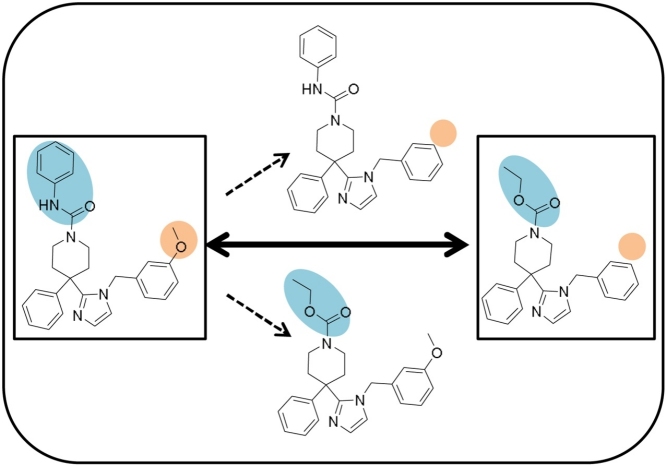
Graphical abstract
Method name: Third generation activity cliff identification
Keywords: Compound structure and activity data, Computational analysis, Molecular similarity, Analog pairs, Potency value distributions, Potency differences, Substitution sites, Single-site activity cliffs, Multi-site activity cliffs, Structure-activity relationships
Abstract
In medicinal chemistry and chemoinformatics, activity cliffs (ACs) are defined as pairs of structurally similar compounds that are active against the same target but have a large difference in potency. Accordingly, ACs are rich in structure-activity relationship (SAR) information, which rationalizes their relevance for medicinal chemistry. For identifying ACs, a compound similarity criterion and a potency difference criterion must be specified. So far a constant potency difference between AC partner compounds has mostly been set, e.g. 100-fold, irrespective of the specific activity (targets) of cliff-forming compounds. Herein, we introduce a computational methodology for AC identification and analysis that includes three novel components:
-
•
ACs are identified on the basis of variable target set-dependent potency difference criteria (a ‘target set’ represents a collection of compounds that are active against a given target protein).
-
•
ACs are extracted from computationally determined analog series (ASs) and consist of pairs of analogs with single or multiple substitution sites.
-
•
For multi-site ACs, a search for analogs with individual substitutions is performed to analyze their contributions to AC formation and determine if multi-site ACs can be represented by single-site ACs.
Specification Table
| Subject area: | Chemistry |
| More specific subject area: | Computational medicinal chemistry |
| Method name: | Third generation activity cliff identification |
| Name and reference of original method: | H. Hu, D. Stumpfe, J. Bajorath, Second-generation activity cliffs identified on the basis of target set-dependent potency difference criteria, Future Med. Chem. 11 (2019) 379-394. D. Stumpfe, H. Hu, J. Bajorath, Introducing a new category of activity cliffs with chemical modifications at multiple sites and rationalizing contributions of individual substitutions, Bioorg. Med. Chem. 27 (2019) 3605-3612. |
| Resource availability: | https://doi.org/10.5281/zenodo.1436584 H. Hu, D. Stumpfe, J. Bajorath, Systematic identification of target set-dependent activity cliffs. Future Sci. OA 5 (2019) FSO363. |
Method details
AC categories
Considering different ways in which the compound similarity and potency difference criterion can be specified and applied for AC definition, the following AC categories are introduced:
First generation ACs: ACs defined on the basis of a constantly applied similarity criterion, i.e., Tanimoto similarity or substructure-based similarity [1], and a constant potency difference criterion [2], irrespective of the target sets under study.
Second generation ACs: ACs defined on the basis of a constant substructure-based similarity criterion and a variable target set-dependent potency difference criterion [3].
Third generation ACs: ACs formed by pairs of structural analogs with single or multiple substitution sites extracted from individual ASs and applying target set-dependent potency difference thresholds [4].
Methodological framework
The method for the identification of third generation ACs combines methodological components from Hu et al. [3] and Stumpfe et al. [4] and makes use of the following computational concepts:
-
i
ASs were identified using the compound-core relationship (CCR) methodology [5]. Following the CCR approach, exocyclic single bonds in test compounds are systematically fragmented according to retrosynthetic combinatorial analysis procedure (RECAP) rules [6] permitting a maximum of five fragmentation sites per compound. Each of these sites represents a possible substitution site. Accordingly, each fragmentation step yields a compound core and a substituent. A core is required to have at least twice the size of the substituent or combined multiple substituents (i.e., twice the number of non-hydrogen atoms). For each test compound, all possible core-fragment combinations with one to five substitution sites are sampled and substituents at each site are replaced by a hydrogen atom to generalize the core representation. Then, all compounds sharing the same core are combined representing an individual AS [5]. It follows that compounds belonging to the same AS are distinguished by modifications at a single or multiple substitution sites. In addition, search calculations for analogs having individual substitutions found in ACs with multiple substitution sites were carried out with the aid of the OpenEye chemistry toolkit [7]
-
ii
As substructure-based similarity criteria for ACs, our group developed a preference for the formation of matched molecular pairs (MMPs) [8,9] with size-restricted substituents [10] (second generation ACs) and pairs of analogs from the same AS [4] (third generation ACs). An MMP is defined as a pair of compounds that only differ by a structural modification (substitution) at a single site [8]. To systematically generate MMPs, single bonds in compounds can be randomly fragmented [9] or on the basis of RECAP rules, yielding retrosynthetic MMPs (RMMPs) [11]. The resulting ACs are termed MMP-cliffs [10] and RMMP-cliffs [12], respectively. The application of MMPs and RMMPs as a similarity criterion yields ACs with only a single substitution site.
