S. Dublin causes economic losses in cattle production, and the bacterium is a public health concern. A surveillance and control program has been in place in Denmark since 2002 with the ultimate goal to eradicate S. Dublin from Danish cattle herds; however, a small proportion of herds have remained positive for many years. In this study, we demonstrate that herds with persistent infection often were infected with the same strain for many years, indicating that internal biosecurity has to be improved to curb the infection. Further, domestic cases of S. Dublin infection in humans were found to be caused both by Danish cattle isolates and by isolates acquired abroad. This study shows the strength of whole-genome sequencing to obtain detailed information on epidemiology of S. Dublin and allows us to suggest internal biosecurity as a main way to control this bacterium in Danish cattle herds.
KEYWORDS: Salmonella, genomics, veterinary epidemiology
ABSTRACT
Salmonella enterica serovar Dublin is a cattle-adapted S. enterica serovar causing both intestinal and systemic infection in its bovine host, and it is also a serious threat to human health. The present study aimed to determine the population structure of S. Dublin isolates obtained from Danish cattle herds and to investigate how cattle isolates relate to Danish human isolates, as well as to non-Danish human and bovine isolates. Phylogenetic analysis of 197 Danish cattle isolates from 1996 to 2016 identified three major clades corresponding to distinct geographical regions of cattle herds. Persistence of closely related isolates within the same herd and their circulation between epidemiologically linked herds for a period of more than 20 years were demonstrated. These findings suggest that a lack of internal biosecurity and, to some extent, also a lack of external biosecurity in the herds have played an important role in the long-term persistence of S. Dublin in Danish cattle herds in the period investigated. Global population analysis revealed that Danish cattle isolates clustered separately from bovine isolates from other countries, whereas human isolates were geographically spread. Resistance genes were not commonly demonstrated in Danish bovine isolates; only the isolates within one Danish clade were found to often harbor two plasmids of IncFII/IncFIB and IncN types, the latter plasmid carrying blaTEM-1, tetA, strA, and strB antibiotic resistance genes.
IMPORTANCE S. Dublin causes economic losses in cattle production, and the bacterium is a public health concern. A surveillance and control program has been in place in Denmark since 2002 with the ultimate goal to eradicate S. Dublin from Danish cattle herds; however, a small proportion of herds have remained positive for many years. In this study, we demonstrate that herds with persistent infection often were infected with the same strain for many years, indicating that internal biosecurity has to be improved to curb the infection. Further, domestic cases of S. Dublin infection in humans were found to be caused both by Danish cattle isolates and by isolates acquired abroad. This study shows the strength of whole-genome sequencing to obtain detailed information on epidemiology of S. Dublin and allows us to suggest internal biosecurity as a main way to control this bacterium in Danish cattle herds.
INTRODUCTION
Salmonella enterica serovar Dublin is a cattle-adapted Salmonella serovar. It is a relatively rare cause of human infections in Denmark (1), but since it is highly pathogenic and is associated with a high mortality rate (2), it is considered a public health concern globally (3, 4). Most human infections are linked to the consumption of contaminated cow milk and beef. Thus, efforts to reduce S. Dublin infection in cattle will benefit human health (5).
S. Dublin causes acute disease in young cattle and abortions in adult cows (6). An important feature of S. Dublin is its ability to persist in the cattle host in a carrier state without showing any clinical signs of infection, which makes it difficult to diagnose. If reactivated, persistent carriers can excrete bacteria in large numbers and cause new infections in the herd (7).
Denmark has had long-term endemic occurrence of S. Dublin in cattle (8). In response to the S. Dublin threat to industry and to human health, a national surveillance program for S. Dublin was initiated in 2002 and further strengthened to a control program in 2007 (9). The program included increased active surveillance for the infection of all cattle herds and both voluntary and mandatory efforts to reduce bacteria in the environment. On the basis of defined serology cutoff values and bacteriology results, cattle herds are categorized into three levels: herds assigned to level 1 are considered free from S. Dublin infection, herds in level 2 must undergo a mandatory plan of reduction and, in addition to this, level 3 herds are put on restrictions with regard to slaughter and trade in order to prevent the transfer of S. Dublin to noninfected herds (10). The program has been updated continuously, and as a result, the prevalence of S. Dublin infection in cattle herds has been reduced from 25 to 7% between 2002 and 2015 (11). However, in some regions of endemicity with large cattle herds and a high cattle density, a herd-level prevalence has remained at approximately 15% and is often associated with long-term infections in this relatively small number of herds. The goal of the Danish national program is to eradicate S. Dublin from the cattle population. To achieve this, additional knowledge is needed to improve the understanding of the S. Dublin epidemiology, especially concerning the persistence of infection in some herds.
Recently, several studies have demonstrated that whole-genome sequencing (WGS) information can be highly valuable in understanding the epidemiology of S. Dublin infections, as well as infections caused by other serovars (12–16). Comparisons of the core genomes of S. Dublin isolates revealed minor single nucleotide polymorphism (SNP) differences between both epidemiologically linked isolates (up to 13 SNPs) and nonlinked isolates (up to 333 SNPs), indicating a stable core genome (12, 13). Furthermore, acquisition of additional region-associated plasmids has been shown to be associated with a global increase in antibiotic resistance in this bacterium and is therefore of concern (4, 15, 17, 18). Recently, it was demonstrated that the evolution of both S. Dublin core and accessory genomes is geographical region dependent, with no influence from a specific host (15).
In the present study, WGS was used to characterize isolates originating from Danish dairy cattle herds tested in the period from 1996 until 2016. In addition, clinical human isolates from 2014 to 2016 were included. The aim was to determine the overall population structure of S. Dublin isolates in Denmark, to determine whether herds that were positive for many years were infected with one or several strains, and to determine the extent to which Danish human infections were caused by Danish cattle isolates. Our thorough comparison of these isolates will potentially contribute to improved control efforts and hence to reducing the occurrence of S. Dublin in cattle, as well as the number of human cases, in Denmark.
