Abstract
Bacteria often exist in polymicrobial communities where they compete for limited resources. Intrinsic to this competition is the ability of some species to inhibit or kill their competitors. This phenomenon is pervasive throughout the human body where commensal bacteria block the colonization of incoming microorganisms. In this regard, molecular epidemiological and microbiota-based studies suggest that species-specific interactions play a critical role in the prevention of nasal colonization of the opportunistic pathogen Staphylococcus aureus. Despite this, S. aureus exists as part of the microbiota of ∼25% of the population, suggesting that the interplay between S. aureus and commensals can be complex. Microbiota studies indicate that several bacterial genera are negatively correlated with S. aureus colonization. While these studies paint a broad overview of bacterial presence, they often fail to identify individual species-specific interactions; a greater insight in this area could aid the development of novel antimicrobials. As a proof of concept study designed to identify individual bacterial species that possess anti-S. aureus activity, we screened a small collection of clinical isolates from the Walter Reed National Military Medical Center for the ability to inhibit multiple S. aureus strains. We found that the majority of the isolates (82%) inhibited at least one S. aureus strain; 23% inhibited all S. aureus strains tested. In total, seven isolates mediated inhibitory activity that was independent of physical contact with S. aureus, and seven isolates mediated bactericidal activity. 16S rRNA based-sequencing revealed that the inhibitory isolates belonged to the Acinetobacter, Agromyces, Corynebacterium, Microbacteria, Mycobacterium, and Staphylococcus genera. Unexpectedly, these included seven distinct Acinetobacter baumannii isolates, all of which showed heterogeneous degrees of anti-S. aureus activity. Defined mechanistic studies on specific isolates revealed that the inhibitory activity was retained in conditioned cell free medium (CCFM) derived from the isolates. Furthermore, CCFM obtained from S. saprophyticus significantly decreased mortality of S. aureus-infected Galleria mellonella caterpillars. While future studies will seek to define the molecular mechanisms of the inhibitory activities, our current findings support the study of polymicrobial interactions as a strategy to understand bacterial competition and to identify novel therapeutics against S. aureus and other pathogens.
Keywords: Staphylococcus aureus, MRSA, polymicrobial interactions, bacterial interaction, clinical isolates
Introduction
Staphylococcus aureus is an opportunistic pathogen that, due to its ability to quickly adapt to harsh conditions and evade the host’s immune system, can colonize virtually any niche throughout the human body. S. aureus causes a variety of diseases, most frequently skin and soft tissue infections, but also systemic and toxin-mediated disease (Otto, 2010). To further exacerbate matters, numerous S. aureus strains are resistant to multiple antibiotics, which subsequently makes treatment more difficult. Even amongst otherwise healthy individuals, the lack of appropriate treatment often leads to more severe morbidity and higher mortality rates (Lowy, 2003). Accordingly, methicillin-resistant S. aureus (MRSA)-mediated disease was responsible for approximately 10,000 deaths from 2005 to 2013 in the United States (Klevens et al., 2007). Furthermore, the worldwide pervasiveness of multidrug-resistant S. aureus strains has led the World Health Organization to designate MRSA as a “high” threat to the global population (WHO, 2017).
Despite the propensity to cause significant morbidity and mortality, S. aureus exists as a part of the microbiota of approximately one-quarter of the population (Sakr et al., 2018); however, colonized individuals are more likely to develop S. aureus-mediated disease (Kluytmans and Wertheim, 2005). In thinking about the dynamics of colonization of the host, S. aureus must interact and compete with the other resident flora as a means to establish itself as a part of the microbiota of a particular niche (Burian et al., 2017). This is undoubtedly a complicated process. However, even the vast amount of currently available microbiota data has not substantially increased our current understanding of the molecular mechanisms underlying the complex interactions between resident flora and incoming pathogens like S. aureus. It is well-established that commensal microbes play a critical role in decreasing and preventing pathogen colonization. A well-known example of this can be found with the ability of fecal transplants from healthy donors to treat patients with recurrent Clostridium difficile infections; restoration of the normal gastrointestinal microbiota eliminates and prevents C. difficile colonization (Buffie et al., 2015).
The ability of commensal bacteria to block pathogen colonization is true at other anatomical locations as well. S. aureus nasal colonization in particular is greatly dependent on molecular interactions with the nasal flora (Brugger et al., 2016; Sakr et al., 2018). Indeed, the nasal cavity is a high salinity and nutrient scarce niche where resident and incoming bacteria compete for limited resources and space in a type of “bacterial warfare” (Krismer et al., 2014). These interactions are often species-specific, and commensal bacterial have been found to use a variety of mechanisms to block pathogen colonization, including the production and secretion of toxic compounds that directly kill or inhibit competitors. For example, various species from within the Streptococcus and Corynebacterium genera are inversely correlated with the presence of S. aureus in the nasal cavity or have been found to directly antagonize S. aureus (Lemon et al., 2010; Bomar et al., 2016). Even other members of the Staphylococcus genus have been found to negatively impact S. aureus viability; several coagulase-negative Staphylococcus (CoNS) species have evolved mechanisms to inhibit S. aureus colonization. Specifically, some S. epidermidis strains secrete a serine protease that is capable of disrupting S. aureus biofilm formation and blocking nasal colonization (Iwase et al., 2010). S. hominis and S. epidermidis both secrete strain-specific antimicrobial peptides that have potent selective bactericidal activity against S. aureus (Nakatsuji et al., 2017). Moreover, lugdunin, a novel cyclic peptide antibiotic produced by S. lugdunensis, has bactericidal properties against several Gram-positive pathogens, including S. aureus, and can prevent S. aureus nasal colonization (Zipperer et al., 2016). It is clear that within the context of the human nose, there is a selective pressure, even amongst closely related commensal species, to block or eliminate S. aureus.
Despite recent advancements detailing the negative molecular interactions that occur between S. aureus and the resident nasal flora, little is known about S. aureus interactions with bacteria isolated from other anatomical locations. Given this deficit and the fact that S. aureus can colonize the human body virtually ubiquitously, as a proof of concept study we set out to characterize S. aureus interactions with clinical bacterial isolates obtained from a variety of body sites from a diverse patient population at the Walter Reed National Military Medical Center. Herein, we show that the majority (82%, 28/34) of clinical isolates possessed some degree of in vitro anti-S. aureus activity when tested against multiple strains of S. aureus, including MRSA. Moreover, eight clinical isolates showed anti-S. aureus activity against all tested strains. Several of the clinical isolates that belonged to the Staphylococcus and Corynebacterium genera mediated contact-independent inhibitory activity against S. aureus. Furthermore, a portion of the clinical isolates (7/28) showed bactericidal activity against S. aureus. Unexpectedly, Acinetobacter baumannii isolates represented the most commonly identified species that produced heterogenous strain-specific anti-S. aureus activity. Finally, analysis of conditioned cell free medium (CCFM) from several isolates revealed that inhibitory activity was often present in the CCFM. Furthermore, CCFM derived from S. saprophyticus was able to reduce mortality of S. aureus-infected Galleria mellonella caterpillars. These findings suggest that S. aureus interactions with other bacteria are far more multifaceted than previously recognized, and strongly support the study of these interactions at the molecular level as a means to reveal novel S. aureus molecular targets or therapeutics.