-
iii
Compound potency distributions in target sets are analyzed in boxplots and the interquartile range (IQR) is determined [12]. The IQR represents the potency range between quartile 1 (Q1) and 3 (Q3) for ∼50 % of the compounds in a set. For AC analysis, only target sets with an IQR of at least one order of magnitude are selected because sets with a smaller IQR value rarely contain ACs [12].
Activity data requirements
To ensure accuracy of AC assignments, only compounds with confirmed specific activity against a given target and numerically defined (assay-independent) equilibrium constants (pKi values) are considered.
Method design
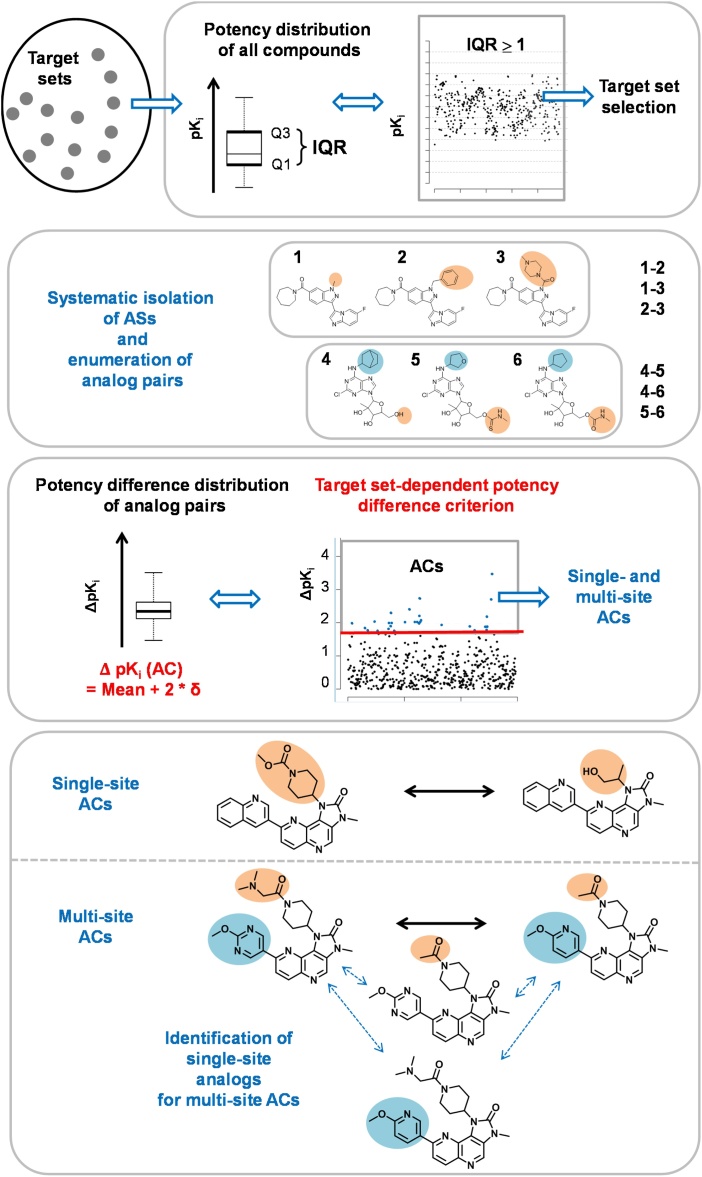
Fig. 1 provides a schematic summary of the different steps involved in computationally identifying and analyzing third generation ACs (from the top to the bottom), as detailed in the following:
Fig. 1.
Summary of the computational methodology for the identification of third generation ACs in target sets. Boxes present different steps of the approach.
1.0. Target sets are pre-selected on the basis of variable compound potency distributions (IQR ≥ 1) that typically yield ACs.
2.1. From a qualifying target set, ASs are systematically extracted. Compounds comprising each AS share a common core and contain single or multiple substitution sites.
2.2. For each AS, all analog pairs are enumerated and the potency difference captured by each pair is calculated. The formation of an analog pair with a single or multiple substitution sites serves as a similarity criterion for AC formation.
3.1. For each target set, all analog pairs (from all ASs) are collected and their potency difference distribution is determined.
3.2. As potency difference criterion for AC formation, the value of the mean of the distribution plus two standard deviations (sigma) is used.
4.1. All single- and multi-site ACs are collected (third generation ACs).
4.2. For multi-site ACs, a search for single-site analogs is carried out that contain individual substitutions and make it possible to study their contributions to AC formation, thus further characterizing multi-site ACs.
