Dental plaque is omnipresent in healthy oral cavities and part of our commensal microbial colonization. At the same time, dental plaque is the cause of the most common human diseases, caries and gum disease. Dental plaque consists of billions of microbes attached to the surface of your teeth. Communication among these microbes is pivotal for development of these complex communities yet poorly studied in dental plaque. In the present study, we show that a specific communication molecule induces changes within the community related to the development of gum disease. This finding suggests that interfering with microbial communication may represent an interesting novel strategy to prevent gum disease that should be further investigated.
KEYWORDS: dental plaque, quorum sensing, periodontitis, acylhomoserine lactone
ABSTRACT
Acylhomoserine lactones (AHLs), the quorum-sensing (QS) signals produced by a range of Gram-negative bacteria, are involved in biofilm formation in many pathogenic and environmental bacteria. Nevertheless, the current paradigm excludes a role of AHLs in dental plaque formation, while other QS signals, such as AI-2 and autoinducer peptides, have been demonstrated to play an important role in biofilm formation and virulence-related gene expression in oral pathogens. In the present work, we have explored the effect of externally added AHLs on in vitro oral biofilm models for commensal, cariogenic, and periodontal dental plaque. While little effect on bacterial growth was observed, some AHLs specifically affected the lactic acid production and protease activity of the biofilms. Most importantly, the analysis of bacterial diversity in the biofilms showed that the addition of C6-homoserine lactone (C6-HSL) results in a shift toward a periodontal bacterial composition profile by increasing the relative presence of the orange-complex bacteria Peptostreptococcus and Prevotella. These results point to a relevant role of AHL-mediated QS in dental plaque formation and might be involved in the development of dysbiosis, the mechanism of which should be further investigated. This finding potentially opens new opportunities for the prevention or treatment of the periodontal disease.
IMPORTANCE Dental plaque is omnipresent in healthy oral cavities and part of our commensal microbial colonization. At the same time, dental plaque is the cause of the most common human diseases, caries and gum disease. Dental plaque consists of billions of microbes attached to the surface of your teeth. Communication among these microbes is pivotal for development of these complex communities yet poorly studied in dental plaque. In the present study, we show that a specific communication molecule induces changes within the community related to the development of gum disease. This finding suggests that interfering with microbial communication may represent an interesting novel strategy to prevent gum disease that should be further investigated.
INTRODUCTION
Biofilms are sessile microbial communities characterized by cells that are irreversibly attached to a substratum or to each other, embedded in a matrix of self-produced extracellular polymers, and exhibit an altered phenotype with respect to growth rate and gene transcription (1). Dental plaque is a complex biofilm of oral microorganisms, mainly bacteria and fungi, attached to the tooth surface. In most cases, oral biofilms are composed of commensal bacteria, creating a harmless or even beneficial microbial community, coexisting with the host in symbiosis. However, in some cases, a shift in ecology and associated functions can result in dysbiosis, eventually leading to a harmful biofilm (2) that is the origin of the development of major human oral diseases, such as caries or periodontitis (3, 4). The complexity of the bacterial community that constitutes the dental plaque is exemplified by the high taxonomic diversity that can be found: between 500 and 1,000 bacterial species have been identified as the core microbiome of the oral cavity, although many of them still remain uncultured (5, 6). Most oral bacteria belong to the bacterial phyla Firmicutes, Proteobacteria, Actinobacteria, Bacteroidetes, and Fusobacteria. Archaea, protozoa, viruses, and fungi are also present in the oral cavity of a healthy adult (7).
Plaque formation is a highly dynamic process of ecological succession, regularly reinitiated by daily oral hygiene measures. Early colonizers adhere to the salivary pellicle, where they influence the environmental conditions, thereby enabling secondary and late colonizers to grow and become part of the biofilm (8). Different types of genes with a wide variety of functions are required for the successful establishment and development of dental plaque. In addition to metabolic and physical interactions, a cell-to-cell communication mechanism known as quorum sensing (QS) is involved. Previous studies have reported the production of QS molecules, such as autoinducer peptides (AIP) and AI-2 signals in monospecific cultures of different oral pathogens (9 to 14). Both Gram-negative and Gram-positive bacteria can synthetize and sense AI-2, known as the “universal” QS signal, which may therefore play a decisive role in multispecies oral biofilm. luxS, the gene responsible for the production of AI-2, is conserved among many species of bacteria, including Aggregatibacter actinomycetemcomitans, Streptococcus mutans, Streptococcus gordonii, Streptococcus oralis, Porphyromonas gingivalis, and other oral pathogens (9–12, 14). Several reports point to an important role of this QS signal in dental plaque formation. AI-2 is essential for mutualistic biofilm growth on saliva as the sole source of nutrients in cocultures of S. oralis and Actinomyces naeslundi (14), neither of which grows well in monoculture. In addition, S. gordonii required the presence of AI-2 to form mixed-species biofilm with the periodontal pathogen P. gingivalis (12). The external addition of partially purified AI-2 from Fusobacterium nucleatum affected biofilm formation in monospecific and multispecies cultures of P. gingivalis, Treponema denticola, and Tannerella forsythia. In the presence of AI-2, biofilms showed higher biomass, greater average depth, and enhanced coaggregation between bacteria (15). In the case of Gram-positive oral pathogens, AIPs, such as competence-stimulating peptide (CSP), have been identified in various oral streptococci, including S. mutans, S. gordonii, and S. intermedius. CSP is involved in biofilm formation, bacteriocin synthesis, stress resistance, and autolysis (13, 16–18).
N-Acylhomoserine lactones (AHLs) are well-studied QS signals produced by Gram-negative bacteria, although the production of AHLs by the Gram-positive bacterium Exiguobacterium strain MPO was described recently (19). AHLs contain a homoserine lactone ring (HSL) linked by an amide bond to a fatty acid (between 4 and 18 carbons). Despite the widespread ability among Gram-negatives to produce AHLs, attempts to establish production of these QS signals by oral bacteria using different bacterial reporter strains have remained unsuccessful (10, 20, 21). AHL-producing bacteria have been isolated from the oral cavity (22–26), although these isolates are not considered members of the core oral microbiome. Additionally, AHLs seem to influence growth and protein expression in P. gingivalis (27, 28). However, despite these reports, it is still generally accepted that AHL-based signaling does not play a major role in dental plaque formation during health and disease.
Due to the key role of QS processes for the expression of pathogenic traits, including biofilm formation, the inhibition of QS, a process general known as quorum quenching (QQ), has been proposed as an alternative approach for antimicrobial therapy and for controlling pathogenic bacterial behavior to treat or prevent infectious diseases (29). QQ strategies do not directly interfere with bacterial growth ability, and hence, the probabilities of inducing resistance or tolerance against these mechanisms are lower (30, 31). Previous studies have reported the successful use of QS inhibitors to control bacterial biofilms (32–37). Since most QS inhibitors described so far act on AHL-mediated QS circuits, the confirmation of a possible role of AHL-type QS signals in dental plaque formation would open new perspectives in the prevention and treatment of oral diseases.
The aim of the present study was to explore a possible role for AHL-based signaling in dental plaque formation by evaluating the effect of externally added AHL on oral biofilms grown in vitro. The effect on pathology-related phenotypes in three different, clinically relevant, in vitro oral biofilm models for commensal, cariogenic, and periodontal biofilms developed recently (38) was assessed. Lactate production capacity and proteolytic activity were determined as indicators of shifts from commensal toward cariogenic biofilms or periodontal biofilms, respectively (38). In addition, for a selection of AHLs, the effect on microbial composition of commensal and periodontal biofilms was determined using 16S rRNA gene sequencing.