Materials and Methods
Strains, Culture and Bacterial Interaction Assays
All deidentified clinical isolates were obtained as a part of a memorandum of understanding (MOU) between the Uniformed Services University of the Health Sciences (USU) and the Walter Reed National Military Medical Center (WRNMMC), Department of Clinical Microbiology. The described studies represent research Not Involving Human Subjects since all isolates were obtained from discarded clinical microbiology plates that contained samples that were obtained during routine diagnostic testing and treatment of WRNMMC patients. Both USU and WRNMMC agree and acknowledge that the activities and projects pursued under the MOU complied with the applicable rules and regulations governing human subjects research within the Department of Defense; the Institutional Review Board at WRNMMC was the IRB of record for the collection of all patient samples. Strains were maintained as −80°C freezer stocks and were cultured under the following conditions unless otherwise noted: Clinical isolates were streaked from frozen glycerol stocks on Brain Heart Infusion (BHI) agar (Becton Dickinson) supplemented with 1% Tween80 (BHIT, Sigma-Aldrich). S. aureus strains were streaked from glycerol stocks on BHI agar. Each isolate was incubated overnight at 37°C. Bacterial interaction assays were performed as previously described (Yan et al., 2013; Hardy et al., 2019). Briefly, 40 mg of S. aureus or a clinical isolate was directly harvested from an agar plate with a sterile inoculating loop and then re-suspended in 200 μL of sterile saline solution (Fisher Chemicals). Eight microliters of the S. aureus cell suspension was inoculated into 15 mL of sterile BHIT agar that had been cooled to 55°C; inoculated agar was poured into a sterile petri dish, and allowed to solidify under sterile conditions. Next, 25 μL of a clinical isolate cell suspension was spotted onto the center of an agar dish (one clinical isolate per a S. aureus-seeded agar plate), and was allowed to dry for 40 min under sterile conditions. The resulting plates were incubated at 28°C, and the formation of a zone of clearance (ZOC) was visually assessed at 24, 72, and 120 h. Images of the ZOC were taken with an Amersham Imager 680 (General Electric). The ZOC was defined as the distance between the edge of the clinical isolate spot and the visible edge of the clearance ring. To measure ZOC length, images were analyzed using ImageJ software (NCBI). Each clinical isolate was assessed in three independent biological replicates against S. aureus strains 2014.N, LAC, and Mu50 (Table 1).
TABLE 1.
Clinical isolates and S. aureus strains assayed.
| Strain | Lab strain designation | Origin | Accession # | Year isolated |
Anti-S. aureus activity4 |
Contact dependent vs. Independent | Bactericidal vs. Bacteriostatic | References | ||
| 2014.N | LAC | Mu50 | ||||||||
| Staphylococcus aureus test strains | ||||||||||
| S. aureus LAC | DSM1485 | Blood | NC_002758.2 | 2005 | N/A | N/A | N/A | Voyich et al. (2005) | ||
| S. aureus 2014.N | DSM1416 | Nose | N/A | 2012 | N/A | N/A | N/A | Hardy et al. (2019) | ||
| S. aureus Mu50 | DSM1633 | Abscess | NC_002758.2 | 1997 | N/A | N/A | N/A | Kuroda et al. (2001) | ||
| Acinetobacter clinical isolates | ||||||||||
| A. baumannii-1 | DSM1675 | Wound | MN175920 | 2016 | Strong | Weak | Weak | Dependent1 | Bactericidal1 | This Study |
| A. baumannii-2 | DSM1676 | Wound | MN175921 | 2016 | Strong | Weak | None | Dependent1 | Bacteriostatic1 | This Study |
| A. baumannii-3 | DSM1923 | Wound | MN175922 | 2016 | Strong | Weak | Weak | Dependent1 | Bacteriostatic1 | This Study |
| A. baumannii-4 | DSM1924 | Wound | MN175925 | 2016 | None | Weak | Weak | Not Tested | Not Tested | This Study |
| A. baumannii-5 | DSM1917 | Blood | MN175926 | 2016 | None | Weak | Strong | Dependent3 | Not Tested∗ | This Study |
| A. baumannii-6 | DSM1762 | Wound | MN175924 | 2016 | None | Weak | Strong | Dependent3 | Bactericidal3 | This Study |
| A. baumannii-7 | DSM1918 | Wound | MN175923 | 2016 | None | Weak | Weak | Not Tested | Not Tested | This Study |
| Corynebacterium clinical isolates | ||||||||||
| C. amycolatum-1 | DSM1914 | Nasal | MN175942 | 2016 | Weak | None | None | Not Tested | Not Tested | This Study |
| C. amycolatum-2 | DSM1567 | Nasal | MN175937 | 2016 | Weak | None | Strong | Independent3 | Bactericidal3 | This Study |
| C. aurimucosum-1 | DSM1560 | Urine | MN175936 | 2016 | Strong | None | Weak | Independent1 | Bacteriostatic1 | This Study |
| C. aurimucosum-2 | DSM1678 | Wound | MN175938 | 2016 | Weak | None | Strong | Independent3 | Bacteriostatic3 | This Study |
| C. aurimucosum-3 | DSM1912 | Wound | MN175945 | 2016 | None | None | Strong | Dependent3 | Not Tested∗ | This Study |
| C. aurimucosum-4 | DSM1913 | Wound | MN175932 | 2016 | None | None | Weak | Not Tested | Not Tested | This Study |
| C. jeikeium | DSM1915 | Would | MN175945 | 2016 | None | Weak | None | Not Tested | Not Tested | This Study |
| C. striatum-1 | DSM1564 | Wound | MN175927 | 2016 | Weak | None | Weak | Not Tested | Not Tested | This Study |
| C. striatum-2 | DSM1566 | Blood | MN175947 | 2016 | Strong | None | None | Independent1 | Bacteriostatic1 | This Study |
| C. tuberculostearicum | DSM1925 | Nasal | MN175944 | 2016 | None | None | Weak | Not Tested | Not Tested | This Study |
| Microbacterium clinical isolates | ||||||||||
| M. paraoxydans-1 | DSM1919 | Nasal | MN175940 | 2016 | None | Weak | Weak | Not Tested | Not Tested | This Study |
| M. paraoxydans-2 | DSM1920 | Wound | MN175935 | 2016 | None | Weak | Weak | Not Tested | Not Tested | This Study |
| Staphylococcus clinical isolates | ||||||||||
| S. epidermidis-1 | DSM1679 | Would | MN175939 | 2016 | Strong | Strong | Weak | Independent1 | Bacteriostatic1 | This Study |
| S. epidermidis-2 | DSM1759 | Wound | MN175929 | 2016 | Strong | Strong | Weak | Dependent1 | Bactericidal1 | This Study |
| S. epidermidis-3 | DSM1760 | Wound | MN175930 | 2016 | Strong | Strong | Weak | Independent1 | Bactericidal1 | This Study |
| S. epidermidis-4 | DSM1922 | Wound | MN175931 | 2016 | Strong | Strong | Weak | Dependent1 | Bactericidal1 | This Study1 |
| S. epidermidis-5 | DSM1761 | Nasal | MN175933 | 2016 | Weak | Weak | None | Not Tested | Not Tested | This Study |
| S. hominis | DSM1916 | Wound | MN175934 | 2016 | Strong | Strong | Weak | Dependent1 | Bactericidal1 | This Study |
| S. saprophyticus | DSM1655 | Urine | MN175941 | 2016 | Weak | Strong | Weak | Independent2 | Bactericidal2 | This Study |
| Other clinical isolates | ||||||||||
| Agromyces sp. 3098BRRJ | DSM1921 | Wound | MN175928 | 2016 | None | None | Weak | Not Tested | Not Tested | This Study |
| Mycobacterium yunnanensis | DSM1677 | Wound | MN175946 | 2016 | Strong | None | Weak | Dependent1 | Bacteriostatic1 | This Study |
Subscript “1” indicates assay was tested against S. aureus 2014.N. Subscript “2” indicates assay was tested against S. aureus LAC. Subscript “3” indicates assay was tested against S. aureus Mu50. Subscript “4” indicates strong anti-S. aureus activity defined as follows: ZOC was completely transparent, ≥2 mm, and a defined edge at 72 h. Weak anti-S. aureus was defined as follows: ZOC was not completely transparent, with a hazy and undefined edge at 72 h. None indicates a ZOC was not present at 72 h. A “∗” indicates that the ZOC was too small to perform Bactericidal vs. Bacteriostatic assays. “N/A” denotes information that was unavailable or not applicable.
