Abstract
Chinese hamster ovary (CHO) cell expression systems have been exquisitely developed for the production of recombinant biotherapeutics (e.g. standard monoclonal antibodies, mAbs) and are able to generate efficacious, multi-domain proteins with human-like post translational modifications at high concentration with appropriate product quality attributes. However, there remains a need for development of new CHO cell expression systems able to produce more challenging secretory recombinant biotherapeutics at higher yield with improved product quality attributes. Amazingly, the engineering of lipid metabolism to enhance such properties has not been investigated even though the biosynthesis of recombinant proteins is at least partially controlled by cellular processes that are highly dependent on lipid metabolism. Here we show that the global transcriptional activator of genes involved in lipid biosynthesis, sterol regulatory element binding factor 1 (SREBF1), and stearoyl CoA desaturase 1 (SCD1), an enzyme which catalyzes the conversion of saturated fatty acids into monounsaturated fatty acids, can be overexpressed in CHO cells to different degrees. The amount of overexpression obtained of each of these lipid metabolism modifying (LMM) genes was related to the subsequent phenotypes observed. Expression of a number of model secretory biopharmaceuticals was enhanced between 1.5-9 fold in either SREBF1 or SCD1 engineered CHO host cells as assessed under batch and fed-batch culture. The SCD1 overexpressing polyclonal pool consistently showed increased concentration of a range of products. For the SREBF1 engineered cells, the level of SREBF1 expression that gave the greatest enhancement in yield was dependent upon the model protein tested. Overexpression of both SCD1 and SREBF1 modified the lipid profile of CHO cells and the cellular structure. Mechanistically, overexpression of SCD1 and SREBF1 resulted in an expanded endoplasmic reticulum (ER) that was dependent upon the level of LMM overexpression. We conclude that manipulation of lipid metabolism in CHO cells via genetic engineering is an exciting new approach to enhance the ability of CHO cells to produce a range of different types of secretory recombinant protein products via modulation of the cellular lipid profile and expansion of the ER.
Keywords: Lipid metabolism, Chinese hamster ovary (CHO) cells, ER expansion, Difficult to express recombinant proteins, Cell line engineering
Highlights
-
•
We have engineered lipid metabolism gene expression, SREBF1 and SCD1, in CHO cells.
-
•
The amount of lipid metabolism gene overexpression is phenotype related.
-
•
Secretory biopharmaceutical yield is enhanced 1.5–9 fold in lipid engineered cells.
-
•
Overexpression of SCD1 and SREBF1 modifies the lipid profile of CHO cells.
-
•
Overexpression of SCD1 and SREBF1 results in an expanded ER.
Abbreviations:
- ACC
acetyl CoA carboxylase
- AMP
adenosine monophosphate
- AMPK
adenosine monophosphate activated protein kinase
- CAB
sodium cacodylate
- CHO
Chinese hamster ovary
- DGAT2
diacylglycerol O-Acyltransferase 2
- DTE
difficult to express
- FcFP
Fc fusion protein
- GS
glutamine synthetase
- INSIG
insulin-induced gene 1
- IVC
integral of viable cells
- LMM
lipid metabolism modifier
- mAb
monoclonal antibody
- MSX
methionine sulfoximine
- MUFA
monounsaturated fatty acid
- PA
phosphatidic acid
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- PS
phosphatidylserine
- S1P
site-1 protease
- S2P
site-2 protease
- SCAP
SREBF cleavage-activating protein
- SCD1
stearoyl CoA desaturase 1
- SFA
saturated fatty acid
- SNAREs
soluble N-ethylmaleimide-sensitive factor attachment protein receptors
- SRE
sterol regulatory element
- SREBF1
sterol regulatory element binding factor 1
- TAG
triacylglycerol
- XBP1
X box binding protein 1
1. Introduction
Chinese hamster ovary (CHO) cells are routinely employed for the production of recombinant biotherapeutics such as monoclonal antibodies (mAbs) (Walsh, 2018; Feary et al., 2017; Mead et al., 2015; Povey et al., 2014). Their endogenous cellular machinery facilitates efficient protein folding and human-like post-translational modification of complex multi-domain, multi-chain molecules and the generation of efficacious therapeutic material. Furthermore, their ability to grow in suspension culture in chemically defined, animal component free media has made them an attractive host for the production of such molecules and CHO cells are now the most commonly employed host for generation of recombinant biotherapeutics. Advances in high throughput technology, cell culture and cell line development processes including vector, expression host and media improvements focused upon expanding transcriptional capacity, sugar/amino acid/chaperone metabolism and manipulation, and avoiding lactate accumulation have enabled manufacturers to regularly achieve concentrations >5 g/L of mAb (Marichal-Gallardo and Álvarez, 2012).
Although CHO cell platforms for standard format mAb production are well refined, the emergence of new, novel format biotherapeutic molecules that are often non-natural has presented new challenges for existing CHO cell hosts rendering such proteins ‘difficult to express’ (DTE). Despite this, the current approach used by industry to create cell lines expressing the desired DTE protein is to force a ‘fit’ with the systems and processes successfully used with mAbs (Laux et al., 2013). These existing host cell lines have not evolved pathways for efficient synthesis, folding, assembly and secretion of DTE next-generation biologics (NGBs) of appropriate quality, resulting in significantly reduced growth and low productivity and product quality (Johari et al., 2015). There is thus a need to develop systems for improved productivity and reduced product quality issues with DTE proteins. Efforts to date have been based around media and process development through high throughput screening applied to current expression platforms (Pybus et al., 2014) and the use of medium additives (e.g. DMSO), host cell line engineering; and modifying expression vector architecture. Others have approached the problem via the manipulation of (a) the unfolded protein response and endoplasmic reticulum (ER) associated chaperones, foldases and protein degradation (Johari et al., 2015; Pybus et al., 2014), (b) via secretion and manipulation of soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) and components of the translational machinery (Le Fourn et al., 2014) or (c) engineering of the target protein itself (protein engineering) (Grote et al., 2012).
Although there have been attempts to manipulate multiple aspects of CHO cell biology to improve their ability to grow and/or synthesize and secrete recombinant biotherapeutics, one area which has received little attention to date is that of lipid biosynthesis. Lipids are the major component of cellular membranes, and are integral in a kaleidoscope of essential cellular activities such as energy metabolism, cell signaling, cell growth and survival, organelle formation and containment, and transport/secretion via vesicle formation and trafficking (Marichal-Gallardo and Álvarez, 2012). It is therefore surprising that lipid metabolism has been little studied in a CHO cell bioprocessing context. One of the few studies in this area compared lipid profiles of CHO, HEK293 and SP2/0-Ag14 cells and concluded that a knowledge of lipid metabolism could help define physiological differences across different mammalian recombinant protein production hosts and help guide metabolic engineering and medium formulation to improve production of biotherapeutics (Zhang et al., 2017).
Glycerophospholipids are the most abundant lipid species in eukaryotic cells and consist of phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), phosphatidic acid (PA) and phosphatidylinositol (PI). Whilst PC is the most abundant lipid species in a mammalian cell, varying ratios of lipid species, in combination with incorporation of different membrane proteins, influences membrane and cellular properties (van Meer et al., 2008). For example, the ER requires a specific membrane fluidity as it is the primary secretory organelle for trafficking of proteins and lipids whilst the plasma membrane requires a more rigid membrane to provide cellular structure (Monje-Galvan and Klauda, 2015; Jackson et al., 2016). The ER is not only the primary site of de novo lipid biogenesis but also the initial organelle involved in vesicle trafficking in the exocytic pathway by which proteins are transported to the Golgi and eventually secreted from the cell. The ER is typically a large organelle contained by a continuous membrane system and lipid turnover in the ER is crucial for optimal ER and, in turn, cellular function. Overall, cellular lipid homeostasis is governed by a balance of de novo biogenesis and membrane trafficking together with the modification of existing lipid species subsequent to their synthesis. These homeostatic pathways can be activated or suppressed in response to specific cellular conditions such as temperature, redox status and cellular sterol levels (Han and Kaufman, 2016). For example, the unfolded protein response (UPR) can be induced by the excessive accumulation of lipids intracellularly and results in the regulation of ER quantity in the cell through synthesis of both proteins and lipids (Han and Kaufman, 2016). X box binding protein 1 (XBP1) is a key regulator of the UPR and processing of XBP1 induces the formation of a specific splice variant which upregulates a cascade of genes including stearoyl CoA desaturase 1 (SCD1), diacylglycerol O-acyltransferase 2 (DGAT2) and acetyl CoA carboxylase (ACC) which serve to modify cellular lipid content. Overexpression of XBP1 has been shown to expand the ER of CHO cells and has subsequently been successfully employed to improve recombinant protein production yields (Tigges and Fussenegger, 2006).
With regard to temperature stress and the homeostatics of lipid biosynthesis, we, and others, have previously shown that at subphysiological temperatures (<37 °C, particularly around 32 °C) cells respond to such sub-optimal temperature by changing the abundance and saturation level of a range of lipids, particularly increasing the level of unsaturated fatty acids (Roobol et al., 2011). Under such culture temperature conditions, secretory recombinant protein yields from CHO cells can be increased (Masterton et al., 2010), and we therefore set out to investigate (1) whether the engineering of CHO cells to manipulate the abundance and unsaturation of lipids would result in changes to the amounts and saturation level of lipids in CHO cells and (2) if this resulted in increased secretory recombinant protein yields, particularly for difficult to express proteins, at physiological temperatures. To achieve this we targeted manipulation of genes/proteins directly involved in generating unsaturated lipids and/or that globally control lipid biosynthesis in CHO cells via engineering of sterol regulatory element binding factor 1 (SREBF1) and SCD1.
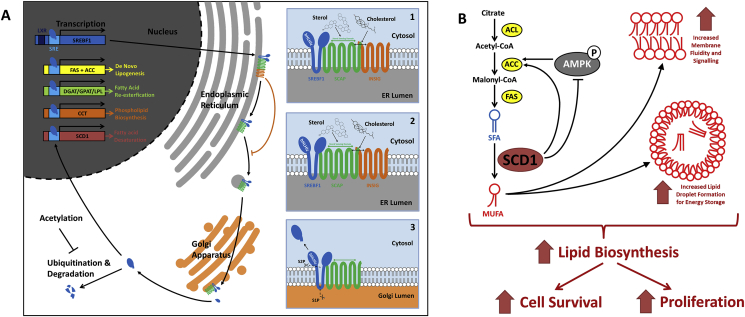
SREBF1 is a global transcriptional regulator capable of activating a plethora of genes involved in lipid metabolism including de novo lipogenesis, fatty acid re-esterification, phospholipid biosynthesis and fatty acid desaturation (Fig. 1). The activity of SREBF1 as a transcriptional activator is governed by its post-translational processing in the cell. Initially, SREBF1 localizes to the ER membrane where it integrates into the phospholipid bilayer and forms a complex with SREBF cleavage-activating protein (SCAP) which can facilitate migration of SREBF1 to the Golgi. However, under high cellular sterol levels (particularly cholesterol) a conformational change in SCAP is induced which aids binding to the membrane integral protein insulin-induced gene 1 (INSIG), inhibiting migration of this complex from the ER. In the absence of sterols, INSIG does not bind to SCAP, allowing migration of the SREBF:SCAP complex to the Golgi. Sequential proteolytic cleavage of SREBF1 occurs in the Golgi mediated by site-1 protease (S1P) and site-2 protease (S2P) proteins liberating the N-terminal basic helix loop helix leucine zipper (bHLHLz) in the cytosol. Lysine residues present on the cleaved SREBF1 are ubiquitinated and degraded by the 26S proteasome, but this ubiquitination can be inhibited through acetylation of the lysine residues, which allows migration to the nucleus. Finally, mature nuclear SREBF1 binds to sterol regulatory element (SRE) sequences upstream of various genes involved in lipid metabolism causing them to be transcriptionally activated (Scaglia et al., 2009; Shimano, 2001).
