Short abstract
TLRs are a family of PRRs that respond to PAMPs or host-derived Danger-Associated Molecular Patterns (DAMPs) to initiate host inflammation and immune responses. TLR dimerization and recruitment of adapter molecules is critical for intracellular signaling and is mediated through intracellular Toll-Interleukin 1 Receptor Resistance (TIR) domain interactions. Human TIR domains, including reported structures of TIR1, TIR2, TIR6, TIR10, TIRAP, and MyD88, contain Cysteine (Cys) interactions or modifications that are disproportionally at, or near, reported biological TIR interfaces, or in close proximity to functionally important regions. Therefore, we hypothesized that intracellular TIR Cys regulation may have greater functional importance than previously appreciated. Expression of mutant TLR4-C747S or treatment of TLR4 reporter cells with a small molecule, Cys-binding inhibitor of TLR4, TAK-242, abrogated LPS signaling in vitro. Using TAK-242, mice were protected from lethal influenza challenge as previously reported for extracellular TLR4 antagonists. Molecular modeling and sequence analysis of the region surrounding TLR4-Cys747 indicate conservation of a WxxxE motif identified among bacterial and NAD+-consuming TIRs, as well as within the TIRs domains of surface TLRs 1, 2, 4, 6, and 10. Together, these data support the hypothesis that critical Cys within the TIR domain are essential for TLR4 functionality.
Keywords: Innate immunity, influenza, Toll-like receptor
Introduction
The ability of TLRs and related molecules to dimerize and to associate with intracellular adapters or co-receptors (e.g., IL-1 accessory protein) is dependent on TIR domain interactions.1,2 A review of TIR domain structures and decoy TIR peptide sequences shown to inhibit TLR signaling revealed that approximately half contained cysteine (Cys) residue(s) that might be integral for this interaction.3 Based on these initial observations, we hypothesized that intercellular TIR Cys modification and/or interaction may have a greater functional importance than currently appreciated. An in-depth bioinformatics examination of human TIR domain structures and their reported biological interfaces, coupled with functionally characterized TIR mutants and inhibitory peptides, identified a highly conserved region involving the receptor TIR domains and Cys residues. Accordingly, we sought to identify inhibitory compounds shown to target specific TIR Cys residues and assess their ability to inhibit TLR-specific functions and specifically, those that target a highly conserved C helix Cys-containing region within TIR domains. In this study, we confirm that TAK-242 (resatorvid), a small molecule TLR4 antagonist that blocks signaling by binding to C747 of the intracellular TIR domain of TLR4, blocks both LPS-induced signaling, as well as protects mice in a lethal model of influenza.
Materials and methods
Reagents
Sterile TAK-242 was purchased from Millipore-Sigma (CAS 243984-11-4). HEK293-Blue™ null, TLR4 and MD2-CD14 cells and reagents were purchased (InvivoGen, San Diego, CA) and used as described. DMEM Media and supplements were purchased from GIBCO-Life-technologies. Mutant plasmid Cys747Ala TLR4 was engineered by quick-change mutagenesis using WT hTLR4 as a template.4 Fugene transfection reagent was purchased from Promega and used as directed.
Molecular visualization and modeling
The crystal structures and reported biological interfaces for TLR1, TLR2, TLR6, TLR10, TIRAP, MyD88, TRIF, and TRAM were examined using Chimera and Pymol programs.5,6 TLR4 TIR, dimer, and ternary complex with TIRAP and MyD88, were modeled based on the reported crystal structures for TLR10 and TLR6 TIR domain, TIRAP, and MyD88, similar to previously described approaches.7–10 Briefly, a least-squares comparison of the TLR4 dimer was performed using reported biological interfaces for TIR 1, 2, 6, 10, TIRAP, and MyD88 aligned to chain B of TLR10 and TLR6. Figures were produced using Pymol.6
Sequence alignments
A sequence alignment of TLR and adapter TIR domains was performed using the corresponding TIR domains of TLR1-13, TIRAP, and MyD88 using Clustal W.11 This sequence alignment was used to plot functionally characterized Cys TIR mutants, inhibitory peptides and reported interfaces.
Mice
Mice (6- to 8-wk-old, WT C57BL/6J) were purchased from The Jackson Laboratory (Bar Harbor, ME). All animal experiments were conducted with institutional IACUC approval from University of Maryland, Baltimore.
Virus
Mouse-adapted H1N1 influenza A/PR/8/34 virus (“PR8”) (ATCC, Manassas, VA) was grown in the allantoic fluid of 10-day-old embryonated chicken eggs as described,12 and was kindly provided by Donna Farber (Columbia University).
Virus challenge and treatment
For survival experiments, C57BL/6J WT mice were infected with mouse-adapted influenza virus, strain A/PR/8/34 (PR8; ∼7500 TCID50, intranasally (i.n.), 25 μL/nares). This dose was found previously to kill ∼90% of PR8-infected mice.13 Two days after infection, mice received either vehicle or TAK-242 (100 μg/mouse in 100 μL, i.p.) once daily for five consecutive days (days 2–6). Mice were monitored daily for survival, mass loss, and clinical signs of illness (e.g., lethargy, piloerection, ruffled fur, hunched posture, rapid shallow breathing, audible crackling) for 14 days. A clinical score ranging from 0 (no symptoms) to 5 (moribund) was ascribed to each mouse daily.13 In some experiments, mice were euthanized at day 7 post-infection to harvest lungs for analysis of gene expression and to assess lung pathology.
