Abstract
Insights into individual differences in gene expression and its heritability (h2) can help in understanding pathways from DNA to phenotype. We estimated the heritability of gene expression of 52,844 genes measured in whole blood in the largest twin RNA-Seq sample to date (1497 individuals including 459 monozygotic twin pairs and 150 dizygotic twin pairs) from classical twin modeling and identity-by-state-based approaches. We estimated for each gene h2total, composed of cis-heritability (h2cis, the variance explained by single nucleotide polymorphisms in the cis-window of the gene), and trans-heritability (h2res, the residual variance explained by all other genome-wide variants). Mean h2total was 0.26, which was significantly higher than heritability estimates earlier found in a microarray-based study using largely overlapping (>60%) RNA samples (mean h2 = 0.14, p = 6.15 × 10−258). Mean h2cis was 0.06 and strongly correlated with beta of the top cis expression quantitative loci (eQTL, ρ = 0.76, p < 10−308) and with estimates from earlier RNA-Seq-based studies. Mean h2res was 0.20 and correlated with the beta of the corresponding trans-eQTL (ρ = 0.04, p < 1.89 × 10−3) and was significantly higher for genes involved in cytokine-cytokine interactions (p = 4.22 × 10−15), many other immune system pathways, and genes identified in genome-wide association studies for various traits including behavioral disorders and cancer. This study provides a thorough characterization of cis- and trans-h2 estimates of gene expression, which is of value for interpretation of GWAS and gene expression studies.
Subject terms: Gene expression, Gene regulatory networks, RNA sequencing
Introduction
Individual differences in RNA expression may result from variation in environmental exposures, stochastic variation, age, sex and genotype differences [1] and thereby may also be involved in the widely observed contribution of DNA, age and sex to the etiology of complex diseases [2–5]. Quantifying human transcriptomic heritability (h2) is of interest for transcriptomic and genomic studies. For example, one possible reason for the association of gene expression with a certain phenotype is that DNA variants influence the amount of gene expression through expression quantitative trait loci (eQTLs). Gene expression can have substantial h2 not explained by eQTLs [6], and therefore the h2 of gene expression and corresponding eQTL findings can be considered complementary to transcriptomic and genomic studies.
The h2 of whole blood gene expression has been established previously for genome-wide transcriptomic data generated by microarray technology and by RNA-Seq. For array technology, Wright et al. [7] found a mean gene expression h2 of 0.10 (SD = 0.14, N = 2752, 18,392 genes) from modeling data assessed in mono- and dizygotic twin pairs. Local identity-by-descent (IBD) analyses, which provide an estimate of variance of gene expression explained by genetic relatedness, resulted in a mean local h2 of 0.03, explaining 23% of the total heritability. A population-based study (N = 2765) by Lloyd-Jones et al. [6] found mean estimates of h2 of microarray-based gene expression of 0.19 with a local h2 of 0.06, resulting in a mean proportion of genetic variance explained by all eQTLs of 31%. Viñuela et al. [8] estimated a mean of blood-derived h2 of 0.23 in a twin-based sample (N = 855) for RNA-Seq-based data.
Here, we analyzed RNA-Seq (Illumina Hiseq2000) data from 459 MZ twin pairs, 150 DZ twin pairs, 24 relatives of twin pairs, and 255 unrelated participants, leading to a total dataset with genotype and expression data of 1497 adult participants (998 females) from the Netherlands Twin Register [9–11] (Supplementary Table 1). Our RNA-Seq dataset contained a large (>60%) sample overlap with the microarray-based dataset from Wright et al. [7], allowing a reliable comparison between microarray and RNA-Seq-based h2 estimates. We first estimated twin-based h2 of gene expression by making use of the classical twin design [12]. Next, we simultaneously estimated the variance explained by a genetic relationship matrix (GRM)-containing SNPs in a 250 kilobase (kb) cis-window of the gene (h2cis), and the variance explained by a second GRM including only closely related individuals (h2res) using GCTA [13]. Together, h2cis and h2res constitute the total heritability (h2total). We performed cis- and trans-eQTL analyses of the same samples and compared h2 and eQTL findings to test for consistency.
Methods
Participants
RNA samples were obtained from 1497 participants from the Netherlands Twin Registry (NTR) and included 459 complete MZ twin pairs and 150 complete DZ twin pairs, 24 relatives of twin pairs, and 255 unrelated participants. NTR is a longitudinal cohort study of twins and their families [10, 14]. The age of the participants ranged from 17.6 to 79.6 years old (mean = 36.7, SD = 14.0), 67% of the sample was female. The data used for this study largely overlap (60%) with those used in an earlier study [7]. See Supplementary Table 1 for a description of the samples.
RNA extraction and sequencing
Venous samples were drawn in the morning after an overnight fast. Heparinized whole blood samples were transferred within 20 min of sampling into PAXgene Blood RNA tubes (Qiagen, Valencia, CA, USA) and stored at −20 °C. Total RNA from whole blood was depleted of globin transcripts using the Ambion GLOBINclear kit and subsequently processed for sequencing using the Illumina TruSeq version 2 library preparation kit. Paired-end sequencing of 2 × 50-bp reads was performed using the Illumina HiSeq 2000 platform, pooling ten samples per lane and aiming for >15 million read pairs per sample. Adapters were identified and clipped, and low-quality read ends were trimmed (min length 25, min quality 20 Read alignment was performed using STAR 2.3.0e against the Genome of the Netherlands (GoNL) reference panel [15]. Expression was using Ensembl v.71 annotation (which corresponds to GENCODE v.16). Overlapping exons (on either of the two strands) were merged into meta-exons, and expression was quantified for the whole meta-exon, resulting in base counts per exon or meta-exon. Gene expression, as base count per gene, was calculated as the sum of the expression values for all exons of each gene (excluding meta-exons). This pipeline is explained in detail in Zhernakova et al. [16].
Gene expression values per gene were ranked and mapped to a normal distribution with mean 0 and SD 1, after which values were corrected for sex, age, and cell counts: monocyte, lymphocyte, eosinophil, basophil, neutrophil and red blood cell counts, and 27 measurement batches. We then performed a principal-component analysis, and values were corrected on only the first principal component, which was not heritable based on comparison of goodness of fit between different twin-based structural equation models (p = 0.74). This PC explained 16.5% of the variance in the data (see Supplementary Figs. 1 and 2). Finally, expression values were centered and subsequently scaled by dividing these values by their respective standard deviations. Analyses of both classical twin modeling-based and identity-by-state (IBS)-based h2 were based on this final dataset.