RESULTS
Salmonella Dublin from Danish cattle.
A total of 197 S. Dublin isolates originating from 58 dairy cattle herds sampled in the period from 1996 to 2016 in Jutland, Denmark (29,767 km2), were compared. Based on the seven-locus multilocus sequence typing (MLST) scheme in EnteroBase, all isolates were assigned to sequence type 10 (ST-10). The raw read mapping of all genomes to the reference genome CT02021853 revealed only 1,184 variable nucleotide positions in the core genome. Of 4,842,908 positions present in the reference genome, 3,803,142 (78.5%) were present in all genomes of Danish cattle isolates under study.
The phylogenetic analysis resulted in isolate clustering into three major clades (I, II, and III). Isolates of clade I differed from isolates of clades II and III with between 232 and 272 SNPs, and there were between 106 to 159 SNP differences between the isolates representing clades II and III (see Fig. 1). SNP variation was similar within all clades, with maximum differences of 26, 61, and 44 SNPs between strains in clades I, II, and III, respectively.
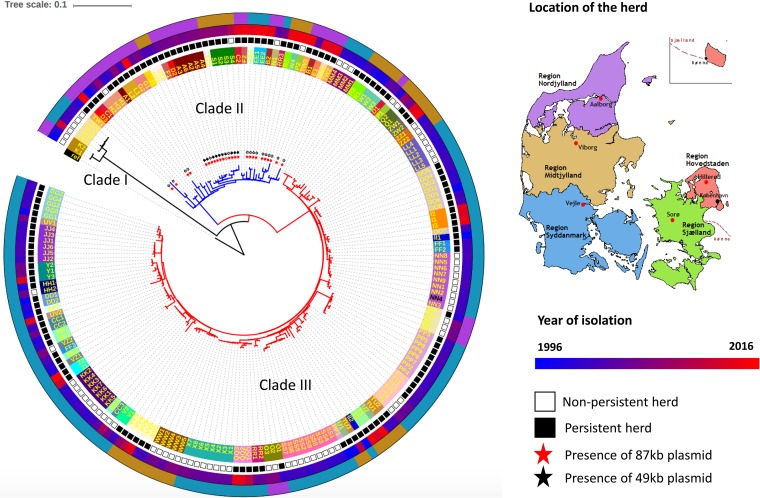
FIG 1.
Core SNP phylogeny of 197 S. Dublin isolates from 58 cattle herds sampled in Denmark in the period from 1996 to 2016. The tips represent herd identification numbers, and isolates from the same herd are colored the same. The map of Denmark (right) is taken from https://upload.wikimedia.org/wikipedia/commons/7/7c/Map_DK_Regions2.png. Colors in the outer ring of the phylogenetic tree correspond to the coloring of north, middle, and south Jutland in this map and indicate the regions where the strains originated. The second outer ring represents the year of S. Dublin isolation as shown on the right side of the figure. All isolates originating from persistently infected herds are labeled with a black square, and those originating from nonpersistent herds are labeled with an empty square in the third ring from the outside. The stars at the tips of the branches indicate S. Dublin isolates containing plasmids, with the filled and empty stars showing the presence or absence of a particular plasmid in the isolate, respectively.
The largest clade, clade III, consisted of 149 isolates, which were predominantly obtained from the southern part of the Jutland peninsula, spanning the full period from 1996 to 2016. Clade II consisted of 42 isolates and included isolates obtained from the years 1997 to 2015, mostly from the northern part of Jutland. Finally, six isolates from the years 1997 to 2012 were placed within clade I. This clade consisted of isolates from both northern and southern Jutland. The isolates from the middle region of mainland Jutland were distributed within both clades II and III (see Fig. 1). Interestingly, clade III isolates were detected in the northern herds only until 2006, while clade II isolates were found in southern herds only from 2005 onward.
Isolates from the same herd (between two and nine isolates per herd, spanning from 1 to 13 years) clustered together in 42 of 58 herds. Thirty-three of the herds where this was observed had been classified as persistently infected at least once based on epidemiological and serological evidence (see Materials and Methods) (Fig. 1). This was observed in all three clades, and the number of SNPs in isolates from the same herd clustering together was <10. Thus, isolates from the same herd were the closest relative to each other among the strains analyzed, showing that herds were generally persistently infected (defined as bacterial strains circulating for more than 1 year).
Closely related isolates (<10 SNPs) were occasionally shown to belong to different herds. We grouped such isolates on the basis of the year of isolation. With this analysis, we identified ten networks. In each of these networks, between 2 and 16 herds shared strains that were closely similar to each other with a difference of fewer than 10 SNPs (see Fig. S1 in the supplemental material).
These networks included 39 out of the 58 herds under analysis. The largest network consisted of 16 herds and included several herds which shared closely related isolates with more than one other herd in the network, suggesting an important role of these herds in the transmissions of S. Dublin (see Fig. S1 in the supplemental material).
Pangenome structure of S. Dublin from cattle in Denmark.
The pangenome analysis of the 197 S. Dublin cattle isolates showed 6,535 coding sequences (CDS). Of these, 4,356 CDS constituted a core genome present in 99 to 100% of the genomes, 143 a soft core present in 95 to 98% of the genomes, and 183 an accessory genome present in 15 to 94% of the genomes, while 1,853 genes were present in <15% of the isolates. The core genome for S. Dublin isolates was calculated to be approximately 94% of the genomic information.
The hierarchic tree based on the accessory genome revealed isolate clustering into the same three major clades, with the same strains in each clade, as revealed by core genome SNPs (see Fig. 2). However, in comparison to phylogeny based on the core genome SNP approach, herd-specific clustering was less frequent in this analysis, indicating possible herd-independent gene gain and loss.
