Although FAM134B-driven ER-phagy is also stimulated by nutrient starvation, how these two ER-phagy pathways, which degrade distinct ER subdomains, cooperate and affect cellular functions in starvation conditions still remains unknown.
ATL3 is a member of Atlastins, which are dynamin-like GTPases involved in ER fusion.36,37) However, Chen et al. reported that ATL3, unlike other Atlastins, functions as an ER-phagy receptor in mammalian cells (Fig. 3).38) Similar to RTN3L, ATL3 promotes autophagic degradation of ER tubules during nutrient starvation. In addition, overproduction of RTN3L in ATL3 knockout cells rescued defects in ER-phagy. Moreover, ATL3 was shown to interact with RTN3L. These results suggested that ATL3 and RTN3L cooperatively act in ER-phagy in starvation conditions. On the other hand, unlike RTN3L, ATL3 specifically interacts with GABARAP among mammalian ATG8 homologs via two LIR motifs (GABARAP interaction motifs or GIMs) (Fig. 3). This unique property of ATL3 may be important for its cooperation with RTN3L. Of note, ATL3 is known to be a gene responsible for hereditary sensory and autonomic neuropathy type I (HSAN I),39,40) and Chen et al. showed that mutations that cause this disease impaired the ATL3-GABARAP interaction and ER-phagy.38)
3. TEX264 — a major receptor for both constitutive and starvation-induced ER-phagy.
Recently, Noboru Mizushima’s group and Wade Harper’s group independently identified TEX264 (testis expressed gene 264) as a new ER-phagy receptor in mammalian cells.41,42) TEX264 is a single membrane-spanning protein with its long C-terminal, cytoplasmic region containing the LIR motif (Fig. 3).41,42) TEX264 is ubiquitously expressed in mouse tissues, and efficiently binds to ATG8 homologs compared to other mammalian ER-phagy receptors.41) Knockdown/knockout analyses suggested that TEX264 makes the largest contribution to ER-phagy among known ER-phagy receptors under both nutrient-replete and starvation conditions.41) Moreover, mass spectrometry estimated that TEX264 is responsible for about 50% of autophagic degradation of ER resident proteins during amino acid starvation.42) These results suggested that TEX264 is a major receptor for both constitutive and starvation-induced ER-phagy.
Chino et al. also revealed an interesting molecular property of TEX264.41) Because the ER (rough ER) associates with a high density of ribosomes, these ribosomes are thought to be an “intermembrane spacer” when TEX264 links the ER to the autophagosomal membrane. To circumvent this issue, TEX264 has a long intrinsically disordered region (IDR) between its transmembrane domain and the LIR motif in the C-terminal cytoplasmic region (Fig. 3). This IDR allows the TEX264 LIR motif to reach ATG8 homologs on the forming autophagosomal membrane even in the presence of ribosomes on the ER membrane.
4. CCPG1 — ER stress-induced ER-phagy.
Simon Wilkinson’s group reported that CCPG1 (cell-cycle progression gene 1) acts as an ER-phagy receptor in cells exposed to ER stress.43) CCPG1 is transcriptionally upregulated upon ER stress and mainly mediates degradation of the tubular regions of the ER. When Ccpg1 expression was repressed in mouse pancreatic cells, which are susceptible to ER stress, ER-resident proteins formed insoluble aggregates, and the unfolded protein response was evoked. In addition, ER swelling and tissue injury associated with inflammatory infiltration were observed. These results suggested that CCPG1-mediated ER-phagy is important for homeostasis of the ER in ER stress conditions.
It is noteworthy that CCPG1 was shown to interact with FIP200, which is likely to be a functional homolog of Atg11 in mammalian cells.43) CCPG1 binds to FIP200 via two FIP200-binding regions, which contain amino acid sequences similar to the Atg11-binding motif found in autophagy receptors in yeast (Fig. 3). In yeast, almost all autophagy receptors in association with Atg11 recruit the autophagy-initiating Atg1 complex to trigger autophagosome formation in the vicinity of degradation targets (Fig. 2).18) By contrast, although mammalian autophagy receptors bind to Atg8-family proteins, their roles in the initiation of autophagosome formation is still poorly understood. FIP200 is a component of the autophagy-initiating complex in mammals (the ULK1 complex).44,45) Thus, in CCPG1-mediated ER-phagy, the recognition of the target and the initiation of its sequestration by the autophagosome proceed in the same manner as those in yeast. Future studies will investigate other mammalian autophagy receptors for their roles in the initiation of selective autophagy.