RESULTS
Presence of AHL-type quorum sensing molecules in saliva samples.
The saliva of a healthy donor was analyzed for the presence of AHLs by high-performance liquid chromatography–mass spectrometry (HPLC-MS). Two saliva samples were taken at a 4-month interval. Using high-sensitivity HPLC-MS techniques, three different AHLs could be unequivocally identified (see Fig.S1 to S3 in the supplemental material) in the first sample, while only one of them was present in the second sample (Table 1). On both occasions, OC8-HSL was identified in the samples at concentrations of 1.35 and 10.71 ng/ml of saliva, respectively (Table 1). C8-HSL and HC10-HSL were identified only in the first sample, indicating variability in the pattern of AHLs present in the saliva with time.
TABLE 1.
Quantification of AHLs present in two saliva samples obtained from a healthy donor at a 4-month interval
| Saliva sample | AHL | AHL concn (ng/ml) |
|---|---|---|
| 1 | OC8-HSL | 1.35 |
| C8-HSL | 197.68 | |
| HC10-HSL | 4.17 | |
| 2 | OC8-HSL | 10.71 |
Effect of exogenous AHLs on oral biofilm formation.
Very small changes were observed in the number of CFU when the different unsubstituted and oxo-substituted AHLs were added to the culture medium in the three oral biofilm models tested, since in no case was the difference observed with the respective control biofilm higher than 1 order of magnitude (Fig. S4). Still, among the 19 AHLs tested, the exogenous addition of the long-chain AHLs C16-HSL, OC16-HSL, and OC18-HSL significantly reduced (P < 0.05) the formation of cariogenic biofilms after 48 h of growth (2.6 × 108 CFU, 2.4 × 108 CFU, and 2.5 × 108 CFU, respectively, versus 4.3 × 108 CFU for the control biofilms) (Fig. S4A). When a commensal biofilm was induced, the signal OC6-HSL (9.0 × 107 CFU) slightly but significantly (P < 0.05) reduced the number of CFU in comparison to that of the control (1.2 × 108 CFU) (Fig. S4B). Lower CFU values were also observed in commensal biofilms when C10-HSLs and C12-HSLs were added, but the difference was not statistically significant in comparison to the respective controls. Periodontal biofilms were the most sensitive to the exogenous addition of AHLs, since both higher and lower numbers of CFU were observed in comparison with the control. In this sense, the short-chain signals C4-HSL, C6-HSL, and OC6-HSL and the very long-chain OC18-HSL significantly promoted biofilm formation (6.8 × 107 CFU, 1.7 × 108 CFU, 9.6 × 107 CFU, and 1.0 × 108 CFU, respectively, versus 5.3 × 107 CFU for the control biofilms) (Fig. S4C). In contrast, C14-HSL clearly decreased the number of CFU (2.6 × 107 CFU versus 7.0 × 107 CFU for the control biofilms) (Fig. S4C).
Effect of exogenous AHLs on lactate production.
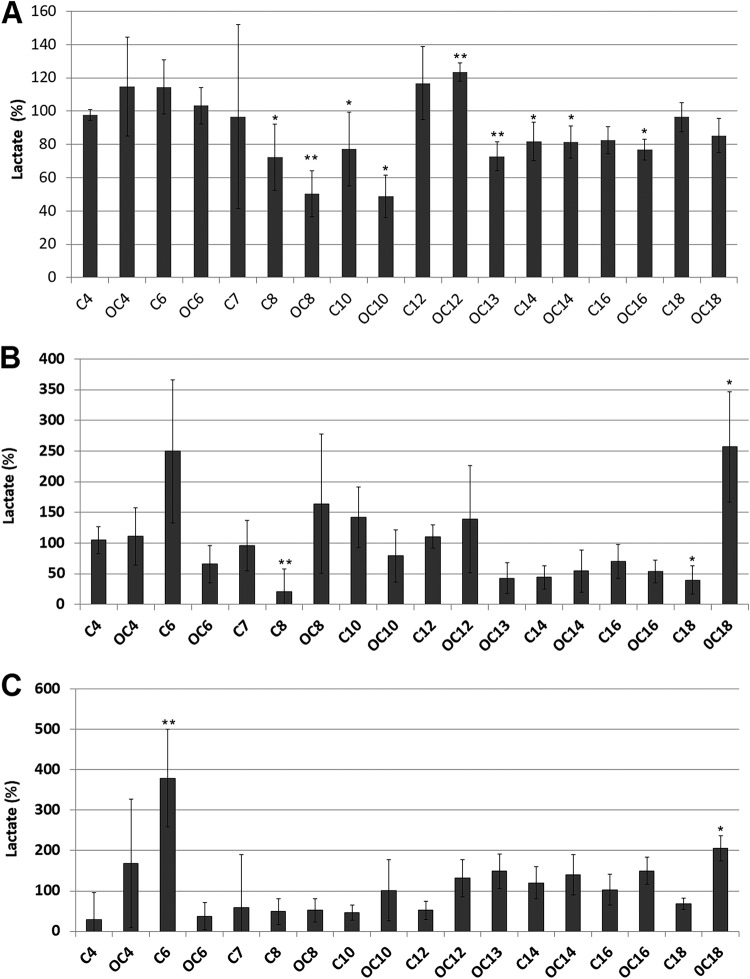
As expected, the cariogenic control biofilms showed the highest levels of lactate production (4.2 to 9.4 mM) in comparison to those of commensal control (0.2 to 2.4 mM) and periodontal control (0.2 to 2.0 mM) biofilms. Significant effects of the addition of AHLs were observed on lactic acid production in the different oral biofilm models when each treated condition was compared to its respective untreated control. The addition of the medium- and long-chain AHLs C8-HSL, OC8-HSL, C10-HSL, OC10-HSL, OC13-HSL, C14-HSL, and OC14-HSL caused a reduction in the production of lactic acid (72.3%, 50.3%, 77.2%, 48.8%, 72.8%, 81.9%, and 81.2%, respectively) in comparison to the control (100%) in cariogenic biofilms. Interestingly, the molecule OC12-HSL caused an increase (123.5%) in the production of lactate (Fig. 1A). Shorter AHLs had no significant effect on lactic acid accumulation in cariogenic biofilms. These effects were not observed in the experiments in which lactic acid production in the control cariogenic biofilms was lower than 7.0 mM (data not shown), indicating that robust fermentative activity is needed in the biofilm in order to observe the effect of the addition of AHLs. In the case of commensal biofilms, the addition of C8-HSL caused a significant decrease in the production of lactate (20.5%) (Fig. 1B). An inhibitory effect on lactic acid production was also observed in commensal biofilms when the long-chain signals between OC13-HSL and C18-HSL were added to the culture medium, although this effect was statistically significant only for the latter. The opposite trend seems to be present in the periodontal biofilm, where, in general, the long-chain AHLs increased lactate accumulation while the short-chain ones decreased lactic acid production (Fig. 1C). The addition of OC18-HSL caused a large increase (256.9%) in lactic acid production in commensal biofilms (Fig. 1B), and the effect was also observed in periodontal biofilms (205.5%) (Fig. 1C). The signal C6-HSL also produced an increase in lactate accumulation in both commensal and periodontal biofilms. Due to the marginal effects of AHLs on cariogenic biofilms, they were not included in the subsequent sequencing analysis.
FIG 1.