DNA Extraction, Amplification, Cloning, and 16S rRNA Gene Sequencing
All clinical isolates that possessed anti-S. aureus activity (28/34) were streaked from frozen glycerol stocks on BHIT agar and incubated overnight at 37°C. Single colonies of each isolate were subcultured in 2 mL of BHIT broth and incubated at 37°C with shaking for 24–48 h. Overnight broth cultures were pelleted by centrifugation and re-suspended in 0.2 mL of Phosphate Buffer Solution (PBS, Fisher Chemicals). Cell suspensions were lysed in a Bullet Blender Homogenizer for 5 min by mechanical disruption in bead-beater tubes that contained 0.1 mm sterile glass beads. Genomic DNA was extracted from lysed cells suspensions with the Wizard Genomic DNA Purification Kit (Promega) according to the manufacturer’s instructions.
Purified genomic DNA from each sample was subjected to PCR amplification of the 16S rRNA gene using the 8F (5′ AGAGTTTGATCCTGGCTCAG 3′) and 1492R (5′ GGTTACCTTGTTACGACTT 3′) primers. PCR mixtures (25 μL) contained 5X Phusion HF buffer, 200 mM of each dNTP, 0.5 μM of each primer, and 0.02 U/μL of Phusion DNA polymerase. PCR amplification was performed with the following reaction conditions: 98°C for 30 s, 30 cycles of 98°C for 5 s, 51°C for 30 s, 72°C for 1 min 30 s, with a final elongation step of 72°C for 5 min. The PCR amplified products were visualized on a 1% agarose gel to confirm the presence of an approximately 1,500 base pair band.
PCR products were purified using the QIAquick PCR purification kit according to the manufacturer’s instructions. Purified PCR products were polyadenylated utilizing the A-tailing procedure; reaction components (10 μL), including PCR-amplified DNA, 10X ThermoPol Buffer, 1mM dATP, and Taq DNA Polymerase, were incubated at 70°C for 30 min. A-tailed PCR products were subsequently cloned into the pGEM-T Easy vector according to the manufacturer’s instructions (Promega). Ligation products were transformed into E. coli TOP10 CaCl2 chemically competent cells. Transformants with the desired insert were isolated via “blue/white” selection on LB (Luria-Bertani) agar supplemented with ampicillin (100 μg/mL), X-gal (40 μg/mL) and IPTG (1 μM). To confirm the presence of the correct insert, colony PCR was performed on at least five white colonies per transformation using the GoTaq Green Master Mix (Promega) and pGEM-T Easy specific T7 (5′ GGGTTTTCCCAGTCACGA 3′) and SP6 (5′ GCACCCCAGGCTTTACAC 3′) primers with the following PCR conditions: 95°C for 3 min, 30 cycles of 95°C for 30 s, 45°C for 30 s, 72°C for 1 min 30 s, with a final elongation step of 72°C for 5 min. White colonies that contained the correct insert were cultured overnight in LB Broth plus ampicillin (100 μg/mL) with shaking. Plasmids were purified using QIAprep Spin Miniprep Kit (Qiagen) according to the manufacturer’s instructions and then used for sequencing.
As previously described (Johnson et al., 2016), to ensure near full-length coverage of the 16S rRNA gene, six individual sequencing reactions were performed on purified plasmids using the following primers: T7, SP6, 8F, 1492R, 515F (5′ GTGYCAGCMGCCGCGGTA 3′), and 806R (5′ AGAGTTTGATCCTGGCTCAG 3′). Sequence reads were manually assembled into a double stranded near full length 16S rRNA gene sequence, and taxonomic information was assigned after comparison with other 16S rRNA gene sequences in the Ribosomal Database Project (RDP)1 and GenBank2 using the Basic Local Alignment Search Tool (BLAST). The 16S rRNA gene sequences of all the strains speciated in this study were deposited in GenBank and assigned accession numbers. Strain descriptions, species identification, and accession numbers can be found in Table 1.
Contact-Dependent Assays
Strongly inhibitory clinical isolates were defined as follows: a ZOC that was visibly transparent, at least 2 mm in length, and with a defined edge. These isolates (17/28) were assayed to determine if anti-S. aureus activity was dependent on direct physical contact between the bacteria; in each case, the activity of each clinical isolate was tested against the S. aureus strain for which the strongest ZOC was obtained in the absence of a filter disk. A sterile 0.2 μm filter disk was placed on top of the BHIT agar that had been seeded with S. aureus; each clinical isolate was then individually spotted on top of the filter disk so that none of the cell suspension physically touched the S. aureus seeded agar plate. Plates were incubated at 28°C and were visually assessed at 24, 72, and 120 h for the absence or presence of a ZOC. The absence of a ZOC in the presence of a filter disk indicates that physical contact is necessary for anti-S. aureus activity against the corresponding most sensitive S. aureus strain. Clinical isolates were assessed in three independent biological replicates.
Recovery of S. aureus From ZOC
To determine if anti-S. aureus activity was bacteriostatic (growth inhibition) or bactericidal (killing), S. aureus survival and growth was monitored as compared to the original inoculum. Immediately after the plates solidified and before a clinical isolate was spotted, five-milligram punches of S. aureus-seeded agar were taken with a sterile pipette tip as a means to enumerate S. aureus colony forming units (CFU) present at T0. Bacterial interaction assays were performed with 15/28 strongly inhibitory clinical isolates as described above. Two isolates that produced a defined and transparent ZOC, but exactly 2 mm in length, were excluded from these experiments as the ZOC produced against S. aureus was too small to accurately extract agar punches. Each strongly inhibitory clinical isolate was tested against the S. aureus strain for which the strongest ZOC was produced. After 48 h (T48) of incubation at 28°C, five-milligram punches of agar directly adjacent to the clinical isolate spot (Inside ZOC) or at the edge of the petri dish (Outside ZOC) were again taken with a sterile pipette tip. To determine the number of S. aureus CFU present in an agar punch, punches were resuspended in 1 mL of BHI broth and heated to 55°C for 10 min. 10-fold serial dilutions of each suspension were prepared in PBS and then plated on Mannitol Salt Agar (MSA, Criterion). Plates were incubated at 37°C overnight, and recovered colonies were quantified. The number of CFU present in the 1 mL original suspension was calculated, and the fold change from T0 was calculated as follows: (Number of CFU present Inside or Outside ZOC at T48/Number of CFU present at T0). Fold change values less than 1 indicate bactericidal activity; S. aureus CFU recovered in an agar punch at T48 was less than the S. aureus CFU recovered in an agar punch at T0. Contact-dependent experiments were completed in three independent biological replicates.