Histology
Lungs were inflated and perfused and fixed with 4% paraformaldehyde. Fixed sections (8 μm) of paraffin-embedded lungs were stained with hematoxylin and eosin (H&E). Slides were randomized, read blindly, and examined for tissue damage and inflammatory cellular infiltration.13
Quantitative real-time PCR
Total RNA isolation and quantitative real-time PCR (qRT-PCR) were performed as previously described.14 Levels of mRNA for specific genes are reported as relative gene expression normalized to mock-infected lungs.
Cell culture and NF-κB activation
HEK293 reporter cells (HEK-Blue™ hTLR4, HEK-Blue™ null, and HEK- Blue™ CD14-MD2 cells InvivoGen, San Diego, CA) were maintained and sub-cultured according to manufacturer’s instructions using DMEM media containing 4.5 g/L Glc, 10% (v/v) FBS, 50 U/mL penicillin, 50 μg/mL streptomycin, 100 μg/mL Normocin™, 2 mM L-glutamine, at 37°C in 5% CO2 and supplemented with appropriate selection antibiotic for each cell type.
SEAP inhibition assay in HEK-Blue™ TLR4 and null cells TLR4-treated cells
The growth media of HEK293-Blue™ cultured cells in a vented T-75 flask at log phase growth 50–80% was removed by pipette and the cells gently rinsed using 10 mL of pre-warmed 1× PBS. Cells were detached by gentle pipetting 2–5 mL of 1× PBS. Re-suspended cells were counted and cell suspension of 1.0 × 105 cells/mL made using prepared HEK-Blue™ Detection media, and immediately added 180 μL to 96-well tissue culture plate wells. Cells were then treated with 20 μL of detection media alone and or in combination with LPS of Escherichia coli K-12 W3110 strain (final concentration 10 ng/mL), TAK-242 [0–50 μΜ] dissolved in DMSO, or vehicle alone (DMSO 0–0.2% final concentration) in triplicate. Tissue culture plates (96-well) were returned for incubation at 37°C in 5% CO2. After 16 h, plate absorbance was measured at 640 nm using a Versa Max Microplate Reader (Molecular Devices Inc., Sunnyvale, CA, USA). Absorbance readings were graphed and statistics performed using Graph Pad PRISM. All samples performed in triplicate and are representative of at least three separate experiments. Purified LPS from E. coli K-12 W3110 strain, was a gift from Robert Ernst.
Statistics
Statistical differences between two groups were determined using an unpaired, two-tailed Student’s t test with significance set at P < 0.05. For comparisons between ≥ 3 groups, analysis was done by one-way ANOVA followed by a Tukey’s multiple comparison test with significance determined at P < 0.05. For survival studies, a Log-Rank (Mantel-Cox) test was used.
Results
Bioinformatic analysis of TIR domain reveals a highly conserved C helix motif among bacterial and mammalian TIR proteins
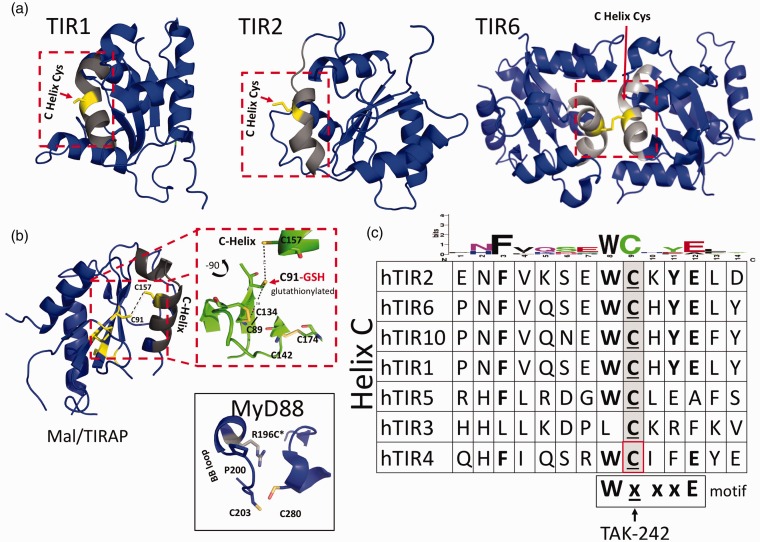
Previous reviews of reported biological interfaces for TIR crystal structures revealed several modified cysteines among receptor and adapter TIR proteins.3,7,8,10,15–18 These include several unique Cys modifications and interactions at suspected TIR–TIR interfaces and functionally important regions in the reported structures of TIR1, TIR2, TIR6, and TIR10, as well as in TIR domain-containing adapters MyD88 and the MyD88 adapter like (Mal) or TIR adapter protein (TIRAP).3,8,10,15,16,18,19 A review of TIR-derived inhibitor peptides at the time also identified a preponderance of functionally characterized inhibitory “decoy peptides” that contain one or more Cys residues as part of their functional sequence (Table 1).3 In contrast, scrambled control antennapedia