Genotype data
Within the NTR, genotype information is available for 15,111 individuals for four different genotyping arrays (Affymetrix 6.0 (N = 11,781), Affymetrix Perlegen 5.0 (N = 1265), Illumina 660 (N = 1439) and Illumina Omni Express 1 M (N = 257), as well as sequence data from the Netherlands reference genome project GONL (BGI full sequence at 12 × (N = 368; [17]). Samples were removed if they had a genotype call rate below 90%, heterozygosity fell outside the range of −0.075 to 0.075, gender and IBS status mismatch occurred, or if the Mendelian error rate was larger than 5 standard deviations from the mean of all samples and for samples measured on Affymetrix 6.0 when the contrast quality control value was smaller than 0.40. Quality control of the SNPs was done for each platform separately, with SNPs being removed when they could only be aligned to the forward strand of the reference panel, the allele frequencies differed more than 10% with the reference allele, minor allele frequency (MAF) was below 0.005, Hardy–Weinberg equilibrium (HWE) test p < 10−12, and a genotype call rate of <0.95. The data of the different genotyping methods, except GONL sequence individuals, were subsequently merged into a single dataset. The missing SNP genotypes between each platform were imputed to the GONL reference data. Filtering of the imputed dataset included the removal of SNPs which were significantly associated with a single genotyping platform, if the allele frequency difference differed more than 10% with the GONL reference set, HWE p < 10−5, Mendelian error rate >mean + 5 SD and if the imputation quality (R2) was below 0.90. After filtering the GONL samples were readded to a cross-platform imputed dataset that includes 1,261,818 SNPs. We did not perform additional genotype quality control after subsetting the individuals with RNAseq.
Statistical methods
We employed two methods for estimating total heritability, a classical twin modeling-based approach, and an IBS-based approach. The classical twin modeling approach requires information on relatedness between subjects, while the IBS-based approach quantifies the genetic relatedness between subjects based on their genome-wide genetic data. For both methods, only autosomal genes were considered.
Classical twin modeling
Based on the resemblance of MZ and DZ twin pairs, the variance of the expression of each gene can be decomposed into additive genetic, common (or shared) environmental and unique environmental variance. Classical twin-based modeling was done in the structural equation modeling (SEM) R-package OpenMx [18, 19]. Models were fitted to decompose the variance of gene expression due to additive genetic (A), shared environmental (C), and unique environmental (E) effects. Additive genetic effects combine all the effects of genetic variants influencing gene expression. Shared environmental variance represents the proportion of variance explained by effects are shared by both members of a twin pair. Unique environmental variance results from environmental effects that are not shared by twins. We used a standard ACE model assuming dizygotic twins have an average IBD sharing of 0.5 across the genome, and monozygotic twins share an IBD of 1. Parameters were estimated by maximum likelihood (ML). We restricted the estimates to be positive. The results of these analyses provide the ML estimates of variance components. The comparison of ACE and CE models gave an estimate for the significance of the A-component.
IBS-based analysis
Techniques to quantify genetic similarity of ‘unrelated’ individuals who are genotyped for a large number of SNPs across the entire genome have been developed to estimate h2 due to SNPs. In software packages such as GCTA [13] the relatedness among individuals based on measured SNPs can be combined with known genetic relatedness in relatives in a two-variance component linear mixed model [20], in which simultaneous estimation of SNP heritability and total h2 is feasible. We used NTR genotype information for the 1497 individuals for whom both expression- and genotype data were available. A GRM was created for each cis-window of a gene, defined by the coordinates of a gene with 250 kb flanking area on each side. This GRM is referred to as the cis-GRM and represents the variance explained by all measured SNPs (and SNPs tagged by these measured SNPs) in a cis-window around the gene of 250 kb. Variance explained by this cis-GRM is referred to as cis-h2 (h2cis). The mean number of SNPs in cis-GRMs was 223.2 (SD = 119.9) (see Supplementary Fig. 3). A second GRM including closely-related individuals (that is, a genetic correlation > 0.05) was created for the autosomes. This GRM is referred to as the residual GRM. This GRM had all off-diagonal elements below 0.05 set to 0, to remove distant relatedness from the matrix. Variance explained by this residual GRM is referred to as residual h2 (h2res). The sum of h2cis and h2res is h2total (h2total = h2cis + h2res). As the cis-GRM is based on a limited number of SNPs there is substantial power to detect the genetic effects in cis [21], the second GRM will absorb all genetic variance not explained by the SNPs in the cis-window, or in high LD with SNPs in the cis-window. Note that due to the presence of a large number of related individuals this GWAS will capture genetic variance tagged by substantial IBD sharing and thus the sum of the two effects will be approximately equal to the heritability estimated in a twin study.
A total of 52,844 genes were analyzed, and subsequently filtered for being protein coding, having read counts above zero in at least 85% samples in each zygosity group (e.g. expressed in in at least 780 MZ twins and 255 DZ twins), a median expression count above 10, and more than 20 SNPs in the cis-window, resulting in an analysis of 11,353 genes (Supplementary Table 2).
Annotation and enrichment
We employed multiple annotation steps to interpret h2 estimates. We tested whether h2 was correlated with gene expression level, gene length, GC content or several loss-of-function scores obtained from Lek et al. [22] using linear regression. Gene locations and lengths were downloaded on 2017-12-28 using the Biomart community portal [23] using data from Ensembl [24]. We investigated genes for which the expression correlates highly (>0.8) with other genes.