Fig. 1.
Schematics illustrating the function of selected genes involved in lipid biosynthesis in eukaryotic cells. Figure A outlines the main regulatory mechanisms of sterol regulatory element binding factor 1 (SREBF1). SREBF1 is initially expressed in the ER as a membrane integrated protein bound to the SCAP/INSIG complex. In the presence of high sterol levels the affinity level of INSIG is high and this complex is unable to migrate away from the ER (panel 1). In the absence of sterols INSIG binds with low affinity and can dissociate from the complex (panel 2). The SREBF1:SCAP complex can then migrate to the Golgi apparatus via vesicles. Once in the Golgi S1P and S2P proteases sequentially cleave the SREBF1 molecule liberating the bHLH domain (panel 3) which can migrate into the nucleus through the cytosol. The liberated SREBF1 molecule is subject to modifications such as ubiquitination and acetylation when in the cytosol which can activate or repress degradation of the molecule respectively. When in the nucleus, nuclear SREBF1 (nSREBF1) is active as a transcriptional activator leading to the expression of a plethora of genes involved in lipid metabolism. Figure B shows the role of stearoyl CoA desaturase 1 (SCD1); an enzyme involved in desaturation of saturated fatty acids (SFA) to monounsaturated fatty acids (MUFA) which results in increased membrane fluidity and lipid droplet formation. Furthermore, SCD1 can also directly and indirectly influence the activity of acetyl CoA carboxylase (ACC) which is a rate limiting step involved in de novo lipogenesis. Overall, the activity of SCD1 can increase the rate of lipid biosynthesis and enhance both cell survival and proliferation rates of the cell.
SCD1 expression is itself partially controlled by SREBF1 mediated transcriptional activation, and is localized to the ER where it catalyzes the conversion of saturated fatty acids (SFA) to monounsaturated fatty acids (MUFA). An increase in the MUFA:SFA ratio results in greater membrane fluidity and cell signaling as well as the formation of triacylglycerol (TAG) containing lipid droplets (Ren et al., 2018; Maulucci et al., 2016; Jaureguiberry et al., 2014). SCD1 also controls the phosphorylation status of adenosine monophosphate (AMP) activated protein kinase (AMPK), consequentially reducing its ability to phosphorylate and inhibit acetyl CoA carboxylase (ACC); a rate-limiting enzyme in the lipid synthesis process. Finally, desaturation of SFA prevents their accumulation, which can cause cell death through lipotoxicity. Together the aforementioned functions of SCD1 result in increased lipid biosynthesis, cell survival and proliferation rates (Igal and Ariel, 2011; Igal, 2016).
In this study we show that overexpression of SCD1 and SREBF1 in the Lonza Biologics’ proprietary CHOK1SV GS-KO™ host cell line; a glutamine synthetase (GS) knockout host cell line, results in enhanced secreted recombinant protein yields of a number of different model biotherapeutic proteins, although this is dependent upon the amount of overexpression of the lipid metabolism modifying (LMM) genes (SCD1 and SREBF1). Their overexpression also influenced cell growth and batch culture longevity. Mass spectrometry based lipidomics analysis confirmed that genetic manipulation of lipid metabolism altered the global lipid profile of engineered cells whilst specific lipids were up or down regulated compared to the host cell in the LMM engineered cells. Electron microscopy imaging revealed substantial changes to the structure and membranes of LMM engineered cells compared to controls. Finally, LMM engineered cells were shown to have an expanded ER compared to control cells that was related to the amount of LMM overexpression and improvement in secretory biotherapeutic protein production, suggesting that expansion of the ER may at least partially underpin the enhanced secretory yields from LMM engineered cells.
2. Methods
2.1. Cloning and construction of vectors used for cell line engineering
A summary of the vectors generated in this study is outlined in the associated Date in Brief (Budge et al., 2019) (Table 1). The process for amplifying and cloning the CHO specific SCD1 gene sequence and the SREBF1 mouse gene sequence is described in the associated Data in Brief (Budge et al., 2019). The SREBF1 CHO cell gene sequence (NCBI accession no. NM_001244003.1) was synthesized by GeneART (Thermo Fisher Scientific, USA) and cloned into a Lonza expression vector where expression was driven by either what was considered to be a strong (high, Fig. 5) or weaker (low, Fig. 5) promoter to drive transcription of the SREBF1 CHO cell gene to relatively low or high levels. The vector also encoded for a glutamine synthetase (GS) metabolic selection marker to isolate cell pools.
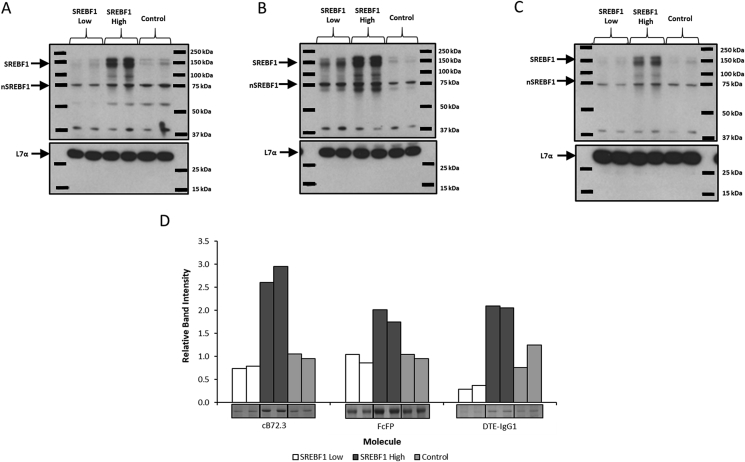
Fig. 5.
Analysis of SREBF1 engineered CHOK1SV GS-KO™ host cells stably expressing a variety of secretory recombinant biotherapeutic molecules. Cell pools were constructed by the simultaneous co-transfection of two expression vectors. The first vector contained the GS gene for selection as well as genes appropriate for production of either cB72.3, FcFP or DTE-IgG1 whilst the second vector contained the SREBF1 gene driven by different promoters to yield either low (SREBF1 Low) or high (SREBF1 High) SREBF1 protein abundance. Control pools (Control) were generated using the appropriate first vector (also generated using GS selection) in conjunction with an empty second vector (i.e. not encoding the SREBF1 gene). Western blot analysis was carried out to determine relative levels of SREBF1 abundance from cell pools constructed to express either cB72.3 (A), FcFP (B) or DTE-IgG1 (C) where SREBF1 Low, SREBF1 High or a Control vector were co-transfected during the generation of pools as highlighted in the figure (L7a was used as a loading control). Figure D shows bands from a Coomassie blue stained SDS-PAGE gel and densitometry analysis of bands corresponding to the recombinant products harvested at 7 days of batch culture from the constructed cell pools. All lanes show bands corresponding to an independent cell pool construction.
Several vectors were constructed which contained the gene(s) for appropriate expression of a range of different format, industrially relevant biotherapeutics to be used as model molecules to assess the performance of the engineering approach implemented and described herein. An in-house Lonza expression vector containing sequences based on chimeric B72.3 (cB72.3) antibody was used as a model IgG4 molecule (considered to be easy to express by the authors) and a second Lonza vector was also utilized which contained sequences for appropriate expression of a model Fc-fusion protein (FcFP). An additional vector for expression of a model IgG1 antibody (DTE-IgG1) which is considered by the authors to be difficult to express was also constructed. All vectors described above also contained the gene sequence of glutamine synthetase (GS) for use as a metabolic selection marker where necessary. Further detail can be found in the associated Data in Brief (Budge et al., 2019).
2.2. Cell culture and cell line construction
The CHOK1SV GS-KO™ host cell line was cultured in CD-CHO medium (ThermoFisher Scientific) supplemented with 6 mM L-glutamine at 37 °C in a 5% CO2 environment and shaking at 140 rpm. Cells were routinely counted using a ViCell (Beckman Coulter) instrument using a 1 ml cell sample to determine viable and total cell concentrations. Culture viability was estimated as the number of viable cells expressed as a proportion of total cells. Cells were sub-cultured every 3–4 days, seeding new cultures at 0.2 × 106 viable cells per ml in 20 ml of culture volume in a 125 ml Erlenmeyer flask (Corning®). Following cell line construction (see below) with vectors containing the GS gene, cells were maintained in CD-CHO medium in the absence of L-glutamine but in the presence of 25 μM L-methionine sulfoximine (MSX). In order to generate and maintain cell pools and lines using vectors derived from pcDNA3.1V5-His/TOPO, G418-disulphate (Melford, UK) was supplemented into media to a final concentration of 750 μg/ml.
The lipid metabolism modified CHOK1SV GS-KO™ cell pools CHO-SCD1POOL, CHO-SREBF1POOL (mouse SREBF1 sequence) were generated using linearized expression vectors whereas the CHO-ControlPOOL cells were generated using the unmodified pcDNA3.1V5-His/TOPO vector. The vectors used were linearized using PvuI restriction enzyme (NEB®). The engineered pools were used to isolate clones via limiting one round of dilutions and are considered to have a high probability of being monoclonal (these clones are referred to as monoclonal lines herein). LMM engineered cell pools and monoclonal lines were transfected with a linearized GS containing vector (linearized with PvuI restriction enzyme (NEB®)) encoding either cB72.3 or the model FcFP. Producer pools were selected for in glutamine-free medium supplemented to a final concentration of 25 μM MSX. In all cases, electroporation was performed with 1 × 107 viable cells with 20 μg of each linearized vector DNA in a BioRad cuvette using a GenePulser Xcell electroporator (Bio-Rad). The DNA/cell mix was electroporated at 300 V and 900 μF in a cuvette with a 0.4 cm electrode gap. Selection agent (either 25 μM MSX or 750 μg/ml G418) was added 24 h post-transfection. Batch cultures were initially seeded with 0.2 × 106 viable cells/ml in 20 ml CD-CHO medium. Fed-batch cultures were also initially seeded at 0.2 × 106 viable cells/ml in 30 ml and efficient feed B (ThermoFisher Scientific) was supplemented at 0 h to comprise 15% of the total culture volume. Further supplementation was carried out on day 3, 6 and 9 in line with condition 3 as described in the manufacturer's instructions.