coupled decoy peptides (RQIKIWFQNRRMKWKK-SLHGRGDPMEAFII) that were biologically inert did not contain cysteines as part of their functional sequence.20–23 To explore the potential role of intracellular TIR Cys in TIR domain function further, we performed a sequence alignment of the available TLR TIR domains and compared this alignment with reported X-ray and NMR structures. This sequence alignment showed several highly conserved Cys residues among various receptor and adapter TIR proteins (Supplemental Figure S1). An in-depth examination of human TIR domain structures and their reported biological interfaces, coupled with functionally characterized TIR mutants and inhibitory peptides, identified a highly conserved region involving the receptor TIR domains and cysteines (Figure 1). Based on this preliminary survey of TIR domain sequences, structures, and inhibitory peptides, we queried whether this region centered around a specific Cys may be playing a larger role in mediating TIR signaling than previously appreciated. A conserved Cys located on the C helix of multiple receptor TIR domains is observed at several reported biological interfaces (Figure 1a). This conserved C helix Cys is located within the WXXXE motif identified among NAD-consuming bacterial TIRs.8,24,25 Residues located within this motif are important for substrate binding and enzymatic function in NAD-consuming bacterial TIRs.8,10,24,26,27 Additionally, several functionally characterized TIR mutants and inhibitory decoy peptides also contain a highly conserved sequence motif, which includes a Cys (Table 1 and Figure 1c). Based on the observation of this highly conserved sequence motif involving NAD active site residues and C-helix containing Cys residue in multiple receptor TIR proteins, as well as functionally characterized inhibitory decoy peptides, we sought to identify compounds reported to bind TIR cysteines selectively.
Table 1.
Summary of Cys-containing inhibitory and decoy TIR peptides.
| Designation | Region | Sequence / Formula | Effect |
|---|---|---|---|
| BB loop | BB | hydrocinnamoyl-1-valyl- | MyD88-IL1-1RI |
| BB loop | BB | RDVLPGT | MyD88-MyD88 |
| MyD88-Ia | BB | RDVLPGTCVNS | MyD88 |
| Decoy peptidea | BB | IVFAEMPCGRLHLQ | TLR4 inhibition |
| Decoy peptidea | BB | VSDRDVLPGTCVWS | TLR4 inhibition |
| Decoy peptide | BB | RDVLPGT | IL-1 inhibition |
| Decoy peptide | BB | LHKRDFVPGKWllD | TLR2,TLR4 inhibition |
| Decoy peptide | BB | LHYRDFIPGVAIAA | TLR2, TLR4 inhibition |
| A46 peptide | Helix 1 | KYSFKLILAEY | TLR4 inhibition |
| Decoy peptide | CC' | LDEDEHGLHTKY | SEFIR inhibition IL17/IL25 |
| TLR4 DPa | TM | AGC KKYSRG ESIYD | TLR4 but not TLR2 |
| TLR4 DPa | AB, βB | EEGVPRFHLC | TLR4 but not TLR2 |
| TLR4 DP | βB,BB, αB | LHYRDFIPGVAIAA | TLR4 but not TLR2 |
| TLR4 DP | αD | LRQQVELYRLLSR | TLR4 but not TLR2 |
| TLR4 DP | αE | HIFWRRLKNALLD | TLR4 but not TLR2 |
| MAL DPa | αA,AB, βB | EGSQASLRCF | TLR4, TLR2/1 |
| MAL DPa | αB,BC | ELQALSRSHCR | TLR4 |
| MAL DPa | CC, C-HELIX | PGFLRDPW CKYQML | TLR4, TLR2/1 |
| MALDP | DD, αD,DE | AAYPPELRFMYYVD | TLR4 |
| MAL DP | αE | GG FYQVKEAVIHY | TLR4 |
| 2Rl PRE TIRa | PRE-TIR | RKPKKAPCRDVCYD | |
| 2R2 | A HELIX | EQDSHWVENLMVQQ | |
| 2R3a | AB LOOP | ENSDPPFKLC | |
| 2R3-4a | B STRAND | PPFKLCLHKRDF | |
| 2RS | B HELIX | NllDSIEKSHKT | |
| 2R6a | C HELIX | ENFVRSEWCKYEL | |
| 2R7a | CD LOOP | FSHFRLFDENNDAA | |
| 2R8 | D STRAND | LLEPIERKAI | |
| 2R9a | DD LOOP | PQRFKLRKIMNT |
aCysteine containing peptide.
Figure 1.