In addition, we tested whether heritable genes are overrepresented in the canonical gene pathways from the molecular signature database (MSIGDB):: KEGG, REACTOME, BIOCARTA pathways downloaded from http://software.broadinstitute.org/gsea/downloads.jsp (c2.cp.v6.1) and genes identified in GWAS for immune diseases, mental or behavioral disorders, cardiovascular diseases, or cancer (extracted from the GWAS catalog [25] (as of September 2018) with the search terms: ‘immune system disease’, ‘mental or behavioral disorder’, ‘cardiovascular disease’ or ‘cancer’, respectively). For each pathway/gene group, a Wilcoxon test was performed between the median heritability of the genes in the pathway and the median heritability of the genes outside the pathway. Comparisons were made with previous analyses of h2 of gene expression from a recent whole blood-based RNA-Seq study by Battle et al. [26] and the GTEx project [27], which published heritability estimates for gene expression in adipose tissue (subcutaneous), tibial artery, heart (left ventricle), lung, muscle (skeletal), tibial nerve, skin (sun-exposed), thyroid, and whole blood.
eQTL analysis
The 1497 gene-level RNA samples with the same preprocessing as described above which were also used for heritability analysis, were also used for cis- and trans-eQTL analysis. For this analysis, the same cross-platform imputed dataset that includes 1,261,818 SNPs as described above, was filtered at MAF >0.01 and HWE <1 × 10−3, resulting in 1,239,670 SNPs.
For cis-eQTL analysis all associations between DNA variants and genes at distance <250 kb were computed, for trans-eQTL analysis all SNP - gene pairs at distance >250 kb. eQTL effects were detected with a mixed linear model approach using fastGWA a implemented in GCTA (https://cnsgenomics.com/software/gcta/#fastGWA, https://www.biorxiv.org/content/10.1101/598110v1).
For fastGWA, first a GRM is built using the –make-grm option in GCTA. Then, a sparse GRM is built using the option –make-bK-sparse 0.05. fastGWA is then run using this sparse GRM, with expression level as the dependent variable and SNP genotype values as independent variable. Correction for multiple testing was done using FDR, for cis- and trans-eQTL analysis separately, resulting in a P-value threshold of 1 × 10−5 for cis-eQTLs, and 1.5 × 10−7 for trans-eQTLs. We are aware that FDR used like this may result in more false positives than 5%, however, we are merely interested in the overlap between heritability and eQTL analysis and do not draw any conclusions on the amount of identified eQTLs.
Simulation
To gain insight in the performance of our models, we simulated twin-based phenotype data with prespecified heritabilities and tested whether our estimations were in accordance. We ran OpenMX and GCTA models using simulated phenotypes and real genotype data, with different values for variance components (see Supplementary Figs. 6 and 7).
Results
The gene-level whole blood RNA-Seq data from 1497 participants, including 459 MZ twin pairs and 150 DZ twin pairs, originally contained 52,844 genes (Supplementary Table 2). After filtering (see “Methods”), 11,409 protein coding genes that were expressed in at least 85% of the samples were left for analysis. In these genes, twin-based h2 for each gene was estimated based on the classical twin design (see Methods). Next, IBS-based methods were applied to compute SNP heritability.
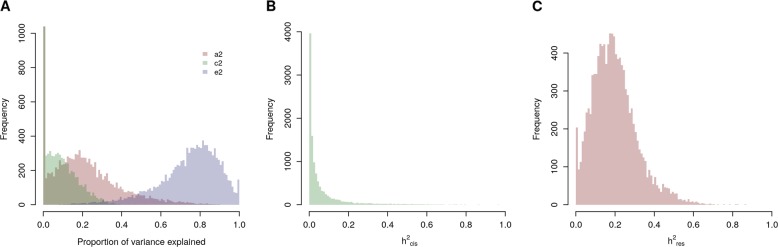
For the twin-based analyses, a genetic structural equation model [28] that included additive genetic, common, and unique environmental factors was fitted to the expression data from each gene. The mean h2 (the standardized estimate of the contribution of additive genetic factors) was estimated at .20, the standardized mean contribution of shared environment at 0.05, and the standardized mean unshared environment at 0.74. There was a considerable spread in estimates, with estimates ranging between 0 and 1, except for the common environment component which had a maximum of 0.51 (Fig. 1 and Table 1).
Fig. 1.
Histograms of estimates of heritability of gene expression. a Classical twin modeling-based estimates of each gene. Based on the resemblance of MZ and DZ twin pairs, the variance of the expression of each gene was decomposed into additive genetic, common (or shared) environmental, and unique environmental variance. A standard ACE model was used, assuming dizygotic twins have an average IBD sharing of 0.5 across the genome, and monozygotic twins share an IBD of 1. Parameters were estimated by maximum likelihood (ML). b IBS-based h2cis estimates of each gene, i.e. variance explained by a GRM created for each cis-window of a gene, defined by the coordinates of a gene with 250 kilobase (kb) flanking area on each side. c IBS-based h2res estimates of each gene, i.e. variance explained by a GRM including closely related individuals for the autosomes with all off-diagonal elements below 0.05 set to 0
Table 1.
Estimates for classical twin modeling-based and IBS-based heritability of gene expression
| Variance component | Min | Median | Mean | Max |
|---|---|---|---|---|
| A | 0 | 0.1793 | 0.2042 | 0.8988 |
| C | 0 | 0 | 0.0532 | 0.5125 |
| E | 0.0800 | 0.7702 | 0.7426 | 1 |
| h2cis | 0.0001 | 0.0212 | 0.0638 | 0.9666 |
| h2res | 0.0001 | 0.1848 | 0.1980 | 0.8655 |
| h2total | 0.0001 | 0.2338 | 0.2618 | 0.9874 |
Genes were filtered for being protein coding, having less than 50 DZ twins with zero expression counts, a median expression count above 10, and more than 20 SNPs in the cis-window, resulting in a total of 11,353 genes tested. Classical twin-based modeling was done in the structural equation modeling (SEM) R-package OpenMx [18, 19]. Models were fitted to decompose the variance of gene expression due to additive genetic (A), shared environmental (C), and unique environmental (E) effects. h2cis is the variance explained by measured SNPs in a cis-window around the gene of 250 kb. h2res is the variance explained by genome-wide closely related individuals
Since estimates for the contribution of common environment on average were low, we proceeded with an IBS-based approach that did not take into account common environment. Gene expression is controlled by both local (cis) eQTLs and non-local (trans) eQTLs. Therefore, when studying the genetic component of expression in terms of heritability, we also make a distinction between the genetic component that is close to the gene (h2cis), and the genetic component that is not close to the gene (h2res). h2cis consisted of the variance explained by SNPs in a 250-kb cis-window of the gene and h2res the variance explained by genome-wide close relatedness. From the IBS approach we obtained heritability estimates for 11,353 out of 11,409 genes. The correlation between IBS-based total h2 (h2cis + h2res) and classical twin modeling-based h2 was 0.98 (p < 10−308, Supplementary Fig. 4). The IBS-based approach resulted in a mean h2res of 0.20 and a mean h2cis of .06 (Fig. 1 and Table 1), summing up to a h2total of 0.26. We found 721 genes to have a Bonferroni-corrected significant h2cis (p < 4.40 × 10−6), all of which had a h2cis larger than 0.16. We found 5636 genes to have a Bonferroni-corrected significant h2res (p < 4.40 × 10−6), all of which had a h2res larger than 0.01 (Supplementary Table 3, Supplementary Fig. 5). The mean contribution of h2cis to the total h2 (mean h2cis/(mean h2cis + mean h2res)) was 20%, with a range from 0 to 100%. The correlation between h2res and h2cis was 0.06 (p = 6.36 × 10−10). We found good performance of our models, slightly underestimating local heritability (see Supplementary Figs. 6 and 7).
