Lactobacillus brevis is a member of the lactic acid bacteria and is often reported as the causative agent of food or beverage spoilage, in particular, that of beer. Bacterial spoilage of beer may result in product withdrawal or recall, with concomitant economic losses for the brewing industry. A very limited number of genes involved in beer spoilage have been identified and primarily include those involved in hop resistance, such as horA, hitA, and horC. However, since none of these genes are universal, it is clear that there are likely (many) other molecular players involved in beer spoilage. Here, we report on the importance of a plasmid-encoded glycosyltransferase associated with beer spoilage by L. brevis that is involved in hop tolerance. The study highlights the complexity of the genetic requirements to facilitate beer spoilage and the role of multiple key players in this process.
KEYWORDS: lactic acid bacteria, plasmid, resistance, HorA, cell wall polysaccharide, phage, beer spoilage, Lactobacillus brevis, microbiology
ABSTRACT
Lactobacillus brevis beer-spoiling strains harbor plasmids that contain genes such as horA, horC, and hitA which are known to confer hop tolerance. The L. brevis beer-spoiling strain UCCLBBS124, which possesses four plasmids, was treated with novobiocin, resulting in the isolation of UCCLBBS124 derivatives exhibiting hop sensitivity and an inability to grow in beer. One selected derivative was shown to have lost a single plasmid, here designated UCCLBBS124_D, which harbors the UCCLBBS124_pD0015 gene, predicted to encode a glycosyltransferase. Hop tolerance and growth in beer were restored when UCCLBBS124_pD0015 was introduced in one of these hop-sensitive derivatives on a plasmid. We hypothesize that this gene modifies the surface composition of the polysaccharide cell wall, conferring protection against hop compounds. Furthermore, the introduction of this gene in trans in L. brevis UCCLB521, a strain that cannot grow in and spoil beer, was shown to furnish the resulting strain with the ability to grow in beer, while its expression also conferred phage resistance. This study underscores how the acquisition of certain mobile genetic elements plays a role in hop tolerance and beer spoilage for strains of this bacterial species.
IMPORTANCE Lactobacillus brevis is a member of the lactic acid bacteria and is often reported as the causative agent of food or beverage spoilage, in particular, that of beer. Bacterial spoilage of beer may result in product withdrawal or recall, with concomitant economic losses for the brewing industry. A very limited number of genes involved in beer spoilage have been identified and primarily include those involved in hop resistance, such as horA, hitA, and horC. However, since none of these genes are universal, it is clear that there are likely (many) other molecular players involved in beer spoilage. Here, we report on the importance of a plasmid-encoded glycosyltransferase associated with beer spoilage by L. brevis that is involved in hop tolerance. The study highlights the complexity of the genetic requirements to facilitate beer spoilage and the role of multiple key players in this process.
INTRODUCTION
Lactobacillus brevis is a major threat for commercial and amateur brewers, as strains of this species are the predominant bacterial contaminants associated with beer spoilage (1). Such L. brevis strains can grow in beer despite the presence of ethanol, low pH, and the depletion of oxygen and nutrients (2). Moreover, hop compounds added to beer for bitter flavor development during the fermentation process also exert antibacterial activity through the presence of iso-α-acids (1, 2). L. brevis beer-spoiling (BS) strains appear to have acquired chromosomally or plasmid-derived genetic content to survive and grow in beer (2). L. brevis resistance to ethanol (up to 10%) and pH lower than the optimal growth conditions (pH 4 to 6) seem to be associated with chromosomal genes, possibly due to the general stressors they represent (3, 4). However, L. brevis BS strains are also known to harbor plasmids that are associated with their beer spoilage phenotype and, more specifically, with hop tolerance (5–8). Plasmid-derived genes that underpin hop resistance in L. brevis include horA, horC, hitA, and orf5ABBC45 (1, 2). The genes horA and horC encode multidrug transporter proteins driven by ATP and proton motive force (PMF), respectively, and were identified as being involved in iso-α-acid extrusion from the bacterial cell (5, 7). The gene hitA encodes a transmembrane protein involved in the transport of divalent cations, such as Mn2+, in the exchange of protons released from hop bitter acids (8). The orf5ABBC45 gene was identified in L. brevis BS strain ABBC45, which was unable to grow in beer after it had lost a plasmid carrying this gene. The orf5ABBC45 gene encodes a predicted transmembrane protein resembling a PMF-dependent multidrug transporter, which is presumed to be responsible for iso-α-acid export (9).
However, these genes are not always indicative of BS ability, as the presence of such genes can be found among L. brevis strains that are unable to grow and consequently spoil beer (here designated NBS strains) (10). Indeed, horA is present in the L. brevis NBS strain UCCLB556 (10). Moreover, genes identified as conferring hop resistance are not always simultaneously present in BS strains, e.g., the BS strain UCCLBBS124 carries plasmids harboring horA and horC; however, it does not possess hitA (10). Analysis of the BS strain L. brevis BSO 464 has highlighted the importance of plasmids and genes on mobile genetic elements for bacterial growth in beer and beer spoilage ability (6). Recently, a gene predicted to encode a glycosyltransferase was identified among BS strains responsible for excess β-glucan formation (11). This gene is also present on the genome of L. brevis BS strain UCCLBBS124, while it is absent in that of BS strain UCCLBBS449 (10). This indicates that beer spoilage is not uniquely governed by the presence of a few genes but rather by a combination of genes acting in concert to confer beer resistance to the strain. It also suggests that other plasmid-borne genes involved in beer spoilage are yet to be discovered.