Effect of the AHLs (1 μM) on lactate accumulation (percent in comparison to control cultures) in cariogenic (A), commensal (B), and periodontal (C) biofilms. The increase observed when the signal OC18-HSL is added to commensal and periodontal model biofilms was confirmed in a second independent experiment. Data are presented as means ± standard deviations (SD) (n = 4). A significant difference in comparison to untreated controls is indicated as follows: *, Welch’s test, P < 0.05; **, Welch´s test, P < 0.01.
Effect of exogenous AHLs on protease activity.
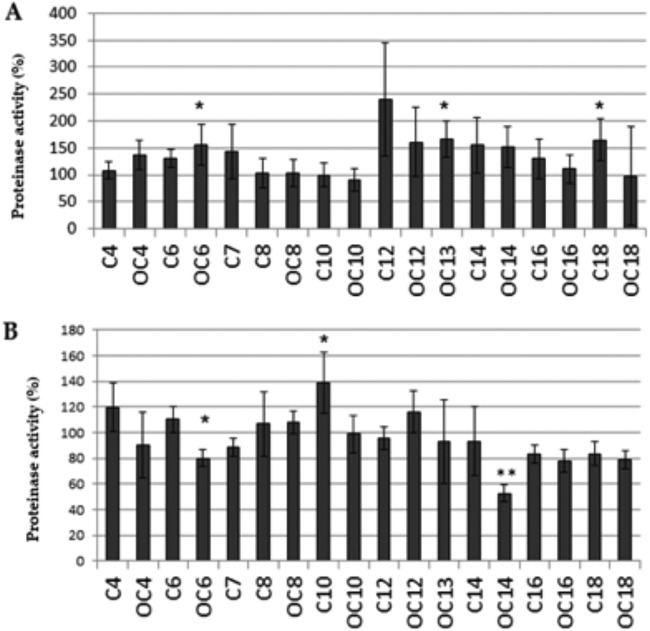
Protease activity was measured only in the commensal and periodontal biofilm models. As expected, the protease activity was higher in periodontal control biofilms (2.3 × 107 to 3.4 × 107 relative fluorescence units [RFU]/min) than in commensal control biofilms (7 × 106 to 2.9 × 107 RFU/min). The addition of several short- and long-chain AHLs caused an increase in protease activity of the commensal biofilm in comparison with that of the respective untreated controls, but this increase was statistically significant only for OC6-HSL, OC13-HSL, and C18-HSL (156%, 166.4%, and 165.1% versus 100% for control biofilms) (Fig. 2A). In contrast, the protease activity was reduced by OC6-HSL and OC14-HSL in the periodontal biofilm (80% and 52.6%) and increased by C10-HSL (138.8%) (Fig. 2B).
FIG 2.
Effect of the addition of AHLs (1 μM) on protease activity (%) in commensal (A) and periodontal (B) biofilms. Data are presented as means ± SD (n = 4). A significant difference in comparisons to untreated controls is indicated as follows: *, Welch´s test, P < 0.05; **, Welch´s test, P < 0.01.
Effect of exogenous AHLs on microbial composition.
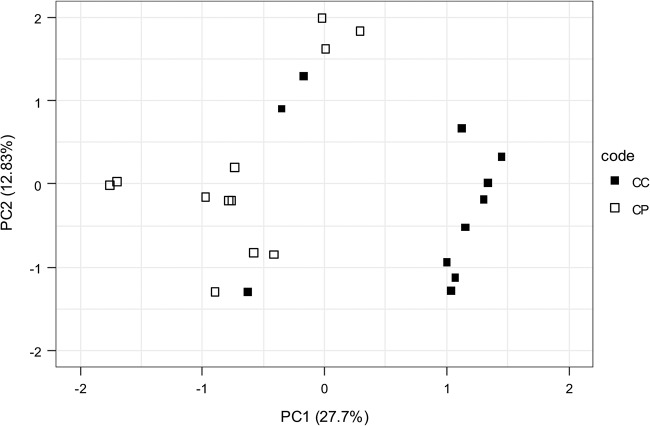
In order to explain the effects observed in the phenotypic assays, the bacterial composition of the biofilms was determined using 16S rRNA gene sequencing (Fig. S5). The control, commensal, and periodontal biofilms differed significantly in composition (permutational multivariate analysis of variance [PERMANOVA], F = 6.08, P = 1e−04) (Fig. 3). The Shannon diversity for the periodontal biofilms was higher than that for commensal biofilms (commensal biofilm mean of 2.21 versus a periodontal biofilm mean of 2.38; P = 0.03). PC1, explaining 27.7% of the variance, separated the commensal and periodontal biofilms and is represented most notably by a differential abundance of OTU5 and OTU69 (both Fusobacterium species) and OTU17 and OTU18 (Prevotella and Alloprevotella, respectively).
FIG 3.
Comparison of species compositions between in vitro commensal (control commensal [CC]) and periodontal (control periodontal [CP]) biofilms from 3 independent experiments performed over a period of 3 weeks. Principal-component analysis of commensal biofilms (filled squares) and periodontal biofilms (open squares) grown without exogenously added AHLs was performed. PC1, explaining 27.7% of the variance, separated the commensal and periodontal biofilms (PERMANOVA, F = 6.08, P = 1e−04).
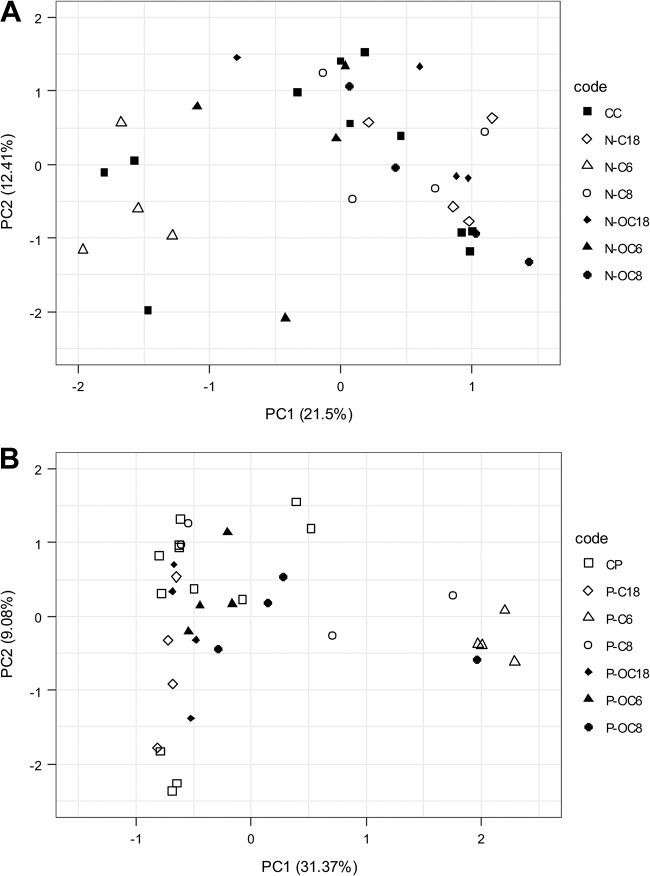
The effects of C6-HSL, OC6-HSL, C8-HSL, OC8-HSL, C18-HSL, and OC18-HSL on the bacterial composition of commensal and periodontal biofilms were determined and compared with those of their respective intraexperimental untreated control biofilms. When 1 μM AHL was added exogenously during growth of commensal biofilms, the community compositions did not shift, in comparison to their respective controls (Fig. 4A). In contrast, in the periodontal biofilms, a clear shift was observed upon addition of C6-HSL along the PC1 axis, explaining 31.37% of the variance (Fig. 4B).