Conditioned Cell Free Medium (CCFM) Preparation and Disk Diffusion Assays
Clinical isolates that produced contact-independent bactericidal anti-S. aureus activity (C. amy-2, S. sap, and S. epi-3) were independently cultured in 10 mL BHIT broth overnight at 37°C with shaking at 190 rpm. Cultures were pelleted by centrifugation, and the supernatant was filter sterilized with a.2 μm filter (Corning). One-milliliter of sterile supernatant was retained, and the remaining supernatant was concentrated (50X) with ammonium sulfate precipitation as previously described (Hardy et al., 2019). For heat-treatments, 50 μL aliquots of unconcentrated or 50X CCFM were incubated at 90°C for 10 min, then allowed to cool. For the disk diffusion assays, the S. aureus strain that was most sensitive to the corresponding inhibitory activity (C. amy-2/Mu50, S. sap/LAC, and S. epi-3/LAC) was cultured on BHI agar overnight at 37°C. The following day, the plate-grown cells were recovered and diluted to 1 × 108 cells/ml (OD600 0.1) in BHI broth. A sterile swab was then used to spread the S. aureus cell suspension on BHIT agar as a lawn. The plate was allowed to dry in a laminar flow hood for 30 min. Next, a sterile 5 mm diffusion disk was placed on top of the S. aureus lawn, and 50 μL of unconcentrated CCFM or 50X CCFM was inoculated onto the disk. Plates were incubated at 28°C, and images were taken after 72 h of incubation. Disk diffusion assays were conducted in three independent biological replicates.
S. aureus Infection and CCFM Treatment of Galleria mellonella Caterpillars
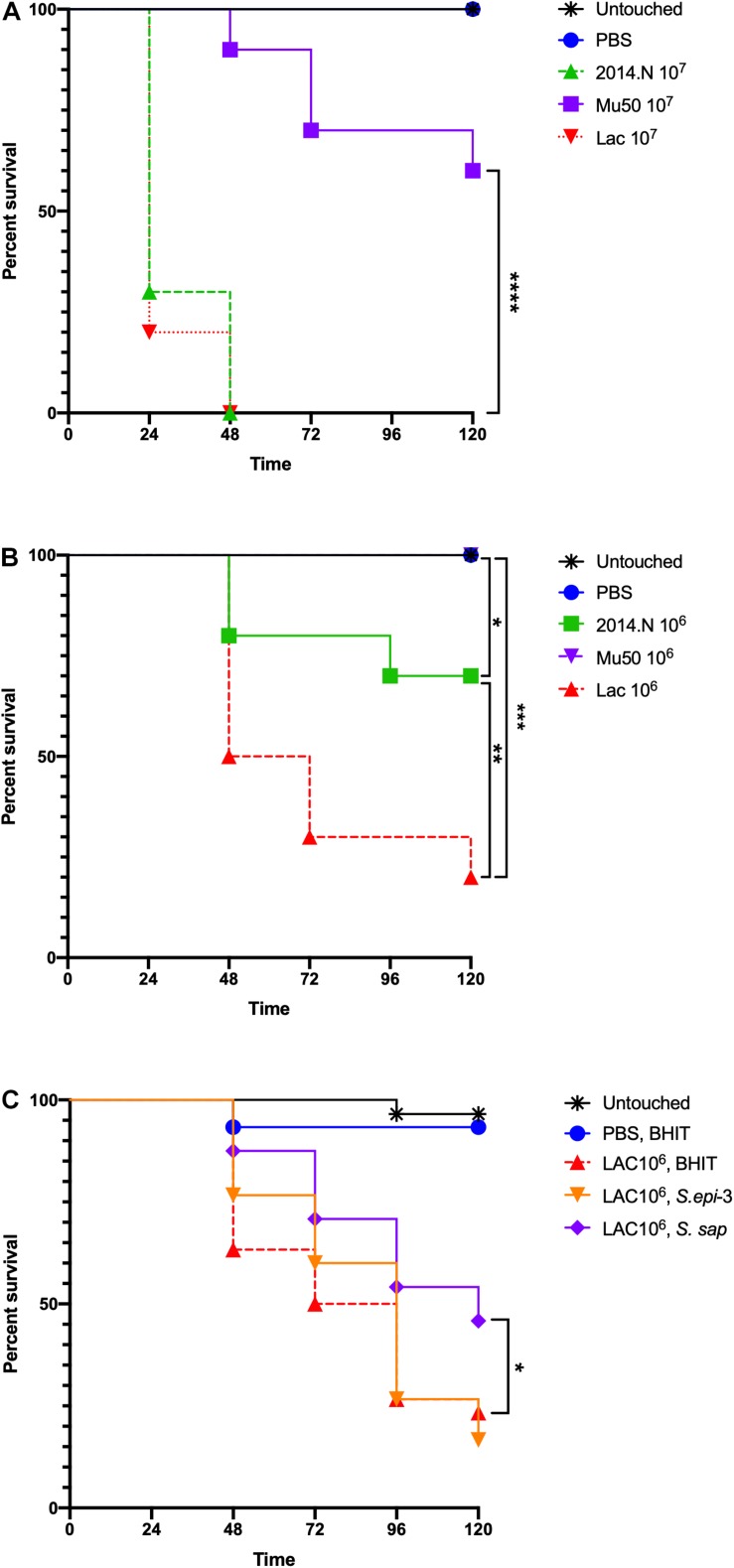
Staphylococcus aureus strains 2014.N, Mu50, and LAC were cultured overnight on BHI agar at 37°C. The following day, S. aureus cells were recovered and diluted to 1 × 108 cells/ml (OD600 0.1) in PBS. Total CFU were then further adjusted to obtain the required doses; i.e., 107 CFU or 106 CFU in 5 μL of PBS + 0.01% bromophenol dye. For infections, Galleria mellonella caterpillars (Vanderhorst Wholesale Inc) were utilized within 1 day of receipt. Caterpillars between 200 and 300 mg were chosen for infection. The injections were carried out as described previously (Desbois and Coote, 2011) with minor adaptations. Briefly, 5 μL of inoculum that contained 107 or 106 total CFU of S. aureus was injected into the last left proleg using a 10 μL glass syringe (Hamilton) fitted with a 31G needle. For caterpillars that were treated with CCFM, the caterpillars were maintained at room temperature for 1 h following the S. aureus injection, then refrigerated at 4°C for 12 min and then injected with 5 μL of freshly prepared 50X CCFM from S. sap or S. epi-3 (treated) or 50X concentrated BHIT (sham treated). These injections were into the last right proleg. All caterpillars were incubated at 37°C, and survival was monitored over 120 h. Untouched, and PBS injected caterpillars were included as controls. Data found in Figures 7A,B represent two completely independent biological replicates (n = 15 caterpillars) performed with different batches of caterpillars. Data found in Figure 7C represent a single batch of caterpillars, but two independently derived batches of CCFM (n = 15 caterpillars)/CCFM preparation. Kaplan–Meier survival curves were compared between groups using the Mantel–Cox test with Holm’s correction for multiple comparisons (excluding Untouched and PBS negative controls). An alpha value of 0.05 was considered statistically significant.