In the present study, we generated plasmid-cured derivatives of L. brevis BS strain UCCLBBS124 using novobiocin. This approach has been successfully employed previously to cure plasmids from isolates of lactic acid bacteria (LAB) (6, 12). Plasmid-cured derivatives were assessed for their ability to grow in the presence of hops and in beer. A derivative that showed inability to grow in beer was selected and analyzed to ascertain which plasmids were responsible for this phenotype. Bioinformatic analysis of the genetic content of such plasmids revealed candidate genes required for growth in beer. These genes were used in transformation experiments to revert the NBS phenotype.
RESULTS AND DISCUSSION
Derivatives with impaired growth in beer reveal a loss of plasmid UCCLBBS124_D.
The beer-spoiling L. brevis strain UCCLBBS124 (here, UCC124) possesses four plasmids carrying genes of interest for bacterial beer spoilage (Table 1). Following exposure to novobiocin, surviving L. brevis UCC124 cells were plated, and 50 isolated colonies (10) were randomly selected for further analysis. Thirty-four of these 50 colonies displayed impaired growth in beer. PCR-based identification of the hop resistance gene horA revealed the loss of this gene, located on plasmid UCCLBBS124_D (here, UCC124_D) in 33 out of the 34 isolates. One derivative, here designated MB569, was selected for genome sequencing, after which its sequence was compared to that of the wild type (WT), confirming that plasmid UCC124_D had been lost from strain MB569.
TABLE 1.
L. brevis UCC124 plasmids and genes of interest for beer spoilage
| UCC124 plasmid | Size (bp) | No. of ORFsa | GenBank accession no. | Gene(s) of interest | Reference(s) |
|---|---|---|---|---|---|
| UCCLBBS124_A (UCC124_A) | 49,560 | 42 | CP031170 | ||
| UCCLBBS124_B (UCC124_B) | 23,078 | 20 | CP031171 | gtf family 2 | 11 |
| UCCLBBS124_C (UCC124_C) | 22,370 | 27 | CP031172 | horB, horC, orf5 | 7, 9 |
| UCCLBBS124_D (UCC124_D) | 20,971 | 16 | CP031173 | horA | 5 |
ORFs, open reading frames.
Tolerance of MB569 to iso-α-acids, ethanol, and low pH.
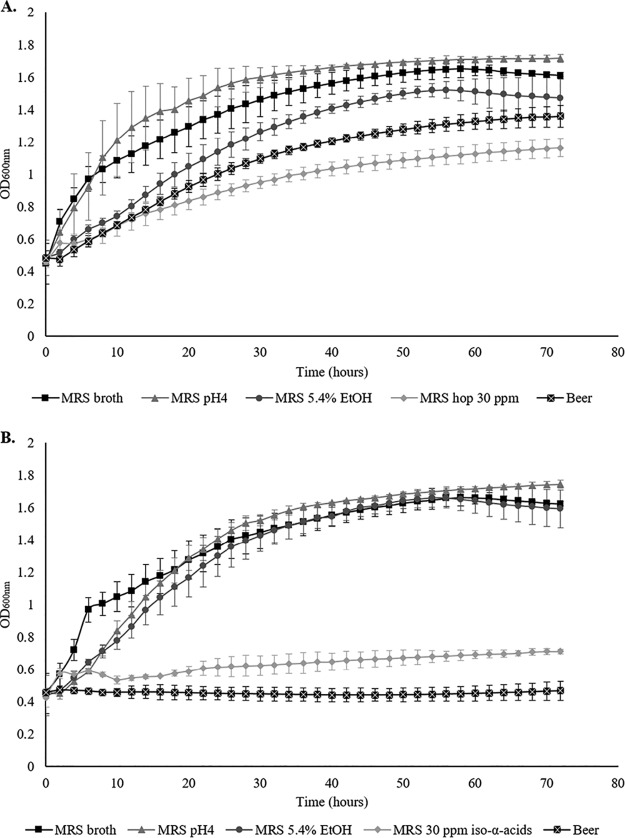
The inability of strain MB569 to grow in beer highlights the apparent importance of plasmid UCC124_D in conferring a beer spoilage phenotype on strain UCC124 (Fig. 1). Beer is a harsh environment incorporating a number of stresses, such as low pH, lack of nutrients, and the presence of ethanol and hop compounds. In order to understand which of these stresses imposed a negative impact on growth of MB569, the WT strain and MB569 were grown in de Man-Rogosa-Sharpe (MRS) broth and mimicking conditions encountered in beer, e.g., pH 4, 5.4% ethanol, and 30 ppm iso-α-acids. Strain MB569 was shown to be capable of growth in MRS broth at neutral pH and at pH 4, while it can also grow in the presence of ethanol comparable to the WT strain (Fig. 1). However, MB569 is incapable of growth in the presence of iso-α-acids, unlike the WT strain UCC124 (Fig. 1). This indicates that the plasmid-cured derivative MB569 has lost the ability to spoil beer due to its sensitivity to the antimicrobial compounds present in hops. Therefore, this phenotype and the finding that MB569 lacks plasmid UCC124_D (compared to its parental strain) indicate that this plasmid is linked to hop tolerance and thus contributes to the ability of strain UCC124 to cause beer spoilage.
FIG 1.
(A and B) Growth of the WT beer-spoiling strain L. brevis UCC124 (A) and its plasmid-cured derivative MB569 (B) in beer, MRS broth, MRS broth at pH 4, and MRS broth supplemented with 5.4% ethanol (EtOH) or 30 ppm iso-α-acids.
Identification and functional annotation of genes present on plasmid UCC124_D.