FIG 4.
Effect of 1 μM exogenously added AHL on species composition of grown commensal (A) and periodontal (B) biofilms. Filled and open squares represent the untreated controls in commensal and periodontal biofilms, respectively. Open symbols represent the C variant of the HSL, while filled symbols represent the OC variant of the HSL. Triangles, C6-HSL and OC6-HSL; circles, C8-HSL and OC8-HSL; diamonds, C18-HSL and OC18-HSL in both panels.
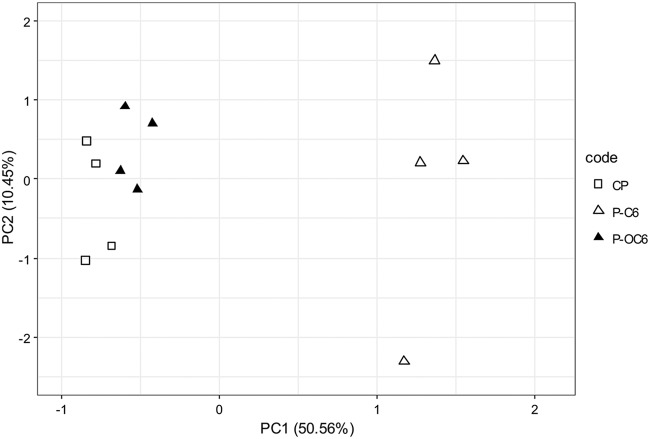
When the effect of C6-HSL was specifically compared to that of the closely related OC6-HSL, a clear and specific C6-derived shift in community composition was observed (Fig. 5). The difference was not significant for the commensal biofilms (data not shown); however, it was significant for the periodontal biofilms (PERMANOVA; F = 9.31, P = 2e−04). A clear shift along PC1 (explaining 50.1% variance) can be observed. PC1 is represented most notably by a Peptostreptococcus species (OTU9), an Alloprevotella species (OTU10), and a Prevotella species (OTU23), indicating that these species were largely responsible for the observed shift. Of importance, these species also differentiate commensal biofilms from biofilms grown in a periodontal model (Fig. S6).
FIG 5.
Comparison of the effects of C6-HSL and OC6-HSL on species composition of periodontal biofilms. A clear shift along PC1, explaining 50.6% variance in periodontal biofilms, can be observed. The three groups differed significantly (PERMANOVA; F = 9.31, P = 2e−04).
DISCUSSION
Cell-to-cell communication is known to play an essential role in microbial pathogenesis, coordinating the physiological behavior, biofilm development, and virulence of several oral pathogens (39). In view of the complexity of the oral microbiota, the characterization of these communication processes is crucial for understanding how oral bacteria interact with each other and for predicting their potential impact on the development of dental plaque-associated oral diseases. Moreover, since the structure and physiological attributes of biofilms confer an inherent resistance to antimicrobial agents (1), the description of the key factors contributing to the formation of pathogenic oral biofilm may allow the identification of novel targets for the development of new antipathogenic strategies. Current intervention strategies attempt to prevent the initial microbial attachment or penetrate the biofilm matrix and kill the associated cells. However, the future approaches to control bacterial biofilms will probably be based on the inhibition of genes involved in biofilm formation (1), in some cases through the disruption of the QS systems (36, 37). Since most QS inhibitors described so far act on AHL-mediated QS circuits, the confirmation of a possible role of AHL-type QS signals in dental plaque formation would open new perspectives in the prevention and treatment of oral diseases.
Despite the fact that the current paradigm excludes a role of the AHL-type QS signals in oral biofilm (39, 40) due to the unsuccessful identification of these molecules in pure cultures of oral pathogenic bacteria, important changes mainly in acid lactic production and protease activity were observed when these QS signals were added. These changes do not seem to be derived from gross quantitative changes in biofilm formation, since the observed differences in CFU counts, smaller than 1 log, should not result in a clinically different outcome in the oral cavity (41). Therefore, changes in biofilm formation and metabolic activity observed in the presence of particular exogenous AHLs do not seem to be caused by growth inhibition of bacteria. The potential antimicrobial activity of each AHL at 1 mg/ml on bacterial growth of S. mutans was analyzed, but no inhibitory effect was observed (data not shown), supporting the hypothesis that the changes generated in biofilm activity and microbial composition are not derived from growth inhibition. Toxicity of OC12-HSL and its tetramic acid degradation product has been reported for Gram-positive bacteria (42) at concentrations around 10-fold higher than those used in this study. Accordingly, no inhibitory activity of oxo-substituted AHL derivatives was observed in the oral biofilm models (see Fig. S4 in the supplemental material).
While only small changes could be observed in CFU numbers, important changes in metabolic activities were found after the addition of specific AHLs in the three oral biofilm models. These phenotypic changes without a concurrent large effect on bacterial growth are typical of a QS-derived gene regulation process. All together, these data strongly support the idea of a potential role of the AHL-type QS molecules in the oral cavity. The wide range of responses to the addition of the signals, with biofilm models responding in opposite directions to the addition of very similar AHL signals, may be derived from the specificity of some of the QS receptors. It is well known that while the cognate AHLs can activate the QS circuits, noncognate AHLs, even of a very similar structure, may act to inhibit the same receptor (43, 44). The signaling molecules C6-HSL, C8-HSL, C10-HSL, and C16-HSL, which generated the most important changes in the model biofilms, have been previously described in bacteria isolated from the oral environment (23–26). In contrast, other signals such as OC6-HSL, C18-HSL, and OC18-HSL, which showed the highest effect on the studied phenotypes, have not been described in the oral environment so far. Moreover, C14-HSL, one of the AHL signals that generated changes in the model oral biofilms, has been previously reported for causing changes in growth and protein expression in P. gingivalis (27). In that case, the concentration used was 2 orders of magnitude higher than the one used in the present work, 1 μM, which is in range with the concentration of QS molecules produced by pathogens (45) and has previously shown effects on the non-AHL-producing food-borne pathogens Escherichia coli and Salmonella enterica (46). Regarding the production of AHLs by oral pathogens, previous studies have been unable to obtain evidence of the production of AHLs by P. gingivalis and other oral pathogens by using the supernatants of monospecific cultures (10, 21). Beyond the limitations of the use of biosensors for the detection of AHLs, it should be taken into account that production of AHLs may change depending on culture medium and conditions (47, 48) and also depends on other factors, such as surface attachment or cell-to-cell adherence, which activates AHL synthesis. For example, the gene A1S_0114 of Acinetobacter baumannii, encoding a small acidic acyl carrier protein (ACP) essential for AHL synthesis, is expressed at high levels in biofilms and downregulated in planktonic cells (49), further linking cell adherence and QS expression. Moreover, AHLs can be detected only in the culture medium of this pathogen under static conditions (48). This could explain that AHLs have not been found so far in pure cultures of oral pathogens.
