Abstract
Background:
The aims of the current study were to determined present status of CL in Shiraz City, identify the causative species of Leishmania and conduct phylogenetic evaluations in detected parasites
Methods:
This study was conducted on 70 individuals with suspected CL that referred to the major health centers of Shiraz (Valfajr), Fars province, Iran, from Sep 2016 to Jul 2017. DNA was extracted from cultured Leishmania promastigotes and PCR-RFLP were performed using ITS1-rDNA gene.
Results:
Overall, 39 male (55.70%) and 31 (44.30%) female were found to be positive microscopically. All of direct examined positive samples were confirmed to be positive for Leishmania spp. DNA. Based upon the PCRRFLP patterns and phylogenetic analysis, 46 (65.72%), 17 (24.28%) and 7 (10%) isolates were clearly identified as L. major, L. tropica and C. fasciculata, respectively.
Conclusion:
The dominat detected species in Shiraz City was L. major and L. tropica, respectively. CL has high prevalence in Shiraz City; therefore, more studies on leishmaniasis in the natural vectors and also reservoirs infection in this region is exceedingly recommended. Skin leisons due to C. fasciculata, was described for the first time in Iran (Shiraz City).
Keywords: ITS1, Leishmania tropica, Leishmania major, Crithidia fasciculata, Iran
Introduction
Leishmaniasis (L) is endemic in 98 countries and is considered as major public health problems over the world (1, 2). There are various forms of leishmaniasis, cutaneous (CL), visceral (VL) and mucocutaneous (MCL), caused by the members of the genus Leishmania (L) (2). According to WHO, CL is the most common form of the disease with approximately 0.6–1 million new cases in the world (1). Based on causative agents, there are two major forms of CL with several differences in clinical symptoms in Iran, including anthroponotic CL (ACL) and zoonotic CL (ZCL) (1).
Although microscopic examination has been considered the major standard method for identification of CL, due to morphological similarity between different Leishmania spp. diagnosis of the main species is not possible (3). Furthermore, identification of causative species is necessary for successful control of CL. Therefore, molecular methods such as restriction fragment length polymorphism PCR (PCR-RFLP) are being used for species identification. Moreover, molecular methods are helpful to evaluate inter-species and intra-species genetic diversity of the genus (4).
The aims of the current study were to determine the present status of CL in Shiraz city, diagnosis of the causative species of disease by using the molecular technique (PCR-RFLP) via ITS1-rDNA gene and conduct phylogenetic evaluations in detected parasites.
Materials and Methods
Study area
Current study is a descriptive study conducted from Sep 2016 to Jul 2017 in Shiraz City (Fig. 1). It is one of the southern cities of Iran, located at approximately 1500 meters above sea level. The weather in this city cold and rainy in winter and moderate to hot dry in summer. The annual mean temperature of the city is 18 °C.
Fig. 1:
Map of Iran indicating the location of sampling site (Shiraz city) in Fars province, Southwest of Iran (Created by Arc GIS version 10.2)
Sampling and parasite culture
Samples were taken from 70 individuals with suspected CL that referred to the major health centers of Shiraz (Valfajr).
Most of the individuals were from urban areas of Shiraz City. After obtaining written informed consent for each participants, demographic information and site of lesion on the body were recorded using a questionnaire (Table 1).