Plasmid UCC124_D is 21 kb in size and is predicted to encompass 16 genes. Interestingly, a 7-kb region of this plasmid contains six genes that are uniquely present among the plasmids of L. brevis BS strains (Table 2) (10). In order to assess the possible role of these genes in beer spoilage, the BS plasmid-specific genes UCCLBBS124_pD0014 (here, UCC124_D14), encoding a predicted cytosine deaminase, UCCLBBS124_pD0015 (renamed gtfD15), encoding a predicted glycosyltransferase, and UCCLBBS124_pD0016 (here designated UCC124_horA), which encodes HorA (Table 2), were individually cloned into plasmid pNZ44 prior their transformation into NZ9000. The resulting plasmids were then introduced into strain MB569 to determine the ability of the obtained recombinant strains to grow in beer (where MB569 itself is unable to do so). Genes with locus tags UCCLBBS124_pD0017 (UCC124_D17), UCCLBBS124_pD0018 (UCC124_D18), and UCCLBBS124_pD0019 (UCC124_D19), encoding acyl-sn-glycerol-3-phosphate acyltransferases and a glycosyltransferase (Table 2), were cloned together as a cluster (as present in plasmid UCC124_D) in pNZ44 prior their introduction into NZ9000 and, subsequently, MB569.
TABLE 2.
Presence and absence of genes of UCCLBBS124_D among L. brevis BS strains
| Gene (abbreviation) | Predicted function | Presence of L. brevis BS strain: |
||||||
|---|---|---|---|---|---|---|---|---|
| UCCLBBS124 | UCCLBBS449 | UCCLB95 | TMW1.2108 | TMW1.2111 | TMW1.2112 | TMW1.2113 | ||
| UCCLBBS124_pD0014 (UCC124_D14) | Cytosine deaminase | + | + | − | + | + | − | + |
| UCCLBBS124_pD0015 (gtfD15) | Glycosyltransferase family 8 | + | + | − | + | + | + | + |
| UCCLBBS124_pD0016 (UCC124_horA) | HorA | + | + | − | + | + | − | + |
| UCCLBBS124_pD0017 (UCC124_D17) | Acyl-sn-glycerol-3-phosphate acyltransferase | + | + | − | + | + | + | + |
| UCCLBBS124_pD0018 (UCC124_D18) | Glycosyltransferase family 8 | + | + | − | + | + | + | + |
| UCCLBBS124_pD0019 (UCC124_D19) | Acyl-sn-glycerol-3-phosphate acyltransferase | + | + | − | + | + | + | + |
The introduction of genes UCC124_D14, UCC124_horA, UCC124_D17, UCC124_D18, and UCC124_D19 in MB569 did not enable any obvious improvement of growth in the presence of iso-α-acid (30 ppm) or beer (compared to strain MB569) (data not shown).
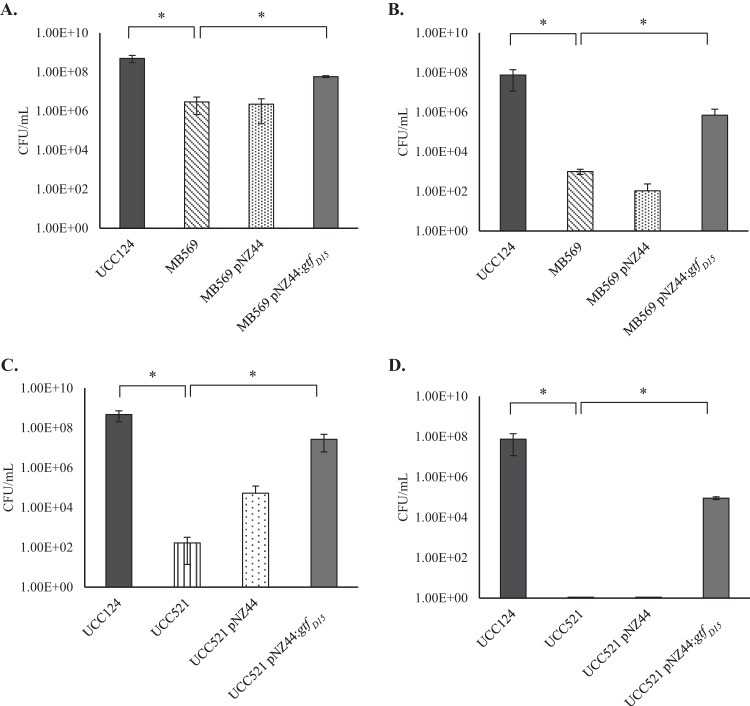
Interestingly, the expression of gtfD15 in MB569 was shown to confer a positive effect on its ability to grow in MRS broth containing 30 ppm iso-α-acids, with a significant (P < 0.05) growth increase after 72 h compared to the noncomplemented strain or MB569 carrying the control plasmid pNZ44 (Fig. 2A). When L. brevis MB569/pNZ44:gtfD15 was cultivated in beer, it also exhibited an ability to grow in beer that was significantly better than that of MB569 itself (P < 0.05) (Fig. 2B). Provision of gtfD15 in trans in MB569 did not restore its growth in beer to the same level as the WT strain (i.e., the strain from which MB569 was derived) but nonetheless allowed survival and growth in beer for this recombinant strain across 96 h. MB569 and MB569/pNZ44 are still able to survive in the presence of iso-α-acids or beer after culture for 72 h (Fig. 2A and B), which might be due to the presence of plasmid UCCLBBS124_C carrying the gene horC (Table 1). The gtfD15 gene is predicted to encode a glycosyltransferase based on BLAST analysis, and HHpred analysis (13) predicted the protein to belong to the glycosyltransferase family 8 associated with cell wall glycosylation (99.9% probability, E value < 10−28). Further sequence scrutiny suggests that the GtfD15 protein is a membrane-associated protein (TMHMM server 2.0 [14]) with a predicted signal peptide in its N terminus that may act as a membrane anchor for the protein (http://phobius.sbc.su.se/ [15]). These predictions suggest that GtfD15 is a cell envelope-associated protein that confers protection against certain environmental stressors, such as hop compounds.