Method validation
Second and third generation ACs were systematically extracted from target set available in ChEMBL [13], the major public repository of compounds and activity data from medicinal chemistry. From ChEMBL release 23, a total of 16,096 target set-dependent RMMP-cliffs were extracted that originated from 212 different target sets [3]. In addition, on the basis of ChEMBL release 24.1, a collection of 13,546 target set-dependent MMP-cliffs and 7995 set-dependent RMMP-cliffs was generated and made publicly available in an open access deposition to enable follow-up investigations of second generation ACs [14]. Furthermore, in ChEMBL release 24.1, a total of 16,454 third generation ACs were detected that originated from 209 target sets. These ACs included 12,249 instances with a single and 4205 instances with multiple substitution sites, 3805 of which were dual-site ACs [4]. Analog search identified both single-site analogs for 297 dual-site ACs that contained individual substitutions and revealed their potency effects. The potency difference of a subset of 141 dual-site ACs was determined by one of two substitutions. By contrast, in 156 cases, both substitutions were found to significantly contribute. These 156 confirmed dual-site ACs made it possible to study substitution-associated potency effects in greater detail, revealing additive, synergistic, and compensatory effects of individual substitutions [4].
Acknowledgements
The authors thank the OpenEye Free Academic Licensing Program for providing a free academic license for their chemistry toolkit. H. Hu is supported by the China Scholarship Council (CSC).
Acknowledgments
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Stumpfe D., Bajorath J. Exploring activity cliffs in medicinal chemistry. J. Med. Chem. 2012;55:2932–2942. doi: 10.1021/jm201706b. [DOI] [PubMed] [Google Scholar]
- 2.Bajorath J. Evolution of the activity cliff concept for structure-activity relationship analysis and drug discovery. Future Med. Chem. 2014;6:1545–1549. doi: 10.4155/fmc.14.94. [DOI] [PubMed] [Google Scholar]
- 3.Hu H., Stumpfe D., Bajorath J. Second-generation activity cliffs identified on the basis of target set-dependent potency difference criteria. Future Med. Chem. 2019;11:379–394. doi: 10.4155/fmc-2018-0299. [DOI] [PubMed] [Google Scholar]
- 4.Stumpfe D., Hu H., Bajorath J. Introducing a new category of activity cliffs with chemical modifications at multiple sites and rationalizing contributions of individual substitutions. Bioorg. Med. Chem. 2019;27:3605–3612. doi: 10.1016/j.bmc.2019.06.045. [DOI] [PubMed] [Google Scholar]
- 5.Naveja J.J., Vogt M., Stumpfe D., Medina-Franco J.L., Bajorath J. Systematic extraction of analog series from large compound collections using a new computational compound-core relationship method. ACS Omega. 2019;4:1027–1032. doi: 10.1021/acsomega.8b03390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewelll X.Q., Judd D.B., Watson S.P., Hann M.M. RECAP -- retrosynthetic combinatorial analysis procedure: a powerful new technique for identifying privileged molecular fragments with useful applications in combinatorial chemistry. J. Chem. Inf. Comput. Sci. 1998;38:511–522. doi: 10.1021/ci970429i. [DOI] [PubMed] [Google Scholar]
- 7.OEChem T.K. OpenEye Scientific Software, Inc.; Santa Fe, NM, USA: 2012. Version 1.7.7. [Google Scholar]
- 8.Griffen E., Leach A.G., Robb G.R., Warner D.J. Matched molecular pairs as a medicinal chemistry tool. J. Med. Chem. 2011;54:7739–7750. doi: 10.1021/jm200452d. [DOI] [PubMed] [Google Scholar]
- 9.Hussain J., Rea C. Computationally efficient algorithm to identify matched molecular pairs (MMPs) in large data sets. J. Chem. Inf. Model. 2010;50:339–348. doi: 10.1021/ci900450m. [DOI] [PubMed] [Google Scholar]
- 10.Hu X., Hu Y., Vogt M., Stumpfe D., Bajorath J. MMP-cliffs: systematic identification of activity cliffs on the basis of matched molecular pairs. J. Chem. Inf. Model. 2012;52:1138–1145. doi: 10.1021/ci3001138. [DOI] [PubMed] [Google Scholar]
- 11.de la Vega de León A., Bajorath J. Matched molecular pairs derived by retrosynthetic fragmentation. Med. Chem. Commun. 2014;5:64–67. [Google Scholar]
- 12.Hu H., Stumpfe D., Bajorath J. Rationalizing the formation of activity cliffs in different compound data sets. ACS Omega. 2018;3:7736–7744. doi: 10.1021/acsomega.8b01188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaulton A., Bellis L.J., Bento A.P., Chambers J., Davies M., Hersey A., Light Y., McGlinchey S., Michalovich D., Al-Lazikani B., Overington J.P. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40(40):D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu H., Stumpfe D., Bajorath J. Systematic identification of target set-dependent activity cliffs. Future Sci. OA. 2019;5 doi: 10.4155/fsoa-2018-0089. FSO363. [DOI] [PMC free article] [PubMed] [Google Scholar]