Table 1:
The characteristics of confirmed CL patients from Shiraz City (Fars Province, Iran) base on personal information and lesion characteristics
| Age Groups (yr) | Sex | Lesion duration (month) | Lesion location | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Male | Female | ≤1 | 1–2 | 3≤ | Head, face and neck | Hand and arm | Feet and leg | Other* | |
| <9 | 9 | 5 | 4 | 7 | 1 | 2 | 5 | 14 | 2 | 3 |
| 10–19 | 14 | 8 | 6 | 5 | 0 | 8 | 13 | 13 | 11 | 0 |
| 20–29 | 12 | 6 | 6 | 5 | 3 | 4 | 15 | 7 | 2 | 3 |
| 30–39 | 18 | 10 | 8 | 8 | 5 | 5 | 7 | 28 | 8 | 0 |
| 40–49 | 9 | 6 | 3 | 5 | 2 | 2 | 9 | 13 | 7 | 0 |
| 50≤ | 8 | 4 | 4 | 2 | 1 | 4 | 4 | 9 | 2 | 1 |
| Total | 70 | 39 | 31 | 32 | 12 | 23 | 53 | 70 | 30 | 4 |
Clinical samples (smears) were taken from the swollen margin of the skin lesions of each patient suspected to have CL by sterile vaccine style or lancet. Giemsa stained slides were prepared and examined by light microscopy. Meanwhile at the same time, samples from each CL lesions were aseptically inoculated and cultured into Novy–McNeal–Nicolle (NNN) medium and immediately transported to department of parasitology, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Informed consent was taken from all patients before taking smear samples. The local Ethics Committee approved the study.
Genomic DNA extraction
Genomic DNA of isolated parasites from confirmed CL patients was extracted from the stationary growth phase of cultured Leishmania promastigotes using the Phenol-chloroform protocol as described elsewhere (5).
PCR–RFLP for ITS1-rDNA amplification
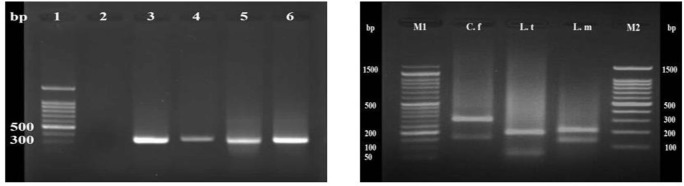
The standard PCR was conducted to identify the Leishmania infection by amplifying ribosomal internal transcribed spacer (ITS1) using previously designed specific primers, LITSR (5`- CTGGATCATTTTCCGATG-3`) as Forward primer and L5.8S (5`TGATACCACTTATCGCACTT-3`) as reverse primer (6). Reference strains of L. tropica (MHOM/IR/02/M39-Khorasan; Accession Number: JN860725) and L. major (MHOM/IR/75/ER; Accession Number: EF653269) were used as positive standard controls (Fig. 2). Next, RFLP was performed on PCR products to determine the parasite species (Fig. 2).
Fig. 2:
(Left) PCR results of ITS1-rDNA for identification of Leishmania sp. Lane 1, 100 bp ladder marker; Lane 2, negative control; Lane 3 and 4, reference strains of Leishmania parasites including: L. tropica (MHOM/IR/02/M39-Khorasan; Accession Number: JN860725) and L. major (MHOM/IR/75/ER; Accession Number: EF653269), respectively; Lanes 5 and 6, positive samples of CL patients due to Leishmania infection. (Right) PCR-RFLP analysis of ITS1-rDNA of Leishmania spp. using the HaeIII restriction enzyme. M1 and M2, 50 bp and 100 bp ladder markers, respectively; C. f, C. fasciculata (fragments 350 bp and 150 bp), L.t, L. tropica (fragments 57 bp, 56 bp and 200 bp) and L.m, L. major (fragments 155 bp and 206 bp) are isolated Leishmania spp. from patients
Sequencing and phylogenetic analyses
Out of the 70 confirmed Leishmania isolates, 15 were randomly selected and purified from the agarose gel by PCR purification kit (Bioneer, South Korea) for sequencing analysis (C. fasciculata (seven isolates), L. major (four isolates) and L. tropica (four isolates)). Hence, the first PCR amplicons were sequenced from both directions (forward and reverse) by targeting the ITS1-rDNA gene, same primers in the PCR, according to the Sanger instructions (Bioneer, South Korea). The sequences alignment was done using BLAST program of GenBank and was compared with those of existing sequences related to Leishmania spp. which was available in the GenBank database. Eight nucleotide sequences are deposited in the GenBank database under the following accession numbers: MG755819, MG755820 and MG755821 for L. major, MG755822, MG755823 and MG755824 for L. tropica and MG755817 and MG755818 for C. fasciculata.
