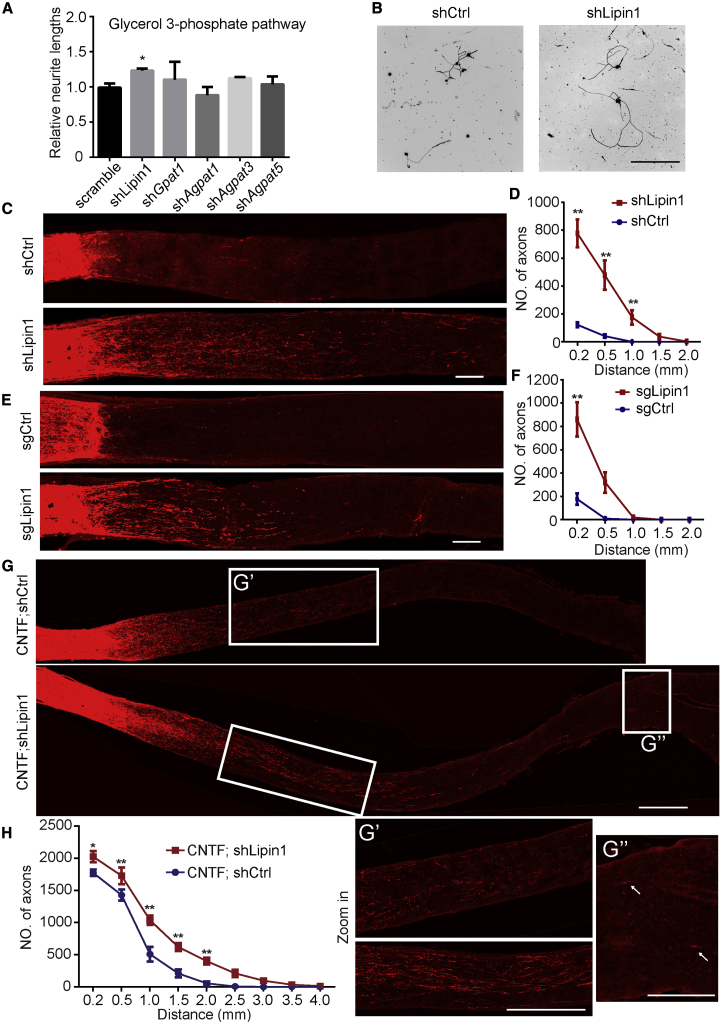
Figure 1.
Lipin1 Depletion Promotes Axon Regeneration
(A) Quantification of the axon elongation by in vitro screening of glycerol-3-phosphate (G3P) metabolic genes in adult DRG neurons. We tested five genes including lipin1, Gpat1, Agpat1, Agpat3, Agpat5. Gpat, Glycerol-3-phosphate acyltransferase. Agpat, 1-acyl-sn-glycerol-3-phosphate acyltransferase. Adult DRG neurons were dissociated and transfected with the plasmids for 3 days. Neurons were then replated and fixed 24 h after replating. DRG neurites were visualized by using Tuj1 staining. Three mice and 10–20 neurons from each mouse were quantified in each group. ∗p ≤ 0.05, ANOVA followed by Dunnett’s test.
(B) Representative images of replated neurons from control shRNA and lipin1 shRNA groups with Tuj1 staining. Scale bar: 400 μm.
(C) Sections of optic nerves from WT mice at 2 weeks post-injury (WPI). The vitreous body was injected with either AAV-control-shRNA or AAV-lipin1-shRNA. Axons were labeled by CTB-FITC. Scale bar: 100 μm.
(D) Number of regenerating axons at indicated distances distal to the lesion site. ∗∗p ≤ 0.01, ANOVA followed by Bonferroni’s test, n = 6 mice.
(E) Sections of optic nerves from Rosa26-Cas9 mice at 2 WPI injected with either AAV-control-sgRNA or AAV-lipin1-sgRNA. Scale bar: 100 μm.
(F) Number of regenerating axons at indicated distances from the lesion site. ∗∗p ≤ 0.01, ANOVA followed by Bonferroni’s test, n = 6 mice.
(G) Sections of optic nerves from WT mice at 2 WPI injected with AAV-CNTF combined with either AAV-control or lipin1-shRNA. Scale bar: 400 μm. Zoomed-in images are shown in the bottom panel. (G′) Zoomed-in images are shown in the bottom panel. Scale bar: 400 μm. (G″) Zoomed-in images of optic chiasm from (G). Arrows indicate regenerating axons in optic chiasm. Scale bar: 200 μm.
(H) Number of regenerating axons at indicated distances distal to the lesion site. ∗∗p ≤ 0.01, ∗p ≤ 0.05, ANOVA followed by Bonferroni’s test, n = 6 mice. Error bars indicate SEM.
See also Figure S1.