5. SEC62 — ER-phagy for recovery from ER stress.
When cells are exposed to ER stress, proteins to cope with perturbations in the ER, such as molecular chaperones and ER-associated degradation (ERAD)-related proteins, are upregulated.46,47) After the removal of the stress, these proteins decrease to the basal level. Maurizio Molinari and colleagues found that SEC62-dependent ER-phagy plays a crucial role in this recovery from ER stress, and termed this type of ER-phagy “recovER-phagy”.48) SEC62 serves as a subunit of the protein translocation machinery in the ER in normal conditions, but it interacts with LC3 via the LIR motif in its C-terminal region during recovery from ER stress (Fig. 3). A mutation in the LIR motif of SEC62 impaired recovER-phagy with its function for protein translocation intact. Overexpression of SEC62 alone or knockout of another subunit of the protein translocation machinery resulted in autophagic degradation of the ER in a manner dependent on the SEC62 LIR motif. Based on these observations, the authors proposed that SEC62 dissociates from the protein translocation machinery upon amelioration of ER stress to act as an ER-phagy receptor.
Concluding remarks
ER-phagy is one of the latest topics in the field of autophagy research. As outlined here, since the first reports on the ER-phagy receptors yeast Atg39 and Atg40 and mammalian FAM134B in 2015, five new receptors have been discovered one after another in mammals. It has also been revealed that these receptors are diverse in terms of when they induce ER-phagy, which ER subdomain they target, and what is the purpose of ER degradation. However, there remain many intriguing issues to be addressed. In mechanistic aspects, it is still unclear how ER fragmentation occurs during ER-phagy. The regulation of ER-phagy also needs further investigation. Given the fact that ER-phagy receptors are all integral membrane proteins but different in their amino acid sequences and structures (Fig. 3), the molecular basis underlying ER fragmentation and ER-phagy regulation may vary among forms of ER-phagy mediated by different receptors. It is also interesting to analyze the mechanistic and functional interplay of ER-phagy receptors in ER degradation.
In physiological and pathological aspects, it is important to know how the impairment of ER-phagy leads to defects in the cellular functions and disease-related phenotypes observed. In addition to the sensory neuropathies HSAN I and II, which are caused by mutations in ATL3 and FAM134B, respectively, ER-phagy can be linked to different human diseases through the regulation of ER functions and homeostasis. For instance, because the ER is a site for the assembly of some viruses, ER-phagy can be involved in this process. Indeed, FAM134B and RTN3 have been reported to be involved in the suppression of the proliferation of viruses, including Ebola, Dengue, and Zika viruses.49–51)
Although this review has focused on macroautophagic degradation of the ER, ER-derived whorl-like structures (ER whorls) are formed and delivered into the vacuole via microautophagy during ER stress.52) ER-phagy research has great potential for further development, and future studies will uncover new aspects of ER-phagy.
Profile
Hitoshi Nakatogawa was born in Kanagawa Prefecture in 1974. He graduated from Chuo University in 1997, then went on to the Graduate School of Science of Kyoto University, and received his Ph.D. degree in 2002 for studies on protein translation and translocation in Escherichia coli under the supervision of Prof. Koreaki Ito at the Institute for Virus Research. In 2004, he joined Prof. Yoshinori Ohsumi’s group at the National Institute for Basic Biology as a JSPS Research Fellow to commence work on autophagy in yeast, which continues to the present. He became an Assistant Professor the next year, and along with Prof. Ohsumi, he moved to the Frontier Research Center, Tokyo Institute of Technology in 2009. He also concurrently served as a PRESTO researcher (Japan Science and Technology Agency) from 2006 to 2010. He moved to the Graduate School of Bioscience and Biotechnology, Tokyo Institute of Technology as an Associate Professor in 2014, and assumed his present position in 2016. He was awarded an Inoue Research Award for Young Scientists in 2003, Young Investigator Award from the Japanese Biochemical Society in 2013, a Young Scientists’ Prize from the Commendation for Science and Technology by MEXT in 2014, and the JSPS Prize in 2017.