Conserved cysteines at TIR–TIR interfaces and C helix. (a) Structures of cytoplasmic TLR1, 2 and 6, and TIR adapter protein TIR domains. Inset boxes show conserved C helix (light grey) and Cys side chains. (b) Structures of MAL/TIRAP and MyD88. Inset box for MAL/TIRAP illustrates C91, which has been shown to require glutathionylation for signaling, as well as unique disulfide bond interactions between C89 and C134, which results in an extended AB loop, a collapsed BB loop, and a noncanonical TIR fold.8,9,18 Inset box for MyD88 illustrates unique crystal packing interactions involving Cys203 and Cys280 and the human mutation Arg to Cys at position 196 (R196C) in patients with deficient MyD88 signaling and a susceptibility to pyogenic infections.45 (c) Sequence alignment of TLRs and TIRAP C helix. Amino acids W, C, and E are conserved within the WxxxE motif observed in bacterial and NADase TIR proteins. TAK-242 binds C747 found on the C helix of TLR4 TIR domain.30
TAK-242 blocks LPS activation of HEK-TLR4 cells, but not HEK-TLR4 Cys747Ala mutant cells
To confirm TLR4 specificity of TAK-242, we compared NF-κB activation stimulated by LPS in the absence or presence of TAK-242 in stably transfected secreted embryonic alkaline phosphatase (SEAP) HEK-293 reporter cell lines, e.g., HEK293-TLR4 (“TLR4”), and in HEK293 cells that do not express TLR4 (“HEK293-null” and “HEK293-CD14-MD2”). TAK-242 abrogated LPS-mediated NF-κB activation in HEK-TLR4 and had no effect on vehicle-treated cells (Supplemental Figure S2). HEK-null, HEK-CD14-MD2 cells (stably expressing MD2 and CD14 coreceptors, but lacking TLR4), or HEK-CD14-MD2 cells transiently transfected with mutant TLR4 (C747S) failed to respond to LPS stimulation when compared with HEK293-CD14-MD2 cells transiently transfected with wildtype (WT) TLR4, further supporting the importance of this residue (Supplemental Figure S3).
Targeting Cys-747 of TLR4 TIR domain protects against lethal influenza infection
TLR4 antagonists that block the extracellular co-receptor, MD-2, e.g., Eritoran13,28 and FP7,29 or bind the extracellular domain of TLR4, e.g., anti-TLR4 Ab,28 have been shown to be protective against lethal mouse influenza infection. To date, all of the TLR4-specific antagonists used to blunt influenza-induced disease have targeted the extracellular domain of TLR4.13,29 The small molecule inhibitor TAK-242 (resatorvid) antagonizes TLR4 by binding to the intracellular TIR domain of TLR4 and has been shown to be specific for intracellular Cys TLR4-C747 (Figures 1 and 2).27–33 Therefore, we sought to determine if specific targeting of the TLR4 TIR Cys-747 with TAK-242 would also protect mice against lethal influenza infection.
Figure 2.
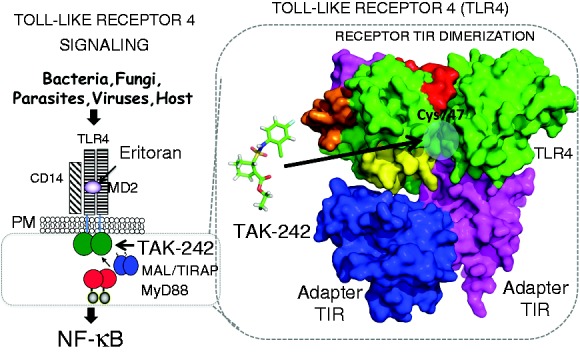
TLR4 Signaling schematic. Ligand-induced TLR4 signaling leads to NF-κB and IRF-3 activation and inflammation. Overview of TLR4 signaling. Extracellular TLR4 antagonist Eritoran acts by blocking LPS binding to MD-2, while TAK-242 targets cytoplasmic TLR4 TIR domain. Shown is a surface representation of cytoplasmic TLR4 receptor and adapter TIR interactions. TAK-242 is represented as ball and stick (green), highlighted surfaces (red, magenta, and orange) depict inhibitory peptides and blue circle indicates location of Cys 747 at predicted interface found on helix C of the TIR domain of TLR4.