We cannot discard that AHL producers and sensors may play a key role in biofilm formation despite representing a minority in the biofilm community. Some bacteria have LuxR homologs, called LuxR orphans, which do not have an associated LuxI autoinducer synthase but can interact with the autoinducers synthetized by other bacteria (47, 50). As an example, although E. coli does not produce AHLs, it possesses a LuxR homolog, called SdiA, and uses various AHLs as switches for its regulatory activity (51). Also, although the AHL-type QS molecules are considered typical of Gram-negative bacteria, previous reports indicate the ability of Gram-positive bacteria not only to produce them (19) but also to respond to them (52, 53). In the genome of Streptococcus mutans, the predicted product of the smu.46 gene showed features of the LuxR family of regulatory proteins, suggesting the existence of a LuxR orphan (53). Indeed, a previous study with Staphylococcus aureus has demonstrated the interaction of this bacterium with the OC12-HSL produced by Pseudomonas aeruginosa in a saturable and specific manner, inhibiting the production of exotoxins and enhancing protein A expression (52). Our results strongly indicate the existence of complex QS networks in the oral biofilm beyond the accepted model in which the oral bacterial communication is mediated by AIPs and AI-2 (39, 40).
The 16S rRNA gene sequencing data indicate that exogenous C6-HSL has the ability to change the microbial composition of in vitro oral biofilms. Interestingly, OC6-HSL does not induce a similar shift in composition. This specificity of C6-HSL points toward the involvement of a specific C6-HSL binding protein (possibly a receptor) that detects the presence of this signal. Surprisingly, the effect of the addition of C6-HSL on the microbial composition of both commensal and periodontal biofilms was accompanied by an increase in lactate production in both models (Fig. 1). In contrast, no significant effect on protease activity was observed (Fig. 2), indicating that this phenotype is not sensitive enough to allow the detection of important shifts in the population. C6-HSL increases the abundance of Alloprevotella, Peptostreptococcus, and Prevotella species in periodontal biofilms, the latter two being important members of the orange complex as defined by Socransky et al. (54). This could be a direct effect, for instance, by increasing the biofilm-forming capacity of these species. Alternatively, the reduction of the fitness of bacterial species that compete with, or have an antagonistic activity toward, Peptostreptococcus and Prevotella species could also explain this effect. Further research on the mechanism of the observed increased abundance is needed to determine this. If C6-HSL induces the presence of certain periodontal pathogens directly, inhibition of C6-HSL sensing could become a viable option to impact the shift from a commensal to a periodontal biofilm. The QQ strategies to prevent the biofilm formation by oral pathogens constitute an attractive alternative, since these approaches do not interfere with bacterial growth, and hence, the probabilities of inducing resistance or tolerance against these mechanisms are lower. In fact, it has been demonstrated that QQ strategies increase the susceptibility to antibiotics in biofilm-forming pathogens (55–57). Further studies are required to assess the possible role of the AHL-type QS molecules in the virulence of oral pathogens and to explore the viability of the application of these strategies in the field of oral health.
The results indicate a potential role of AHLs in the development of dysbiosis related to periodontal diseases. Further studies are required to confirm the role of the AHL-type QS molecules in dental plaque formation in vivo, which would open the possibility of applying QQ strategies to prevent the development of dysbiosis in oral biofilms leading to the development of oral diseases.
MATERIALS AND METHODS
Extraction and identification of AHLs by HPLC-MS.
The identification of AHLs was done in different acidified saliva samples from the same healthy donor who provided a written informed consent that was approved by the Comité Ético de Investigación Clínica de Galicia (protocol 2009/319 modified in July 2017). The extraction of the sample (approximately 0.5 ml) was performed twice in an appropriate volume of organic solvents, ethyl acetate, and dichloromethane, and then it was evaporated to dryness at 40°C. AHLs present in the oral samples were reconstituted in acetonitrile and quantified by HPLC-MS methodology (45). Pure AHLs covering the whole range of AHL lengths (from C4-HSL to C18-HSL), both substituted and unsubstituted, were obtained from Sigma-Aldrich and from the University of Nottingham and used as external standards for quantification.
Test compounds.
QS AHL type molecules were used at 1 μM for this study. They included N-butanoyl-l-homoserine lactone (C4-HSL), N-oxobutanoyl-l-homoserine lactone (OC4-HSL), N-hexanoyl-l-homoserine lactone (C6-HSL), N-oxohexanoyl-l-homoserine lactone (OC6-HSL), N-heptanoyl-l-homoserine lactone (C7-HSL), N-octanoyl-l-homoserine lactone (C8-HSL), N-oxooctanoyl-l-homoserine lactone (OC8-HSL), N-decanoyl-l-homoserine lactone (C10-HSL), N-oxodecanoyl-l-homoserine lactone (OC10-HSL), N-dodecanoyl-l-homoserine lactone (C12-HSL), N-oxododecanoyl-l-homoserine lactone (OC12-HSL), N-oxotridecanoyl-l-homoserine lactone (OC13-HSL), N-tetradecanoyl-l-homoserine lactone (C14-HSL), N-oxotetradecanoyl-l-homoserine lactone (OC14-HSL), N-hexadecanoyl-l-homoserine lactone (C16-HSL), N-oxohexadecanoyl-l-homoserine lactone (OC16-HSL), N-octadecanoyl-l-homoserine lactone (C18-HSL), and N-oxooctadecanoyl-l-homoserine lactone (OC18-HSL). The compounds were obtained from Sigma-Aldrich (St. Louis, MO) and the University of Nottingham.
Biofilm formation.
Saliva was collected on ice from a single healthy donor, as previously described (38). The saliva was diluted 2-fold with sterile glycerol, aliquoted, and stored at –80°C.
Biofilms were grown in the Amsterdam active attachment model (AAA model) (58) assembled with round glass coverslips (diameter, 12 mm). The model was differentiated toward commensal, cariogenic, or periodontal biofilms by using one of three culture media (38): (i) for commensal biofilms, buffered semidefined McBain medium (59), containing 2.5 g/liter mucin, 2 g/liter Bacto peptone, 2 g/liter Trypticase peptone, 1 g/liter yeast extract, 0.35 g/liter NaCl, 0.2 g/liter KCl, 0.2 g/liter CaCl2, 1 mg/ml hemin, and 2 mg/liter vitamin K1 with PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] at pH 7.0; (ii) for cariogenic biofilms, buffered semidefined McBain medium with 0.2% sucrose; and (iii) for periodontal biofilms, buffered semidefined McBain medium with 10% fetal calf serum (FCS).
All biofilms were inoculated for 8 h at 37°C under anaerobic conditions using a 1:50 dilution of saliva in the model-specific medium. Cariogenic biofilm was grown in the presence of sucrose for a total of 48 h, with twice daily (at 8 and 16 h) medium refreshments as described previously (38). Commensal and periodontal biofilms were grown for a total of 9 days. After an initial 8-h inoculation, the medium was refreshed every 24 h, except for the weekends.
The respective AHLs were added during all phases of biofilm formation, including the initial 8-h inoculation step.
Acid production assay.
Lactic acid production by the biofilms was determined prior to harvesting to estimate the cariogenic phenotype (38, 58). The biofilms on coverslips were placed in a 24-well plate containing 1.5 ml of buffered peptone water (BPW) with 0.2% sucrose in each well. Acid formation was allowed for 3 h at 37°C under anaerobic conditions. The amount of lactic acid produced was analyzed using the colorimetric assay described previously (60).
Protease activity.
To quantify the protease activity of the biofilms, the fluorescence resonance energy transfer (FRET) assay was used (61, 62). The probe used in the present study was PEK-054 (63), a probe for total protease activity (kindly provided by F. J. Bikker, Department of Oral Biochemistry, Academic Centre for Dentistry Amsterdam, The Netherlands). The spent medium was filter sterilized using 0.2-μm-pore-size filters and stored at –20°C until needed. Wells of clear-bottom 96-well plates were filled with 100 μl of Tris-buffered saline (TBS; 50 mM Tris, 150 mM NaCl, pH 7.6) and 100 μl of sterile spent medium. Four microliters of substrate PEK-054 (800 μM) was added to each well to a final concentration of 16 μM. Sterile fresh culture broth was used as the control. The fluorescence of each well was read for 2 h with 5-min intervals on a fluorescence microplate reader with an excitation wavelength of 485 nm and an emission wavelength of 530 nm. Relative fluorescence (RF) values were obtained after correction against the culture broth control. The protease activity was defined in RFU per minute. The protease activity was measured in commensal and periodontal biofilms.