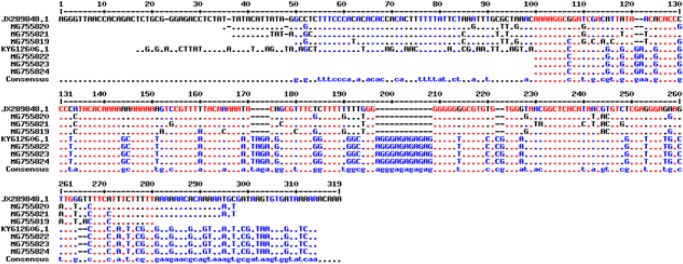
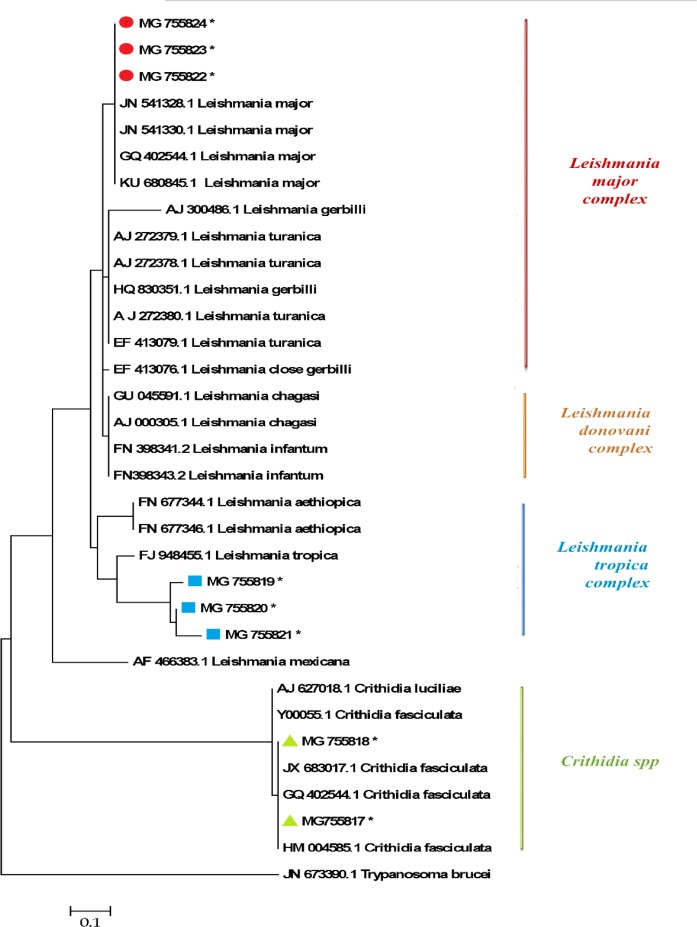
After extraction of the related genes forms Gen-Bank, the multiple sequence alignment was performed using online Multalin software (http://multalin.toulouse.inra.fr/multalin/) (Fig. 3 and 4). In this regard, overlapped sequences (contigs) were edited at each consensus positions by using Sequencher Tmv.4.1.4 Software (Gene Codes Corporation). For demonstrating the precise taxonomic status of all isolates, a maximum likelihood phylogenetic tree was constructed by using the MEGA 5.05 software based on kimura two-parameter model (Fig. 5).
Fig. 3:
The multiple sequence alignment of ITS1-rDNA gene using online Multalin software (http://multalin.toulouse.inra.fr/multalin/) for comparison of L. major and L. tropica. Six sequenced Leishmania ITS1-rDNA genes in the current study are MG755819, MG755820 MG755821, MG755822, MG755823 and MG755824. L. tropica (JX289848.1) isolate MHOM/IR/12/Bam4 and L. major isolate MLM-IR88 were used for comparison of similarity and differences between Leishmania spp.