2.3. Western blotting
Western blotting was undertaken essentially as described previously (Roobol et al., 2014). Details of primary and secondary antibody conjugates are outlined in Data in Brief (Budge et al., 2019) (Table 2).
2.4. Extraction of lipids from cells and subsequent mass spectrometry analysis
Between 1.0 and 1.5 × 106 viable cells were pelleted by centrifugation in a bench top centrifuge (Hereaus Biofuge pico for 5 min at 400 g), frozen immediately on dry ice and stored at −80 °C until lipids were extracted. Each pellet was extracted by adding 3 ml of a 2:1 chloroform:methanol solution and 0.5 ml of water to the cell pellet. This was then centrifuged in a bench top centrifuge (Thermo Megafuge 16R for 5 min at 3000 rpm, 1500 g at 4 °C) and then the lower chloroform layer removed and dried under a nitrogen stream. Pellets were resuspended in 150 μl of chloroform:methanol (2:1) and then diluted 1:1 with a isopropanol:acetonitrile:water (2:1:1) solution. Separation of the extracted lipids was performed on a Acquity UPLC CSH C18 column (2.1 × 100 mm, 1.7 μm) using buffer A (acetonitrile:water (3:2) containing 10 mM ammonium formate and 0.1% formic acid) and buffer B (isopropanol:acetonitrile (9:1) containing 10 mM ammonium formate and 0.1% formic acid v/v) and a flow rate of 0.4 ml/min. A gradient from 60% buffer A to 1% buffer A over 18 min was used for separation of lipids with the system being coupled to the mass spectrometer. Mass spectrometry analysis of the material eluting from the liquid chromatography system was performed on a Synapt G2Si (Waters) mass spectrometer set to negative ionization mode with MSE acquisition over 100 to 1200 m/z mass range with a scan time of 0.5 s using a leucine encephalin LockSpray solution (Waters) and the following instrument settings; collision energy 20–30 eV, capillary voltage 1.0 kV, cone voltage 40 V, desolvation temperature 400 °C, desolvation gas 800 l/h and source temperature 100 °C. The data was analyzed using the Waters software UNIFI searching a Waters Lipid Maps database. UNIFI was linked to EZInfo software to generate the principal component analysis (PCA) plots.
2.5. Preparation of samples for immunofluorescence analysis and capture of images
In order to adhere suspension cells to coverslips, coverslips were first submerged in a poly-L-lysine solution (molecular weight 70,000–150,000 (P4707), Sigma Aldrich, USA) and incubated for 15 min at room temperature. Coverslips were then removed and left to dry in a sterile environment before being transferred to a 24 well plate. Cells were added to the wells at a concentration of 2 × 105 viable cells per well and incubated in a static incubator for a minimum of 1 h before further processing. Samples were fixed and subsequently permeabilized using 4% paraformaldehyde in PBS (w/v) followed by 0.1% Triton-X100 in PBS (v/v) respectively. Samples were blocked using 3% BSA in PBS (w/v) and exposed to the appropriate primary antibodies overnight at 4 °C and secondary antibodies for 2 h at room temperature. The details of the antibodies used are described in Data in Brief (Budge et al., 2019) (Table 2). Finally, samples were exposed to DAPI (10 mg/ml) before being mounting in ProLong™ Gold Antifade Mountant (ThermoScientific). Images were captured using a Zeiss LSM 880/Elyra/Axio Observer Z1 confocal microscope instrument.
2.6. Fluorescence microscopy image analysis for ER/Nuclear quantification
Images for analysis were captured on a Zeiss LSM 880/Elyra/Axio Observer Z1 confocal microscope as described above. Image analysis for quantification of ER size was performed using Zeiss Zen Blue software. The “spline contour” tool was used to manually outline the outer perimeter of the ER (highlighted using calnexin antibody Abcam ab22595) and the nucleus (highlighted by DNA staining with DAPI) and the area of both were determined for each cell image. The outer area of the ER defines both the ER and nuclear area and therefore the area determined for the nucleus was subtracted from that of the area enclosed by the ER to calculate the area of ER. The calculated ER area was then divided by the area of nucleus to establish the ratio of ER area in relation to the area of the nucleus through a cross section of each individual cell. Care was taken to ensure that the z-position of each of the images ran approximately through the equator of the cells (determined via the diameter of the nuclear stain). Since the estimated area of ER may be dependent on the z-position, the nucleus was used to determine the ratio to compensate for drift from the equator. Between 30-68 individual cells were analyzed per sample.
2.7. Electron microscopy analysis
Cells attached to poly-L-lysine coated Aclar membrane (Agar Scientific) were fixed for 2 h in 2.5% glutaraldehyde (w/v) in 100 mM sodium cacodylate (CAB) buffer pH 7.2. Samples were washed twice for 10 min in 100 mM CAB and then post-fixed in 1% osmium tetroxide (w/v) in 100 mM CAB for 1 h before being dehydrated using an ethanol series of 50%, 70%, 90% (v/v) and 3 times with 100% ethanol for 10 min per step. The samples were then placed into propylene oxide, for 10 min, and following this into a 1:1 mixture of propylene oxide and Agar LV resin (Agar Scientific) for 30 min. Following this, samples were embedded in freshly prepared Agar LV resin twice for 2 h before being placed in shallow aluminium moulds with the cells facing up and were polymerized at 60 °C for 24 h before being examined with a dissecting microscope to identify areas confluent with cells. These areas were cut out with a jig saw and attached to polymerized resin blocks with superglue, and once attached, the Aclar membrane was peeled off, exposing a monolayer of cells in the block face. Sections of 70 nm were cut on a Leica EM UC7 ultramicrotome using a diamond knife (Diatome) and were collected on 400 mesh copper grids. Sections were counterstained in 4.5% uranyl acetate (w/v) in 1% acetic acid (v/v) for 45 min and in Reynolds’ lead citrate for 7 min. Samples were viewed in a Jeol 1230 transmission electron microscope and images were captured with a Gatan One View 16mp camera.
2.8. Octet® measurement of recombinant protein concentrations
Recombinant molecule concentrations in cell culture supernatants were determined using an Octet® instrument (ForteBio) with IgG calibrators and protein A biosensors. Product concentration and viable cell concentrations determined using a ViCell instrument (Beckman Coulter) were analyzed across at least three timepoints of culture to calculate cell specific productivity values (Qp). Qp is equal to the gradient of the line equation where the average product concentration was plotted on the y axis and the average integral of viable cells (IVC) was plotted on the x axis. IVC was calculated as shown in the equation below where is the viable cell number recorded at a timepoint, is the viable cell number recorded at the previous timepoint, t is the time (in hours) that the timepoint was taken and is the time (in hours) of the previous timepoint. was the IVC recorded at the previous timepoint:
2.9. Relative mRNA expression analysis by qRT-PCR
qRT-PCR was undertaken using Qiagen QuantiFast SYBR Green RT-PCR kits essentially as previously described (Mead et al., 2015) using FcFP specific primers. The amounts of the FcFP transcript was normalized to that of the house keeping genes glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and β-actin as endogenous controls, and the relative levels of expression of each sample calculated using the ΔΔCt method.
3. Results
3.1. SCD1 and SREBF1 proteins can be overexpressed in CHO cells and localize to the expected cellular location
To determine if manipulation of lipid metabolism in CHO cells could be used to enhance cell growth and secretory recombinant protein capacity, we set out to engineer the expression of the transcription factor SREBF1, and the desaturase SCD1. Respectively, these control the expression of multiple genes involved in lipid biosynthesis (SREBF1) and the conversion of saturated fatty acids to monounsaturated fatty acids (SCD1). We initially developed CHOK1SV GS-KO™ cell pools and clonal cell lines engineered to overexpress each of these genes. In order to develop stably expressing engineered cell pools, we initially used the pcDNA3.1V5-His/TOPO vector (Thermo Fisher Scientific) which bears a neomycin phosphate transferase selection marker (used with G418 sulfate (Melford) as the selection agent) to produce two vectors for overexpression of either the CHO SCD1 or mouse SREBF1 genes under the control of a CMV promoter and inclusive of a 3’ V5 tag. Polyclonal cell pools which overexpress either SCD1 (CHO-SCD1POOL) or SREBF1 (CHO-SREBF1POOL) genes were generated using the pcDNA3.1V5-His/TOPO derived vectors. A control cell pool was also generated using the unmodified pcDNA3.1V5-His/TOPO vector and the resulting polyclonal pool was named CHO-ControlPOOL. Clones which overexpressed the LMM molecules were isolated from these original pools and western blot analysis was undertaken on these clones, using antibodies specific for SCD1 and SREBF1, in order to assess and rank relative amounts of expression of the exogenous lipid modifying genes in the isolated clones (data not shown). Three clones showing varying levels of LMM expression were selected from each pool for further analysis. No significant difference in SCD1 or SREBF1 protein amount was evident in clones isolated from the CHO-ControlPOOL, as determined by western blot, so the clones taken forward were selected at random.
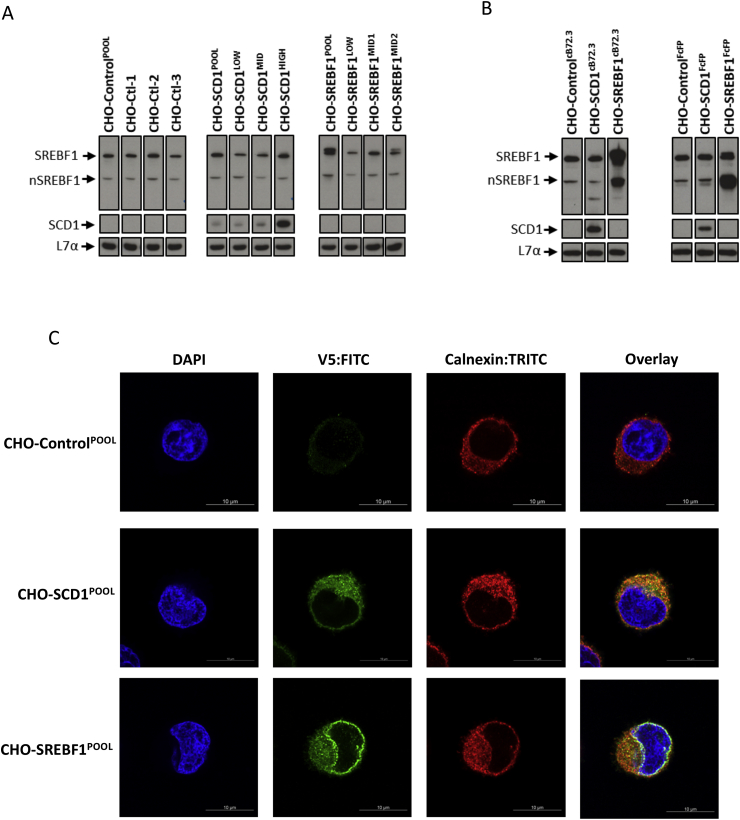
Fig. 2A shows western blot analysis of the cell pools and isolated clones to establish amounts of lipid metabolism modifying proteins (SCD1 and SREBF1) with L7α being used as a loading control (Roobol et al., 2009, 2014). The three clonal lines isolated from the CHO-ControlPOOL were termed CHO-Ctl-1, CHO-Ctl-2 and CHO-Ctl-3. Isolated clones from the CHO-SCD1POOL were named CHO-SCD1LOW, CHO-SCD1MID and CHO-SCD1HIGH based on their relative SCD1 abundance as confirmed in the blots presented in Fig. 2A. Whilst endogenous levels of SCD1 were not detectable in control or SREBF1 samples at the exposure shown, all SCD1 overexpressing cells had elevated levels compared to the control. Densitometry analysis (Data in Brief (Budge et al., 2019) Fig. 1A) of the western blots showed that the low SCD1 (CHO-SCD1LOW) clone had lower overexpression compared to that observed in the polyclonal pool whilst the middle SCD1 overexpressing clone (CHO-SCD1MID) levels were approximately 1.5-fold higher than that in the CHO-SCD1POOL and the high clone (CHO-SCD1HIGH) had nearly a 7-fold higher SCD1 expression (Data in Brief (Budge et al., 2019) Fig. 1A). Both the pool and clones all had much higher SCD1 expression than the controls.