Correlation of gene expression h2 between tissues
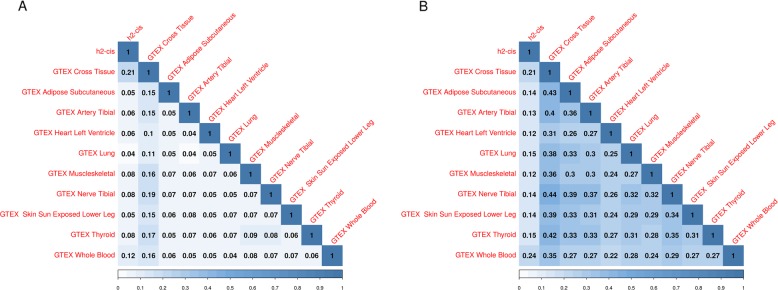
A whole blood-based RNA-Seq study by Battle et al. [26] published heritability estimates due to regulatory genetic variation (N = 922). Estimates of heritability due to local genetic variation from this study correlated 0.81 with h2cis from our study (p < 10−308, Spearman correlation, Supplementary Fig. 8). eQTL studies have shown that gene expression in different tissues is regulated by DNA by partially overlapping, and partially unique QTLs (https://science.sciencemag.org/content/348/6235/648). To study the extent to which the total local genetic component of gene expression is similar between tissues, we looked at h2 estimates of gene expression in The Genotype-Tissue Expression (GTEx). The GTEx consortium, (N = 422) [29] reported h2 estimates for gene expression from RNA-Seq in adipose tissue (subcutaneous), tibial artery, heart (left ventricle), lung, muscle (skeletal), tibial nerve, skin (sun-exposed), thyroid, and whole blood, estimating local h2 both unique for a tissue (tissue specific) and heritability shared between tissues (tissue wide). Our estimates of h2cis showed significant correlations with local h2 in every measured tissue, in both tissue-wide (ρ > 0.12) and tissue-specific heritability (ρ > 0.03) estimates. The strongest correlation was found between h2cis and tissue-wide heritability of whole blood-derived gene expression (ρ = 0.24, p = 1.99 × 10−144, Fig. 2, Supplementary Table 4). Heritability estimates reported in the GTEx study correlated <.44 between tissues in tissue-wide estimates and <0.19 in tissue-specific estimates.
Fig. 2.
Spearman correlations between h2cis and estimates of GTEx per-tissue heritabilities [27]. a Tissue-specific heritabilities. b Tissue-wide heritabilities
Correcting h2cis for median read count only showed marginal effects on correlations. There were 614 genes that were highly expressed in all tissues in GTEx (above the 8th decile). These genes showed decreased h2cis (p = 5.58 × 10−12, mean = 0.04, median = 0.02) and increased h2res (p = 7.77 × 10−14, mean = 0.23, median = 0.22) compared to the full set of tested genes.
Pathway analyses
To annotate gene expression heritability, we studied if certain gene pathways have higher average heritability than expected. In order to so we tested for enrichment of h2res and h2cis in canonical gene pathways covering a broad range of biological pathways that are well curated (KEGG, REACTOME, BIOCARTA). The expression of genes identified in GWAS is likely to be under genetic control: to test if this is the case for gene expression in blood, we also tested if h2res and h2cis is enriched in genes identified in genome-wide association studies (GWAS) for immune diseases, mental and behavioral disorders, cardiovascular diseases, or cancer (extracted from the GWAS catalog [25]) to cover GWAS findings for a broad range of diseases. Enrichment analysis were performed before and after correcting h2 for median gene expression per gene.
We observed significantly higher h2res (false discovery rate (FDR) <0.05) in 343 canonical pathways (top hit: KEGG cytokine-cytokine interaction, p = 4.22 × 10−15), and in genes identified in GWAS for immune diseases (p = 4.99 × 10−13), mental disorders (p = 1.79 × 10−7), cancer (p = 5.53 × 10−5) and cardiovascular diseases (p = 1.06 × 10−9). After correction for mean gene expression, h2res was significantly higher in 125 canonical pathways (top hit: KEGG cytokine-cytokine interaction, p = 6.66 × 10−15) and in genes identified in GWAS for immune diseases (p = 2.33 × 10−10), mental disorders (p = 3.63 × 10−10), cancer (p = 7.10 × 10−6) and cardiovascular diseases (p = 9.69 × 10−10).
We found significantly higher h2cis (FDR <0.05) in 6 canonical pathways (top hit: KEGG lysosome, p = 1.41 × 10−7), and in genes identified in GWAS for immune diseases (p = 6.71 × 10−4), mental disorders (p = 7.13 × 10−4), cancer (p = 1.09 × 10−7) and cardiovascular diseases (p = 3.24 × 10−5). After correction for mean gene expression, h2res was significantly higher in 10 canonical pathways (top hit: KEGG lysosome, p = 5.79 × 10−9) and in genes identified in GWAS for immune diseases (p = 9.13 × 10−5), for mental disorders (p = 2.36 × 10−3), cancer (p = 3.15 × 10−7) and cardiovascular diseases (p = 1.16 × 10−5) (see Table 2 and Supplementary Table 5).
Table 2.