References
- 1).Yang Z., Klionsky D.J. (2010) Eaten alive: A history of macroautophagy. Nat. Cell Biol. 12, 814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Ohsumi Y. (2014) Historical landmarks of autophagy research. Cell Res. 24, 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Oku M., Sakai Y. (2018) Three distinct types of microautophagy based on membrane dynamics and molecular machineries. BioEssays 40, 1800008. [DOI] [PubMed] [Google Scholar]
- 4).Kaushik S., Cuervo A.M. (2018) The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Aizawa S., Contu V.R., Fujiwara Y., Hase K., Kikuchi H., Kabuta C., et al. (2017) Lysosomal membrane protein SIDT2 mediates the direct uptake of DNA by lysosomes. Autophagy 13, 218–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Aizawa S., Fujiwara Y., Contu V.R., Hase K., Takahashi M., Kikuchi H., et al. (2016) Lysosomal putative RNA transporter SIDT2 mediates direct uptake of RNA by lysosomes. Autophagy 12, 565–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009) Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467. [DOI] [PubMed] [Google Scholar]
- 8).Mizushima N., Yoshimori T., Ohsumi Y. (2011) The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132. [DOI] [PubMed] [Google Scholar]
- 9).Lamb C.A., Yoshimori T., Tooze S.A. (2013) The autophagosome: Origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14, 759–774. [DOI] [PubMed] [Google Scholar]
- 10).Dikic I., Elazar Z. (2018) Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349–364. [DOI] [PubMed] [Google Scholar]
- 11).Levine B., Kroemer G. (2019) Biological functions of autophagy genes: A disease perspective. Cell 176, 11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Ashford T.P., Porter K.R. (1962) Cytoplasmic components in hepatic cell lysosomes. J. Cell Biol. 12, 198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Clark S.L., Jr. (1957) Cellular differentiation in the kidneys of newborn mice studies with the electron microscope. J. Biophys. Biochem. Cytol. 3, 349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).de Duve C., Wattiaux R. (1966) Functions of lysosomes. Annu. Rev. Physiol. 28, 435–492. [DOI] [PubMed] [Google Scholar]
- 15).Kirkin V. (2019) History of the selective autophagy research: How did it begin and where does it stand today? J. Mol. Biol. doi:10.1016/j.jmb.2019.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Johansen T., Lamark T. (2019) Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J. Mol. Biol. doi:10.1016/j.jmb.2019.07.016. [DOI] [PubMed] [Google Scholar]
- 17).Gatica D., Lahiri V., Klionsky D.J. (2018) Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 20, 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Zientara-Rytter K., Subramani S. (2019) Mechanistic insights into the role of Atg11 in selective autophagy. J. Mol. Biol. doi:10.1016/j.jmb.2019.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Noda N.N., Ohsumi Y., Inagaki F. (2010) Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584, 1379–1385. [DOI] [PubMed] [Google Scholar]
- 20).Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., et al. (2000) A ubiquitin-like system mediates protein lipidation. Nature 23, 488–492. [DOI] [PubMed] [Google Scholar]
- 21).Kabeya Y. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T. (2004) LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117, 2805–2812. [DOI] [PubMed] [Google Scholar]
- 23).Hamasaki M., Noda T., Baba M., Ohsumi Y. (2005) Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic 6, 56–65. [DOI] [PubMed] [Google Scholar]
- 24).Mochida K., Oikawa Y., Kimura Y., Kirisako H., Hirano H., Ohsumi Y., et al. (2015) Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 522, 359–362. [DOI] [PubMed] [Google Scholar]
- 25).Noda T., Ohsumi Y. (1998) Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273, 3963–3966. [DOI] [PubMed] [Google Scholar]
- 26).Voeltz G.K., Rolls M.M., Rapoport T.A. (2002) Structural organization of the endoplasmic reticulum. EMBO Rep. 3, 944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Cui Y., Parashar S., Zahoor M., Needham P.G., Mari M., Zhu M., et al. (2019) A COPII subunit acts with an autophagy receptor to target endoplasmic reticulum for degradation. Science 365, 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Park Y.-E., Hayashi Y.K., Bonne G., Arimura T., Noguchi S., Nonaka I., et al. (2009) Autophagic degradation of nuclear components in mammalian cells. Autophagy 5, 795–804. [DOI] [PubMed] [Google Scholar]