Fig. 4:
The multiple sequence alignment of ITS1-rDNA gene using online Multalin software (http://multalin.toulouse.inra.fr/multalin/) for comparison of C. fasciculata. Two sequenced Crithidia ITS1-rDNA genes in the current study are MG755817 and MG755818. C. fasciculata (JX683017.1) isolate MHOM/IR/90/M42 was used for comparison of similarity and differences between Crithidia spp.
Fig. 5:
A maximum likelihood phylogenetic tree of Leishmania and Crithidia spp. based on ITS1-rDNA gene compared with other trypanosomatidae family. Evolutionary analyses were conducted in MEGA 5.05. L. mexicana (MNYC/BZ/62/M379; Accession number: AF466383) and Trypanosoma brucei (TS07112; Accession number: JN673390) were utilized as out-groups. Sequenced Leishmania and Crithidia ITS1-rDNA genes in the current study are illustrated in colored symbols with stars
Results
Seventy patients with confirmed CL lesions, who lived in the CL endemic regions of Shiraz city, were recruited for the current study (Fig. 1). CL diagnosis from each isolates was confirmed by demonstration of parasite amastigotes in the skin smears, culture of isolated promastigotes in RPMI-1640 medium and PCR (Fig. 2). All of the 70 suspected CL patients were found to be positive using microscopic examination and PCR. Species of isolated parasites were identified using PCR-RFLP. Of this, PCR products of ITS1-rDNA gene were digested using BsuRI (HaeIII) restriction enzyme. Therefore, of 70 confirmed CL lesions, 46 (65.72%) and 17 (24.28%) were identified as L. major (fragments 155 bp and 206 bp) and L. tropica (fragments 57 bp, 56 bp, and 200 bp) respectively, and the rest (seven isolates; 10%) were C. fasciculata (fragments 350 bp and 150 bp) (Fig. 2). Notably, no mix-infection was found in identified isolates.
Out of the 70 confirmed CL patients recruited in this study, 30 (44.29%) were female and 39 (55.71%) were male, with the age ranges of 1–65 yr. Most of the patients were belong to 30–39 and 10–19 yr’ age group. Total of 70 lesions were observes on the hands and arms; 53 on the head, face and neck; 30 on the feet and leg; and 4 on the back, belly and waist. Most of the lesions were seen on the hands (Table 1). The lesions duration ranged from 15 d to eight months. None of the patients have undergone any CL treatment prior sampling. Complete description of patients and lesions characteristics including age, sex, number of lesions and duration are illustrated in Table 1.
The multiple sequence alignment using ITS1-rDNA gene for L .major, L. tropica and C. fasciculata is illustrated in Fig. 3 and 4. Of 15 randomly sequenced ITS1-rDNA genes of Leishmania and Crithidia, eight sequences were chosen and directly compared with extracted GenBank sequences of three Leishmania and Crithidia reference species including L. tropica (JX289848.1) isolate MHOM/IR/12/Bam4, L. major isolate MLMIR88 and C. fasciculata isolate MHOM/IR/90/M42 Fig. 3 and 4. Totally 33 ITS1-rDNA gene sequences of Leishmania and Crithidia spp. were prepared for analyzing in order to create the phylogenetic tree including eight sequenced isolates in the current study which registered currently in the GenBank, 18 and five sequences belonging to distinct Leishmania complexes and Crithidia spp.
Respectively, and two sequences were used as outgroups (L. mexicana and Trypanosoma brucei). The constructed maximum likelihood phylogenetic tree elucidated that the sequenced Leishmania ITS1-rDNA genes in the current study were distributed into two min complexes including L. major and L. tropica; and C. fasciculata distributed into own genus, Crithidia (Fig. 5). Interestingly, three sequenced L. major from Shiraz City were categorized into the first group close to Iran sequences, and two isolated C. fasciculata were grouped in the Iran sequences (Fig. 5). The topology of constructed maximum likelihood phylogenetic tree showed that the L. tropica and L. major were grouped with the bootstrap value of higher than 60% in their specific complex. The phylogenetic relationship among 31 Leishmania and Crithidia parasites and two outgroup species is presented in Fig. 5.