Gene pathway enrichment of heritable genes in certain gene pathways (KEGG, REACTOME, BIOCARTA) and genes identified in GWAS for immune diseases, mental or behavioral disorders, cardiovascular diseases, or cancer (extracted from the GWAS catalog [25])
| h2res enrichment p-value | h2cis enrichment p-value | |
|---|---|---|
| All immune-related GWAS genes | 4.99 × 10−13 | 6.71 × 10−4 |
| All immune-related GWAS genesa | 2.33 × 10−10 | 9.13 × 10−5 |
| All psychiatric-related GWAS genes | 1.79 × 10−7 | 7.13 × 10−4 |
| All psychiatric-related GWAS genesa | 3.63 × 10−10 | 2.36 × 10−3 |
| All cancer-related GWAS genes | 5.53 × 10−5 | 1.09 × 10−7 |
| All cancer-related GWAS genesa | 7.10 × 10−6 | 3.15 × 10−7 |
| All cardiovascular GWAS genes | 1.06 × 10−9 | 3.24 × 10−5 |
| All cardiovascular GWAS genesa | 9.69 × 10−10 | 1.16 × 10−5 |
| KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION | 4.22 × 10−15 | |
| KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTIONa | 6.66 × 10−15 | |
| KEGG_LYSOSOME | 1.41 × 10−7 | |
| KEGG_LYSOSOMEa | 5.79 × 10−9 |
ah2 corrected for median read count. See also Supplementary Table 5.
Gene expression h2 correlations
In order to identify physiological gene properties that are correlated with gene expression heritability, we evaluated the correlation between heritability and gene expression level, gene length, GC content and several loss-of-function scores, see Table 3). We found a significant association of h2res and h2cis with median read count (P = 6.81 × 10−276, P = 2.15 × 10−2, respectively) and GC content of a gene (P < 1.80 × 10−115, P = 4.02 × 10−26, respectively). After correcting for median read count, GC content was still significantly correlated with h2res and h2cis (P = 2.27 × 10−3, P = 6.11 × 10−28, respectively). The length of a gene was significantly correlated with h2res, with longer genes having a slightly higher h2res (P = 4.94 × 10-8). Gene length did not influence h2cis (P = .50). A high intolerance to LoF or high probability of loss-of-function (pLI) did not significantly influence h2 estimates.
Table 3.
Predictors of h2
| h2res | h2cis | |||||
|---|---|---|---|---|---|---|
| Predictor | Estimate | P-value | r-squared | Estimate | P-value | r-squared |
| Median Read count | 5.21 × 10−2 | 6.81 × 10−276 | 1.16 × 10−1 | −3.76 × 10−3 | 2.15 × 10−2 | 8.16 × 10−4 |
| NA | NA | NA | NA | NA | NA | |
| GC content | 3.10 × 10−3 | 1.80 × 10−115 | 4.81 × 10−2 | 1.58 × 10−3 | 4.02 × 10−26 | 8.85 × 10−3 |
| GC contenta | 2.27 × 10−3 | 1.07 × 10−69 | 2.83 × 10−2 | 1.64 × 10−3 | 6.11 × 10−28 | 9.85 × 10−3 |
| LOF score | 7.24 × 10−3 | 8.16 × 10−18 | 3.14 × 10−2 | −3.19 × 10−3 | 8.52 × 10−5 | 2.55 × 10−3 |
| LOF scorea | −1.65 × 10−4 | 8.04 × 10−1 | 3.95 × 10−6 | 6.10 × 10−4 | 4.08 × 10−1 | 6.18 × 10−5 |
| pLI score | 3.54 × 10−2 | 7.09 × 10−19 | 2.47 × 10−2 | −2.37 × 10−2 | 1.27 × 10−9 | 7.41 × 10−3 |
| pLI scorea | −1.73 × 10−3 | 5.98 × 10−1 | 1.50 × 10−5 | −3.00 × 10−3 | 4.13 × 10−1 | 1.29 × 10−4 |
| pNull | −4.59 × 10−2 | 2.86 × 10−17 | 2.54 × 10−2 | 1.72 × 10−2 | 6.30 × 10−4 | 1.90 × 10−3 |
| pNulla | −4.31 × 10−3 | 2.99 × 10−1 | 7.50 × 10−5 | −3.54 × 10−3 | 4.45 × 10−1 | 7.75 × 10−5 |
| pRec | −1.30 × 10−2 | 3.25 × 10−3 | 1.43 × 10−3 | 1.58 × 10−2 | 1.96 × 10−4 | 3.34 × 10−3 |
| pReca | −1.64 × 10−3 | 6.50 × 10−1 | 5.74 × 10−5 | 7.37 × 10−3 | 6.62 × 10−2 | 4.33 × 10−4 |
| Gene length | 1.80 × 10−8 | 8.10 × 10−2 | 2.12 × 10−4 | 9.54 × 10−9 | 3.86 × 10−1 | 2.30 × 10−4 |
| Gene lengtha | 4.94 × 10−8 | 4.13 × 10−7 | 2.26 × 10−3 | 7.48 × 10−9 | 4.96 × 10−1 | 1.59 × 10−4 |
We tested whether h2 could be predicted by gene expression level, gene length, GC content or several loss-of-function scores obtained from Lek et al. [22]
LOF loss-of-function, PLI probability of loss-of-function, pNull completely tolerant of loss-of-function, pRec intolerant of two loss-of-function variants
ah2 corrected for median read count
Relation of h2cis with strength of cis-eQTLs
Gene expression can have substantial h2 not explained by eQTLs [6]. In order to study the overlap between heritability and eQTL results, we performed eQTL analysis in the same sample (see Methods) and found 5249 genes with a significant cis-eQTL (p-value threshold 1.5 × 10−7 for a FDR of 5%). In addition, we found a significant association between h2cis and the beta of the corresponding top cis-eQTLs (ρ = 0.7644, p < 10−308). We also tested the correlation between h2cis and the presence of cis-eQTLs in results from Zhernakova et al. [16], who performed RNA-Seq-based eQTL analysis in an independent sample (N = 2116 unrelated adults). There was a strong correlation between h2cis and the Z-score of the strongest eQTL (ρ = 0.75, p < 10−308) (Supplementary Fig. 9).