FIGURE 7.
Staphylococcus aureus-infected Galleria mellonella. Galleria mellonella caterpillars were divided into the indicated groups and were monitored for survival over 120 h at 37°C after the indicated inoculations. (A,B) Untouched and PBS injected Galleria served as negative controls and the indicated doses of 2014.N, LAC and Mu50 were tested. (C) For the treatment experiments, untouched caterpillars were maintained. Additionally, PBS injected caterpillars were treated 1-h post S. aureus infection with CCFM prepared from BHIT in which no bacteria had been cultured to ensure no effect of dual injection on survival. Caterpillars that were injected with 106 CFU of S. aureus strain LAC were treated 1-h post infection with BHIT CCFM (sham treatment) or S. epi-3 or S. sap derived CCFM. Kaplan-Meier survival curves were compared (excluding negative controls) using the Mantel–Cox test with Holm’s correction for multiple comparisons. In (A,B), the three S. aureus strains were compared to identify statistically significant differences in Galleria mortality; untouched and PBS dosed caterpillars were not included in the comparisons. In (A), Mu50 was significantly different than both 2014.N and LAC. In (B), significant differences between the various groups are indicated. In (C), BHIT treated caterpillars were compared to CCFM treated (S. epi-3 or S. sap) Galleria to identify difference; only S. sap was significantly different. Asterisks signifying the P value as follows: ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
Results
Activity of Clinical Bacterial Isolates Against S. aureus
Polymicrobial interactions within the human host are complex and dynamic. Numerous studies have shown that several genera that inhabit the skin and nasal cavity prevent the colonization of opportunistic pathogens (Jarraud et al., 2002; Bomar et al., 2016). However, these studies often focus on specific anatomical locations and do not represent the host as one environmental niche. Given this, we questioned whether bacterial species isolated from a diverse patient population and a variety of body sites would display antagonistic interactions against S. aureus. To this end, we obtained a collection of clinical isolates (Table 1) from the WRNMMC Clinical Microbiology Lab and assayed in vitro anti-S. aureus activity utilizing a bacterial interaction assay (Hardy et al., 2019). As prior studies have shown that antagonistic polymicrobial interactions are often strain-specific and because we previously showed that Corynebacterium pseudodiphtheriticum, a common skin and nasal commensal microbe, mediates heterotypic bactericidal activity against specific S. aureus strains (Hardy et al., 2019), we assayed anti-S. aureus activity against three phenotypically different S. aureus strains: S. aureus LAC (Community-Acquired, MRSA), S. aureus Mu50 (Hospital-Acquired, MRSA) and 2014.N (Methicillin-Sensitive S. aureus), a recently acquired nasal isolate (Table 1). To this end, 34 individual clinical isolates were assessed against each S. aureus strain in the bacterial interaction assays; appearance of a visible zone of clearance (ZOC) around the clinical isolate was considered a positive indicator of anti-S. aureus activity. While we found that six clinical isolates showed no anti-S. aureus activity, the majority (28/34, 82%) of tested clinical isolates possessed inhibitory activity against at least one of the S. aureus strains (Figure 1A). Furthermore, eight of the clinical isolates were able to inhibit the growth of all tested S. aureus strains. As expected, many of the clinical isolates mediated inhibitory activity in a S. aureus strain-specific manner: three clinical isolates only inhibited 2014.N, five only inhibited Mu50, and two only inhibited LAC (Figure 1A).
FIGURE 1.
Bacterial species isolated from patients at the Walter Reed National Military Medical Center inhibit Staphylococcus aureus growth. (A) Venn Diagram of anti-S. aureus activity: thirty-four clinical isolates were screened for anti-S. aureus activity in an in vitro bacterial interaction assays against S. aureus strains 2014.N, LAC and Mu50. 28/34 isolates tested possessed anti-S. aureus activity against at least one S. aureus strain. 6/34 possessed no activity against S. aureus. 8/34 inhibited growth of all S. aureus strains tested while the remaining 20 strains showed activity against one or two of the S. aureus strains as indicated in the Venn Diagram. (B) Representative Bacterial Interaction Assays: C. aurimucosum (C. aur-1), M. yunnanensis (M. yun), or S. saprophyticus (S. sap) were spotted onto an agar plate that had been seeded with the indicated S. aureus strains (2014.N, LAC, or Mu50). Plates were incubated at 28°C, and images were taken at 24, 72, and 120 h (24 and 120 h images are shown). Images are representative of three independent biological replicates. Scale bar = 10 mm and is the same in the corresponding 24 h and 120 images; in some cases the 120 h spots appear larger than the 24 h spots due to growth of the bacteria within the spots.
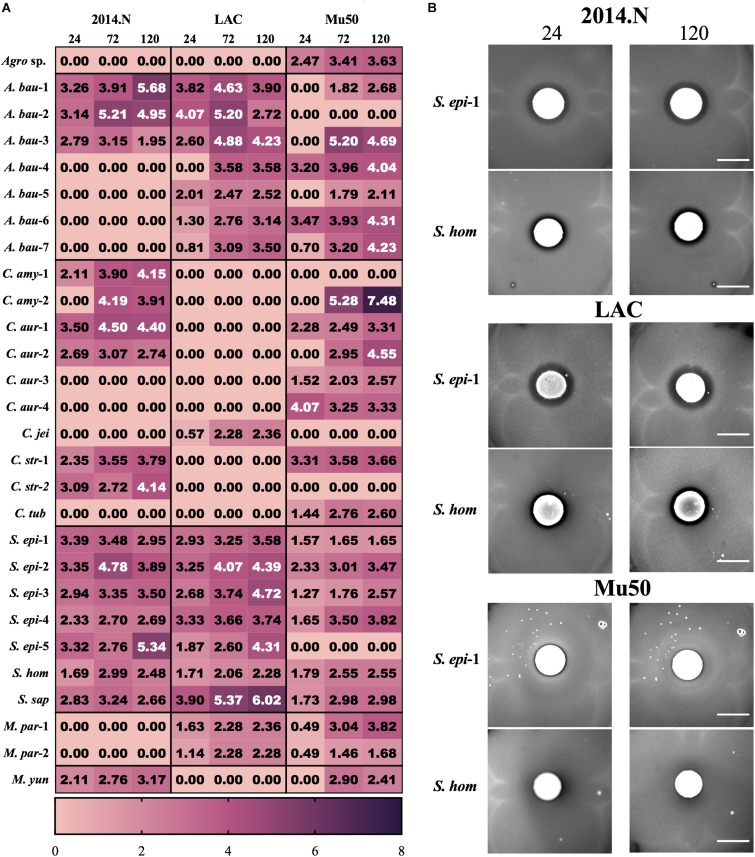
The species of the 28 isolates that exhibited anti-S.aureus activity were next identified via cloning and sequencing of the 16S rRNA gene; sequences were deposited into GenBank and accession numbers are available in Table 1. Analysis of the species information combined with the bacterial interaction assays revealed several types of ZOCs that developed over time (Figure 1B). For example, co-incubation of Corynebacterium aurimucosum (C. aur-1) or Mycobacterium yunnanensis (M. yun) with S. aureus 2014.N or Mu50 resulted in a diffused and moderately sized ZOC; a ZOC did not develop upon co-incubation with S. aureus LAC for either clinical isolate (Figure 1B). In contrast, co-incubation of Staphylococcus saprophyticus (S. sap) with S. aureus LAC resulted in a defined and transparent ZOC, while only a modest and hazy ZOC was produced against S. aureus 2014.N and Mu50.