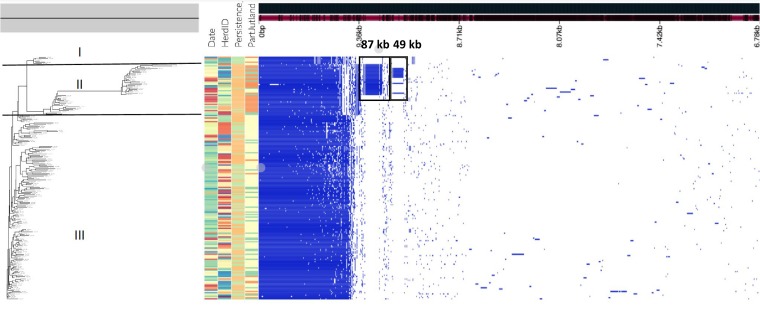
FIG 2.
Pangenome of 197 S. Dublin isolates from 58 cattle herds sampled in Denmark from 1996 to 2016. The hierarchic tree was generated based on gene presence/absence in the genomes. I to III indicate the three clades of S. Dublin as defined by core genome SNP phylogeny. The top row represents the calculated pangenome of S. Dublin, and blue and white squares represent the presence and absence, respectively, of genes in each S. Dublin genome. The black squares indicate CDS blocks representing the 49- and 87-kb plasmids.
The heat map for presence of genes showed two DNA regions to be specifically present in the isolates belonging to clade II, and additional analysis revealed the presence of statistically significant clade II-specific genes (n = 129), whereas there were no genes found to be specific for the isolates located in clade I or III.
Evidence for recombination in the sequences.
Recombination analysis of 197 isolates of S. Dublin from cattle identified potential regions of recombination occurring throughout the chromosome (see Fig. S2 in the supplemental material). Only a very small fraction of bases in the whole chromosome (on average 0.1%) was affected by recombination (minimum = 0, maximum = 25,023 SNPs, and average = 3,925) (see Table S1). A mean of 2.3 recombination events was detected on each branch, and the mean ratio of SNPs introduced by recombination to those introduced by point mutations was 0.24. The clustering of S. Dublin isolates in the phylogeny generated by Gubbins was not altered in comparison to their phylogenetic relationship determined by using the CSI phylogeny tool, where mapping to a reference genome was used to identify the SNPs.
Plasmid characterization in S. Dublin genomes from cattle.
The list of clade II-specific genes was subjected to local blastN analysis against the genomes belonging to clade II. The genes were found to be located on single contigs of either 49 or 87 kb. The 49-kb contig was found in 10 isolates and the 87-kb contig was found in 25 isolates of 42 assigned to clade II (seen as gene blocks in Fig. 2). The contigs were found to be similar to two plasmids present in the nonredundant nucleotide database. The 49-kb region was homologous (coverage = 99%, identity = 100%) to an IncN plasmid of S. enterica serovar Stanleyville (GenBank accession no. CP017725), and the 87-kb region was most similar (coverage = 60%, identity = 99%) to an IncFII/IncFIB plasmid of Escherichia coli (GenBank accession no. KJ484628). Neither of these plasmids had been previously characterized in S. Dublin.
To confirm the presence of these plasmids, as well as their correct size, plasmid profiling was performed on selected isolates from all three clades (n = 6 [clade I], 31 [clade II], and 27 [clade III]). The analysis showed the presence of one to five plasmids in the S. Dublin isolates (see Fig. S3 in the supplemental material). One 83-kb plasmid corresponded to the virulence plasmid of S. Dublin and was present in all isolates. The presence of additional 49- and 87-kb plasmids was only detected in strains from clade II, an observation in agreement with the findings in silico. Two small plasmids of approximately 4 and 6 kb were randomly distributed across the isolates and may represent previously described plasmids of S. Dublin, which did not indicate any influence on S. Dublin resistance or virulence phenotype (19).
To analyze the gene content of these plasmids, we used blastN to compare them to the plasmids to which they had shown homology. The gene content of the 49-kb plasmid was exactly the same as that of the IncN plasmid of S. enterica serovar Stanleyville (GenBank accession no. CP017725), with the exception of one gene encoding isochorismatase, which was not present in the S. Dublin plasmid, and one gene encoding a hypothetical protein found only in the plasmid of S. Dublin. The plasmid harbored type IV secretion system genes (virB1, virB4 to virB6, and virB9 to virB11) and several antibiotic resistance genes (blaTEM-1, tetA, strA, and strB) (see Fig. S4 in the supplemental material). All isolates from clade II were subjected to antimicrobial susceptibility testing, and those containing the 49-kb plasmid were found to be phenotypically resistant to ampicillin, amoxicillin-clavulanic acid, and tetracycline. The rest of the isolates were susceptible to all antibiotics tested.
The 87-kb plasmid was only partly homologous to a plasmid found in E. coli (GenBank accession no. KJ484628) (see Fig. S5 in the supplemental material). BLASTP of the proteins from this part of the plasmid showed a minimum of 97% amino acid identity and 100% length coverage to plasmid replicons of the IncFII and IncFIB type and to 21 Tra and 8 Trb conjugal transfer proteins. The rest of the predicted plasmid did not show homology to any specific plasmid in GenBank. This part consisted of 58 mostly uncharacterized proteins but included the type IV secretion proteins ImpA and ImpC, colicin-1b, and the Enterobacteriaceae toxin-antitoxin proteins VapC and VapB as well.
Comparison of S. Dublin from cattle and humans in Denmark with global isolates from the period from 2001 to 2016.