Fig. 2.
Overexpression and cellular localization of SCD1 and SREBF1 in engineered cells. Western blot analysis of CHOK1SV GS-KO™ cells engineered to overexpress either SCD1 or SREBF1 genes where controls have been generated with an unmodified pcDNA3.1V5-His/TOPO vector. CHO-Ctl-1, CHO-Ctl-2 and CHO-Ctl-3 are monoclonal lines derived from the CHO-ControlPOOL; CHO-SCD1LOW, CHO-SCD1MID and CHO-SCD1HIGH are monoclonal cell lines isolated from CHO-SCD1POOL and CHO-SREBF1LOW, CHO-SREBF1MID1 and CHO-SREBF1MID2 are monoclonal lines isolated from CHO-SREBF1POOL. The pools which had been engineered were used to generate cells expressing cB72.3 (CHO-ControlcB72.3, CHO-SCD1cB72.3 and CHO-SREBF1cB72.3) or an Fc fusion protein (FcFP; CHO-ControlFcFP, CHO-SCD1FcFP and CHO-SREBF1FcFP). Lysate samples were generated from these cells harvested at day 6 of culture from either host cells (A) or cells stably producing the recombinant products cB72.3 or FcFP where specified (B). A primary antibody with specificity for L7α was used as a loading control. (C) Cellular localization of overexpressed SCD1 and SREBF1 proteins in CHO-ControlPOOL, CHO-SCD1POOL and CHO-SREBF1POOL cell pools as determined by immunofluorescent detection using an anti-V5 antibody conjugated with a FITC secondary antibody. An anti-calnexin antibody conjugated with a TRITC secondary antibody was used to highlight the position of the ER. Images were obtained using confocal microscopy (see Data in Brief (Budge et al., 2019) Table 2 for full details of antibodies used).
Overexpression of SREBF1 was clearly evident in the polyclonal pool (CHO-SREBF1POOL) engineered to overexpress the protein with both the full length and nuclear SREBF1 (nSREBF1) levels higher than observed in the controls (Fig. 2A and Data in Brief (Budge et al., 2019) Fig. 1B). Both full length and nSREBF1 levels were determined for each sample as only nSREBF1 is effective as a transcriptional activator and the engineering process reported here is dependent on the processing of this molecule. Interestingly, when the SREBF1 polyclonal pool (CHO-SREBF1POOL) was cloned, resulting clones did not have higher amounts of SREBF1 expression than observed in the controls or the polyclonal pool (CHO-SREBF1POOL). Nevertheless, a low (CHO-SREBF1LOW) and two clones termed mid-SREBF1 expressers (CHO-SREBF1MID1 and CHO-SREBF1MID2) were taken forward for further study.
The CHO-ControlPOOL, CHO-SCD1POOL and CHO-SREBF1POOL pools were also transfected with expression vectors for either the model chimeric IgG mAb cB72.3 or a model Fc Fusion Protein (FcFP). The resulting pools were termed CHO-ControlcB72.3, CHO-SCD1cB72.3 and CHO-SREBF1cB72.3 in those where the recombinant cB72.3 was stably expressed and CHO-ControlFcFP, CHO-SCD1FcFP and CHO-SREBF1FcFP in those where the model FcFP was stably expressed. Fig. 2B shows western blot analysis of the resulting cell pools for SREBF1 (CHO-SREBF1cB72.3 and CHO-SREBF1FcFP) and SCD1 (CHO-SCD1cB72.3 and CHO-SCD1FcFP) expression. The SCD1 and SREBF1 proteins were overexpressed in the appropriate lipid modified cells with high amounts of SCD1 and SREBF1 expression observed compared to the control cells (CHO-ControlcB72.3 and CHO-ControlFcFP). In the case of the SREBF1 overexpressing cells, both the full-length (SREBF1, band expected at 127 kDa) and nuclear SREBF1 (nSREBF1, band expected at 70 kDa) species were expressed at higher amounts than in the control cells in both the CHO-SREBF1cB72.3 and CHO-SREBF1FcFP pools (Fig. 2B). The presence of an aberrant, lower molecular weight SREBF1 band (42 kDa) was observed in the CHO-SCD1cB72.3 and CHO-SREBF1cB72.3 pools which was absent in the CHO-ControlcB72.3 and all the FcFP pool lysates (Fig. 2B).
In addition to western blotting, immunofluorescence analysis was undertaken to confirm overexpression of the target exogenous LMM proteins and to determine the localization of the expressed exogenous LMM proteins (Fig. 2C and Data in Brief (Budge et al., 2019) Fig. 1C). An antibody against the V5 tag was used to detect the exogenous lipid modifying proteins in the pools of engineered cells whilst an anti-calnexin antibody, an established ER marker (Bergström et al., 2014; Roobol et al., 2015), was used to define the ER and determine any co-localization of the LMM proteins to the ER. The images shown in Fig. 2C and the Data in Brief (Budge et al., 2019) Fig. 1C are representative of all images obtained and confirm exogenous SCD1 and SREBF1 protein expression. Overlay of the V5 signal with the calnexin staining showed co-localization suggesting that both proteins are localized to the ER, consistent with previous literature reports (Sato et al., 1994; Man et al., 2006). As described in the introduction, SREBF1 is initially localized to the ER but translocated to the Golgi where it is processed by the cellular machinery to liberate an N-terminal active transcription factor that is able to migrate to the nucleus where it acts as a transcriptional activator. However, since the V5 tag on the exogenous proteins was present on the C-terminus of SREBF1, the mature N-terminal nuclear species will not be detected using the V5 staining approach. The immunostaining of the full length SREBF1 protein yielded intense fluorescent signal at the peri-nucleus, likely due to the presence of an abundance of intracellular cholesterol which prevents migration of SREBF1 subsequent to its initial localization, upon production, in the ER (Rintoul et al., 1978).
3.2. CHO cells overexpressing the lipid metabolism modifying genes SCD1 and SREBF1 show different batch culture growth profiles compared to control host cells
We next determined if the overexpression of SCD1 or SREBF1 had any impact on cell growth. Defining the growth profiles was of particular interest since this attribute has been shown to be affected by lipid metabolism imbalances (Rintoul et al., 1978; McGrew et al., 1998). Batch-cultures were setup in biological triplicate for each cell pool and clonal cell line and samples taken daily to monitor the viable cell concentration and culture viability (Data in Brief (Budge et al., 2019) Fig. 2). There was no obvious impact on the growth characteristics of exogenous SCD1 expression on CHO cell growth in the CHO-SCD1POOL when compared to the CHO-ControlPOOL (Data in Brief (Budge et al., 2019) Fig. 2A). However, clonal cells engineered to overexpress SCD1 had different growth profiles dependent on the relative SCD1 expression levels with CHO-SCD1LOW cells reaching the highest maximum viable cell concentration of any culture (Data in Brief (Budge et al., 2019) Fig. 2C). However, whilst the CHO-SCD1MID cells had a similar growth profile to the CHO-SCD1POOL, the CHO-SCD1HIGH cells initially grew in a similar fashion but began to decline slowly in both viable cell number and culture viability at an earlier timepoint (6 days, Data in Brief (Budge et al., 2019) Fig. 2C). CHO-SCD1cB72.3 cells, expressing the cB72.3 antibody, obtained a higher maximum viable cell concentration compared to the CHO-ControlcB72.3 cultures at a later timepoint and began to decline later than the control (Data in Brief (Budge et al., 2019) Fig. 2E).
In comparison, the CHO-SREBF1POOL did not attain the same maximum viable cell concentration as the CHO-ControlPOOL or CHO-SCD1POOL, suggesting that expression of endogenous SREBF1 negatively impacted on the cells ability to attain higher viable cell concentrations (Data in Brief (Budge et al., 2019) Fig. 2A). A similar pattern was observed when the pools were engineered to express either cB72.3 (Data in Brief (Budge et al., 2019) Fig. 2E) or the FcFP (Data in Brief (Budge et al., 2019) Fig. 2F) molecules in SREBF1 overexpressing cell pools. SREBF1 engineered overexpressing clones also showed variations in growth profiles (Data in Brief (Budge et al., 2019) Fig. 2D). Whilst all cells initially grew in a similar fashion, CHO-SREBF1MID2 cells obtained the highest maximum viable cell concentration compared to other SREBF1 engineered cells and the CHO-ControlPOOL. However, CHO-SREBF1MID2 exhibited a decline in viable cell concentration from the peak concentration to almost zero within 24 h between days 8 and 9 of batch culture (Data in Brief (Budge et al., 2019) Fig. 2D). CHO-SREBF1MID1 had a similar growth profile to the control and CHO-SREBF1LOW began to decline early in culture (6 days) than the other SREBF1 engineered cells (Data in Brief (Budge et al., 2019) Fig. 2D).