FIG 2.
(A and B) Number of viable bacteria (CFU per milliliter) of the WT BS strain L. brevis UCC124, the derivative MB569 with and without the empty plasmid pNZ44, and MB569 carrying the gene gtfD15 after growth in MRS broth containing 30 ppm iso-α-acids for 72 h (A) and beer for 96 h (B) (P < 0.05). (C and D) CFU per milliliter of the WT BS strain L. brevis UCC124, the NBS UCC521 with and without the empty plasmid pNZ44, and the NBS UCC521 carrying the gene gtfD15 after growth in MRS broth containing 30 ppm iso-α-acids (C) and beer for 96 h (D) (P < 0.05).
Introduction of gtfD15 in NBS L. brevis strains allows growth in beer.
The introduction of gtfD15 in MB569 was shown to significantly improve growth of the strain in MRS broth containing hop compounds (30 ppm iso-α-acids) and in beer, indicating the importance of this gene for beer spoilage by L. brevis strain UCC124. In order to assess the potential growth-promoting effect of this gene for an NBS strain when inoculated in beer, gtfD15, when cloned into pNZ44 (pNZ44:gtfD15), was introduced into the NBS L. brevis strain UCCLB521 (here renamed UCC521) (Table 3). Remarkably, the presence of pNZ44:gtfD15 in the NBS strain L. brevis UCC521 permitted the strain to grow significantly better (P < 0.05) in MRS broth containing 30 ppm iso-α-acids and in beer than in the strain carrying an empty plasmid which is incapable of survival or growth in these environments (Fig. 2C and D). These observations reinforce our results described above and highlight the significance of the gtfD15 gene in hop tolerance and beer spoilage. An alternative, though in our opinion a less likely explanation, is that strains for which we obtained no or reduced CFU had entered a viable but nonculturable (VBNC) state, as has been previously observed for beer-passaged L. brevis strains (16).
TABLE 3.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description (isolation location) | Reference or source |
|---|---|---|
| Strains | ||
| L. brevis | ||
| UCCLBBS124 (UCC124) | Beer-spoiling strain isolated from spoiled beer keg (Singapore) | 10 |
| UCCLBBS449 | Beer-spoiling strain isolated from unpasteurized spoiled beer (The Netherlands) | 10 |
| MB569 | Non-beer-spoiling strain derivative of UCCLBBS124 | This study |
| UCCLB521 (UCC521) | Non-beer-spoiling strain isolated from a brewery environment (The Netherlands) | 10 |
| MB569/pNZ44 | MB569 carrying pNZ44 | This study |
| MB569/pNZ44:gtfD15 | MB569 carrying pNZ44 with gtfD15 | This study |
| UCCLB521/pNZ44:gtfD15 | UCCLB521 carrying pNZ44 with gtfD15 | This study |
| L. lactis | ||
| NZ9000 | Transformation host | 34 |
| Plasmids | ||
| pNZ44 | Transformation vector, chloramphenicol resistance gene | |
| pNZ44:gtfD15 | pNZ44 harboring gtfD15 | This study |
The introduction of pNZ44:gtfD15 into ATCC 367, another NBS strain, did not allow improved survival or growth in the presence of hop compounds or in beer (data not shown). This suggests that a strain-specific mechanism and involvement of other genes that are absent in ATCC 367 are responsible for increased hop tolerance. Among beer spoilage-related genes, UCC521 possesses the orf5ABBC45 gene previously identified as involved in hop tolerance (9), unlike ATCC 367, which does not harbor known genes involved in beer spoilage. Moreover, UCC521, although a non-beer-spoiling strain, was isolated from the brewery environment, unlike ATCC 367, which was isolated from silage (10). UCC521 may have acquired genes (such as orf5ABBC45) or plasmids (UCC521 harbors five plasmids) throughout its presence in the brewery environment which confer hop tolerance when combined with gtfD15. This scenario has previously been observed, indicating that L. brevis strains can only survive and grow in beer when multiple beer spoilage-related genes are present in a particular combination (17). Selection pressures of the beer environment determine the genetic content of beer-spoiling strains. The identification of diagnostic marker genes (DMGs) are important in distinguishing BS from NBS strains, as well as predicting the ability of a given strain to grow in beer (17, 18). In a study from Bergsveinson and Ziola, proposed DMGs were not related to hop tolerance, and no genes encoding glycosyltransferases were identified as DMGs. However, a glycosyltransferase-encoding gene located on the plasmid of the BS L. brevis strain BSO 464 which showed more than 99% nucleotide (nt) similarity to gtfD15 was described as unique compared to an NBS L. brevis strain KB290 and the BS strain Pediococcus damnosus Pc344T (17). From these observations and knowing that gtfD15 is highly prevalent in BS L. brevis strains (Table 2), we propose to include this gene as a DMG to assess the beer-spoiling potential of L. brevis strains.
Effect on phage sensitivity.