Discussion
This study is the first documented report in the Shiraz city, southern Iran done to identify Leishmanis species including L. major, L. tropica and C. fasciculata by molecular methods and confirm them by using sequencing and phylogenetic analysis in clinical samples of CL patients in this city, as one of the main endemic areas in Iran.
Our findings by using ITS-based PCR-RFLP showed that 70 confirmed CL lesions, 46 and 17 were identified as Leishmania spp. including L. major and L. tropica respectively, and seven isolates belonged to Crithidia genus, C. fasciculata. Moreover, the findings of phylogenetic analysis based upon ITS1-rDNA gene sequences plainly showed close relationship of distinct Leishmania and Crithidia species, in the constructed maximum likelihood phylogenetic tree.
The dominant species in Shiraz City was L. major. However, the results are in agreement with those of other studies in Shiraz Province, in that, the major causative agent of CL in three villages of rural regions of Shiraz is L. major (7). Furthermore, another molecular study was done in Fasa City (southeast of Shiraz) which confirmed that L. major was predominant species of Leishmania in CL patients. Our results about the predominance of L. major is in accordance with some of previous studies conducted in different parts of Iran (8). Moreover, unlike rural region of Shiraz, L. tropica is the predominant species in urban region of the city (9). The main effectors which cause active transmission in endemic areas can be the following factors: agricultural and herbal planting development, building clay houses and dumping the building and garbage waste products near to residential areas which end to the attraction of sand fly vectors and rodents reservoirs close to the Shiraz city (7).
One of the interesting and challenging findings of the current study was molecular identification of C. fasiculate from seven samples of the CL patients. Studies have shown that C. fasiculate, as an insects parasite, is non-pathogenic to mammals (10); however, coinfection of C. fasiculate and L. major or L. tropica was reported in Khuzestan Province (southwest of Iran) (11). Crithidia in the lesion of right hand, L. major was isolated in left hand and L. tropica in face of CL patient (11). However, we report here in some patients only Crithidia without coinfection with other pathogenic Leishmania spp. It could be very important because lower trypanosomatid such as Crithidia in some situations such as immunosuppression caused diffuse cutaneous lesions (12). Moreover, Crirhidia could inter dermal mouse fibroblasts and stay inside these cells and could resist lysis by the complement system (12). All of the isolated samples were cultured and DNA was extracted from cultured samples, one of the possible hypothesis that why Crithidia was reported in the current study could be its ability to strongly develop and rapidly multiplicate inside culture media and destroy immediately other Leishmania parasites in coinfection situations. Therefore, when we isolated the parasite from active lesions both nonpathogenic Crithidia and pathogenic Leishmania spp. were present, but in culture, Crithidia was probably dominant parasite and killed other species.
Furthermore, the highest frequency was observed in 30–39 yr old, it can be caused by more outdoor activity of this group (1). The most affected part of the patient’s body was belonged to hand and face respectively, which has also been observed in the other distinct studies in various parts of the country (6, 7).
Conclusion
Both L. major and L. tropica are circulating in Shiraz City as an endemic foci with high infection rates; however, L. major is predominant species of Leishmania in this region. Isolation of Crithidia in different sites of body can approve the existence of this nonpathogenic insect’s parasite in confirmed CL patients. Along with microscopic method, molecular and phylogenetically methods are necessary to determine the exact causative species of CL in endemic areas.
Ethical considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Acknowledgements
This work was financially supported by Shahid Beheshti University of Medical Sciences with grant number 10511.Thanks to Dr. Zahra Asadgol for her technical assistance in designing the GIS map.
Footnotes
Conflicts of interest
The authors declare that there is no conflict of interest.