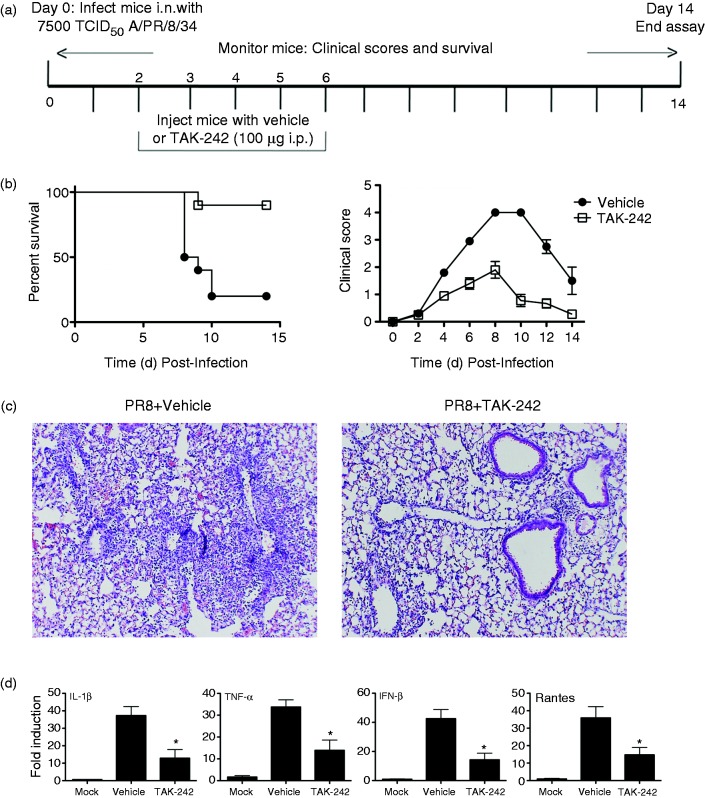
Briefly, C57BL/6J mice were infected i.n. with a dose of influenza A/PR/8/34 (“PR8”) previously determined to kill ∼90% of mice (∼LD90).13 Two days later, TAK-242 was administered once daily for 5 consecutive days (100 μg/mouse; i.p.; days 2-6) (Figure 3a). Each mouse was monitored daily for survival and clinical symptoms (e.g., lethargy, piloerection, ruffled fur, hunched posture, etc.)13 for 14 days. TAK-242-treated mice were significantly (P < 0.006) protected from PR8-induced lethality, while 80% of mice administered vehicle succumbed to infection (Figure 3b, left panel). Clinical scores were also more severe (Figure 3b, right panel) in vehicle-treated mice than in mice treated with TAK-242. To examine the effect TAK-242 has on influenza-induced lung pathology, groups of mice were infected with PR8 and treated with vehicle or TAK-242 as in Figure 3a, and were euthanized 7 days post-infection. Extensive lung damage was observed in mice infected with PR8 and treated with vehicle (Figure 3c, left panel), while lung sections from mice infected with PR8 and treated with TAK-242 exhibited less inflammatory infiltrates and lung damage (Figure 3c, right panel). Influenza infection potently induces expression of inflammatory genes that contribute to the overall inflammatory response.34,35 To assess whether TAK-242 could mitigate PR8-induced gene expression, lung homogenates from the same samples used for histopathology were also used to measured gene expression. TAK-242-treated mice had significantly lower gene expression of NF-κB- and IRF-3-dependent genes (IL-1β, TNF-α, and IFN-β, RANTES, respectively) compared with vehicle-treated mice (Figure 3d). Taken together, our observations that TAK-242 treatment blocks TLR4-mediated, influenza-induced acute lung injury support the central hypothesis that targeting TIR Cys interactions may be a viable therapeutic approach for influenza.
Figure 3.
TAK-242 therapy improves survival in a mouse model of influenza. (a) Basic protocol for in vivo survival assay of mice infected with PR8. WT mice infected with PR8 (∼7500 TCID50; i.n.) on day “0”. Mice received TAK-242 (100 µg/mouse i.p.) or vehicle (saline and 0.001% DMSO) once daily for 5 days (days 2–6). (b) Survival was monitored daily for 2 wk. TAK-242 reduces influenza-induced lethality in mice. WT C57BL/6 mice (6–8 wk old) were infected on day 0 with PR8 (7500 TCID50) and treated with vehicle (saline and 0.001% DMSO) or TAK-242 (100 µg/mouse i.p.) starting on day 2 daily for 5 consecutive days. (b) Survival and clinical score (i.e., 0 (no symptoms) to 5 (moribund), including mass loss, piloerection, lethargy, and lung “crackling,”) were assessed daily for 2 wk. Data shown are combined results of two assays (n = 5 mice/treatment group/experiment). (c and d) Mice were treated as in (a) and euthanized on day 7 post-infection for lung extraction. Lungs were stained for histopathology (c). Photomicrographs of representative sections were taken at 40X. (d) Total RNA was extracted to measure gene expression by qRT-PCR. Data shown are mean ± SEM. n = 5 mice/treatment group/experiment. *P < 0.05.
Discussion
Conserved C helix Cys interactions have been identified in several reported crystal structures of mammalian TIR proteins including TIR1, TIR2, and TIR6. Based on observed Cys interactions, modifications, sequence conservation, as well as Cys inclusion in multiple inhibitory TIR decoy peptides, we sought to test whether specific targeting of intracellular TIR cysteines would have a functional effect on TLR4 signaling. After an extensive literature search, we identified TAK-242, a small molecule inhibitor previously characterized to interact selectively with intracellular Cys 747 found on the C helix of the cytoplasmic TIR domain of TLR4. The novelty of this report stems not from using TAK-242 to antagonize TLR4 signaling, but rather, in the bioinformatics and structural analyses showing proof of principle for targeting intracellular TIR domains via Cys (an environment typically thought to be reducing) as an effective, if not more efficient, therapeutic approach. Approaches targeting intracellular TIR domain signaling have several advantages over extracellular approaches, including multiplicity of receptor pathways affected (e.g., Toll-like, Interleukin-1 and 18 receptors and TIR domain containing adaptor molecules) and no requirement for a priori knowledge of activating ligand, resulting in decoupling of receptor-ligand activation signals. Thus, approaches targeting intracellular TIR domains may be effective against previously unidentified, multiple and high concentrations of activating ligands. However, at present, our understanding and targeting of intracellular TIR signaling remains poorly underdeveloped in comparison to approaches targeting extracellular TLRs.