- 29).Shoji J.Y., Kikuma T., Arioka M., Kitamoto K. (2010) Macroautophagy-mediated degradation of whole nuclei in the filamentous fungus Aspergillus oryzae. PLoS One 5, e15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Papandreou M.E., Tavernarakis N. (2019) Nucleophagy: From homeostasis to disease. Cell Death Differ. 26, 630–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Khaminets A., Heinrich T., Mari M., Grumati P., Huebner A.K., Akutsu M., et al. (2015) Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 522, 354–358. [DOI] [PubMed] [Google Scholar]
- 32).Kurth I., Pamminger T., Hennings J.C., Soehendra D., Huebner A.K., Rotthier A., et al. (2009) Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat. Genet. 41, 1179–1181. [DOI] [PubMed] [Google Scholar]
- 33).Voeltz G.K., Prinz W.A., Shibata Y., Rist J.M., Rapoport T.A. (2006) A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124, 573–586. [DOI] [PubMed] [Google Scholar]
- 34).Bhaskara R.M., Grumati P., Garcia-Pardo J., Kalayil S., Covarrubias-Pinto A., Chen W., et al. (2019) Curvature induction and membrane remodeling by FAM134B reticulon homology domain assist selective ER-phagy. Nat. Commun. 10, 2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35).Grumati P., Morozzi G., Hölper S., Mari M., Harwardt M.I., Yan R., et al. (2017) Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. eLife 6, e25555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Hu J., Shibata Y., Zhu P.P., Voss C., Rismanchi N., Prinz W.A., et al. (2009) A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 138, 549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Orso G., Pendin D., Liu S., Tosetto J., Moss T.J., Faust J.E., et al. (2009) Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 460, 978–983. [DOI] [PubMed] [Google Scholar]
- 38).Chen Q., Xiao Y., Chai P., Zheng P., Teng J., Chen J., et al. (2019) ATL3 is a tubular ER-phagy receptor for GABARAP-mediated selective autophagy. Curr. Biol. 29, 846–855.e6. [DOI] [PubMed] [Google Scholar]
- 39).Fischer D., Schabhüttl M., Wieland T., Windhager R., Strom T.M., Auer-Grumbach M. (2014) A novel missense mutation confirms ATL3 as a gene for hereditary sensory neuropathy type 1. Brain 137, e286. [DOI] [PubMed] [Google Scholar]
- 40).Kornak U., Mademan I., Schinke M., Voigt M., Krawitz P., Hecht J., et al. (2014) Sensory neuropathy with bone destruction due to a mutation in the membrane-shaping atlastin GTPase 3. Brain 137, 683–692. [DOI] [PubMed] [Google Scholar]
- 41).Chino H., Hatta T., Natsume T., Mizushima N. (2019) Intrinsically disordered protein TEX264 mediates ER-phagy. Mol. Cell 74, 909–921.e6. [DOI] [PubMed] [Google Scholar]
- 42).An H., Ordureau A., Paulo J.A., Shoemaker C.J., Denic V., Harper J.W. (2019) TEX264 is an endoplasmic reticulum-resident ATG8-interacting protein critical for ER remodeling during nutrient stress. Mol. Cell 74, 891–908.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43).Smith M.D., Harley M.E., Kemp A.J., Wills J., Lee M., Arends M., et al. (2018) CCPG1 is a non-canonical autophagy cargo receptor essential for ER-phagy and pancreatic ER proteostasis. Dev. Cell 44, 217–232.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J.-L., et al. (2008) FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181, 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Ganley I.G., Lam D.H., Wang J., Ding X., Chen S., Jiang X. (2009) ULK1·ATG13·FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284, 12297–12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46).Mori K. (2009) Signalling pathways in the unfolded protein response: Development from yeast to mammals. J. Biochem. 146, 743–750. [DOI] [PubMed] [Google Scholar]
- 47).Hwang J., Qi L. (2018) Quality control in the endoplasmic reticulum: Crosstalk between ERAD and UPR pathways. Trends Biochem. Sci. 43, 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Fumagalli F., Noack J., Bergmann T.J., Cebollero E., Pisoni G.B., Fasana E., et al. (2016) Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat. Cell Biol. 18, 1173–1184. [DOI] [PubMed] [Google Scholar]
- 49).Chiramel A.I., Dougherty J.D., Nair V., Robertson S.J., Best S.M. (2016) FAM134B, the selective autophagy receptor for endoplasmic reticulum turnover, inhibits replication of Ebola virus strains Makona and Mayinga. J. Infect. Dis. 214, S319–S325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Lennemann N.J., Coyne C.B. (2017) Dengue and Zika viruses subvert reticulophagy by NS2B3-mediated cleavage of FAM134B. Autophagy 13, 322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51).Wu M.J., Ke P.Y., Hsu J.T.A., Yeh C.T., Horng J.T. (2014) Reticulon 3 interacts with NS4B of the hepatitis C virus and negatively regulates viral replication by disrupting NS4B self-interaction. Cell. Microbiol. 16, 1603–1618. [DOI] [PubMed] [Google Scholar]
- 52).Schuck S., Gallagher C.M., Walter P. (2014) ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J. Cell Sci. 127, 4078–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]