Harvesting of biofilms.
All biofilms were harvested by transferring the glass coverslips into 2 ml of phosphate-buffered saline (PBS), as previously described (38). Biofilms were sonicated to disperse following the phenotypic assays, to allow CFU determination and subsequent DNA isolation for sequencing analysis.
Biofilm biomass estimation.
The numbers of CFU were determined to estimate biofilm formation (38, 58). Serial dilutions in cysteine peptone water (CPW) of the dispersed biofilms were plated on tryptic soy agar blood plates (TSA-b). The plates were incubated anaerobically for 96 h at 37°C, and the CFU were counted. Since not all AHLs could be tested simultaneously for each model, the results are expressed as a percentage with respect to the corresponding control. Average CFU values for control cultures were 6.47 × 107 ± 2.57 × 107 for cariogenic, 8.59 × 107 ± 4 × 107 for commensal, and 1 × 108 ± 0.8 × 108 for periodontal biofilms.
DNA isolation and microbiome analysis.
Commensal and periodontal biofilms were used for microbiome analysis. Total DNA was isolated and purified as previously described (38). Bacterial DNA concentration was determined by quantitative PCR (qPCR), using a universal primer-probe set targeting the 16S rRNA gene (64). Next, 1 ng of DNA was used to amplify the V4 hypervariable region of the 16S rRNA gene, as described previously (65) except that 33 amplification cycles were performed. The amplicons were pooled equimolarly and purified from agarose gel (Illustra; GE Healthcare, United Kingdom). Paired-end reads of 251 nt were generated using the Illumina MiSeq platform and Illumina MiSeq reagent kit v3 (Illumina, Inc., San Diego, CA, USA) at the VUmc Cancer Center Amsterdam (Amsterdam, the Netherlands). The sequence data were processed as described previously (66); however, 10% mismatches in the overlap here translates to a maximum of 25 mismatches. Seventy-five samples (including controls), represented by 1.87 million read pairs, were processed; of these, 72.3% passed the merging and quality-filtering steps (maximum expected error, 0.5). Of these quality-filtered reads, 91.0% mapped to the final OTU table.
Sequencing data analysis.
The OTU table was randomly subsampled at 11,000 reads/sample and in R v3.5.1 (67) using the microbiome v1.4.0 (68), phyloseq v1.26.0 (69), and vegan v2.5-3 (70) packages from R. The OTU table was log2 transformed to ordinate the data by principal-component analysis (PCA) into two dimensions. The PCA coordinates were eigenvalue scaled. Plots were generated with ggplot2 v.3.1.0 (71).
Statistical analysis.
Due to the large number of conditions obtained, it was not possible to test all of them in a single experiment. Therefore, several batches of experiments, all with their own untreated controls, were performed using the same frozen saliva inoculum. A Welch’s t test was performed to determine the statistical significance of the effect of the addition of each AHL on biofilm formation (CFU), lactic acid production, and protease activity between each treated well and its respective untreated control. Significant differences were determined at P values of <0.05 and <0.01.
Permutational multivariate analysis of variance (PERMANOVA) using the Bray-Curtis distance was performed in R on the transformed OTU table using 9,999 permutations. Differences in Shannon diversities were tested in R using the Wilcoxon rank sum (Mann-Whitney) test.
Data availability.
The data are available in the NCBI BioProject database under accession number PRJNA573890.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the grant Axudas do Programa de Consolidación e Estructuración de Unidades de Investigación Competitivas (GPC) from the Consellería de Cultura, Educación e Ordenación Universitaria, Xunta de Galicia (ED431B2017/53). A.M. was supported by a predoctoral fellowship from the Consellería de Cultura, Educación e Ordenación Universitaria, Xunta de Galicia (ED481A-2015/311), and by the Real Academia Galega de Ciencias (Premio á Transferencia de Tecnoloxía en Galicia 2016). B.P.K. was supported by a UvA focal point grant on oral infection and inflammation.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–193. doi: 10.1128/cmr.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R, Romero H, Simón-Soro A, Pignatelli M, Mira A. 2012. The oral metagenome in health and disease. ISME J 6:46–56. doi: 10.1038/ismej.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marsh PD. 1994. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res 8:263–271. doi: 10.1177/08959374940080022001. [DOI] [PubMed] [Google Scholar]
- 4.Hajishengallis G, Darveau RP, Curtis MA. 2012. The keystone-pathogen hypothesis. Nat Rev Microbiol 10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. 2005. Defining the normal bacteria flora of the oral cavity. J Clin Microbiol 43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaura E, Keijser BJ, Huse SM, Crielaard W. 2009. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol 9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolenbrander PE, Palmer RJ, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. 2006. Bacterial interactions and successions during plaque development. Periodontol 2000 42:47–49. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 9.Fong KP, Chung WO, Lamont RJ, Demuth DR. 2001. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect Immun 69:7625–7634. doi: 10.1128/IAI.69.12.7625-7634.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burgess NA, Kirke DF, Williams P, Winzer K, Hardie KR, Meyers NL, Aduse-Opoku J, Curtis MA, Cámara M. 2002. LuxS-dependent quorum sensing in Porphyromonas gingivalis modulates protease and haemagglutinin activities but is not essential for virulence. Microbiology 148:763–772. doi: 10.1099/00221287-148-3-763. [DOI] [PubMed] [Google Scholar]
- 11.Merritt J, Qi F, Goodman SD, Anderson MH, Shi W. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect Immun 71:1972–1979. doi: 10.1128/iai.71.4.1972-1979.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNab R, Ford SK, El-Sabaeny A, Barbieri B, Cook GS, Lamont RJ. 2003. Lux-S based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J Bacteriol 185:274–284. doi: 10.1128/jb.185.1.274-284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petersen FC, Pecharki D, Scheie AA. 2004. Biofilm mode of growth of Streptococcus intermedius favored by a competence-stimulating signaling peptide. J Bacteriol 186:6327–6331. doi: 10.1128/JB.186.18.6327-6331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rickard AH, Palmer RJ, Blehert DS, Campagna SR, Semmelhack MF, Egland PG, Bassler BL, Kolenbrander PE. 2006. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol Microbiol 60:1446–1456. doi: 10.1111/j.1365-2958.2006.05202.x. [DOI] [PubMed] [Google Scholar]