To investigate the relationship between Danish and non-Danish S. Dublin isolates, we performed a phylogenetic analysis based on the SNPs in the core genomes of 185 selected genomes from Denmark (human = 46, cattle = 139) and 103 genomes from various sources from abroad. A subset of Danish cattle isolates included in this data set was selected to span the years of isolation of Danish human and international S. Dublin isolates (2001 to 2016). In total, 2,571 SNPs were identified among all the isolates, and the isolates were separated into nine clades, including three Danish clades (DK/I to DK/III), each dominated by isolates from specific geographic locations around the globe (see Fig. 3).
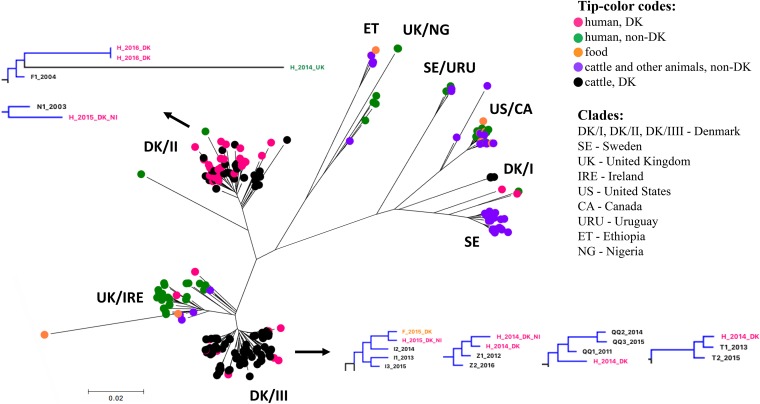
FIG 3.
Core genome SNP phylogeny of 288 genomes from Danish human isolates (n = 46), Danish cattle isolates (n = 139), and publicly available genomes representing a global collection of S. Dublin isolates (n = 103). DK/I, DK/II, and DK/III were named according to the clades determined in the phylogeny of only Danish cattle isolates. Subtrees with branches in blue show closely related Danish human and cattle isolates at the SNP threshold of <10 SNPs.
Cattle isolates from Denmark were distributed within the clades DK/I, DK/II, DK/III resembling cattle clades I, II, and III described above. Most of the clinical human isolates from Denmark (41 of 46) were found within these clades together with Danish cattle isolates.
Interestingly, one lineage within clade DK/III included a beef isolate from 2015 from Denmark, which clustered together (2-SNP difference) with three 2013-2015 cattle isolates from the same herd and one 2015 human isolate from Denmark with no travel information available. This demonstrates a possible transmission of S. Dublin to humans through the meat produced from a specific cattle herd persistently infected with S. Dublin.
A total of 35 Danish clinical human isolates found within clades DK/II and DK/III were separated by a minimum of 14 core SNPs from the Danish cattle isolates. Among these were three clinical human isolates from ill people who reported traveling to Spain, Canada, and the Canary Islands in the period just before the isolation. Two human isolates from the United Kingdom and Ireland, which differed from Danish isolates by between 27 and 87 SNPs, were included in clade DK/II.
Three Danish clinical human isolates were found within clade UK/IRE; however, the SNP difference with isolates from United Kingdom and Ireland in this clade was >40 SNPs. One of the Danish isolates was obtained from a person who was travelling to Malta, and the two others represented cases with no travel information available. Two more Danish clinical human isolates, with no travel information available, were placed between clades SE and DK/I, together with one human isolate from the United Kingdom. These isolates differed from each other and from all the isolates by a minimum of 40 SNPs.
The raw read mapping of S. Dublin isolates from human revealed the presence of the 49- and 87-kb plasmids in 4 and 11 isolates, respectively. All of these isolates clustered with cattle isolates within clade DK/II. Interestingly, the two plasmids were also detected in one human isolate from the United Kingdom (accession no. SRR1968923), which was a part of clade DK/II as well.
DISCUSSION
In the present study, clinical isolates from cattle and human in Denmark were studied using draft whole-genome analysis and compared to genomes of a collection of global isolates of S. Dublin.
The findings from the phylogenetic analysis of cattle isolates revealed the presence of three distinct populations of S. Dublin in Denmark. Further analysis, including clinical human isolates from Denmark, as well as international isolates, confirmed these clades to be specific to the Danish cattle population. Two possible scenarios can be proposed to explain S. Dublin epidemics in Denmark. The first scenario is the presence of three overlapping S. Dublin epidemics in Denmark in the period under study, and the second is the occurrence of several clones in the perpetual epidemics of S. Dublin in cattle. However, the second scenario is unlikely due to a high number of SNPs (>106) separating the three different populations of S. Dublin and the fact that the three different populations were isolated in different periods: clade I consisted of older isolates from the years 1997 to 2012, clade II consisted of isolates from the years 1997 to 2015, and clade III consisted of isolates from the years 1996 to 2016. A herd-specific clustering of S. Dublin isolates was obvious in all three phylogenetic clades obtained; however, the clustering of isolates did not correlate with the year of isolation or with the persistence status of the herd. This strongly indicates that the main feature of long-term-infected herds is persistence of the same strain within the herd, causing infections either through contaminated environment or feed or by recurrent cow-to-cow infections, and the observation suggests that internal biosecurity has to be improved in such herds. This might include the identification of persistently infected carrier animals for hygienic management or culling (7). Geographic location-dependent clustering was also observed in the global tree of S. Dublin, which indicates limited transmission of this bacterium between countries.