As discussed above, gtfD15 was observed to play a role in hop and beer tolerance and is predicted to encode a glycosyltransferase. Since the protein is predicted to be involved in biosynthesis or modification of a cell surface-associated saccharidic polymer, the possible role of this protein in bacteriophage infection was investigated. L. brevis strain UCC521 is sensitive to L. brevis phages 3-521 and 521B (19). Plaque assays employing these phages and L. brevis UCC521 harboring the empty vector pNZ44, or strain UCC521 containing pNZ44:gtfD15, displayed similar efficiency of plaquing (EOP) values with no significant difference from the WT (Table 4). However, notable differences in plaque morphology were observed, where plaques were faint and hard to distinguish on the bacterial lawn of UCC521 pNZ44:gtfD15. Moreover, overnight incubation of the different strains with the two phages led to complete lysis in broth of UCC521 and UCC521 containing pNZ44 with an approximately 1,000-fold increase of phage titer after overnight propagation (Table 4). In contrast, UCC521/pNZ44:gtfD15 did not show visible lysis and was able to grow after overnight incubation with just a 10-fold increase in phage numbers after overnight propagation (Table 4). These results reinforce the role of the protein GtfD15 in bacterial protection against diverse environmental hazards such as hop compounds or bacteriophages.
TABLE 4.
Effect of phages 3-521 and 521B on L. brevis strain UCC521 and derivatives
| Phage | Parametera | Data for L. brevis strain: |
||
|---|---|---|---|---|
| UCC521 | UCC521/pNZ44 | UCC521/pNZ44:gtfD15 | ||
| 521B | EOP | 1.00 | 0.58 ± 0.29 | 0.64 ± 0.21 |
| Plaque morphology | Small clear plaques | Small clear plaques | Faint plaques | |
| Phage titer after O/N propagation (PFU/ml) | 2.90E+09 | 2.30E+09 | 3.00E+07 | |
| 3-521 | EOP | 1.00 | 1.52 ± 0.20 | 1.19 ± 0.19 |
| Plaque morphology | Small clear plaques | Small clear plaques | Faint plaques | |
| Phage titer after O/N propagation (PFU/ml) | 4.30E+09 | 1.80E+09 | 4.80E+07 | |
Overnight (O/N) propagation was realized with a starting phage titer of 106 PFU/ml (results are average of triplicate assays). EOP, efficiency of plaquing.
Conclusions.
In this study, we identified a novel genetic component required for beer spoilage and, more specifically, for hop tolerance. This gene is located on plasmid UCC124_D of L. brevis BS strain UCC124, validating the importance of plasmids to confer a beer spoilage phenotype. Moreover, this gene had been highlighted previously as common among BS strains (10). Genes required for hop tolerance have all been identified on plasmids (5, 7, 8), reinforcing the importance of such mobile genetic elements in adaptation to the specific hurdles imposed by the beer environment.
A derivative of UCC124, MB569, showed impaired growth in beer after the loss of plasmid UCC124_D and despite the presence of plasmids UCC124_B and UCC124_C, which carry several genes of interest in beer spoilage. The introduction of gtfD15 in strain MB569 restored the hop tolerance phenotype of the strain, which ultimately allowed it to grow in beer. Similar results were observed when the gene was introduced into an NBS strain, confirming the notion that gtfD15 is required for the development of hop tolerance and beer spoilage. Furthermore, this gene impacts the phage sensitivity of its host. This gene seems to be a unique trait shared among BS strains of L. brevis, and we propose gtfD15 as a DMG for the detection of potential bacterial contamination of beer.
The gene is predicted to encode a glycosyltransferase, and analysis of its topology suggests that it is a membrane-anchored protein involved in the biosynthesis or modification of a cell surface-associated saccharidic polymer. BS strains of L. brevis have been shown to increase higher-molecular-weight lipoteichoic acids (LTA) in their cell wall, in the presence of hop bitter acids, thus believed to confer resistance to the bacteria by enhancing the barrier functions of the cell wall and preventing the intrusion of hop compounds (20, 21). Moreover, lipoteichoic acids have been described as phage receptors among lactobacillus phages as seen for Lactobacillus delbrueckii phages LL-H and JCL1032 (22) but also for Lactobacillus plantarum ATCC 8014-B2 (23). Therefore, we speculate that this glycosyltransferase is involved in replacing alanine residues with sugar residues on teichoic acids, thereby changing their charge and not allowing iso-α-acids to penetrate the membrane, as well as affecting phage adsorption and/or DNA injection. This predicted glycosyltransferase shows only limited similarity (36% amino acid similarity in 20% query cover) with the glycosyltransferase identified in a previous study as responsible for β-glucan formation (11) and is thus believed to play a different role in beer spoilage.
Future studies will focus on defining the mechanism that underpins hop tolerance and on determining how the genes identified to date (5, 7, 8) are linked to each other. Moreover, located on the same plasmid as gtfD15 are genes predicted to encode a glycosyltransferase and acyltransferases (Table 2) suggesting a common action on teichoic acids with the acyltransferases involved in the acylation of alanine residues or the lipid moiety of the lipoteichoic acids (24). Follow-up work may therefore focus on determining the precise function of the glycosyltransferase (and other associated genes) in the modification of the cell wall and/or cell surface. Another question to be addressed is if and how hop tolerance is enhanced when these genes are present in a certain combination and how such tolerance is influenced by their expression level.
MATERIALS AND METHODS
Bacterial strains and cultivation media.
The bacterial strains used in this study are listed in Table 3. L. brevis strains were grown in MRS broth (Oxoid Ltd., UK) at 30°C, while Lactococcus lactis NZ9000 was grown in M17 broth (Oxoid Ltd.) supplemented with 0.5% glucose. Chloramphenicol at 5 μg/ml (Cm5) was added to the medium when indicated.
Plasmid curing and content analysis.