- 15.Jang YJ, Choi YJ, Lee SH, Jun HK, Choi BK. 2013. Autoinducer 2 of Fusobacterium nucleatum as a target molecule to inhibit biofilm formation of periodontopathogens. Arch Oral Biol 58:17–27. doi: 10.1016/j.archoralbio.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 16.Senadheera D, Cvitkovitch DG. 2008. Quorum sensing and biofilm formation by Streptococcus mutans. Adv Exp Med Biol 631:178–188. doi: 10.1007/978-0-387-78885-2_12. [DOI] [PubMed] [Google Scholar]
- 17.Perry JA, Cvitkovitch DG, Levesque CM. 2009. Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol Lett 299:261–266. doi: 10.1111/j.1574-6968.2009.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Senadheera D, Krastel K, Mair R, Persadmehr A, Abranches J, Burne RA, Cvitkovitch DG. 2009. Inactivaction of VicK affects acid production and acid survival of Streptococcus mutans. J Bacteriol 191:6415–6424. doi: 10.1128/JB.00793-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biswa P, Doble M. 2013. Production of acylated homoserine lactone by gram-positive bacteria isolated from marine water. FEMS Microbiol Lett 343:34–41. doi: 10.1111/1574-6968.12123. [DOI] [PubMed] [Google Scholar]
- 20.Whittaker CJ, Klier CM, Kolenbrander PE. 1996. Mechanisms of adhesion by oral bacteria. Annu Rev Microbiol 50:513–552. doi: 10.1146/annurev.micro.50.1.513. [DOI] [PubMed] [Google Scholar]
- 21.Frias J, Olle E, Alsina M. 2001. Periodontal pathogens produce quorum sensing signal molecules. Infect Immun 69:3431–3434. doi: 10.1128/IAI.69.5.3431-3434.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin WF, Purmal K, Chin S, Chan KY, Chan KG. 2012. Long chain N-acyl homoserine lactone production by Enterobacter sp. isolated from human tongue surfaces. Sensors (Basel) 12:14307–14314. doi: 10.3390/s121114307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin WF, Purmal K, Chin S, Chan KY, Koh CL, Sam CK, Chan KG. 2012. N-Acyl homoserine lactone production by Klebsiella pneumoniae isolated from human tongue surface. Sensors (Basel) 12:3472–3483. doi: 10.3390/s120303472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen JW, Chin S, Tee KK, Yin WF, Choo YM, Chan KG. 2013. N-Acyl homoserine lactone-producing Pseudomonas putida strain T2-2 from human tongue surface. Sensors (Basel) 13:13192–13203. doi: 10.3390/s131013192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goh SY, Tan WS, Khan SA, Chew HP, Kasim NHA, Yin WF, Chan KG. 2014. Unusual multiple production of N-acylhomoserine lactones a by Burkholderia sp. strain C10B isolated from dentine caries. Sensors (Basel) 14:8940–8949. doi: 10.3390/s140508940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goh SY, Khan SA, Tee KK, Kasim NHA, Yin WF, Chan KG. 2016. Quorum sensing activity of Citrobacter amalonaticus L8A, a bacterium isolated from dental plaque. Sci Rep 6:20702. doi: 10.1038/srep20702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komiya-Ito A, Ito T, Yamanaka A, Okuda K, Yamada S, Kato T. 2006. N-Tetradecanoyl homoserine lactone, signaling compound for quorum sensing, inhibits Porphyromonas gingivalis growth. Res J Microbiol 1:353–359. doi: 10.3923/jm.2006.353.359. [DOI] [Google Scholar]
- 28.Asahi Y, Noiri Y, Igarashi J, Asai H, Suga H, Ebisu S. 2010. Effects of N-acyl homoserine lactone analogues on Porphyromonas gingivalis biofilm formation. J Periodontal Res 45:255–261. doi: 10.1111/j.1600-0765.2009.01228.x. [DOI] [PubMed] [Google Scholar]
- 29.Romero M, Mayer C, Muras A, Otero A. 2015. Silencing bacterial communication through enzymatic quorum sensing inhibition, p 219–236. In Kalia VC. (ed), Quorum sensing vs quorum quenching: a battle with no end in sight. Springer, New Delhi, India. [Google Scholar]
- 30.García-Contreras R, Maeda T, Wood TK. 2016. Can resistance against quorum-sensing interference be selected? ISME J 10:4–10. doi: 10.1038/ismej.2015.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guendouze A, PLener L, Bzdrenga J, Jacquet P, Rémy B, Elias M, Lavigne JP, Daudé D, Chabriêre E. 2017. Effect of quorum quenching lactonase in clinical isolates of Pseudomonas aeruginosa and comparison with quorum sensing inhibitors. Front Microbiol 8:227. doi: 10.3389/fmicb.2017.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeon KM, Lee CH, Kim J. 2009. Magnetic enzyme carrier for affective biofouling control in a membrane bioreactor based on enzymatic quorum quenching. Environ Sci Technol 43:7403–7409. doi: 10.1021/es901323k. [DOI] [PubMed] [Google Scholar]
- 33.Kim JH, Choi DC, Yeon KM, Kim SR, Lee CH. 2011. Enzyme-immobilized nanofiltration membrane to mitigate biofouling based on quorum quenching. Environ Sci Technol 45:1601–1607. doi: 10.1021/es103483j. [DOI] [PubMed] [Google Scholar]
- 34.Oh HS, Yeon KM, Yang CS, Kim SR, Lee CH, Park SY, Han JY, Lee JK. 2012. Control of membrane biofouling in MBR for wastewater treatment by quorum quenching bacteria encapsulated in microporous membrane. Environ Sci Technol 46:4877–4884. doi: 10.1021/es204312u. [DOI] [PubMed] [Google Scholar]
- 35.Lade H, Paul D, Kweon JH. 2014. Quorum quenching mediated approaches for control of membrane biofouling. Int J Biol Sci 10:550–565. doi: 10.7150/ijbs.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanova K, Fernandes MM, Francesko A, Mendoza E, Guezguez J, Burnet M, Tzanov T. 2015. Quorum-quenching and matrix-degrading enzymes in multilayer coatings synergistically prevent bacterial biofilm formation on urinary catheters. ACS Appl Mater Interfaces 7:27066–27077. doi: 10.1021/acsami.5b09489. [DOI] [PubMed] [Google Scholar]
- 37.Muras A, Mayer C, Romero M, Camino T, Ferrer MD, Mira A, Otero A. 2018. Inhibition of Streptococcus mutans biofilms formation by extracts of Tenacibaculum sp. 20J, a bacterium with wide-spectrum quorum quenching activity. J Oral Microbiol 10:1429788. doi: 10.1080/20002297.2018.1429788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janus MM, Keijser BJF, Bikker FJ, Exterkate RAM, Crielaard W, Krom BP. 2015. In vitro phenotypic differentiation towards commensal and pathogenic oral biofilms. Biofouling 31:503–510. doi: 10.1080/08927014.2015.1067887. [DOI] [PubMed] [Google Scholar]
- 39.Guo L, He X, Shi W. 2014. Intercellular communications in multispecies oral microbial communities. Front Microbiol 5:328. doi: 10.3389/fmicb.2014.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jakubovics NS, Kolenbrander PE. 2010. The road to ruin: the formation of disease-associated oral biofilms. Oral Dis 16:729–739. doi: 10.1111/j.1601-0825.2010.01701.x. [DOI] [PubMed] [Google Scholar]
- 41.Janus MM, Crielaard W, Zaura E, Keijser BJ, Brandt BW, Krom BP. 2016. A novel compound to maintain a healthy oral plaque ecology in vitro. J Oral Microbiol 8:32513. doi: 10.3402/jom.v8.32513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaufmann GF, Sartorio R, Lee SH, Rogers CJ, Meijler MM, Moss JA, Clapham B, Brogan AP, Dickerson TJ, Janda KD. 2005. Revisiting quorum sensing: discovery of additional chemical and biological functions for 3-oxo-N-acylhomoserine lactones. Proc Natl Acad Sci U S A 102:309–314. doi: 10.1073/pnas.0408639102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McClean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Cámara M, Daykin M, Lamb JH, Swift S, Bycroft B, Gordon SA, Stewart GS, Williams P. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 44.McLean RJ, Pierson LS III, Fuqua C. 2004. A simple screening protocol for the identification of quorum signal antagonists. J Microbiol Methods 58:351–360. doi: 10.1016/j.mimet.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 45.Romero M, Muras A, Mayer C, Buján N, Magariños B, Otero A. 2014. In vitro quenching of fish pathogen Edwardsiella tarda AHL production using marine bacterium Tenacibaculum sp. strain 20J cell extracts. Dis Aquat Organ 108:217–225. doi: 10.3354/dao02697. [DOI] [PubMed] [Google Scholar]