Temporal quantification of ZOC length additionally revealed distinct patterns of interactions between each clinical isolate and each S. aureus strain. For the majority of the isolates, the ZOC length either remained constant or increased over time (Figure 2A). In support of the literature that suggests that some members of the Corynebacterium genus promote negative interactions with S. aureus (Yan et al., 2013; Hardy et al., 2019), numerous inhibitory isolates were speciated to be members of the Corynebacterium genus. These isolates tended to show anti-S. aureus activity selectively against strains 2014.N and Mu50; only one Corynebacterium isolate, C. jeikeium (C. jei), inhibited S. aureus LAC growth, but neither 2014.N or Mu50. Previous reports have also shown that several CoNS prevent S. aureus colonization by inhibiting growth or by direct killing (Iwase et al., 2010; Zipperer et al., 2016; Nakatsuji et al., 2017). In support of this, numerous Staphylococcal isolates were identified and possessed activity against S. aureus. These isolates generally mediated robust activity against S. aureus 2014.N and LAC, but only modest anti-S. aureus activity against Mu50 (Figures 2A,B). For example, Staphylococcus epidermidis (S. epi-1) and S. hominis (S. hom) produced defined and transparent ZOCs against 2014.N and LAC, but a comparatively small ZOC was produced against Mu50. Taken together, these results support the current hypothesis that antagonistic interactions with S. aureus are often strain-specific. As it would account for the differences in sensitivity amongst the various S. aureus strains, this may indicate that the S. aureus molecular target(s) of each inhibitory isolate is strain-specific and/or differentially expressed between the various S. aureus strains.
FIGURE 2.
Clinical isolates mediate strain-specific anti-S. aureus activity. (A) Heat map showing the activity of the indicated clinical isolate against the indicated S. aureus strains: The Zone of Clearance (ZOC) was defined as the distance between the edge of the clinical isolate spot to the visible edge of the S. aureus ring of clearance. The ZOC was measured using ImageJ software (NCBI) and each value within a box represents the arithmetic mean (in millimeters) of three independent biological replicates measured temporally (24, 72, and 120 h). Clinical isolate species names have been abbreviated as follows: C. aurimucosum (C. aur), C. striatum (C. str), C. amycolatum (C. amy), C. jeikeium (C. jei), C. tuberculostearicum (C. tub), S. epidermidis (S. epi), S. saprophyticus (S. sap), S. hominis (S. hom), A. baumannii (A. bau), M. paraoxydans (M. par), M. yunnanensis (M. yun), and Agromyces sp. 3098BRRJ (Agro sp.). Additionally, a number preceded by a – is used to indicate an independent isolate of the indicated species. (B) Example of strain-specific activity: S. epidermidis (S. epi-1) and S. hominis (S. hom) were co-incubated with agar plates seeded with S. aureus strains 2014.N, LAC or Mu50. Images of the ZOC were taken after 24 and 120 h of incubation at 28°C and are representative of three independent biological replicates. Scale bar = 10 mm and is the same in the corresponding 24 h and 120 images; in some cases the 120 h spots appear larger than the 24 h spots due to growth of the bacteria within the spots.
In addition to the expected members of the Corynebacterium and Staphylococcus genera, several clinical isolates that are not typically associated with the human microbiota were found to have anti-S. aureus activity. For example, there are few reports of the clinical isolation of Microbacterium species (Laffineur et al., 2003). However, M. paraoxydans, a pathogen of various fish species (Soto-Rodriguez et al., 2013), was recovered from two separate patients and both isolates possessed anti-S. aureus activity against strains LAC and Mu50 (Figure 2A). Similarly, Agromyces, a common soil microbe, also mediated anti-S. aureus activity against Mu50 (Figure 2A). These data indicate that antagonistic interactions with S. aureus are not limited to conventional members of the human microbiota that would have been under evolutionary pressure to evolve mechanisms to compete with S. aureus.
Heterotypic Inhibitory Activity of A. baumannii Against S. aureus
While Acinetobacter baumannii and S. aureus have been frequently co-isolated from wounds (Furuno et al., 2008; Castellanos et al., 2019), to our knowledge there is no published evidence that A. baumannii possesses any inhibitory activity against S. aureus. Thus, we were surprised that A. baumannii isolates represented ∼20% (7/34) of the clinical isolates that showed anti-S. aureus activity (Figures 2A, 3). Though not certain, this large representation of A. baumannii clinical isolates may be a result of the “wounded warrior” patient population that is often treated at WRNMMC. Of the seven A. baumannii isolates, two (A. bau-1 and A. bau-3) possessed inhibitory activity against all tested S. aureus strains. The remaining five A. baumannii mediated anti-S. aureus activity against at least two strains (Figure 2A).
FIGURE 3.
Independent Acinetobacter baumannii isolates produce strain-specific anti-S. aureus activity. Individual A. baumannii (A. bau-2 and A. bau-6) isolates were spotted onto an agar plate that had been seeded with various S. aureus strains (2014.N, LAC, or Mu50). Plates were incubated at 28°C, and images were taken at 24, 72, and 120 h (24 and 120 h images are shown). Images are representative of three independent biological replicates. Scale bar = 10 mm and is the same in the corresponding 24 h and 120 images; in some cases the 120 h spots appear larger than the 24 h spots due to growth of the bacteria within the spots.
The type of ZOC produced by A. baumannii varied and was largely dependent on the S. aureus strain being tested. For example, A. bau-2 produced a large and defined ZOC against 2014.N, a large and hazy ZOC against LAC, and no ZOC against Mu50 (Figure 3). In contrast, A. bau-6 produced a moderately sized and very defined ZOC against Mu50, a small and hazy ZOC against LAC, and no ZOC against 2014.N. Taken together, these data indicate that A. baumannii possesses heterogeneous strain-specific anti-S. aureus activity. This may in turn indicate that A. baumannii utilizes multiple independently evolved mechanisms to compete with S. aureus or that the target(s) of anti-S. aureus activity are differentially expressed between S. aureus strains.