The overall experimental approach is presented in Fig. S1 in the supplemental material. Plasmid curing of the BS strain UCC124 was achieved using novobiocin treatment (25). A 1% inoculum of a WT strain overnight culture was used to inoculate 10 ml MRS broth containing 0.25 μg/ml novobiocin. Cultures were incubated at 26°C for 72 h. After incubation, cells were diluted and plated on MRS agar. After 3 days of incubation at 26°C, isolated colonies were randomly selected, and derivatives with impaired growth in beer (no growth observed after 72 h) were checked for the presence or loss of hop resistance genes horA, horC, and orf5ABBC45 (Table 5). A derivative showing loss of a hop resistance gene was selected and sequenced using Illumina sequencing technology. Paired-end sequence reads were generated using an Illumina Hi Seq 2500 system (read length, 2 × 250 bp). FASTQ sequence files were generated using the Illumina Casava pipeline v1.8.3. After Illumina sequencing, the obtained sequences were mapped back against the WT reference sequence to detect mutations by single nucleotide polymorphism (SNP) or plasmid content loss. SNP analysis was performed by aligning Illumina raw reads against a reference sequence using Bowtie2 v2.3.5 (26). The reads were then sorted using SAMtools (27), and VarScan v2.3.9 was applied for the detection of variants (28). A minimum allelic variation frequency cutoff of 0.25 was applied.
TABLE 5.
PCR primers used in this study
| Primer name | Sequence (5′–3′)a | Target | GenBank accession no. |
|---|---|---|---|
| horAF | cgcaactgaggctaacttct | horA gene in UCCLBBS124 | CP031173 |
| horAR | ggcttgctatgctaggata | horA gene in UCCLBBS124 | CP031173 |
| horCF | gtatgcctaagtgacgt | horC gene in UCCLBBS124 | CP031172 |
| horCR | cattctctgcctctatac | horC gene in UCCLBBS124 | CP031172 |
| orf5F | ctggattgaggtgaggg | orf5 gene in UCCLBBS124 | CP031172 |
| orf5R | gctgtaaagggtagtgattg | orf5 gene in UCCLBBS124 | CP031172 |
| pNZ44F | aacaattgtaacccatac | pNZ44 promoter | |
| pNZ44R | gaacgtttcaagccttgg | pNZ44 MCS | |
| pD14F | aaaaaaCTGCAGgtccgaacagcgttcggatt | Gene UCCLBBS124_pD0014 in UCCLBBS124_D | CP031173 |
| pD14R | aaaaaaTCTAGAttaatcttcgaaatagtt | Gene UCCLBBS124_pD0014 in UCCLBBS124_D | CP031173 |
| pD15F | aaaaaaCCATGGgcggtttggatattttatact | Gene UCCLBBS124_pD0015 in UCCLBBS124_D | CP031173 |
| pD15R | aaaaaaTCTAGAtcactcagttttcaattccc | Gene UCCLBBS124_pD0015 in UCCLBBS124_D | CP031173 |
| pD16F | aaaaaaCTGCAGaggcttgctatgctagg | Gene UCCLBBS124_pD0016 in UCCLBBS124_D | CP031173 |
| pD16R | aaaaaaTCTAGAtcacccgttgctcgt | Gene UCCLBBS124_pD0016 in UCCLBBS124_D | CP031173 |
| pD17-19F | aaaaaaCCATGGggggtagaatggttctgtt | Gene UCCLBBS124_pD0017-19 in UCCLBBS124_D | CP031173 |
| pD17-19R | aaaaaaTCTAGAttattgataatgaccagcaa | Gene UCCLBBS124_pD0017-19 in UCCLBBS124_D | CP031173 |
Incorporated restriction sites are indicated by uppercase letters.
Construction of plasmid vectors.
Genes of interest were amplified by PCR (Table 5) and cloned into the expression vector pNZ44 (29). PCR products and pNZ44 plasmid DNA were digested with the appropriate enzymes (Roche, USA) at 37°C for at least 4 h, following the manufacturer’s instructions (Table 5). A ratio of 3:1 was applied for the ligation of the PCR product with pNZ44 using T4 DNA ligase (Promega, USA). The mixture was incubated at room temperature for at least 4 h prior to electrotransformation into L. lactis NZ9000 competent cells.
Preparation of competent cells and electrotransformation.
Competent cells of L. lactis NZ9000 were prepared as previously described (30). Competent cells of L. brevis UCC124 were prepared using an adapted version of a previously described protocol (31): An overnight culture was transferred (1% inoculum) to 10 ml MRS broth containing 1% glycine and incubated overnight at 30°C. Five milliliters of the overnight culture was transferred to fresh MRS broth containing 1% glycine (50 ml final volume), and cells were grown to an optical density at 600 nm (OD600) of 0.6. Cells were harvested by centrifugation at 4,000 × g for 15 min at 4°C and washed in ice-cold wash buffer (0.5 M sucrose, 10% glycerol). The wash step was repeated twice, and the cells were finally resuspended in 200 μl wash buffer prior to storage at −80°C and/or electroporation (see below). All constructs were generated using L. lactis NZ9000 as the cloning host and verified by sequencing after PCR amplification using the primers pNZ44F and pNZ44R (Table 5) prior to their transfer into L. brevis strains. Electrotransformation was performed using freshly prepared competent cells, as described above, where 45 μl of cells and 5 μl of plasmid construct were mixed into a prechilled 2-mm electroporation cuvette (Cell Projects, Kent, England) and subjected to electroporation at 1.5 kV (L. brevis) or 2.0 kV (L. lactis), 200 Ω, and 25 μF. Following electroporation, 950 μl recovery broth was added (MRS broth supplemented with 0.5 M sucrose and 0.1 M MgCl2 [L. brevis] or GM17 broth supplemented with 20 mM MgCl2 and 2 mM CaCl2 [L. lactis]). Cells were recovered at 30°C for 3 h (L. brevis) or 2 h (L. lactis) prior to spread plating on MRS (L. brevis) or GM17 (L. lactis) agar supplemented with Cm5. The presumed transformants were purified on MRS agar plus Cm5, and colonies were checked by sequencing after PCR amplification using the primers pNZ44F and pNZ44R (Table 5) and applied to growth assays as described below.