Previous WGS-based studies on the epidemiology of S. Dublin have used different SNP thresholds to define closely related S. Dublin isolates. Mohammed and Cormican (13) investigated isolates from a human outbreak and identified them to be related by a difference of one to nine SNPs, though these isolates were also closely related to a historical isolate of S. Dublin (15 SNPs), with no link between those isolates identified. In another study, a maximum number of 13 SNPs was detected between epidemiologically linked isolates when cattle outbreak isolates from different regions in Sweden were compared (12). Although different pipelines were used in the studies, our results agreed with these reports. The majority of isolates from the same cattle herds with no gaps in the year of isolation clustered at the threshold of <10 SNPs, whereas those which had a several-year gap with no S. Dublin isolation were related by a minimum of 14 SNPs. Our previous findings using cattle herd contact data revealed that cattle herds with epidemiological links can share S. Dublin isolates related by up to 15 SNPs (11). However, it is not clear whether these isolates represent true transmission of identical S. Dublin strains, thus belonging to the same outbreak, or whether the contact occurred in the past. More-distant isolates may be the same strains which evolved over time. Based on previous reports and observations of our own, we used a strict threshold of <10 SNPs to define closely related isolates of S. Dublin. It should be noted that the selected threshold may not be suitable for the analyses of S. Dublin when genomes are processed with another pipeline or even another sample set, since it is recommended that thresholds on strain identity should also be based on epidemiological information (20). However, we performed additional analysis of our data sets using other pipelines, and different information from the genomes did not change the main findings of the study (see Fig. S6 to S9 in the supplemental material), making the conclusion on strain relation strong.
Using the <10-SNP threshold, a number of herds sharing closely related S. Dublin isolates were identified in two of the three clades of S. Dublin, and specific herds were identified as potential sources of S. Dublin infections in other herds (see Fig. S1 in the supplemental material). Overall, these results suggest that the endemicity of S. Dublin in Denmark has been sustained due to both the long-term infections with the same strain within the persistently infected herds and transmissions of strains between cattle herds, even though strict limitations for movement of cattle from infected to noninfected herds have been in place (11). This might be due to misclassifications of the herds in the surveillance program or alternative transmission routes, such as vehicles, manure, or shared pastures. On the other hand, the fact that the smaller networks showed an absence of a relationship with other herds could be due to sampling. This underpins the importance of frequent sampling/testing and good coverage of herds for tracing the sources of infections in cattle herds. Interestingly, this analysis also demonstrated the presence of two unrelated S. Dublin populations within the same cattle herds (FF, R, and W) but not in the same year, suggesting the dominance of one S. Dublin strain at a given time point.
Eight human isolates were found to be closely related to Danish cattle isolates, and they had caused infection in humans in the same year as the cattle isolates were identified in the cattle herds. In four cases human and cattle isolates were reported to be associated geographically, suggesting an epidemiological link between the isolates. Moreover, a direct link between human infection, a specific cattle herd, and a beef isolate was demonstrated. It was not possible to obtain information about travel inside Denmark for the cases, and further, selling of food of cattle origin is not restricted to the region of Denmark where it is produced. Thus, we cannot conclude that Danish cattle are always the direct source of human infections in Denmark. The latter example also demonstrated that the inclusion of food isolates in WGS comparison allows the prediction of the source of infection in a food chain perspective, and it would have been beneficial to include more food isolates in the present study.
In agreement with previous studies (12, 13), we found a limited sequence variation between strains of S. Dublin. Large SNP differences of up to 272 SNPs were found between isolates of clade I and isolates of clades II and III. This high distance between clades was also seen between the clades of Danish cattle isolates when they were included in the analysis of a global collection of isolates. The results suggest that clade DK/I is the oldest Danish clade and, according to global phylogeny, it was more related to Swedish cattle isolates (96 to 113 SNPs) than to the Danish cattle isolates (175 to 208 SNPs). Accordingly, clade DK/I may represent a population of S. Dublin which had circulated in the past but apparently had not been sampled since 2012. Clades DK/II and DK/III were closely related to each other in both analyses, and the DK/III clade is further related to the UK/IRE clade. The reason for such a close relationship between these clades is unknown, but it may be an indication of S. Dublin transmissions between countries in the past. However, this was not further investigated in the present study.
The current clustering based on phylogeny indicates national S. Dublin populations which evolve mostly by point mutations in the whole genome and by single gene gain and loss. We tested whether there is a pattern for gene gain and loss between the S. Dublin isolates from persistently and nonpersistently infected herds, but significant differences were not detected (data not shown). Overall, the herd-specific clustering over the span of up to 13 years in parallel to an indication of development of new populations may represent a situation similar to that observed in Salmonella Typhi (21).
Although S. Dublin is a highly clonal bacterium with a very conserved genome, it may be able to acquire variation due to acquisition of plasmids carrying additional virulence and resistance genes, thus likely providing the opportunity for S. Dublin to more readily adapt to changing environments, as reported in studies of other bacteria (22–24).
The adaptation ability is supported by multiple reports of several resistance plasmids in S. Dublin from cattle and humans in Germany (25), the United States (15, 26, 27), Canada (16), and Peru (17), for example. These reports indicate that the resistance of S. Dublin is increasing and can lead to the emergence of antibiotic resistance in this bacterium. S. Dublin carrying multiple resistance genes is more likely than susceptible types to result in bloodstream infections, hospitalization, and death (4). An interesting finding in our study was the presence of 49- and 87-kb plasmids in S. Dublin isolates within clade II. These plasmids were not found in clades representing other countries, and yet their presence was detected in both Danish cattle and Danish clinical human isolates. The 49-bp plasmid was almost identical to a pSARB26_02 plasmid of Salmonella enterica Stanleyville (28), as well as to several other plasmids of similar size present in an uncultured bacterium (GenBank accession no. JN102344; 52,809 bp) isolated in a wastewater treatment plant in Germany (29), in Salmonella enterica (GenBank accession no. CP028173; 50,905 bp) from turkey in Germany, and in Salmonella enterica (GenBank accession no. CP026053; 47,793 bp) from humans in the United States. Recently, we also reported the presence of the homologous plasmid in the genomes of S. Enteritidis in clinical human isolates in Ghana (30). The Danish cattle and human isolates possessing this plasmid were isolated in the period between 2004 and 2014, indicating that this plasmid has been circulating in S. Dublin populations already for a decade or more. It may even have circulated for a longer period, since a plasmid of a very similar size (48 kb), carrying resistance genes for streptomycin and tetracycline, was reported in S. Dublin isolates from Denmark between 1983 and 1987 (19), though with no insertion of the blaTEM-1 gene. The replicon type of the S. Dublin 48-kb plasmid was not investigated, but it was shown that resistance genes from this plasmid were transferred to E. coli K-12 by conjugation (19). The presence of the multidrug resistance plasmid of the IncN type in human, cattle, and environmental S. Dublin isolates from Denmark shows that even in countries with low antimicrobial use in the cattle population, such plasmids can apparently be stably maintained in the population. It is phylogenetically related to plasmids of the IncN type in E. coli (29), and it may have been gained from related Enterobacteriaceae and maintained in the presence of antibiotic pressure.