Characterization of Contact-Dependent and Bactericidal Anti-S. aureus Activity
Commensal bacteria utilize a wide variety of molecular mechanisms to compete with other microbes; these include both contact dependent and independent mechanisms (Brugger et al., 2016). Thus, we sought to determine whether the observed anti-S. aureus activity of the clinical isolates required physical bacterial interaction. Of the 28 strains that displayed activity, we focused our efforts on the 17 clinical isolates that showed strong inhibitory activity; these strains produced a defined and transparent ZOC against S. aureus that was greater than or equal to 2 mm. To this end, bacterial interaction assays were repeated, but the clinical isolate was separated from the S. aureus seeded agar with a 0.2 μm filter disk. A ZOC still formed for 41% (7/17) of the tested clinical isolates (Figure 4A), indicating that anti-S. aureus activity was contact-independent. The clinical isolates that mediated contact-independent anti-S. aureus activity were restricted to the Staphylococcus and Corynebacterium genera (Figure 4B and data not shown). For example, two independently recovered S. epidermidis isolates (S. epi-1 and S. epi-3) and S. saprophyticus (S. sap) mediated robust contact-independent inhibitory activity (Figure 4B and data not shown). In addition, four Corynebacterium species (C. amy-2, C. aur-1, C. aur-2, and C. str-1) produced moderate inhibitory activity in the presence of a filter disk (Figure 4B and data not shown). Taken together, these data indicate that the various isolates can use both contact-dependent and contact-independent mechanisms as a means to inhibit S. aureus growth.
FIGURE 4.
Select clinical isolates mediate contact-independent anti-S. aureus activity. (A) Strongly inhibitory clinical isolates (17/28) were defined as follows: visibly transparent ZOC of at least 2 mm with a defined edge. (B) A 0.2 μm filter was placed on top of BHIT agar plates seeded with S. aureus (S. epi-1 and S. sap were incubated with S. aureus LAC, C. amy-2 was incubated with S. aureus Mu50, and C. aur-1 was incubated with S. aureus 2014.N). A clinical isolate (as described above) was then spotted on top of the filter paper such that the clinical isolate and the S. aureus seeded agar plate were physically separated. Images of the ZOC were taken after 120 h of incubation at 28°C. Images are representative of three independent biological replicates. Scale bar = 10 mm.
Commensal bacteria can compete with other bacteria using mechanisms that either inhibit bacterial growth (bacteriostatic) or directly kill (bactericidal) the competitor. To further characterize the anti-S. aureus activities of the strongly inhibitory clinical isolates, the number of S. aureus CFU were determined from within the ZOC, directly adjacent to the clinical isolate spot (Inside ZOC), and outside of the ZOC, on the edge of the petri dish (Outside ZOC), after 48 h (T48) of incubation. These numbers were then compared to the number of S. aureus CFU seeded within a comparable area of the agar plate at the initiation of the experiment (T0). Of the 17 strongly inhibitory isolates, 15 developed a ZOC that was large enough (greater than 2 mm) to take accurate agar punches that fell fully within the ZOC. Of these 15 isolates, 7 mediated bactericidal activity against S. aureus. Most of these isolates belonged to the Staphylococcus genus (4/7), followed by A. baumannii (2/7), and Corynebacterium (1/7, Figure 5). Combined with the contact dependence assays, a total of 3 clinical isolates (C. amy-2, S. sap, and S. epi-3) produced anti-S. aureus activity that was independent of direct contact and was also bactericidal. This strongly suggests that these isolates directly kill S. aureus via the secretion of toxic compound(s).
FIGURE 5.
Select clinical isolates mediate bacteriostatic or bactericidal activity against S. aureus. The number of S. aureus CFU seeded within the agar (T0) was compared to the number of S. aureus found after 48 h of incubation. S. aureus CFU numbers were determined from directly adjacent to the clinical isolate spot (Inside ZOC) or from outside the ZOC and the fold change from T0 was calculated. Each box represents the data from three independent biological replicates; the horizontal line is plotted at the arithmetic mean and the length of the box represents the range. A dotted line is indicated at 1, which would represent a purely bacteriostatic interaction where the numbers of S. aureus at T0 and T48 were unchanged. Values below the line indicate a decrease in S. aureus at the 48-h time point, which indicates bactericidal activity.
Basic Mechanistic Characterization of Contact-Independent Bactericidal Activity
We hypothesized that clinical isolates that produced contact-independent bactericidal anti-S. aureus activity would do so via a secreted compound(s) that would be present in culture supernatants. To test this hypothesis, C. amy-2, S. sap, and S. epi-3 were independently cultured in BHIT broth, and sterile conditioned cell free medium (CCFM) was prepared. Unconcentrated CCFM and 50X concentrated CCFM were then tested in a disk diffusion assay (Figure 6 and data not shown) against the S. aureus strain for which they showed the most robust bactericidal activity (C. amy-2/Mu50, S. sap/LAC, and S. epi-3/LAC). Each of the 50X concentrated CCFM samples produced a ZOC against the tested S. aureus strain (Figure 6). In addition, unconcentrated CCFM derived from S. sap and S. epi-3 produced a small ZOC against S. aureus LAC (data not shown). To determine the thermostability of the compound(s) found in the concentrated CCFM, aliquots of CCFM were also subjected to heat treatment prior to testing for anti-S. aureus activity. In all cases anti-S. aureus activity was maintained after heat treatment (Figure 6).
FIGURE 6.
Anti-S. aureus activity is retained in Conditioned Cell Free Medium. S. aureus strains LAC or Mu50 were spread on the BHI agar surface and a sterile disk was placed in the center of the plate. Fifty μl of concentrated CCFM prepared from C. amy-2; (incubated with Mu50), S. sap (incubated with LAC), and S. epi-3 (incubated with LAC) that was Heat-Treated (H+) or maintained at room temperature (H–) prior to use was inoculated onto the disk and allowed to dry. Images of the ZOC were taken after 72 h of incubation. Images are representative of three independent biological replicates. Scale bar = 10 mm.
To examine the therapeutic potential of the compound(s) found within the CCFM, we next examined the ability of CCFM to rescue S. aureus-infected Galleria mellonella caterpillars. G. mellonella have been established as a simple infection model for several pathogens, including S. aureus (Desbois and Coote, 2011; Tsai et al., 2016), and have also been used to test the efficacy of antimicrobials (Desbois and Coote, 2011). Despite the usefulness of this model, little is understood about the relative virulence of different S. aureus strains in G. mellonella. We previously found that in vitro gene expression of important virulence factors broadly varied amongst S. aureus strains 2014.N, LAC, and Mu50; 2014.N expresses the highest levels followed by LAC and then Mu50 (Hardy et al., 2019). Thus, we first tested the ability of these various strains to induce G. mellonella mortality at various doses. The overall virulence in this model revealed that LAC induced the highest level of death, followed by 2014.N and Mu50. Indeed, infection with LAC or 2014.N killed significantly more G. mellonella than Mu50 at the tested doses (Figures 7A,B). These data support the notion that though in vitro defined virulence factor expression profiles may be helpful, they do not always directly correlate with virulence in every in vivo model.