Growth assays.
The growth profiles of the wild-type strain and its derivative were obtained by transferring an overnight culture (1% inoculum) to MRS broth or MRS broth supplemented with 30 ppm iso-α-acids or beer (fresh Heineken lager 5% ethanol [pH 4], 23 ppm iso-α-acids). Cultures were incubated at 30°C for 72 h. One milliliter of culture was retrieved after 24, 48, 72, and 96 h, diluted in Ringer’s solution, and plated on MRS agar plates. Plates were incubated at 30°C anaerobically for 48 h prior to colony counting. The number of viable bacteria of each strain was assessed after calculation of the CFU per milliliter. Noninoculated controls were used in all of the experiments as blank measurements. These measurements were then subtracted from each experimental condition to produce the values represented on growth curves. Statistical differences were calculated using an unpaired t test method (32).
Phage activity against L. brevis strains and transformants.
To assess the phage sensitivity of L. brevis strains, transformants carrying genes of interest were compared to the wild-type (WT) strain using plaque assays, as previously described (33). A 10-μl volume of the appropriate phage dilution and 200 μl of L. brevis culture were added to 4 ml of soft agar supplemented with 10 mM CaCl2, mixed, and poured onto an MRS agar plate supplemented with 10 mM CaCl2 and 0.5% glycine. Plates were incubated at 30°C overnight, and the resulting plaques were enumerated. The phage titer was determined as PFU per milliliter. The ability of phages to propagate and multiply within the host cell was also tested. L. brevis strains were grown to early exponential phase (OD600, ∼0.25), at which point phages were added to the culture (time zero [T0]) at a multiplicity of infection (MOI) of 1, along with 10 mM CaCl2. The mix was further incubated at 30°C overnight (time 1 [T1]). The number of phages present in the medium (i.e., following removal of bacterial cells by centrifugation) at T1 was then determined using a plaque assay. Phage propagation efficiency on a given host was then determined by dividing the phage titer at T1 by the phage titer at T0.
Data availability.
The GenBank accession numbers for the strain genome sequences are as follows: L. brevis UCCLBBS124, CP031169; L. brevis UCCLBBS124_A, CP031170; L. brevis UCCLBBS124_B, CP031171; L. brevis UCCLBBS124_C, CP031172; and L. brevis UCCLBBS124_D, CP031173.
Supplementary Material
ACKNOWLEDGMENTS
M.F. performed the experiments and genomic analysis. D.V.S., J.M., T.O., and V.B. provided materials and strains. M.F., J.M., T.O., and D.V.S. were involved in project design and wrote the manuscript. All authors read and approved the final manuscript.
V.B. and T.O. are employees of Heineken.
M.F. is the recipient of an Irish Research Council Enterprise Partnership Scheme postgraduate scholarship (reference no. EPSPG/2015/7). D.V.S. is supported by a principal investigator award (reference no. 450 13/IA/1953) through Science Foundation Ireland (SFI). J.M. is in receipt of a Starting Investigator Research Grant (SIRG) (reference no. 15/SIRG/3430) funded by SFI.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Sakamoto K, Konings WN. 2003. Beer spoilage bacteria and hop resistance. Int J Food Microbiol 89:105–124. doi: 10.1016/s0168-1605(03)00153-3. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki K, Iijima K, Sakamoto K, Sami M, Yamashita H. 2006. A review of hop resistance in beer spoilage lactic acid bacteria. J Inst Brew 112:173–191. doi: 10.1002/j.2050-0416.2006.tb00247.x. [DOI] [Google Scholar]
- 3.Wang C, Cui Y, Qu X. 2018. Mechanisms and improvement of acid resistance in lactic acid bacteria. Arch Microbiol 200:195–201. doi: 10.1007/s00203-017-1446-2. [DOI] [PubMed] [Google Scholar]
- 4.Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee J-H, Díaz-Muñiz I, Dosti B, Smeianov V, Wechter W, Barabote R, Lorca G, Altermann E, Barrangou R, Ganesan B, Xie Y, Rawsthorne H, Tamir D, Parker C, Breidt F, Broadbent J, Hutkins R, O'Sullivan D, Steele J, Unlu G, Saier M, Klaenhammer T, Richardson P, Kozyavkin S, Weimer B, Mills D. 2006. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A 103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakamoto K, Margolles A, van Veen HW, Konings WN. 2001. Hop resistance in the beer spoilage bacterium Lactobacillus brevis is mediated by the ATP-binding cassette multidrug transporter HorA. J Bacteriol 183:5371–5375. doi: 10.1128/jb.183.18.5371-5375.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergsveinson J, Baecker N, Pittet V, Ziola B. 2015. Role of plasmids in Lactobacillus brevis BSO 464 hop tolerance and beer spoilage. Appl Environ Microbiol 81:1234–1241. doi: 10.1128/AEM.02870-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iijima K, Suzuki K, Ozaki K, Yamashita H. 2006. HorC confers beer-spoilage ability on hop-sensitive Lactobacillus brevis ABBC45cc. J Appl Microbiol 100:1282–1288. doi: 10.1111/j.1365-2672.2006.02869.x. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi N, Ito M, Horiike S, Taguchi H. 2001. Molecular cloning of a putative divalent-cation transporter gene as a new genetic marker for the identification of Lactobacillus brevis strains capable of growing in beer. Appl Microbiol Biotechnol 55:596–603. doi: 10.1007/s002530100600. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki K, Sami M, Kadokura H, Nakajima H, Kitamoto K. 2002. Biochemical characterization of horA-independent hop resistance mechanism in Lactobacillus brevis. Int J Food Microbiol 76:223–230. doi: 10.1016/s0168-1605(02)00016-8. [DOI] [PubMed] [Google Scholar]