3.3. CHO cells overexpressing SCD1 and SREBF1 have enhanced stable secretory recombinant protein production capacity
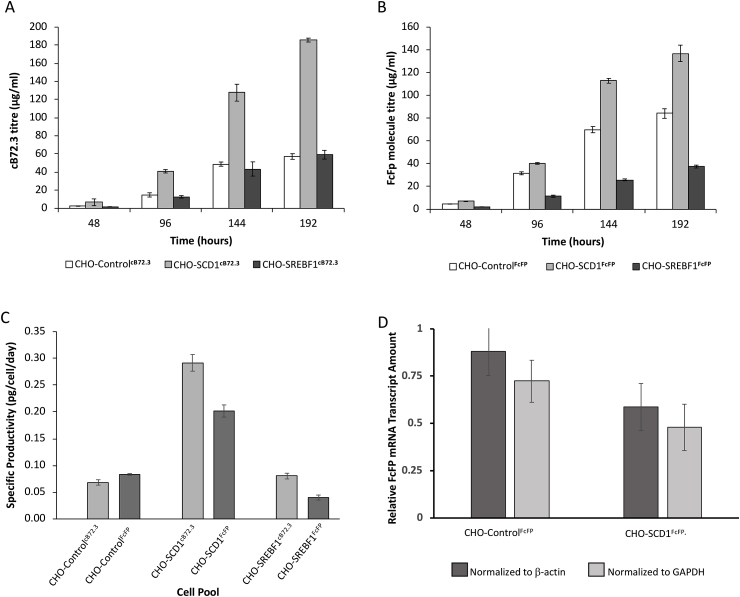
A major aim of this study was to determine if engineering of lipid metabolism in CHO cells impacted on secreted recombinant protein concentration. We therefore next assessed whether stable engineering of SCD1 or SREBF1 in the CHOK1SV GS-KO™ host cell line affected the concentration of secreted protein. Stable lipid engineered pools were transfected to generate stably expressing secreted recombinant protein pools using the stable CHO-ControlPOOL, CHO-SCD1POOL or CHO-SREBF1POOL hosts and vectors containing the recombinant protein cB72.3 or FcFP gene(s) with a GS selection marker. Culture supernatant from the resulting pools were then analyzed using an Octet® instrument to determine the profiles of cB72.3 or FcFP accumulation over 192 h (8 days) of batch culture (Fig. 3). Cells overexpressing SCD1 achieved higher concentrations of both cB72.3 (Fig. 3A) and FcFP (Fig. 3B), compared to the appropriate controls and across all timepoints measured. We also determined the relative transcript (mRNA) amounts of the FcFP transcript normalized to the house keeping genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin in the CHO-ControlFcFP and CHO-SCD1FcFP cell pools to confirm that the observed increase in FcFP secretory productivity was not brought about by changes in mRNA amounts in the engineered cells compared to the controls. As shown in Fig. 3D, the FcFP transcript amounts were not elevated in CHO-SCD1FcFP expressing pools compared to the control suggesting that the observed increased secretory amounts were not due to changes at the transcript level.
Fig. 3.
Productivity of model recombinant products in lipid engineered cell pools. cB72.3 and FcFP concentrations in supernatant of batch-cultured CHO-ControlcB72.3, CHO-SCD1cB72.3 and CHO-SREBF1cB72.3 pools as well as CHO-ControlFcFP, CHO-SCD1FcFP and CHO-SREBF1FcFP pools. These cell pools were generated by stably transfecting host pools engineered to overexpress SCD1 or SREBF1 proteins. The control was constructed using the original pcDNA3.1V5-His/TOPO vector. cB72.3 (A) and FcFP (B) concentrations were measured using an Octet® from supernatants harvested at 48, 96, 144 and 192 h of culture. Specific productivity (C) was calculated for cB72.3 (B) and FcFP (D) using these concentration values and corresponding cell growth measurements from the same time points. (D) shows the relative amount of FcFP mRNA transcript in CHO-SCD1FcFP cells compared to CHO-ControlFcFP expressing cells from samples collected after 72 h of batch culture relative to the two house keeping genes β-actin or GAPDH. n = 3 for each data point and error bars show ± one standard deviation.
Whilst the SCD1 engineered cells showed increased secretory recombinant protein productivity, cells overexpressing the mouse derived SREBF1 gene did not produce higher cB72.3 concentrations than the control and FcFP concentrations were lower (Fig. 3A and B). Specific productivity in cells overexpressing SCD1 was increased approximately 4-fold for cB72.3 and 2.4-fold for FcFP (Fig. 3C) compared to the controls under batch culture conditions. The CHO-SREBF1cB72.3 cell specific productivity was similar to the CHO-ControlcB72.3 cells whilst the CHO-SREBF1FcFP cell specific productivity was approximately half of that of the CHO-ControlFcFP cells (Fig. 3C).
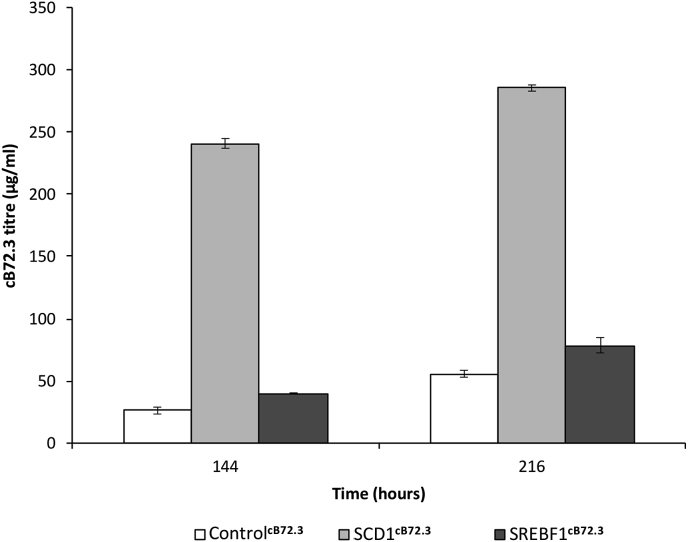
After assessing the performance of the LMM engineered CHO cell pools for stable expression of the model recombinant proteins under batch culture conditions, we investigated if fed-batch conditions affected the observed increase in secreted protein yield. To assess this we took samples at 144 and 216 h (day 6 and 9) during fed-batch culture of the SCD1 and SREBF1 engineered cell pools that were stably expressing the model mAb cB72.3 and determined the amount of secreted recombinant product in the cell culture supernatant. The resulting analysis showed that the CHOK1SV GS-KO™ pools engineered to overexpress SCD1 had a 9-fold and 5-fold increase in cB72.3 concentration after 144 and 216 h (6 and 9 days) of fed-batch culture respectively compared to the controls (Fig. 4). Pools engineered to overexpress mouse SREBF1 showed a more modest increase in secreted product concentration of approximately 1.5-fold above that of the control at both timepoints (Fig. 4).
Fig. 4.
Monoclonal antibody production in fed-batch conditions using lipid engineered cells. cB72.3 concentrations in fed batch cultures where supernatants were harvested from CHO-ControlcB72.3, CHO-SCD1cB72.3 and CHO-SREBF1cB72.3 cultures at 144 and 216 h (6 and 9 days) of culture and feeds were added on day 0, 3 and 6 where samples were harvested before feeding on day 6. n = 3 for each data point and error bars show ± one standard deviation.
As described above, the CHO-SREBF1POOL engineered cell pools had elevated amounts of SREBF1 protein expression compared to the controls but the resulting SREBF1 engineered clonal lines did not have elevated SREBF1 amounts compared to CHO-SREBF1POOL (see Fig. 2A and Data in Brief (Budge et al., 2019) Fig. 1B). We therefore investigated if we could use the CHO specific SREBF1 gene sequence under the control of promoters of different strengths to generate engineered cells with differing intracellular levels of SREBF1, and then make stable cell lines by co-transfection of the appropriate SREBF1 vectors with a second vector containing the GS selection marker and the genes for the model product. To achieve this we used vectors with promoters to drive either relatively high or low expression of SREBF1 and a second vector containing the genes for either of the mAb cB72.3 or DTE-IgG1 (a model IgG1 mAb considered by the authors to be difficult to express), or the FcFP alongside the GS gene as the selectable marker. Western blot analysis of SREBF1 expression in the resulting pools confirmed that varying levels (either high or low) were achieved from the different promoters utilized as intended. In the “high” SREBF1 pool there were elevated levels of both full length SREBF1 and nuclear SREBF1 species in all pools (Fig. 5A–C). Coomassie blue staining of model proteins on an SDS-PAGE revealed that the amount of product in the supernatant in batch culture samples after 7 days of culture was 2-3-fold higher in the high SREBF1 expressing pools compared to the control and low SREBF1 expressing pools (Fig. 5D). This suggests that the amount of SREBF1 is integral to the success of the engineering approach and that overexpression of the CHO specific SREBF1 gene sequence improved recombinant titres to a greater degree than the mouse derived gene. Collectively these data show that SCD1 and SREBF1 engineering of CHO cell hosts can be used to generate hosts that yield enhanced stable secretory recombinant product concentrations under batch and fed-batch conditions, including standard monoclonal antibodies and DTE un-natural molecules (e.g. those that do not exist in nature).
3.4. CHO cells overexpressing SCD1 and SREBF1 show enhanced transient secretory recombinant protein production capacity
We also investigated whether the impact of stable overexpression of SCD1 or SREBF1 in the CHOK1SV GS-KO™ host cell had an impact on accumulation of model secreted recombinant proteins in a transient expression system. To determine this, the engineered host pools (CHO-ControlPOOL, CHO-SCD1POOL and CHO-SREBF1POOL) were transfected with vectors containing the necessary genes for expression of a model IgG mAb, cB72.3, or the difficult to express FcFP and grown under batch culture conditions with supernatant samples taken over a 96 h period post-transfection for analysis of secreted product (Data in Brief (Budge et al., 2019) Fig. 3). Both the CHO-SCD1POOL and CHO-SREBF1POOL lipid modified cell pools produced increased concentrations of both cB72.3 and the FcFP (Data in Brief (Budge et al., 2019) Fig. 3A & C) compared to the control (CHO-ControlPOOL). By 96 h post-transfection, both the CHO-SCD1POOL and CHO-SREBF1POOL cell pools showed an approximate 60% increase in cB72.3 and 40% increase in FcFP concentration compared to the CHOK1SV GS-KO™ control pool. Western blot analysis was also undertaken on supernatant samples from the cB72.3 and FcFP (Data in Brief (Budge et al., 2019) Fig. 3B & D) transfected pools and showed more intense bands for the respective products in the CHO-SCD1POOL and CHO-SREBF1POOL engineered pool derived samples than from CHO-ControlPOOL.
Finally, we investigated whether the transient engineering of LMM CHO cells could deliver enhanced transient product yields of a further DTE secreted recombinant protein molecule. Being able to rapidly produce higher yields of DTE molecules would facilitate rapid material generation for early stage screening of such potential therapeutic molecules. For this purpose, a vector containing the three genes required for expression of this further model DTE antibody fusion (IL2-F) and either the SCD1 or SREBF1 gene (or no LMM gene in the control), were transfected (on the same vector) by electroporation into the CHOK1SV GS-KO™ host cell line. The Data in Brief (Budge et al., 2019) Fig. 4 shows analysis of supernatant samples harvested from transient batch cultures at 96 h post-transfection with the aforementioned IL2-F vectors. Octet® analysis of the supernatant samples showed that there was an approximate 2.5-fold increase in transient IL2-F expression when simultaneously expressed with the SCD1 gene compared to the control whilst simultaneous expression with the SREBF1 gene produced 1.5-fold more IL2-F compared to the control (Data in Brief (Budge et al., 2019) Fig. 4). Western blot analysis of reduced supernatant samples confirmed increased transient production of both the standard and fused heavy chain molecules required for expression of IL2-F at 48 and 96 h post-transfection when SCD1 or SREBF1 were simultaneously expressed (Data in Brief (Budge et al., 2019) Fig. 4B).