The 87-kb plasmid was present in Danish human and cattle isolates, as well as in one human isolate from the United Kingdom, isolated in the period from 1997 to 2016. In contrast to the 49-kb plasmid, no homologous plasmid was detected in GenBank. According to the blastP analysis, the proteins encoded on this plasmid were mostly of unknown function, except for those corresponding to tra and trb regions, as well as the IncFII- and IncFIB-type plasmid replicon. The function of this plasmid remains unclear and needs to be further investigated. All S. Dublin isolates analyzed in this study contained the virulence plasmid pSDV of the IncFII type. This plasmid carries the five spvRABCD genes, some of which are essential for systemic infections in cattle (31).
Conclusions.
Overall, the analysis revealed the presence of three distinct populations of S. Dublin in Danish cattle herds. Of the three Danish populations of S. Dublin, two shared ancestries with global isolates. The clades seem to evolve independently, specifically by the accumulation of a low number of random SNPs within persistently infected herds. In addition, a few gene gains and losses and occasional spread between herds contribute to the evolution. As expected, human strains were cattle related, whether acquired domestically or in relation to travel. These findings suggest that, in order to eradicate S. Dublin infection from cattle in Denmark, biosecurity within herds should be improved and methods to identify persistent carriers should be a priority, in addition to the current focus on control measures to prevent spread between herds.
MATERIALS AND METHODS
Bacterial strains.
This study included 197 S. Dublin isolates, collected from 1996 to 2016 from 58 Danish dairy cattle herds during various activities (national surveillance program, research projects, and demonstration projects). Isolates had been stored at –80°C at the National Veterinary Institute (Denmark). To investigate the relation to the persistence status of the herd, isolates were selected to represent herds where persistent infection was suspected (P; defined as herds where the bulk-tank milk test status had been above the national cutoff value for at least 4 years after the first isolation of S. Dublin bacteria) and herds where the infection had been cleared following the first isolation (nonpersistent [NP]; the antibody levels in bulk-tank milk went down below the national cutoff within 4 years after the first isolation). The herd distributions and their details are shown in Table S2.
Isolates and patient information from all human clinical Salmonella infections were sent to the Statens Serum Institut (SSI) from the local clinical laboratories for the national laboratory-based surveillance. Forty-six S. Dublin isolates received from 2014 to 2016 were selected for this study (see Table S3). Isolates were sequenced at the SSI by extracting DNA and preparation using a Promega Wizard genomic DNA purification kit (Promega, Madison, WI) and a Nextera XT v2 DNA library preparation kit (Illumina, San Diego, CA) according to the manufacturers’ protocols. WGS was performed using an Illumina MiSeq with 250-bp paired-end technology.
In addition to genome sequences obtained in the present study, raw reads of previously sequenced S. Dublin isolates were downloaded from Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) using a fastq-dump 2.9.0 script. The list and metadata for 103 public genomes analyzed in the study are presented in Table S4.
Phylogenetic analysis.
The phylogenetic relationship between the S. Dublin isolates included in the study was determined using the CSI phylogeny tool (32), available on the server of the Centre for Genomic Epidemiology (Technical University of Denmark [DTU]; https://cge.cbs.dtu.dk/services/CSIPhylogeny/). For the analysis, the pair-end reads were mapped to the reference genome of S. Dublin strain CT02021853. Genomic positions containing an SNP in at least one of the isolates and meeting quality-filtering criteria in 100% of the isolates are included in the SNP matrix. The SNPs were excluded from the analyses if the depth at the SNP position was less than 10× or at least 10% of the average depth for the particular genome mapping. SNPs were filtered out if mapping quality was below 25 or the SNP quality was below 30. The recombinant regions were excluded by filtering out SNPs separated by <10 bp. All of the identified SNP positions were concatenated per isolate, and their alignment was used to construct the SNP matrix for phylogenetic analysis. All the obtained trees were visualized using either FigTree v.1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/) or iTOL (33). EnteroBase was used to identify the MLST types of the isolates (34).
Pangenome.
Prokka 1.12-beta (35) was used to obtain the genome annotations, and .gff files were used as an input for the pangenome analysis with Roary 3.7.0 (36). The binary presence/absence data of accessory genes produced in Roary were used for the construction of an accessory binary tree, as well as for univariate and multivariate analyses with Scoary (37), and were visualized using Phandango (39).
Recombination assessment in the sequences.
The core gene alignment of 197 S. Dublin isolates from cattle was generated with Roary. The alignment was analyzed by using the Gubbins algorithm (Genealogies Unbiased By recomBinations In Nucleotide Sequences), which iteratively identifies loci containing elevated densities of base substitutions while concurrently constructing a phylogeny based on the putative point mutations outside these regions (38). For the analysis, the default parameters of Gubbins v.2.3.4 were applied. The output from Gubbins was visualized using Phandango (39).
Antimicrobial susceptibility testing.