- 10.Feyereisen M, Mahony J, Kelleher P, Roberts RJ, O'Sullivan T, Geertman J-MA, van Sinderen D. 2019. Comparative genome analysis of the Lactobacillus brevis species. BMC Genomics 20:416. doi: 10.1186/s12864-019-5783-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraunhofer ME, Geissler AJ, Wefers D, Bunzel M, Jakob F, Vogel RF. 2018. Characterization of beta-glucan formation by Lactobacillus brevis TMW 1.2112 isolated from slimy spoiled beer. Int J Biol Macromol 107:874–881. doi: 10.1016/j.ijbiomac.2017.09.063. [DOI] [PubMed] [Google Scholar]
- 12.Karthikeyan V, Santosh SW. 2010. Comparing the efficacy of plasmid curing agents in Lactobacillus acidophilus. Benef Microbes 1:155–158. doi: 10.3920/BM2009.0038. [DOI] [PubMed] [Google Scholar]
- 13.Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 15.Käll L, Krogh A, Sonnhammer E. 2007. Advantages of combined transmembrane topology and signal peptide prediction–the Phobius Web server. Nucleic Acids Res 35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Deng Y, Soteyome T, Li Y, Su J, Li L, Li B, Shirtliff ME, Xu Z, Peters BM. 2018. Induction and recovery of the viable but nonculturable state of hop-resistance Lactobacillus brevis. Front Microbiol 9:2076–2076. doi: 10.3389/fmicb.2018.02076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergsveinson J, Ziola B. 2017. Comparative genomic and plasmid analysis of beer-spoiling and non-beer-spoiling Lactobacillus brevis isolates. Can J Microbiol 63:970–983. doi: 10.1139/cjm-2017-0405. [DOI] [PubMed] [Google Scholar]
- 18.Behr J, Geissler AJ, Schmid J, Zehe A, Vogel RF. 2016. The identification of novel diagnostic marker genes for the detection of beer spoiling Pediococcus damnosus strains using the BlAst Diagnostic Gene findEr. PLoS One 11:e0152747. doi: 10.1371/journal.pone.0152747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feyereisen M, Mahony J, Lugli GA, Ventura M, Neve H, Franz C, Noben J-P, O’Sullivan T, Dv S. 2019. Isolation and characterization of Lactobacillus brevis phages. Viruses 11:E393. doi: 10.3390/v11050393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behr J, Gänzle MG, Vogel RF. 2006. Characterization of a highly hop-resistant Lactobacillus brevis strain lacking hop transport. Appl Environ Microbiol 72:6483–6492. doi: 10.1128/AEM.00668-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yasui T, Yoda K. 1997. Purification and partial characterization of an antigen specific to Lactobacillus brevis strains with beer spoilage activity. FEMS Microbiol Lett 151:169–176. doi: 10.1111/j.1574-6968.1997.tb12566.x. [DOI] [PubMed] [Google Scholar]
- 22.Räisänen L, Schubert K, Jaakonsaari T, Alatossava T. 2004. Characterization of lipoteichoic acids as Lactobacillus delbrueckii phage receptor components. J Bacteriol 186:5529–5532. doi: 10.1128/JB.186.16.5529-5532.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douglas LJ, Wolin MJ. 1971. Cell wall polymers and phage lysis of Lactobacillus plantarum. Biochemistry 10:1551–1555. doi: 10.1021/bi00785a007. [DOI] [PubMed] [Google Scholar]
- 24.Kiriukhin MY, Neuhaus FC. 2001. d-Alanylation of lipoteichoic acid: role of the d-alanyl carrier protein in acylation. J Bacteriol 183:2051–2058. doi: 10.1128/JB.183.6.2051-2058.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruiz-Barba JL, Piard JC, Jiménez-Díaz R. 1991. Plasmid profiles and curing of plasmids in Lactobacillus plantarum strains isolated from green olive fermentations. J Appl Bacteriol 71:417–421. doi: 10.1111/j.1365-2672.1991.tb03810.x. [DOI] [PubMed] [Google Scholar]
- 26.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGrath S, Fitzgerald GF, van Sinderen D. 2001. Improvement and optimization of two engineered phage resistance mechanisms in Lactococcus lactis. Appl Environ Microbiol 67:608–616. doi: 10.1128/AEM.67.2.608-616.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Pijkeren J-P, Britton RA. 2012. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res 40:e76. doi: 10.1093/nar/gks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahrné S, Molin G, Axelsson L. 1992. Transformation of Lactobacillus reuteri with electroporation: studies on the erythromycin resistance plasmid pLUL631. Curr Microbiol 24:199–205. doi: 10.1007/BF01579282. [DOI] [Google Scholar]
- 32.Kim TK. 2015. T test as a parametric statistic. Korean J Anesthesiol 68:540–546. doi: 10.4097/kjae.2015.68.6.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svensson U, Christiansson A. 1991. Methods for phage monitoring. Bull Int Dairy Fed 263:29–39. [Google Scholar]
- 34.Kuipers OP, de Ruyter P, Kleerebezem M, de Vos WM. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol 64:15–21. doi: 10.1016/S0168-1656(98)00100-X. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GenBank accession numbers for the strain genome sequences are as follows: L. brevis UCCLBBS124, CP031169; L. brevis UCCLBBS124_A, CP031170; L. brevis UCCLBBS124_B, CP031171; L. brevis UCCLBBS124_C, CP031172; and L. brevis UCCLBBS124_D, CP031173.