3.5. CHO cells stably engineered to overexpress the lipid metabolism modifying genes SCD1 and SREBF1 have altered lipid profiles and cellular structure
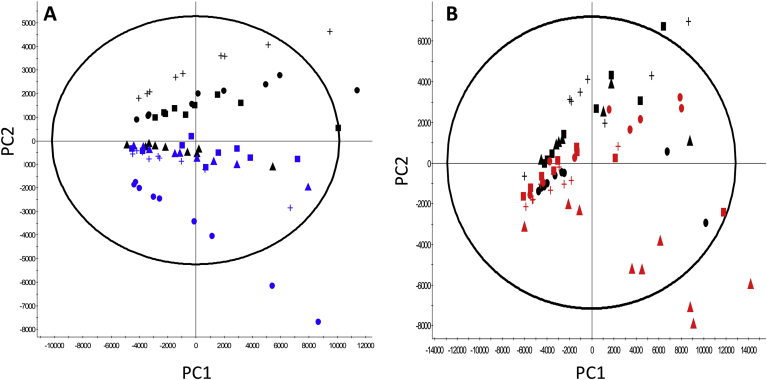
Having shown that overexpression of the LMM proteins SCD1 and SREBF1 could enhance secretory yields of various recombinant biotherapeutic proteins from CHO cells, we next set out to determine if this resulted in changes to the lipid profiles/makeup of engineered cells and their structure. We investigated this by mass spectrometry based lipid profiling of whole cell lipid extracts from triplicate biological replicate batch cultures harvested on day 6 of culture (with each sample analyzed in technical triplicate) and by electron microscopy of the engineered pools and three clonal lines of the engineered and control cells. Mass spectrometry data from the analysis of the lipids from control and engineered cells were initially subjected to principal component analysis (PCA) (Fig. 6A and B). These data show that the lipid profile of the CHO-SCD1HIGH clonal cell line clustered separately from the other SCD1 engineered clones and polyclonal pool and from the control samples (Fig. 6A). The CHO-SCD1MID cell line data points tended to cluster between the CHO-SCD1HIGH and CHO-SCD1LOW cell lines suggesting a link between SCD1 expression levels and the change in the lipid profile. PCA of the SREBF1 data showed that the CHO-SREBF1LOW clone had a distinct lipid profile from the other SREBF1 engineered clones, CHO-SREBF1POOL and the controls (Fig. 6B). CHO-SREBF1MID1 and CHO-SREBF1MID2 clones clustered separately from each other and the controls and CHO-SREBF1POOL engineered pools. Collectively these data confirm that there are changes to the global lipid profile of SCD1 and SREBF1 engineered cells compared to that in the control cells.
Fig. 6.
Analysis of cellular lipid content in control and LMM engineered CHO cells using mass spectrometry. (A) and (B) show Principal Component Analysis (PCA) of the mass spectrometry derived lipid profiles extracted from cells overexpressing either SREBF1 or SCD1 genes highlighting differences in lipidomic profiles of engineered CHOK1SV GS-KO cell lines harvested after 6 days of batch culture. Figure A shows PCA of control samples and samples engineered to overexpress SCD1; black data points represent controls (CHO-Ctl-1 = ●, CHO-Ctl-2 = ▲, CHO-Ctl-3 = +, CHO-ControlPOOL = ■) whilst blue data points represent SCD1 overexpressing cells (CHO-SCD1LOW =  , CHO-SCD1MID = +, CHO-SCD1HIGH =
, CHO-SCD1MID = +, CHO-SCD1HIGH =  , CHO-SCD1POOL =
, CHO-SCD1POOL =  ). Figure B shows analysis of control samples and samples engineered to overexpress SREBF1; black data points represent controls (CHO-Ctl-1 = ●, CHO-Ctl-2 = ▲, CHO-Ctl-3 = +, CHO-ControlPOOL = ■) whilst red data points represent SREBF1 overexpressing cells (CHO-SREBF1LOW =
). Figure B shows analysis of control samples and samples engineered to overexpress SREBF1; black data points represent controls (CHO-Ctl-1 = ●, CHO-Ctl-2 = ▲, CHO-Ctl-3 = +, CHO-ControlPOOL = ■) whilst red data points represent SREBF1 overexpressing cells (CHO-SREBF1LOW =  , CHO-SREBF1MID1 =
, CHO-SREBF1MID1 =  , CHO-SREBF1MID2 =
, CHO-SREBF1MID2 =  , CHO-SREBF1POOL =
, CHO-SREBF1POOL =  ). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We then analyzed individual lipid m/z mass peaks to identify specific lipids that changed in the SCD1 engineered cell lines and polyclonal pool compared to the control. We identified specific lipids that were either upregulated (Data in Brief (Budge et al., 2019) Fig. 5A and B) or downregulated (Data in Brief (Budge et al., 2019) Fig. 5D) in all samples from the CHO-SCD1HIGH clone compared to the control and the other SCD1 engineered clones and pool. This is consistent with the PCA analysis of the total lipid profiles as determined by mass spectrometry analysis whereby the CHO-SCD1HIGH clone was distinct from the others and the protein analysis data (Fig. 2A and Data in Brief (Budge et al., 2019) Fig. 1A), which showed that SCD1 expression was elevated in the high clone to a much greater extent than in any of the other samples. There were also many lipid ion peaks that were unchanged across all samples (see Data in Brief (Budge et al., 2019) Fig. 5C for example). Collectively, these data show that SCD1 and SREBF1 engineering of CHO cells changes the lipid profile of the cell and this is related to the amount of overexpression of the individual LMM genes.
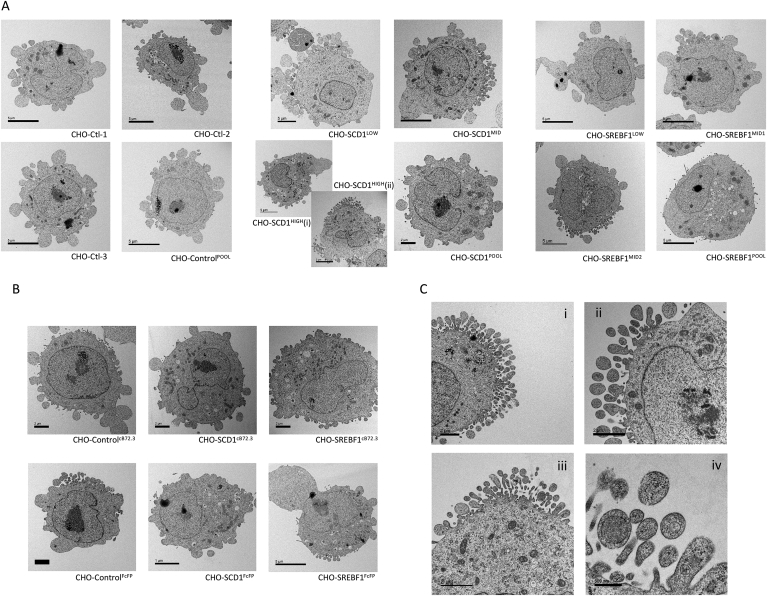
We also undertook electron microscopy analysis to determine if changing the lipid profile of CHO cells gave any observable differences to the membrane and cellular structures in lipid modified cells (Fig. 7). The images shown in Fig. 7 are representative of all images acquired for each sample and are broadly separated into CHO-Control, CHO-SCD1 and CHO-SREBF1 cells where a single representative image for each of the monoclonal or pool samples are displayed. The most striking difference between these samples was the difference in the plasma membrane and cell morphology of SCD1 overexpressing cells. Whilst the plasma membrane of all cells imaged exhibited protrusions, this feature was particularly exaggerated in SCD1 overexpressing cells and Fig. 7C shows images from CHO-SCD1POOL (Fig. 7Ci) and CHO-SCD1HIGH (Fig. 7Cii, iii and iv) in which cells have an abundance of vesicle like protrusions. This attribute was particularly common in CHO-SCD1HIGH cells and is in alignment with an increased membrane fluidity brought about through the activity of SCD1. Additionally, a greater number of membrane bound features such as vesicles were observed in SCD1 and SREBF1 overexpressing cells in comparison to the relevant controls in both host (Fig. 7A) and recombinant protein producing (Fig. 7B) cells.
Fig. 7.
Images obtained by electron microscopy of cells engineered to overexpress lipid modifying genes. Samples were taken at day 6 of batch culture and observed using a Jeol 1230 transmission electron microscope and images were captured using a Gatan One View 16mp camera. Monoclonal and host cell pools (A) and cB72.3 and FcFP expressing cells (B) were imaged and representative images are shown for all cells included in this study. Figure (C) shows images from CHO-SCD1POOL (i) and CHO-SCD1HIGH (ii, iii and iv) of membrane protrusions observed frequently in these cells.
3.6. Engineering of the lipid metabolism modifying genes SCD1 and SREBF1 in CHO cells results in an expansion of the ER
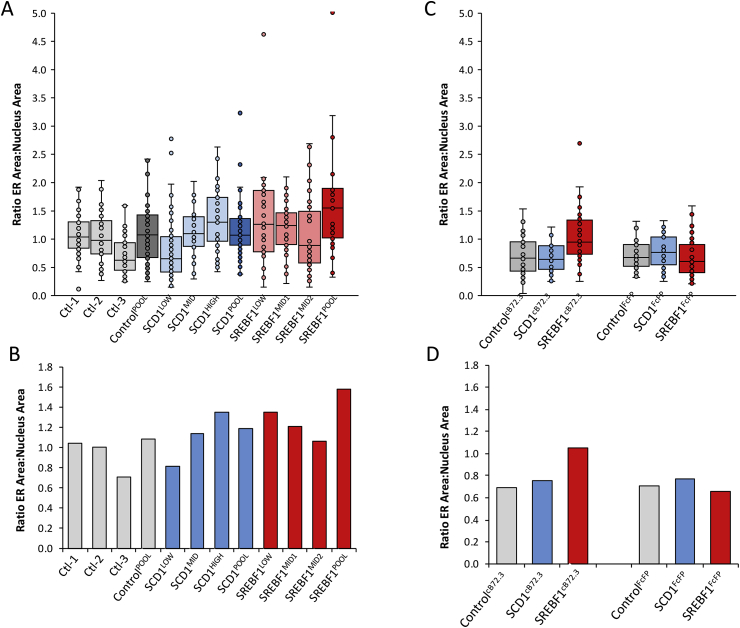
The ER is an organelle which requires high lipid turnover due to the large abundance of continuous membrane required to contain it and is also the major site of de novo lipogenesis. Induced ER expansion approaches have previously been successful in improving recombinant protein productivity in CHO cells (Tigges and Fussenegger, 2006). In consideration of this, ER area was estimated to determine if overexpression of SCD1 and SREBF1 affected ER size. ER size was quantified on day 6 of batch culture relative to the nucleus, to ascertain whether modifying cellular lipid content has an impact on ER expansion as described in the methods section. Fig. 8 reports the data obtained from this analysis and box and whisker plots show a wide range in the ratio of ER to nucleus area in the control and engineered cells (Fig. 8A and C). This can be explained by the general heterogeneity of each culture analyzed. Attributes such as cell cycle stage and natural variation within a population can account for the range of values attained but clear patterns between different cell lines were revealed through this analysis.
Fig. 8.