References
- 1.Mirzapour A, Spotin A, Behniafar H, Azizi H, Maleki B, Shakeraminia H, Seyyed Tabaei SJ. (2019). Intra-Species Diversity of Leishmania major and L. tropica from Clinical Isolates of Cutaneous Leishmaniasis in Southwest Iran Inferred by ITS1-rDNA. Iran J Public Health, 48:893–901. [PMC free article] [PubMed] [Google Scholar]
- 2.Badirzadeh A, Mohebali M, Sabzevari S, Ghafoori M, Arzamani K, Seyyedin M, Hashemi SA. (2018). Case Report: First Coinfection Report of Mixed Leishmania infantum/Leishmania major and Human Immunodeficiency Virus–Acquired Immune Deficiency Syndrome: Report of a Case of Disseminated Cutaneous Leishmaniasis in Iran. Am J Trop Med Hyg, 98:122–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muñoz EB, Santander S, Rojas-Silva P, Cardenas PA, Fornasini M, Cifuentes SC, Salvador D, Baldeón ME. (2016). Diagnostic Efficacy of Molecular Techniques for Detection and Identification of Leishmania Species in Human Whole Blood and Skin Samples from Ecuador. Am J Trop Med Hyg, 95:803–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hajjaran H, Mohebali M, Mamishi S, Vasigheh F, Oshaghi MA, Naddaf SR, Teimouri A, Edrissian GH, Zarei Z. (2013). Molecular identification and polymorphism determination of cutaneous and visceral leishmaniasis agents isolated from human and animal hosts in Iran. Biomed Res Int, 2013:789326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hajjaran H, Mahdi M, Mohebali M, Samimi-Rad K, Ataei-Pirkooh A, Kazemi-Rad E, Naddaf SR, Raoofian R. (2016). Detection and molecular identification of Leishmania RNA virus (LRV) in Iranian Leishmania species. Arch Virol, 161:3385–3390. [DOI] [PubMed] [Google Scholar]
- 6.Hezari F, Niyyati M, Seyyed Tabaei SJ, Mohebali M, Moin Vaziri V, Behniafar H, Azargashb E, Taghipour N. (2016). Frequency of Cutaneous Leishmaniasis and Species Identification in Suspected Individuals from Golestan Province, Northern Iran in 2014. Iran J Public Health, 45:1348–1354. [PMC free article] [PubMed] [Google Scholar]
- 7.Razmjou S, Hejazy H, Motazedian MH, Baghaei M, Emamy M, Kalantary M. (2009). A new focus of zoonotic cutaneous leishmaniasis in Shiraz, Iran. Trans R Soc Trop Med Hyg, 103:727–730. [DOI] [PubMed] [Google Scholar]
- 8.Kheirandish F, Sharafi C, Kazemi B, Mohebali M, Sarlak A, Tarahi MJ, Holakouee K, Hajaran H. (2013). Identification of Leishmania species using PCR assay on giemsa-stained slides prepared from cutaneous leishmaniasis patients. Iran J Parasitol, 8:382–88. [PMC free article] [PubMed] [Google Scholar]
- 9.Izadi S, Mirhendi H, Jalalizand N, Khodadadi H, Mohebali M, Nekoeian S, Jamshidi A, Ghatee MA. (2016). Molecular epidemiological survey of cutaneous leishmaniasis in two highly endemic metropolises of Iran, application of FTA cards for DNA extraction from Giemsa-stained slides. Jundishapur J Microbiol, 9:e32885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alcolea PJ, Alonso A, García-Tabares F, Toraño A, Larraga V. (2014). An Insight into the Proteome of Crithidia fasciculata Choanomastigotes as a Comparative Approach to Axenic Growth, Peanut Lectin Agglutination and Differentiation of Leishmania spp. Promastigotes. PLoS One, 9:e113837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spotin A, Rouhani S, Parvizi P. (2014). The associations of Leishmania major and Leishmania tropica aspects by focusing their morphological and molecular features on clinical appearances in Khuzestan province, Iran. Biomed Res Int, 2014:913510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santos DO, Bourguignon SC, Castro HC, Silva JS, Franco LS, Hespanhol R, Soares MJ, Corte-Real S. (2004). Infection of mouse dermal fibroblasts by the monoxenous trypanosomatid protozoa Crithidia deanei and Herpetomonas roitmani. J Eukaryot Microbiol, 51:570–574. [DOI] [PubMed] [Google Scholar]