Antimicrobial susceptibility of a subset of S. Dublin isolates was determined by the agar disc diffusion method on Mueller-Hinton agar (CM0337; Oxoid, Ltd., England, UK) according to the protocol and guidelines of the European Committee on Antibiotic Susceptibility Testing (EUCAST). The strains were screened for their susceptibility to the following antimicrobials: ampicillin (10 μg), gentamicin (10 μg), amoxicillin-clavulanic acid (30 μg + 10 μg), tetracycline (30 μg), chloramphenicol (30 μg), trimethoprim (5 μg), sulfamethoxazole (240 μg), nalidixic acid (30 μg), and ciprofloxacin (5 μg) (Oxoid). E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used for quality control. EUCAST breakpoints (http://www.eucast.org/clinical_breakpoints/) (40) were used to interpret zone diameters.
Plasmid analysis.
Plasmid isolation from a subset of S. Dublin isolates was performed as described previously (41). The sizes of the detected plasmids were determined by using reference plasmids of E. coli 39R61 and E. coli V517 as markers of plasmid mobility on the agarose gel (19).
Data availability.
The draft genome sequences of S. Dublin isolated from cattle and from human clinical cases in Denmark used in this study are available in the DDBJ/ENA/NCBI databases under study accession numbers PRJEB26501 and PRJEB33058, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the Cattle Levy Fund. Part of the simulations described here were performed using the DeiC National Life Science Supercomputer at DTU.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Anonymous. 2018. Annual report on zoonoses in Denmark 2017. National Food Institute, Technical University of Denmark, Lyngby, Denmark. [Google Scholar]
- 2.Helms M, Vastrup P, Gerner-Smidt P, Mølbak K. 2003. Short- and long-term mortality associated with foodborne bacterial gastrointestinal infections: registry based study. BMJ 326:357. doi: 10.1136/bmj.326.7385.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Schaik G, Klinkenberg D, Veling J, Stegeman A. 2007. Transmission of Salmonella in dairy herds quantified in the endemic situation. Vet Res 38:861–869. doi: 10.1051/vetres:2007036. [DOI] [PubMed] [Google Scholar]
- 4.Harvey RR, Friedman CR, Crim SM, Judd M, Barrett KA, Tolar B, Folster JP, Griffin PM, Brown AC. 2017. Epidemiology of Salmonella enterica serotype Dublin infections among humans, United States, 1968–2013. Emerg Infect Dis 23:1493–1501. doi: 10.3201/eid2309.170136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Funke S, Anker JC, Ethelberg S. 2017. Salmonella Dublin patients in Denmark and their distance to cattle farms. Infect Dis (Lond) 49:208–216. doi: 10.1080/23744235.2016.1249024. [DOI] [PubMed] [Google Scholar]
- 6.Henderson K, Mason C. 2017. Diagnosis and control of Salmonella Dublin in dairy herds. In Practice 39:158–168. doi: 10.1136/inp.j1160. [DOI] [Google Scholar]
- 7.Nielsen LR, Schukken YH, Gröhn YT, Ersbøll AK. 2004. Salmonella Dublin infection in dairy cattle: risk factors for becoming a carrier. Prev Vet Med 65:47–62. doi: 10.1016/j.prevetmed.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen LR, Dohoo I. 2013. Time-to-event analysis of predictors for recovery from Salmonella Dublin infection in Danish dairy herds between 2002 and 2012. Prev Vet Med 110:370–378. doi: 10.1016/j.prevetmed.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen LR, Rattenborg E. 2011. Active surveillance and control programme for Salmonella Dublin in cattle: alternatives to acceptance of endemic infection with poor control options. Epidemiologie and Santé Animale Proceedings of the International Conference on Animal Health Surveillance 2011:210–212. [Google Scholar]
- 10.Nielsen LR. 2013. Within-herd prevalence of Salmonella Dublin in endemically infected dairy herds. Epidemiol Infect 141:2074–2082. doi: 10.1017/S0950268812003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Knegt LV, Kudirkiene E, Rattenborg E, Sørensen G, Denwood MJ, Olsen JE, Nielsen LR. 2018. Combining Salmonella Dublin genome information and contact-tracing to substantiate a new approach for improved detection of infectious transmission routes in cattle populations. Prev Vet Med 2018:S0167-5877(18)30357X. doi: 10.1016/j.prevetmed.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Ågren EC, Wahlström H, Vesterlund-Carlson C, Lahti E, Melin L, Söderlund R. 2016. Comparison of whole-genome sequencing typing results and epidemiological contact information from outbreaks of Salmonella Dublin in Swedish cattle herds. Infect Ecol Epidemiol 6:31782. doi: 10.3402/iee.v6.31782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohammed M, Cormican M. 2016. Whole-genome sequencing provides insights into the genetic determinants of invasiveness in Salmonella Dublin. Epidemiol Infect 144:2430–2439. doi: 10.1017/S0950268816000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gymoese P, Sørensen G, Litrup E, Olsen JE, Nielsen EM, Torpdahl M. 2017. Investigation of outbreaks of Salmonella enterica serovar Typhimurium and its monophasic variants using whole-genome sequencing, Denmark. Emerg Infect Dis 23:1631–1639 doi: 10.3201/eid2310.161248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenske GJ, Thachil A, McDonough PL, Glaser A, Scaria J. 2019. Geography shapes the population genomics of Salmonella enterica Dublin. Genome Biol Evol 11:2220–2231. doi: 10.1093/gbe/evz158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mangat CS, Bekal S, Avery BP, Côté G, Daignault D, Doualla-Bell F, Finley R, Lefebvre B, Bharat A, Parmley EJ, Reid-Smith RJ, Longtin J, Irwin RJ, Mulvey MR, on behalf of the Canadian Integrated Program for Antimicrobial Resistance Surveillance Public Health Partnership. 2019. Genomic investigation of the emergence of invasive multidrug-resistant Salmonella enterica serovar Dublin in humans and animals in Canada. Antimicrob Agents Chemother 63:e00108-19. doi: 10.1128/AAC.00108-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohammed M, Le Hello S, Leekitcharoenphon P, Hendriksen R. 2017. The invasome of Salmonella Dublin as revealed by whole-genome sequencing. BMC Infect Dis 17:544. doi: 10.1186/s12879-017-2628-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mangat CS, Bekal S, Irwin RJ, Mulvey MR. 2017. A novel hybrid plasmid carrying multiple antimicrobial resistance and virulence genes in Salmonella enterica serovar Dublin. Antimicrob Agents Chemother 61:e02601-16. doi: 10.1128/AAC.02601-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olsen JE, Baggesen DL, Nielsen BB, Larsen HE. 1990. The prevalence of plasmids in Danish bovine and human isolates of Salmonella Dublin. APMIS 98:735–740. doi: 10.1111/j.1699-0463.1990.tb04994.x. [DOI] [PubMed] [Google Scholar]