Over the past 5 yr, Shirey and colleagues have demonstrated that multiple TLR4 antagonists mitigate influenza-induced morbidity and mortality.13,28,29 Specifically, when Eritoran and FP7, lipid A analog antagonists that act by binding to the TLR4 co-receptor MD-2, and anti-TLR4 Abs that bind to the extracellular domain of TLR4, were administered therapeutically to mice infected with PR8, lethality and lung pathology were ameliorated, and accompanied by a decrease in cytokine production.13,29 Since influenza does not express known TLR4 PAMPs, it was hypothesized that a host-derived TLR4 agonist released upon influenza-induced tissue damage might underlie this apparent TLR4 dependency. Eritoran and the monosaccharide lipid A analog, FP7, blocked influenza-induced release of High Mobility Group Box-1 (HMGB1), a host-derived, TLR4-activating DAMP in mice and in cotton rats.28,29,36 To test the hypothesis that HMGB1 was the primary TLR4 DAMP induced by influenza infection, P5779, a small molecule inhibitor of HMGB1 that acts by preventing binding of HMGB1 to MD-2,37 was shown to be as effective as Eritoran in mitigating influenza-induced disease.28 Therefore, TAK-242 was tested to provide proof of concept for the hypothesis that specific targeting of the intracellular TLR4 Cys-747 would mimic the action of extracellular TLR4 antagonists. Our data show that TAK-242 protected against lethal PR8-induced influenza infection and lung injury similar to extracellular TLR4 antagonists such as Eritoran.13
The ability to modulate TLR signaling selectively has been highly sought after for vaccine development, and for controlling inflammation, autoimmunity, and cancer. Most studies have focused on targeting extracellular TLR domains using identified naturally occurring and synthetic ectodomain agonists and antagonists. Conversely, select targeting of intracellular TIR domains has focused primarily on developing inhibitory peptides and peptidomimetics based on peptide dissection of TIR domains and resulting functional characterization in signaling assays, resulting in the identification of several BB loops and TIR-derived “decoy” peptides that inhibit TLR4 signaling.3,22 However, TIR-based inhibitory peptides and peptidomimetics have encountered typical peptide challenges, including the need for high effective concentrations and the requirement for adding specific sequences to increase permeability across the cell membrane, as well for stability and solubility.22 Importantly, many of the functionally characterized decoy peptides contain a Cys as part of their functional sequence. Under reducing conditions, decoy peptides do not bind respective TIR targets.3,10,20–22 However, in vitro binding experiments may not fully recapitulate conditions within the cell or this may indicate potential localized redox environment or modification as has been reported.18,38 Reports to identify and develop TIR-specific small molecule inhibitory compounds from peptidomimetics, in silico compound library screening, or chemical synthesis have been met with limited success, including recently developed MyD88 small molecule inhibitors.39–44 Thus, the use of TAK-242 to block influenza-induced disease supports the hypothesis that specifically targeting the highly conserved C helix intracellular Cys-747 of the cytoplasmic TLR4-TIR domain may represent an important new approach for influenza therapy.
Bioinformatics analysis of reported bacterial and mammalian TIR structures show that the highly conserved TLR4-C747 targeted by TAK-242 is contained within the functionally important WXC747XXE motif identified in bacterial TIR-domain-containing proteins (Supplemental Figure S4). This motif contains a catalytic glutamic acid (E) at the carboxy-terminus that is essential for enzymatic function of NAD+ consuming bacterial and human TIR proteins (e.g., SARM). Bacterial and mammalian TIR domain-containing proteins have homology with a family of nucleotidases, which also contain a similar highly conserved catalytic glutamic acid (E) that is essential for enzymatic function.27 It remains to be seen if mammalian TIR domain-containing proteins other than SARM utilize the conserved WxxxE motif for enzymatic function or binding of NAD+ and NAD-like compounds. Additionally, it is unknown if recently identified TLR signaling inhibitors using methyl-piperidinio-pyrazole and scaffold analogs target this region containing the highly conserved C helix Cys and WxxxE motif.44 Finally, it remains to be determined if other compounds like TAK-242 also target the conserved WxcxxE motif.
Explicit targeting of the WxxxE motif and, potentially, conserved cryptic dinucleotide binding pocket in the TIR family members, may be an effective therapeutic approach. To our knowledge, targeting within conserved human receptor and bacterial TIR WXC747-Tak-242XXE motifs, conserved active site E, and potential NADase-like pockets have not been explicitly identified or correlated for potential small molecule development. Recent reports showing that TIR protein family members (bacterial and human) are an ancient family of NAD-consuming enzymes with NADase activity that retain a highly conserved active site glutamic acid residue located with the WxxxE motif.
Supplemental Material
Supplemental material, INI846281 Supplemental Material for Select targeting of intracellular Toll-interleukin-1 receptor resistance domains for protection against influenza-induced disease by Kari Ann Shirey, Wendy Lai, Lindsey J Brown, Jorge C G Blanco, Robert Beadenkopf, Yajing Wang, Stefanie N Vogel, Greg A Snyder, in Innate Immunity
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by NIH grants R21CA191726 (GAS) AI125215, AI123371 (to SNV and JCGB), T32AI095190 (LJB).