Relation of h2res with strength of trans-eQTLs
We performed eQTL analysis in the same sample for SNPs outside a cis-window of 250 kb around each gene, resulted in 2433 genes with a significant trans-eQTL (p-value threshold 1.5 × 10−7 for a FDR of 5%). Estimates of h2res correlated with the beta of the corresponding top trans-eQTLs (ρ = .04, p = 1.89 × 10−3). The mean p-value of trans-eQTL of genes with h2res > 0.4 was 5.51 × 10−9 (median = 1.44 × 10−9).
Comparing RNA-Seq and micro-arrays: relation of mean expression levels with heritability
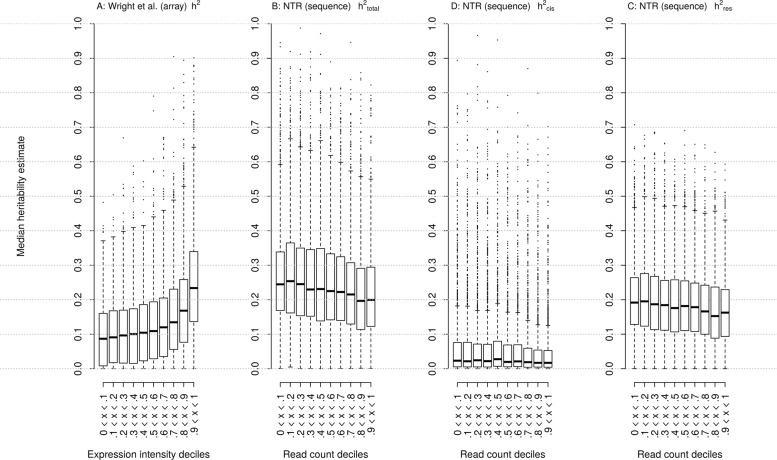
The difference in heritability estimates between RNAseq and array data results, especially with gene expression intensity as a factor, is a valuable comparison to distinguish noise or bias that is inherent to a specific technique. As RNAseq is a more expensive method, it is informative to know if and in which situations this technique offers advantages to answer a particular research question. From our original results on 52,844 genes, we selected the 12,070 genes that were measured both with Affymetrix expression arrays in the study by Wright et al. [7] and with RNA-Seq with read counts above zero in at least 85% of samples in each zygosity group. In contrast to previous analyses, we did not apply any additional filtering (i.e. minimum median read count per gene, protein coding genes only, minimum of SNPs per GRM to allow for an unbiased comparison. In the RNA-Seq data, the Spearman correlation of mean expression with h2total was −0.13 after correcting for covariates (see Methods) (p = 7.49 × 10−45). In the array-based results from Wright et al. [7] the Spearman correlation between mean expression and h2 was 0.28 (p = 2.08 × 10−219) after correcting for covariates.
Using this selection of 12,070 genes, mean h2res, h2cis, h2total, and h2total results from Wright et al. [7] were 0.19, 0.06, 0.25, and 0.14, respectively. When we partitioned mean expression levels into 10 deciles for both microarray and RNA-Seq data, we saw that h2res estimates were also higher in RNA-Seq data for almost all deciles (see Fig. 3 and Supplementary Table 6), with the difference being largest in the lowest deciles (p = 1.06 × 10−72). In the highest decile of expression, the h2 of array-based expression was higher (p = 2.00 × 10−6). This suggests that the resolution of RNA-Seq is better able to capture variation in low to moderately expressed genes. Genes measured by both array and RNA-Seq were mostly in the same or nearest decile (see Supplementary Fig. 10). Estimates of h2cis showed a slight negative correlation with median expression level in the RNA-Seq data eQTLs (ρ = −0.04, p < 5.79 × 10−5).
Fig. 3.
Heritability estimates for genes divided into groups based on gene expression levels per decile. a Total heritability estimates from Wright et al. [7] b IBS-based h2total estimates. c IBS-based h2cis estimates. d IBS-based h2res estimates
Discussion
The present study estimated the h2 of gene expression in RNA-Seq-based expression data by making use of the different genetic relatedness of mono- and dizygotic twins and an IBS approach. The mean of total gene expression h2 (0.26) was substantially higher than found with the microarray-based study by Wright et al. with largely overlapping RNA samples. This was also the case with a direct comparison of 12,070 genes that were measured both with Affymetrix expression arrays in the study by Wright et al. [7] and with RNA-Seq, where mean RNA-Seq-based h2total was 0.25 and mean microarray-based h2total was 0.14 (p < 10−308). Heritability estimates in RNA-Seq did not increase with gene expression level, as opposed to the results from microarray data. This suggests RNA-Seq measurements are less noisy, in particular in genes with low expression, as compared to microarrays measures.
We estimated h2 of gene expression at 0.20 for mean h2res, and at 0.06 for mean h2cis. This resulted in a relative contribution of h2cis to h2total of 20%. This is in line with earlier findings by Wright et al. [7] (relative contribution of local IBS-driven h2 of 23%) and Lloyd-Jones et al. [6] (proportion of h2 explained by cis-eQTL of 0.31). Since local variants in the cis-window of a gene only explain 20% on average of the total h2, and the number of genes for which the majority of heritability stems from genetic relatedness in the cis-window is very low, our findings indicate that loci outside the cis-window of a gene or rare local variants explain a significant proportion of total h2 of gene expression. This is strengthened by our finding that the strength of a trans-eQTL is correlated with h2res.
The h2cis estimates correlated strongly with estimates from an independent sample (ρ = 0.81, whole blood, N = 922) [26], but much less so in the smaller GTEx dataset [27] (ρ = 0.24, whole blood, N = 449). This shows that reasonably large sample sizes are needed for accurate h2 estimation. However, even with the small sample size used in GTEx we found significant correlations between h2cis in whole blood and h2cis in the other tissues, which is in line with the finding that cis-eQTLs are partially shared between tissues [29].
We found a significantly higher h2res in 125 canonical pathways, with the strongest enrichment for genes in the KEGG cytokine-cytokine interaction pathway, and many other immune system pathways (including KEGG Innate Immune system (p = 8 × 10−12), REACTOME interferon signaling (p = 2 × 10−8) and KEGG natural killer cell cytotoxicity (p = 2 × 10−7)) [30].