- 20.Schürch AC, Arredondo-Alonso S, Willems RJL, Goering RV. 2018. Whole-genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin Microbiol Infect 24:350–354. doi: 10.1016/j.cmi.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 21.Achtman M. 2008. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol 62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- 22.Rychlik I, Gregorova D, Hradecka H. 2006. Distribution and function of plasmids in Salmonella enterica. Vet Microbiol 112:1–10. doi: 10.1016/j.vetmic.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 23.Chu C, Feng Y, Chien AC, Hu S, Chu CH, Chiu CH. 2008. Evolution of genes on the Salmonella virulence plasmid phylogeny revealed from sequencing of the virulence plasmids of S. enterica serotype Dublin and comparative analysis. Genomics 92:339–343. doi: 10.1016/j.ygeno.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 24.Moreno Switt AI, den Bakker HC, Cummings CA, Rodriguez-Rivera LD, Govoni G, Raneiri ML, Degoricija L, Brown S, Hoelzer K, Peters JE, Bolchacova E, Furtado MR, Wiedmann M. 2012. Identification and characterization of novel Salmonella mobile elements involved in the dissemination of genes linked to virulence and transmission. PLoS One 7:e41247. doi: 10.1371/journal.pone.0041247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frech G, Schwarz S. 1998. Tetracycline resistance in Salmonella enterica subsp. enterica serovar Dublin. Antimicrob Agents Chemother 42:1288–1289. doi: 10.1128/AAC.42.5.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han J, Lynne AM, David DE, Nayak R, Foley SL. 2012. Sequencing of plasmids from a multi-antimicrobial resistant Salmonella enterica serovar Dublin strain. Food Res Int 45:931–934. doi: 10.1016/j.foodres.2011.04.016. [DOI] [Google Scholar]
- 27.Hsu CH, Li C, Hoffmann M, McDermott P, Abbott J, Ayers S, Tyson GH, Tate H, Yao K, Allard M, Zhao S. 2019. Comparative genomic analysis of virulence, antimicrobial resistance, and plasmid profiles of Salmonella Dublin isolated from sick cattle, retail beef, and humans in the United States. Microb Drug Resist 25:1238–1249. doi: 10.1089/mdr.2019.0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timme RE, Pettengill JB, Allard MW, Strain E, Barrangou R, Wehnes C, Van Kessel JS, Karns JS, Musser SM, Brown EW. 2013. Phylogenetic diversity of the enteric pathogen Salmonella enterica subsp. enterica inferred from genome-wide reference-free SNP characters. Genome Biol Evol 5:2109–2123. doi: 10.1093/gbe/evt159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eikmeyer F, Hadiati A, Szczepanowski R, Wibberg D, Schneiker-Bekel S, Rogers LM, Brown CJ, Top EM, Pühler A, Schlüter A. 2012. The complete genome sequences of four new IncN plasmids from wastewater treatment plant effluent provide new insights into IncN plasmid diversity and evolution. Plasmid 68:13–24. doi: 10.1016/j.plasmid.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Kudirkiene E, Andoh LA, Ahmed S, Herrero-Fresno A, Dalsgaard A, Obiri-Danso K, Olsen JE. 2018. The use of a combined bioinformatics approach to locate antibiotic resistance genes on plasmids from whole-genome sequences of Salmonella enterica serovars from humans in Ghana. Front Microbiol 9:1010. doi: 10.3389/fmicb.2018.01010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallis TS, Paulin SM, Plested JS, Watson PR, Jones PW. 1995. The Salmonella Dublin virulence plasmid mediates systemic but not enteric phases of salmonellosis in cattle. Infect Immun 63:2755–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. 2014. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alikhan NF, Zhou Z, Sergeant MJ, Achtman M. 2018. A genomic overview of the population structure of Salmonella. PLoS Genet 14:e1007261. doi: 10.1371/journal.pgen.1007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 36.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pangenome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. 2016. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol 17:238. doi: 10.1186/s13059-016-1108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole-genome sequences using Gubbins. Nucleic Acids Res 43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hadfield J, Croucher NJ, Goater RJ, Abudahab K, Aanensen DM, Harris SR. 2017. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics 34:292–293. doi: 10.1093/bioinformatics/btx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.EUCAST. 2013. Clinical breakpoints—breakpoints and guidance EUCAST, Basel, Switzerland: http://www.eucast.org/clinical_breakpoints/. [Google Scholar]
- 41.Kado CI, Liu ST. 1981. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol 145:1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The draft genome sequences of S. Dublin isolated from cattle and from human clinical cases in Denmark used in this study are available in the DDBJ/ENA/NCBI databases under study accession numbers PRJEB26501 and PRJEB33058, respectively.