Supplemental material
Supplemental material is available online for this article.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell 2006; 124: 783–801. [DOI] [PubMed] [Google Scholar]
- 2.Gay NJ, Symmons MF, Gangloff M, et al. Assembly and localization of Toll-like receptor signalling complexes. Nat Rev Immunol 2014; 14: 546–558. [DOI] [PubMed] [Google Scholar]
- 3.Snyder GA, Sundberg EJ. Molecular interactions in interleukin and Toll-like receptor signaling pathways. Curr Pharm Des 2014; 20: 1244–1258. [DOI] [PubMed] [Google Scholar]
- 4.Rallabhandi P, Bell J, Boukhvalova MS, et al. Analysis of TLR4 polymorphic variants: new insights into TLR4/MD-2/CD14 stoichiometry, structure, and signaling. J Immunol 2006; 177: 322–332. [DOI] [PubMed] [Google Scholar]
- 5.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 2004; 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- 6.Schrödinger LLC. The PyMOL molecular graphics system, Version 2.0 Schrödinger, LLC.
- 7.Nyman T, Stenmark P, Flodin S, et al. The crystal structure of the human toll-like receptor 10 cytoplasmic domain reveals a putative signaling dimer. J Biol Chem 2008; 283: 11861–11865. [DOI] [PubMed] [Google Scholar]
- 8.Snyder GA, Deredge D, Waldhuber A, et al. Crystal structures of the Toll/Interleukin-1 receptor (TIR) domains from the Brucella protein TcpB and host adaptor TIRAP reveal mechanisms of molecular mimicry. J Biol Chem 2014; 289: 669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valkov E, Stamp A, Dimaio F, et al. Crystal structure of Toll-like receptor adaptor MAL/TIRAP reveals the molecular basis for signal transduction and disease protection. Proc Natl Acad Sci USA 2011; 108: 14879–14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snyder GA, Cirl C, Jiang J, et al. Molecular mechanisms for the subversion of MyD88 signaling by TcpC from virulent uropathogenic Escherichia coli. Proc Natl Acad Sci USA 2013; 110: 6985–6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 12.Teijaro JR, Njau MN, Verhoeven D, et al. Costimulation modulation uncouples protection from immunopathology in memory T cell responses to influenza virus. J Immunol 2009; 182: 6834–6843. [DOI] [PubMed] [Google Scholar]
- 13.Shirey KA, Lai W, Scott AJ, et al. The TLR4 antagonist Eritoran protects mice from lethal influenza infection. Nature 2013; 497: 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirey KA, Cole LE, Keegan AD, et al. Francisella tularensis live vaccine strain induces macrophage alternative activation as a survival mechanism. J Immunol 2008; 181: 4159–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tao X, Xu Y, Zheng Y, et al. An extensively associated dimer in the structure of the C713S mutant of the TIR domain of human TLR2. Biochem Biophys Res Commun 2002; 299: 216–221. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y, Tao X, Shen B, et al. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature 2000; 408: 111–115. [DOI] [PubMed] [Google Scholar]
- 17.Jang TH, Park HH. Crystal structure of TIR domain of TLR6 reveals novel dimeric interface of TIR-TIR interaction for toll-like receptor signaling pathway. J Mol Biol 2014; 426: 3305–3313. [DOI] [PubMed] [Google Scholar]
- 18.Hughes MM, Lavrencic P, Coll RC, et al. Solution structure of the TLR adaptor MAL/TIRAP reveals an intact BB loop and supports MAL Cys91 glutathionylation for signaling. Proc Natl Acad Sci USA 2017; 114: E6480–E6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan JA, Brint EK, O'Neill LA, et al. Crystal structure of the Toll/interleukin-1 receptor domain of human IL-1RAPL. J Biol Chem 2004; 279: 31664–31670. [DOI] [PubMed] [Google Scholar]
- 20.Piao W, Shirey KA, Ru LW, et al. A decoy peptide that disrupts TIRAP recruitment to TLRs is protective in a murine model of influenza. Cell Rep 2015; 11: 1941–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Couture LA, Piao W, Ru LW, et al. Targeting Toll-like receptor (TLR) signaling by Toll/interleukin-1 receptor (TIR) domain-containing adapter protein/MyD88 adapter-like (TIRAP/Mal)-derived decoy peptides. J Biol Chem 2012; 287: 24641–24648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toshchakov VY, Vogel SN. Cell-penetrating TIR BB loop decoy peptides a novel class of TLR signaling inhibitors and a tool to study topology of TIR–TIR interactions. Expert Opin Biol Ther 2007; 7: 1035–1050. [DOI] [PubMed] [Google Scholar]
- 23.Toshchakov VU, Basu S, Fenton MJ, et al. Differential involvement of BB loops of Toll-IL-1 resistance (TIR) domain-containing adapter proteins in TLR4- versus TLR2-mediated signal transduction. J Immunol 2005; 175: 494–500. [DOI] [PubMed] [Google Scholar]