Both h2cis and h2res were higher in all genes identified in GWAS for immune diseases, mental disorders, and cardiovascular diseases, although interestingly the enrichment was much stronger for h2res. This indicates that for genes associated with a disease through GWAS, expression in blood is not only locally regulated but also enriched with genome-wide SNP signal as reflected in the high average h2res, suggesting that SNPs found in GWAS are influencing expression of genes outside the cis-window of the gene. A high intolerance to loss-of-function (LoF) or high probability of loss-of-function (pLI) did not significantly influence h2 estimates, suggesting that increased mutational load in a gene increases the genetic variation as much as it influences variation in gene expression and therefore does not influence h2.
The comparison of mean h2 of genes across different RNA measurement techniques can be viewed as a proxy for the comparison of the measurement error between techniques. An increase in measurement error always implies a decrease in h2 (because measurement error introduces random divergence within twin pairs). Consider for example the h2 of probes conditioned on the median expression level. When comparing the h2res estimates obtained based on RNA-Seq with those obtained based on Affymetrix expression arrays [7], it becomes apparent that micro-array-based estimates of gene expression heritability are associated with gene expression levels. The fact that differential measurement error conditional on expression level plays less of a role for RNA-Seq data ensures that variation across genes reflects biological signal. The slight negative correlation we found of h2cis with median expression level in the RNA-Seq data eQTLs (ρ = −0.04, p < 5.79 × 10−5) is counterintuitive, difficult to interpret and presumably not meaningful.
If, or when, researchers eventually examine the h2 of RNA expression levels in single cells, or nuclei, an inspection of the relationship between median expression levels and h2 can be used to detect this source of differential measurement error.
In summary, this study shows possible advantages of h2res-informed trans-eQTL discovery, reproducibility of h2cis, and the benefits of using RNA-Seq for estimating heritability of low-expressed genes.
Supplementary information
Acknowledgements
We very warmly thank all participants in the study. This study makes use of data in the Netherlands Twin Register with prof. D.I. Boomsma as principle investigator.
Funding
This work was performed within the framework of the BBMRI - NL Consortium, a research infrastructure financed by the Dutch government (NWO, nos. 184.021.007 and 184.033.111). Genotyping was made possible by grants from NWO/SPI 56-464-14192, Genetic Association Information Network (GAIN) of the Foundation for the National Institutes of Health, Rutgers University Cell and DNA Repository (NIMH U24 MH068457-06), the Avera Institute, Sioux Falls (USA) and the National Institutes of Health (NIH R01 HD042157-01A1, MH081802, Grand Opportunity grants 1RC2 MH089951 and 1RC2 MH089995) and European Research Council (ERC-230374). DIB acknowledges her KNAW Academy Professor Award (PAH/6635).
BIOS Consortium
Bastiaan T. Heijmans12, Peter A. C. ’t Hoen13, Joyce van Meurs14, Aaron Isaacs15, Rick Jansen16, Lude Franke17, Dorret I. Boomsma18, René Pool18, Jenny van Dongen18, Jouke J. Hottenga18, Marleen M. J. van Greevenbroek19, Coen D. A. Stehouwer19, Carla J. H. van der Kallen19, Casper G. Schalkwijk19, Cisca Wijmenga17, Lude Franke17, Sasha Zhernakova17, Ettje F. Tigchelaar17, P. Eline Slagboom12, Marian Beekman12, Joris Deelen12, Diana van Heemst20, Jan H. Veldink21, Leonard H. van den Berg21, Cornelia M. van Duijn15, Bert A. Hofman22, Aaron Isaacs15, André G. Uitterlinden14, Joyce van Meurs14, P. Mila Jhamai14, Michael Verbiest14, H. Eka D. Suchiman12, Marijn Verkerk14, Ruud van der Breggen12, Jeroen van Rooij14, Nico Lakenberg12, Hailiang Mei23, Maarten van Iterson12, Michiel van Galen13, Jan Bot24, Dasha V. Zhernakova17, Rick Jansen16, Peter van’t Hof23, Patrick Deelen17, Irene Nooren24, Peter A. C. ’t Hoen13, Bastiaan T. Heijmans12, Matthijs Moed12, Lude Franke17, Martijn Vermaat14, Dasha V. Zhernakova17, René Luijk12, Marc Jan Bonder17, Maarten van Iterson12, Patrick Deelen17, Freerk van Dijk25, Michiel van Galen13, Wibowo Arindrarto23, Szymon M. Kielbasa26, Morris A. Swertz25, Erik. W van Zwet26, Rick Jansen16, Peter-Bram ’t Hoen13, Bastiaan T. Heijmans12
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The NTR study was approved by the Central Ethics Committee on Research Involving Human Subjects of the VU University Medical Center, Amsterdam (institutional review board [IRB] number IRB-2991 under Federal wide Assurance 3703; IRB/institute code NTR 03-180). All participants provided written informed consent.
Footnotes
Members of the BIOS Consortium are listed at the end of the paper.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Klaasjan G. Ouwens, Email: klaasjan.ouwens@vu.nl
BIOS Consortium:
Bastiaan T. Heijmans, Peter A. C. ’t Hoen, Joyce van Meurs, Aaron Isaacs, Rick Jansen, Lude Franke, Dorret I. Boomsma, René Pool, Jenny van Dongen, Jouke J. Hottenga, Marleen M. J. van Greevenbroek, Coen D. A. Stehouwer, Carla J. H. van der Kallen, Casper G. Schalkwijk, Cisca Wijmenga, Lude Franke, Sasha Zhernakova, Ettje F. Tigchelaar, P. Eline Slagboom, Marian Beekman, Joris Deelen, Diana van Heemst, Jan H. Veldink, Leonard H. van den Berg, Cornelia M. van Duijn, Bert A. Hofman, Aaron Isaacs, André G. Uitterlinden, Joyce van Meurs, P. Mila Jhamai, Michael Verbiest, H. Eka D. Suchiman, Marijn Verkerk, Ruud van der Breggen, Jeroen van Rooij, Nico Lakenberg, Hailiang Mei, Maarten van Iterson, Michiel van Galen, Jan Bot, Dasha V. Zhernakova, Rick Jansen, Peter van’t Hof, Patrick Deelen, Irene Nooren, Peter A. C. ’t Hoen, Bastiaan T. Heijmans, Matthijs Moed, Lude Franke, Martijn Vermaat, Dasha V. Zhernakova, René Luijk, Marc Jan Bonder, Maarten van Iterson, Patrick Deelen, Freerk van Dijk, Michiel van Galen, Wibowo Arindrarto, Szymon M. Kielbasa, Morris A. Swertz, Erik. W van Zwet, Rick Jansen, Peter-Bram ’t Hoen, and Bastiaan T. Heijmans
Supplementary information
The online version of this article (10.1038/s41431-019-0511-5) contains supplementary material, which is available to authorized users.