- 46.Bai JA, Rai VR. 2016. Effect of small chain N acyl homoserine lactone quorum sensing signals on biofilms of food-borne pathogens. J Food Sci Technol 53:3609–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muras A, López-Pérez M, Mayer C, Parga A, Amaro-Blanco J, Otero A. 2018. High prevalence of quorum-sensing and quorum-quenching among cultivable bacteria and metagenomic sequences in the Mediterranean Sea. Genes 9:100. doi: 10.3390/genes9020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mayer C, Muras A, Romero M, Rumbo-Leal S, López Días M, Tomás M, Otero A. 2018. Multiple quorum quenching enzymes are active in the nosocomial pathogen Acinetobacter baumannii ATCC17978. Front Cell Infect Microbiol 8:310. doi: 10.3389/fcimb.2018.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rumbo-Feal S, Gómez MJ, Gayoso C, Álvarez-Fraga L, Cabral MP, Aransay AM, Rodríguez-Ezpeleta N, Fullaondo A, Valle J, Tomás M, Bou G, Poza M. 2013. Whole transcriptome analysis of Acinetobacter baumannii assessed by RNA-sequencing reveals different mRNA expression profiles in biofilm compared to planktonic cells. PLoS One 8:e72968. doi: 10.1371/journal.pone.0072968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patankar AV, González JE. 2009. Orphan LuxR regulators of quorum sensing. FEMS Microbiol Rev 33:739–756. doi: 10.1111/j.1574-6976.2009.00163.x. [DOI] [PubMed] [Google Scholar]
- 51.Yao Y, Martinez-Yamout MA, Dickerson TJ, Brogan AP, Wright PE, Dyson HJ. 2006. Structure of the Escherichia coli quorum sensing protein SdiA: activation of the folding switch by acyl homoserine lactones. J Mol Biol 355:262–273. doi: 10.1016/j.jmb.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 52.Qazi S, Middleton B, Muharram SH, Cockayne A, Hill P, O'Shea P, Chhabra SR, Cámara M, Williams P. 2006. N-Acylhomoserine lactones antagonize virulence gene expression and quorum sensing in Staphylococcus aureus. Infect Immun 74:910–919. doi: 10.1128/IAI.74.2.910-919.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wen ZT, Nguyen AH, Bitoun JP, Abranches J, Baker HV, Burne RA. 2011. Transcriptome analysis of LuxS-deficient Streptococcus mutans grown in biofilms. Mol Oral Microbiol 26:2–18. doi: 10.1111/j.2041-1014.2010.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol 25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 55.Brackman G, Cos P, Maes L, Nelis HJ, Coenye T. 2011. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob Agents Chemother 55:2655–2661. doi: 10.1128/AAC.00045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simonetti O, Cirioni O, Cacciatore I, Baldassarre L, Orlando F, Pierpaoli E, Lucarini G, Orsetti E, Provinciali M, Fornasari E, Di Stefano A, Giacometti A, Offidani A. 2016. Efficacy of the quorum sensing inhibitor FS10 alone and in combination with tigecycline in an animal model of staphylococcal infected wound. PLoS One 11:e0151956. doi: 10.1371/journal.pone.0151956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roudashti S, Zeighami H, Mirshahabi H, Bahari S, Soltani A, Haghi F. 2017. Synergistic activity of sub-inhibitory concentrations of curcumin with ceftzidime and ciprofloxacin against Pseudomonas aeruginosa quorum sensing related genes and virulence traits. World J Microbiol Biotechnol 33:50. doi: 10.1007/s11274-016-2195-0. [DOI] [PubMed] [Google Scholar]
- 58.Exterkate RA, Crielaard W, Ten Cate JM. 2010. Different response to amine fluoride by Streptococcus mutans and polymicrobial biofilms in a novel high-throughput active attachment model. Caries Res 44:372–379. doi: 10.1159/000316541. [DOI] [PubMed] [Google Scholar]
- 59.McBain AJ, Sissons C, Ledder RG, Sreenivasan PK, De Vizio W, Gilbert P. 2005. Development and characterization of a simple perfused oral microcosm. J Appl Microbiol 98:624–634. doi: 10.1111/j.1365-2672.2004.02483.x. [DOI] [PubMed] [Google Scholar]
- 60.van Loveren C, Buijs JF, ten Cate JM. 2000. The effect of triclosan toothpaste on enamel deminelization in a bacterial demineralization model. J Antimicrob Chemother 45:153–158. doi: 10.1093/jac/45.2.153. [DOI] [PubMed] [Google Scholar]
- 61.Kaman WE, Hulst AG, van Alphen PT, Roffel S, van der Schans MJ, Merkel T, van Belkum A, Bikker FJ. 2011. Peptide-based fluorescence resonance energy transfer protease substrates for the detection and diagnosis of Bacillus species. Anal Chem 83:2511–2517. doi: 10.1021/ac102764v. [DOI] [PubMed] [Google Scholar]
- 62.Kaman WE, Galassi F, de Soet JJ, Bizzarro S, Loos BG, Veerman ECI, van Belkum A, Hays JP, Bikker FJ. 2012. Highly specific protease-based approach for detection of Porphyromonas gingivalis in diagnosis of periodontitis. J Clin Microbiol 50:104–112. doi: 10.1128/JCM.05313-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cummings RT, Salowe SP, Cunningham BR, Wiltsie J, Park YW, Sonatore LM, Wisniewski D, Douglas CM, Hermes JD, Scolnick EM. 2002. A peptide-based fluorescence resonance energy transfer assay for Bacillus anthracis lethal factor protease. Proc Natl Acad Sci U S A 99:6603–6606. doi: 10.1073/pnas.062171599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ciric L, Pratten J, Wilson M, Spratt D. 2010. Development of a novel multi-triplex qPCR method for the assessment of bacterial community structure in oral populations. Environ Microbiol Rep 2:770–774. doi: 10.1111/j.1758-2229.2010.00183.x. [DOI] [PubMed] [Google Scholar]
- 65.O'Donnell LE, Robertson D, Nile CJ, Cross LJ, Riggio M, Sherriff A, Bradshaw D, Lambert M, Malcolm J, Buijs MJ, Zaura E, Crielaard W, Brandt BW, Ramage G. 2015. The oral microbiome of denture wearers is influenced by levels of natural dentition. PLoS One 10:e0137717. doi: 10.1371/journal.pone.0137717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koopman JE, Buij MJ, Brandt BW, Keijser BJ, Crielaard W, Zaura E. 2016. Nitrate and the origin of saliva influence composition and short chain fatty acid production of oral microcosms. Microb Ecol 72:479–492. doi: 10.1007/s00248-016-0775-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- 68.Lahti L. 2017. Bioconductor. Tools for microbiome analysis in R. Microbiome package version 1.4.0. https://microbiome.github.io/tutorials/.
- 69.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2018. vegan: community ecology package. R package version 2.4-6. https://CRAN.R-project.org/package=vegan.
- 71.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are available in the NCBI BioProject database under accession number PRJNA573890.