- 24.Felix C, Kaplan Turkoz B, Ranaldi S, et al. The Brucella TIR domain containing proteins BtpA and BtpB have a structural WxxxE motif important for protection against microtubule depolymerisation. Cell Commun Signal 2014; 12: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alaidarous M, Ve T, Casey LW, et al. Mechanism of bacterial interference with TLR4 signaling by Brucella Toll/interleukin-1 receptor domain-containing protein TcpB. J Biol Chem 2014; 289: 654–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Essuman K, Summers DW, Sasaki Y, et al. The SARM1 Toll/interleukin-1 receptor domain possesses intrinsic NAD(+) cleavage activity that promotes pathological axonal degeneration. Neuron 2017; 93: 1334–1343 e1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Essuman K, Summers DW, Sasaki Y, et al. TIR domain proteins are an ancient family of NAD(+)-consuming enzymes. Curr Biol 2018; 28: 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shirey KA, Lai W, Patel MC, et al. Novel strategies for targeting innate immune responses to influenza. Mucosal Immunol 2016; 9: 1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perrin-Cocon L, Aublin-Gex A, Sestito SE, et al. TLR4 antagonist FP7 inhibits LPS-induced cytokine production and glycolytic reprogramming in dendritic cells, and protects mice from lethal influenza infection. Sci Rep 2017; 7: 40791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takashima K, Matsunaga N, Yoshimatsu M, et al. Analysis of binding site for the novel small-molecule TLR4 signal transduction inhibitor TAK-242 and its therapeutic effect on mouse sepsis model. Br J Pharmacol 2009; 157: 1250–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawamoto T, Ii M, Kitazaki T, et al. TAK-242 selectively suppresses Toll-like receptor 4-signaling mediated by the intracellular domain. Eur J Pharmacol 2008; 584: 40–48. [DOI] [PubMed] [Google Scholar]
- 32.Sha T, Sunamoto M, Kitazaki T, et al. Therapeutic effects of TAK-242, a novel selective Toll-like receptor 4 signal transduction inhibitor, in mouse endotoxin shock model. Eur J Pharmacol 2007; 571: 231–239. [DOI] [PubMed] [Google Scholar]
- 33.Ii M, Matsunaga N, Hazeki K, et al. A novel cyclohexene derivative, ethyl (6R)-6-[N-(2-Chloro-4-fluorophenyl)sulfamoyl]cyclohex-1-ene-1-carboxylate (TAK-242), selectively inhibits toll-like receptor 4-mediated cytokine production through suppression of intracellular signaling. Mol Pharmacol 2006; 69: 1288–1295. [DOI] [PubMed] [Google Scholar]
- 34.Mishin VP, Hayden FG, Gubareva LV. Susceptibilities of antiviral-resistant influenza viruses to novel neuraminidase inhibitors. Antimicrob Agents Chemother 2005; 49: 4515–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taubenberger JK, Morens DM. The pathology of influenza virus infections. Annu Rev Pathol 2008; 3: 499–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel MC, Shirey KA, Boukhvalova MS, et al. Serum high-mobility-group box 1 as a biomarker and a therapeutic target during respiratory virus infections. MBio 2018; 9: pii: e00246-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang H, Wang H, Ju Z, et al. MD-2 is required for disulfide HMGB1-dependent TLR4 signaling. J Exp Med 2015; 212: 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stottmeier B, Dick TP. Redox sensitivity of the MyD88 immune signaling adapter. Free Radic Biol Med 2016; 101: 93–101. [DOI] [PubMed] [Google Scholar]
- 39.Olson MA, Lee MS, Kissner TL, et al. Discovery of small molecule inhibitors of MyD88-dependent signaling pathways using a computational screen. Sci Rep 2015; 5: 14246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fekonja O, Avbelj M, Jerala R. Suppression of TLR signaling by targeting TIR domain-containing proteins. Curr Protein Pept Sci 2012; 13: 776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loiarro M, Capolunghi F, Fanto N, et al. Pivotal advance: inhibition of MyD88 dimerization and recruitment of IRAK1 and IRAK4 by a novel peptidomimetic compound. J Leukoc Biol 2007; 82: 801–810. [DOI] [PubMed] [Google Scholar]
- 42.Kissner TL, Moisan L, Mann E, et al. A small molecule that mimics the BB-loop in the Toll interleukin-1 (IL-1) receptor domain of MyD88 attenuates staphylococcal enterotoxin B-induced pro-inflammatory cytokine production and toxicity in mice. J Biol Chem 2011; 286: 31385–31396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pollock JA, Sharma N, Ippagunta SK, et al. Triaryl pyrazole Toll-like receptor signaling inhibitors: structure-activity relationships governing pan- and selective signaling inhibitors. ChemMedChem 2018; 13: 2208–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ippagunta SK, Pollock JA, Sharma N, et al. Identification of Toll-like receptor signaling inhibitors based on selective activation of hierarchically acting signaling proteins. Sci Signal 2018; 11(543). pii: eaaq1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.von Bernuth H, Picard C, Jin Z, et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science 2008; 321: 691–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, INI846281 Supplemental Material for Select targeting of intracellular Toll-interleukin-1 receptor resistance domains for protection against influenza-induced disease by Kari Ann Shirey, Wendy Lai, Lindsey J Brown, Jorge C G Blanco, Robert Beadenkopf, Yajing Wang, Stefanie N Vogel, Greg A Snyder, in Innate Immunity