Quantification of ER area in relation to nuclear area calculated using immunofluorescence confocal microscopy. ER and nuclear features which were highlighted using an anti-calnexin antibody (an established ER marker (Roobol et al., 2009)) followed by conjugation with a TRITC secondary antibody and DAPI respectively and images were taken at the approximate equator of the cell on the z axis, as determined by the intensity and diameter of the DAPI stain. Values obtained were measured using the “spline contour” tool of Zen blue software to outline the ER and nuclear features of the cells. All cells were adhered and fixed to coverslips on day 6 of batch culture and all images analyzed were from this timepoint. A & B show values from pools and monoclonal host CHO cells engineered to overexpress SCD1 or SREBF1 and control CHO cells, generated using the original pcDNA3.1V5-His/TOPO vector whilst C & D show cells stably expressing either cB72.3 or the FcFP generated from control, SCD1 or SREBF1 host pools. Further, A & C show box and whisker plots generated from values obtained where all individual recorded data points are represented by dots and B & D show mean values obtained using these data points (n ≥ 30). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The CHO-Ctl-1, CHO-Ctl-2, and CHO-ControlPOOL yielded similar distributions in the ratio of ER:nucleus area whilst the latter showed a greater range of values which is not unexpected in a polyclonal pool compared to clonal cell lines (Fig. 8A). Conversely, CHO-Ctl-3 cells showed lower ratios highlighting the variation that naturally arises between different clones but the range of each was within those observed in the other controls. SCD1 modified cell lines showed differences in ER size relative to the nucleus that was dependent on the relative overexpression levels. CHO-SCD1LOW cells had the smallest ER:nucleus ratio values, CHO-SCD1MID show values similar to the CHO-ControlPOOL and CHO-SCD1HIGH cells showed increased ER:nuclear area values. Thus, the high expressing SCD1 clone had an increased ER area compared to the nucleus indicative of a general expansion of the ER on average in the cells in this clone. Alternatively, lower overexpression of SREBF1 had higher ER:nuclear area ratios; CHO-SREBF1LOW cells had increased ER:nuclear area in comparison to controls whilst CHO-SREBF1MID1 and CHO-SREBF1MID2 cells show sequentially decreasing average values (Fig. 8A and B). Interestingly, the CHO-SREBF1POOL had the highest ER:nuclear area ratio overall. Whilst box and whisker plots show the entire range of values obtained, mean values calculated from these data are also presented within the box blot and individually (Fig. 8B).
When this analysis was undertaken on the engineered cell pools stably expressing secretory recombinant proteins, the ER:nuclear ratio was generally lower than the corresponding non-producing counterparts. However, the trend observed across these was similar to the hosts alone whereby the CHO-SCD1cB72.3 pools had a small increase in ER compared to the CHO-ControlcB72.3 pools, whilst the CHO-SREBF1cB72.3 pool had an approximate 40% increase in ER area on average (Fig. 8C and D). Interestingly, analysis of FcFP expressing cells shows that CHO-SCD1FcFP cells also had a small increase in ER area compared to the CHO-ControlFcFP but that the CHO-SREBF1FcFP cells did not appear to show any indication of ER expansion compared to the control (Fig. 8C and D).
4. Discussion
The data presented here report the first studies to directly modify lipid metabolism in CHO cells by engineering the overexpression of proteins involved in lipid biosynthesis. CHO cells are the established workhorse for the synthesis of biotherapeutic proteins and decades of process development have been successfully implemented to enhance the capacity of existing CHO hosts for production of a variety of recombinant molecules, but particularly mAbs (Walsh, 2018). Previous cellular engineering strategies have focused on cell metabolism, cell cycle control, protein secretion/chaperone manipulation, apoptosis and post-translational modification of recombinant proteins (Yiping et al., 2010). However, as lipids are essential in many of the processes that underpin secretory recombinant protein production from mammalian cell systems, it is somewhat surprising that there have been few efforts to understand and manipulate lipid biogenesis in CHO cells with a view to enhancing their ability to produce such proteins. We therefore hypothesized that the manipulation of lipid metabolism pathways could bring about desired attributes for biotherapeutic production in CHO cells.
SREBF1 was overexpressed in CHO cells with the aim to increase the overall rate of lipid turnover and modify existing lipid species by transcriptionally activating a plethora of genes involved in lipid metabolism. The functionality and survival of eukaryotic cells is reliant on tightly regulated lipid homeostasis and the accumulation of excessive lipids can induce lipotoxicity leading to apoptosis (Kliewer et al., 1997; van Herpen and Schrauwen-Hinderling, 2008). It is likely that a large increase in de novo lipogenesis promoted by high SREBF1 overexpression could have a detrimental effect on CHO host survival. This theory is supported by the fact that only a modest increase in SREBF1 is observed in engineered cells, suggesting that high expressing cells were unable to survive in the recovered population. Furthermore, stable expression of SREBF1 at higher levels appeared to have an adverse effect on cell growth in culture. This was the case for both host and cB72.3 or FcFP producing cells, which did not grow to the same viable cell concentration as the relevant controls. However, clonal hosts overexpressing SREBF1 to lower levels than the polyclonal pool (which expressed the highest amount) showed less dramatic changes in growth profiles. The CHO-SREBF1MID2 cell line obtained a higher maximum viable cell number but perished more quickly than other SREBF1 engineered cell pools and lines.
The data presented in this study suggest that there is an ‘optimal’ level of SREBF1 in CHO host lines to achieve desired attributes and that tuning of SREBF1 expression to different extents could either improve or negatively impact on growth and productivity characteristics of CHO cells. Lower levels of SREBF1 overexpression also showed the greatest increase in ER size of the monoclonal CHO-SREBF1 cells whilst higher expression levels showed progressively lower average ER size compared to the nucleus. Interestingly, CHO-SREBF1POOL cells showed the greatest increase in ER size overall suggesting that overexpression of SREBF1 can induce ER expansion, perhaps since its expression is endogenously upregulated in the UPR and contributes to the tuning of the abundance of lipids required for ER expansion (Han and Kaufman, 2016; Basseri and Austin, 2012). Interestingly, cells that show increased ER size also show compromised growth profiles, which may be a result of either cell stress or a limit in resources and energy availability.
ER size was considerably lower in CHO cells expressing recombinant products than in non-expressing counterparts. This is likely due to differences in growth profiles, brought about by the introduction of the recombinant molecules, where impacts on cell cycle progression can considerably affect organelle morphology (van Meer et al., 2008; English and Voeltz, 2013; Heald and Cohen-Fix, 2014). Interestingly, cells producing the IgG4 molecule cB72.3 showed a similar trend, in terms of ER size, to their host counterparts i.e. SREBF1 overexpressing cells show an increased ER area compared to the control for both the host pool and producing cell pools. Conversely, CHO-SREBF1FcFP cells had ER sizes more similar to the relevant control. It has previously been demonstrated that bottlenecks in secretory recombinant protein product yields are dependent on both the molecule and cell line used for expression (Mayrhofer et al., 2015; Mason et al., 2012). Whilst SREBF1 overexpression was able to transiently boost cB72.3, FcFP and antibody fusion expression (Budge et al., 2019), it did not initially exhibit the same effect in a stable scenario where the increases in cB72.3 produced were more modest. However, when higher expressing stable SREBF1 pools were generated by co-transfection of the CHO cell specific SREBF1 with three different recombinant molecules the higher expressing SREBF1 pools had a 2-3-fold increase in the stable expression of the secreted recombinant molecules (Fig. 5), providing further evidence that the amount of SREBF1 overexpression is important in ‘dialing in’ the desired effect on CHO cell phenotype. It is also noteworthy that stable overexpression of the CHO specific SREBF1 gene sequence generated higher titres of model recombinant molecules than when the mouse derived gene was employed suggesting that the specific SREBF1 sequence may be important in the success of this engineering approach. Overexpression of SCD1 increased secretory recombinant yields of all molecules evaluated and was shown to be effective in both transient and stable scenarios. This shows that SCD1 is able to modify CHO cells such that cellular processes involved in recombinant protein production are enhanced.
A combination of membrane lipid composition, the presence of membrane integral proteins, cellular scaffolds and cytoskeletal assembly can influence mechanisms of membrane curvature during vesicle budding processes (McMahon and Gallop, 2005). It has been suggested that a decrease in membrane rigidity can lower the energy required for vesicle formation (Settles et al., 2010). Since SCD1 increases membrane fluidity through the conversion of SFA to MUFA, it is likely that overexpression of SCD1 increases the propensity for vesicle formation. This is supported by the data presented in Fig. 7 where CHO-SCD1 cells showed higher numbers of vesicles on the surface, and in some cases intracellularly, of engineered cells. Furthermore, the inhibition of SCD1 in huh7 cells has been shown to decrease the size of lipid droplets (Ben M'barek et al., 2017). Conversely, large vesicles were frequently observed when SCD1 was overexpressed in CHO cells in the current study. Elongated protrusions at the plasma membrane were also observed in SCD1 overexpressing cells likely due to an increase in membrane fluidity. Whilst the implications this may have on recombinant protein production is unknown, this phenomenon is likely to increase the surface area to volume ratio increasing the contact surface that these hosts have with external medium and increasing the potential for secretion via fusion of cell membranes. An increase in vesicle budding and trafficking may increase the secretory capacity of the CHO hosts and the SCD1 engineered cells consistently had improved recombinant concentrations. In addition, an increase in lipid droplet formation or size may increase energy storage and lipid availability and this process has been shown to protect cells against lipotoxicity in CHO cells (Plötz et al., 2016).
In conclusion, we show for the first time that cellular engineering of lipid metabolism can be employed to enhance desired attributes and cellular processes, increasing both transient and stable secretory biotherapeutic production from cultured CHO cells. Excitingly, this study provides proof-of-concept of an entirely new approach to engineer CHO cells for enhanced bioprocessing attributes that may allow further enhancement of existing and new CHO expression systems to generate hosts that allow improved production of a range of more easy and DTE proteins. Importantly, we have also shown that engineering of these targets in other host CHO cells delivers a similar impact on product yields to that observed in the host described here (data not shown). However, in order to obtain the maximum enhanced attributes from CHO cells engineered to overexpress either SCD1, involved in formation of MUFAs, or SREBF1, involved in more global control of lipid biogenesis, it will be necessary to tune the amount of expression of these LMM genes in engineered CHO cells. As such, development and commercialization of new lipid engineered host cell lines will require the generation of stably expressing cell lines with specific SCD1 or SREBF1 profiles that may differ depending on the target secretory molecule to be expressed. Finally, the success of this approach highlights the importance of lipids in mammalian cell culture processes for the production of secreted biopharmaceuticals in general and provides further evidence that lipidomics should be applied to the understanding and enhancement of expression systems in the bioprocessing industry as suggested by others (Zhang et al., 2017).
Declaration of competing interest
AD, ST, CMJ, RJY and AJR are employed by Lonza Biologics, who developed and license the GS Gene Expression System®.
Lonza is the assigned owner of, and CMS, JDB, TJK, and RYJ are named inventors on, the filed patent ‘Modulation of lipid metabolism for protein production’, patent number WO2017191165A1.
The authors declare no other financial or commercial conflict of interest.