References
- 1.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–35. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryois J, Buil A, Ferreira PG, Panousis NI, Brown AA, Viñuela A, et al. Time-dependent genetic effects on gene expression implicate aging processes. Genome Res. 2017;27:545–52. doi: 10.1101/gr.207688.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jansen R, Batista S, Brooks AI, Tischfield JA, Willemsen G, van Grootheest G, et al. Sex differences in the human peripheral blood transcriptome. BMC Genomics. 2014;15:33. doi: 10.1186/1471-2164-15-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meder B, Backes C, Haas J, Leidinger P, Stähler C, Großmann T, et al. Influence of the confounding factors age and sex on microRNA profiles from peripheral blood. Clin Chem. 2014;60:1200–8. doi: 10.1373/clinchem.2014.224238. [DOI] [PubMed] [Google Scholar]
- 5.Tower J. Sex-specific gene expression and life span regulation. Trends Endocrinol Metab. 2017;28:735–47. doi: 10.1016/j.tem.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lloyd-Jones LR, Holloway A, McRae A, Yang J, Small K, Zhao J, et al. The genetic architecture of gene expression in peripheral blood. Am J Hum Genet. 2017;100:228–37. doi: 10.1016/j.ajhg.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright FA, Sullivan PF, Brooks AI, Zou F, Sun W, Xia K, et al. Heritability and genomics of gene expression in peripheral blood. Nat Genet. 2014;46:430–7. doi: 10.1038/ng.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vinuela A, Brown AA, Buil A, Tsai PC, Davies MN, Bell JT, et al. Age-dependent changes in mean and variance of gene expression across tissues in a twin cohort. Hum Mol Genet. 2018;27:732–41. doi: 10.1093/hmg/ddx424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vink JM, Jansen R, Brooks A, Willemsen G, van Grootheest G, de Geus E, et al. Differential gene expression patterns between smokers and non‐smokers: cause or consequence? Addiction Biol. 2017;22:550–60. doi: 10.1111/adb.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willemsen G, de Geus EJ, Bartels M, van Beijsterveldt CE, Brooks AI. Estourgie-van Burg GF et al: The Netherlands Twin Register biobank: a resource for genetic epidemiological studies. Twin Res Hum Genet. 2010;13:231–45. doi: 10.1375/twin.13.3.231. [DOI] [PubMed] [Google Scholar]
- 11.Willemsen G, Vink JM, Abdellaoui A, den Braber A, van Beek JH, Draisma HH, et al. The adult netherlands twin register: twenty-five years of survey and biological data collection. Twin Res Hum Genet. 2013;16:271–81. doi: 10.1017/thg.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boomsma D, Busjahn A, Peltonen L. Classical twin studies and beyond. Nat Rev Genet. 2002;3:872–82. doi: 10.1038/nrg932. [DOI] [PubMed] [Google Scholar]
- 13.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willemsen G, Vink JM, Abdellaoui A, den Braber A, van Beek JH, Draisma HH, et al. The Adult Netherlands Twin Register: twenty-five years of survey and biological data collection. Twin Res Hum Genet. 2013;16:271–81. doi: 10.1017/thg.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genome of the Netherlands C. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet. 2014;46:818–25. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 16.Zhernakova DV, Deelen P, Vermaat M, van Iterson M, van Galen M, Arindrarto W, et al. Identification of context-dependent expression quantitative trait loci in whole blood. Nat Genet. 2017;49:139–45. doi: 10.1038/ng.3737. [DOI] [PubMed] [Google Scholar]
- 17.Boomsma DI, Wijmenga C, Slagboom EP, Swertz MA, Karssen LC, Abdellaoui A, et al. The Genome of the Netherlands: design, and project goals. Eur J Hum Genet. 2014;22:221–7. doi: 10.1038/ejhg.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R Development Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2014.
- 19.Neale MC, Hunter MD, Pritikin JN, Zahery M, Brick TR, Kirkpatrick RM, et al. OpenMx 2.0: extended structural equation and statistical modeling. Psychometrika. 2016;81:535–49. doi: 10.1007/s11336-014-9435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaitlen N, Kraft P, Patterson N, Bogdan P, Gaurav, Samuela P, et al. Using extended genealogy to estimate components of heritability for 23 quantitative and dichotomous traits. PLoS Genet. 2013;9:e1003520. doi: 10.1371/journal.pgen.1003520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Visscher PM, Hemani G, Vinkhuyzen AA, Chen GB, Lee SH, Wray NR, et al. Statistical power to detect genetic (co) variance of complex traits using SNP data in unrelated samples. PLoS Genet. 2014;10:e1004269. doi: 10.1371/journal.pgen.1004269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smedley D, Haider S, Durinck S, Pandini L, Provero P, Allen J, et al. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic acids Res. 2015;43:W589–W598. doi: 10.1093/nar/gkv350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinsella R. J., Kahari A., Haider S., Zamora J., Proctor G., Spudich G., Almeida-King J., Staines D., Derwent P., Kerhornou A., Kersey P., Flicek P. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011;2011(0):bar030–bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Battle A, Mostafavi S, Zhu X, Patash JB, Weissman MM, McCormick C, et al. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consortium GT, Laboratory DA, Coordinating Center -Analysis Working G, Enhancing GTEx (eGTEx) groups, NIH Common Fund, NIH/NCI, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–13. [DOI] [PMC free article] [PubMed]
- 28.Franić S, Dolan CV, Borsboom D, Boomsma DI. Structural equation modeling in genetics. In: Hoyle RH (ed) Handbook of structural equation modeling. New York: Guilford Press; 2012. pp 617–35.
- 29.Wheeler HE, Shah KP, Brenner J, Garcia T, Aquino-Michaels K, GTEX Consortium et al. Survey of the heritability and sparse architecture of gene expression traits across human tissues. PLoS Genet. 2016;12:e1006423. doi: 10.1371/journal.pgen.1006423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2016;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.