Acknowledgements
This worked was funded via the Biotechnology and Biological Sciences Research Council (BBSRC) via Industrial CASE PhD studentships (JDB and TJK) and grant BB/N023501/1 (to CMS, Kent) and InnovateUK (to AJR, Lonza Biologics, grant No. 102621). We thank Mr Kevin Howland for help with lipid mass spectrometry analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ymben.2019.11.007.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Basseri S., Austin R.C. Endoplasmic reticulum stress and lipid metabolism: mechanisms and therapeutic potential. Biochem. Res. Int. 2012;2012:841362. doi: 10.1155/2012/841362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben M'barek K. ER membrane phospholipids and surface tension control cellular lipid droplet formation. Dev. Cell. 2017;41:591–604. doi: 10.1016/j.devcel.2017.05.012. [DOI] [PubMed] [Google Scholar]
- Bergström J.H. AGR2, an endoplasmic reticulum protein, is secreted into the gastrointestinal mucus. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budge, J. D. et al. Data for engineering lipid metabolism of Chinese hamster ovary (CHO) cells for enhanced recombinant protein production. Data in Brief, in press (2019). [DOI] [PMC free article] [PubMed]
- English A.R., Voeltz G.K. Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a013227. a013227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feary M., Racher A.J., Young R.J., Smales C.M. Methionine sulfoximine supplementation enhances productivity in GS-CHOK1SV cell lines through glutathione biosynthesis. Biotechnol. Prog. 2017;33:17–25. doi: 10.1002/btpr.2372. [DOI] [PubMed] [Google Scholar]
- Le Fourn V., Girod P.-A., Buceta M., Regamey A., Mermod N. CHO cell engineering to prevent polypeptide aggregation and improve therapeutic protein secretion. Metab. Eng. 2014;21:91–102. doi: 10.1016/j.ymben.2012.12.003. [DOI] [PubMed] [Google Scholar]
- Grote M., Haas A.K., Klein C., Schaefer W., Brinkmann U. Bispecific antibody derivatives based on full-length IgG formats. Methods Mol. Biol. 2012;901:247–263. doi: 10.1007/978-1-61779-931-0_16. [DOI] [PubMed] [Google Scholar]
- Han J., Kaufman R.J. The role of ER stress in lipid metabolism and lipotoxicity. J. Lipid Res. 2016;57:1329–1338. doi: 10.1194/jlr.R067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R., Cohen-Fix O. Morphology and function of membrane-bound organelles. Curr. Opin. Cell Biol. 2014;26:79–86. doi: 10.1016/j.ceb.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Herpen N.A., Schrauwen-Hinderling V.B. Lipid accumulation in non-adipose tissue and lipotoxicity. Physiol. Behav. 2008;94:231–241. doi: 10.1016/j.physbeh.2007.11.049. [DOI] [PubMed] [Google Scholar]
- Igal R.A. Stearoyl CoA desaturase-1: new insights into a central regulator of cancer metabolism. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2016;1861:1865–1880. doi: 10.1016/j.bbalip.2016.09.009. [DOI] [PubMed] [Google Scholar]
- Igal R.A., Ariel R. Roles of stearoyl CoA desaturase-1 in the regulation of cancer cell growth, survival and tumorigenesis. Cancers. 2011;3:2462–2477. doi: 10.3390/cancers3022462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C.L., Walch L., Verbavatz J.-M. Lipids and their trafficking: an integral part of cellular organization. Dev. Cell. 2016;39:139–153. doi: 10.1016/j.devcel.2016.09.030. [DOI] [PubMed] [Google Scholar]
- Jaureguiberry M.S. Role of plasma membrane lipid composition on cellular homeostasis: learning from cell line models expressing fatty acid desaturases. Acta Biochim. Biophys. Sin. 2014;46:273–282. doi: 10.1093/abbs/gmt155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johari Y.B., Estes S.D., Alves C.S., Sinacore M.S., James D.C. Integrated cell and process engineering for improved transient production of a “difficult-to-express“ fusion protein by CHO cells. Biotechnol. Bioeng. 2015;112:2527–2542. doi: 10.1002/bit.25687. [DOI] [PubMed] [Google Scholar]
- Kliewer S.A. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux H. Generation of genetically engineered CHO cell lines to support the production of a difficult to express therapeutic protein. BMC Proc. 2013;7:P1. [Google Scholar]
- Man W.C., Miyazaki M., Chu K., Ntambi J. Colocalization of SCD1 and DGAT2: implying preference for endogenous monounsaturated fatty acids in triglyceride synthesis. J. Lipid Res. 2006;47:1928–1939. doi: 10.1194/jlr.M600172-JLR200. [DOI] [PubMed] [Google Scholar]
- Marichal-Gallardo P.A., Álvarez M.M. State-of-the-art in downstream processing of monoclonal antibodies: process trends in design and validation. Biotechnol. Prog. 2012;28:899–916. doi: 10.1002/btpr.1567. [DOI] [PubMed] [Google Scholar]
- Mason M., Sweeney B., Cain K., Stephens P., Sharfstein S.T. Identifying bottlenecks in transient and stable production of recombinant monoclonal-antibody sequence variants in Chinese hamster ovary cells. Biotechnol. Prog. 2012;28:846–855. doi: 10.1002/btpr.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterton R.J., Roobol A., Al-Fageeh M.B., Carden M.J., Smales C.M. Post-translational events of a model reporter protein proceed with higher fidelity and accuracy upon mild hypothermic culturing of Chinese hamster ovary cells. Biotechnol. Bioeng. 2010;105:215–220. doi: 10.1002/bit.22533. [DOI] [PubMed] [Google Scholar]
- Maulucci G. Fatty acid-related modulations of membrane fluidity in cells: detection and implications. Free Radic. Res. 2016;50:S40–S50. doi: 10.1080/10715762.2016.1231403. [DOI] [PubMed] [Google Scholar]
- Mayrhofer P. Identification of bottlenecks in antibody expression using targeted gene integration. BMC Proc. 2015;9:P7. [Google Scholar]
- McGrew J.T., Richards C.L., Smidt P., Dell B., Price V. New Developments and New Applications in Animal Cell Technology 205–207. Kluwer Academic Publishers; 1998. Lipid requirements of a recombinant Chinese hamster ovary cell line (CHO) [Google Scholar]
- McMahon H.T., Gallop J.L. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- Mead E.J. Biological insights into the expression of translation initiation factors from recombinant CHOK1SV cell lines and their relationship to enhanced productivity. Biochem. J. 2015;472:261–273. doi: 10.1042/BJ20150928. [DOI] [PubMed] [Google Scholar]
- van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje-Galvan V., Klauda J.B. Modeling yeast organelle membranes and how lipid diversity influences bilayer properties. Biochemistry. 2015;54:6852–6861. doi: 10.1021/acs.biochem.5b00718. [DOI] [PubMed] [Google Scholar]
- Plötz T., Hartmann M., Lenzen S., Elsner M. The role of lipid droplet formation in the protection of unsaturated fatty acids against palmitic acid induced lipotoxicity to rat insulin-producing cells. Nutr. Metab. 2016;13:16. doi: 10.1186/s12986-016-0076-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povey J.F. Rapid high-throughput characterisation, classification and selection of recombinant mammalian cell line phenotypes using intact cell MALDI-ToF mass spectrometry fingerprinting and PLS-DA modelling. J. Biotechnol. 2014;184:84–93. doi: 10.1016/j.jbiotec.2014.04.028. [DOI] [PubMed] [Google Scholar]
- Pybus L.P. Model-directed engineering of “difficult-to-express” monoclonal antibody production by Chinese hamster ovary cells. Biotechnol. Bioeng. 2014;111:372–385. doi: 10.1002/bit.25116. [DOI] [PubMed] [Google Scholar]
- Ren C. Scd1 contributes to lipid droplets formation in GMEC via transcriptional regulation of Tip47 and Adrp. Eur. J. Lipid Sci. Technol. 2018;120:1700238. [Google Scholar]
- Rintoul D.A., Sklar L.A., Simonin R.D. Membrane lipid modification of Chinese hamster ovary cells thermal properties of membrane phospholipids. J. Biol. Chem. 1978;253:7447–7452. [PubMed] [Google Scholar]
- Roobol A., Carden M.J., Newsam R.J., Smales C.M. Biochemical insights into the mechanisms central to the response of mammalian cells to cold stress and subsequent rewarming. FEBS J. 2009;276:286–302. doi: 10.1111/j.1742-4658.2008.06781.x. [DOI] [PubMed] [Google Scholar]
- Roobol A. ATR (ataxia telangiectasia mutated- and Rad3-related kinase) is activated by mild hypothermia in mammalian cells and subsequently activates p53. Biochem. J. 2011;435:499–508. doi: 10.1042/BJ20101303. [DOI] [PubMed] [Google Scholar]
- Roobol A. The chaperonin CCT interacts with and mediates the correct folding and activity of three subunits of translation initiation factor eIF3: b, i and h. Biochem. J. 2014;458:213–224. doi: 10.1042/BJ20130979. [DOI] [PubMed] [Google Scholar]
- Roobol A. p58IPK is an inhibitor of the eIF2α kinase GCN2 and its localization and expression underpin protein synthesis and ER processing capacity. Biochem. J. 2015;465:213–225. doi: 10.1042/BJ20140852. [DOI] [PubMed] [Google Scholar]
- Sato R. Assignment of the membrane attachment, DNA binding, and transcriptional activation domains of sterol regulatory element-binding protein-1 (SREBP-1) J. Biol. Chem. 1994;269:17267–17273. [PubMed] [Google Scholar]
- Scaglia N., Chisholm J.W., Igal R.A. Inhibition of stearoylCoA desaturase-1 inactivates acetyl-CoA carboxylase and impairs proliferation in cancer cells: role of AMPK. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settles E.I., Loftus A.F., McKeown A.N., Parthasarathy R. The vesicle trafficking protein Sar1 lowers lipid membrane rigidity. Biophys. J. 2010;99:1539–1545. doi: 10.1016/j.bpj.2010.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H. Sterol regulatory element-binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes. Prog. Lipid Res. 2001;40:439–452. doi: 10.1016/s0163-7827(01)00010-8. [DOI] [PubMed] [Google Scholar]
- Tigges M., Fussenegger M. Xbp1-based engineering of secretory capacity enhances the productivity of Chinese hamster ovary cells. Metab. Eng. 2006;8:264–272. doi: 10.1016/j.ymben.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Walsh G. Biopharmaceutical benchmarks 2018. Nat. Biotechnol. 2018;36:1136–1145. doi: 10.1038/nbt.4305. [DOI] [PubMed] [Google Scholar]
- Yiping L. Engineering mammalian cells in bioprocessing – current achievements and future perspectives. Biotechnol. Appl. Biochem. 2010;55:175–189. doi: 10.1042/BA20090363. [DOI] [PubMed] [Google Scholar]
- Zhang Y. High-throughput lipidomic and transcriptomic analysis to compare SP2/0, CHO, and HEK-293 mammalian cell lines. Anal. Chem. 2017;89:1477–1485. doi: 10.1021/acs.analchem.6b02984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.