To examine the therapeutic potential of the compound(s) found within the CCFM, we next tested the ability of CCFM to rescue S. aureus-infected G. mellonella. As we found Mu50 to be essentially avirulent in this model (Figures 7A,B), we focused our efforts on CCFM derived from S. sap and S. epi-3, which was most active against S. aureus LAC (Figure 2A). Treatment with S. sap CCFM, but not S. epi-3 CCFM, 1-h post infection with 106 S. aureus LAC significantly reduced mortality of infected G. mellonella compared to sham treated controls (Figure 7C). Taken together, our results indicate that anti-S. aureus activity mediated by the various bacterial species is diverse and suggest that secreted compound(s) derived from S. saprophyticus may have possible future therapeutic value.
Discussion
Humans serve as an incredibly complicated and dynamic environmental niche for microorganisms. Our understanding of this fact has been greatly enhanced by the Human Microbiome Project, which has revealed that most anatomical locations are colonized with dozens, if not hundreds of bacterial species that must compete with each other for limited nutrients (Turnbaugh et al., 2007). While large microbiota-based epidemiological studies have identified the presence of these microbes, they often fail to elucidate the molecular interactions that occur between the resident flora and how these interactions may impact incoming pathogens. In addition, interactions with opportunistic pathogens are difficult to study in particular as the factors that promote commensalism vs. pathogenesis are often ill-defined. This is especially true for S. aureus, which asymptotically colonizes one-quarter of the population at any given time (Kluytmans and Wertheim, 2005; Wertheim et al., 2005), while simultaneously maintaining the ability to cause severe disease. It is well-established that the composition of the host microbiota heavily influences S. aureus carriage (Burian et al., 2017). This is particularly true in the nasal cavity, which serves as a primary reservoir for S. aureus colonization (Sakr et al., 2018). As such, many microbiota studies have focused on S. aureus interactions with the nasal flora. However, little is known about how S. aureus may interact with bacteria commonly found at other anatomical sites. Thus, in a proof of concept study we set out to characterize the basic interactions of S. aureus with bacterial isolates obtained from various sites (wound, blood, urine, and the nasal cavity) from patients at the WRNMMC. By taking a reductionist approach, we found that the majority of clinical isolates we screened displayed some form of in vitro anti-S. aureus activity.
En masse, in vitro bacterial interaction assays against three phenotypically different S. aureus strains revealed that the majority of tested clinical isolates were able to inhibit S. aureus to some degree (Figure 1, 2). Most of the inhibitory isolates were members of the Corynebacterium genera (10/28), which supports well-established findings that show that the Corynebacterium genus heavily impacts S. aureus colonization and viability (Yan et al., 2013; Hardy et al., 2019). For example, we previously showed that C. pseudodiphtheriticum, an important community determinant of S. aureus nasal colonization, mediates potent strain-specific bactericidal activity against S. aureus via production of a secreted factor(s) (Hardy et al., 2019). The results described herein indicate that related Corynebacterium species (C. aurimucosum, C. amycolatum, C. striatum, C. jeikeium, and C. tuberculostearicum) also possess some level of anti-S. aureus activity. Despite this finding, it is not possible to generalize that all Corynebacterium species negatively impact S. aureus. For example, C. accolens has been shown to actually promote S. aureus nasal colonization by reducing competition from other opportunistic pathogens (Yan et al., 2013; Bomar et al., 2016). In our screen, C. accolens possessed no anti-S. aureus activity (Figure 1A). In addition, recent work from Stubbendieck et al. (2019) showed that some Corynebacterium species can inhibit CoNS growth through the production of siderophores that enable these species to out-compete the CoNS for available iron, and thusly influence S. aureus viability. Therefore, individual Corynebacterium species appear to have evolved independent mechanisms that allow them to either cooperate or compete with S. aureus. Overall, our results combined with the growing body of literature suggest that the relationships observed in microbiota-based studies can be translated into in vitro phenotypes, and that the Corynebacterium genus in particular greatly impacts S. aureus viability and thusly colonization.
Culture independent-identification methods have revealed that wound infections, rather than being caused by a single species, are often polymicrobial in nature (Bowler et al., 2001; Peters et al., 2012; Tay et al., 2016). Moreover, microbiota-based studies have shown that wounds that are infected with multiple bacterial species tend to have worse outcomes as compared to wounds that are infected with a single species (Dalton et al., 2011; Pastar et al., 2013). It is worth noting that bacteria within wounds have to compete for resources and must contend with the host’s immune system. To aid these processes, bacteria that commonly infect wounds have evolved multiple mechanisms that help in these responses. For example, Pseudomonas aeruginosa and S. aureus are often co-isolated from wounds (Giacometti et al., 2000; Dowd et al., 2008). P. aeruginosa has been found to limit S. aureus growth by sensing the presence of S. aureus peptidoglycan (Korgaonkar et al., 2013; Pastar et al., 2013). P. aeruginosa responds by producing pyocyanin and elastase; both of these molecules have anti-S. aureus properties (Korgaonkar et al., 2013). Similarly, S. aureus and A. baumannii are also commonly co-isolated from wounds. However, to our knowledge there are no published reports of cooperative or competitive interactions between these two species. Thus, we were surprised that our initial screen revealed that A. baumannii was the most frequently isolated species possessing anti-S. aureus activity (7/28, Figures 2, 3). Moreover, the various A. baumannii isolates displayed a wide range of anti-S. aureus activities that were dependent upon both the A. baumannii and S. aureus strains. Future studies that seek to understand these interactions at a molecular level will be of great interest.
In thinking about the types of inhibition that we observed, contact-dependent inhibition can be mediated by variety of different mechanisms. For example, Type VI Secretion Systems (T6SS), which are found in many Gram-negative species, require physical contact and involve injection of toxic compounds directly into competitor cells (Coulthurst, 2019). Similarly, though mechanistically divergent from the T6SS, the Esx secretion pathway, which is broadly distributed amongst Gram-positive bacteria, also requires physical contact between competing bacterial species to mediate growth inhibition via toxic compounds (Whitney et al., 2017). In both these examples, only target cells that are physically touching the inhibitory cells are negatively impacted. In contrast contact-independent growth inhibition is typically mediated by toxic compounds that are synthesized and then secreted by the inhibitory species as a means to kill/prevent the growth of a competitor; no cell-to-cell contact between the two species is required. This approach is a common mechanism that is used by various microbes across multiple ecological niches and these compounds can include bacteriocins, secondary metabolites, and other small molecules (Zipperer et al., 2016; Terra et al., 2018). Finally, it is worth noting that some antagonistic interactions are more complex and can involve both contact-independent and dependent mechanisms. For example, initial physical contact between Streptococcus pneumoniae and S. aureus induces S. pneumoniae to generate and secrete hydrogen peroxide that can then kill S. aureus (Khan et al., 2016; Wu et al., 2019). Similarly, P. aeruginosa can physically senses S. aureus, which leads to global changes in transcription, resulting in the secretion of multiple compounds that have anti S. aureus activity (Korgaonkar et al., 2013; Filkins et al., 2015). Thus, of the 10 strongly inhibitory clinical isolates that produced a ZOC against S. aureus that was dependent on direct contact (Figure 4), some of these may require initial physical contact with S. aureus as a way to stimulate production of a toxic compound(s) or a secondary metabolite into the surrounding agar that can alter the pH or other environmental conditions in such a way as to impact S. aureus viability in that region. Undoubtedly, various species and strains utilize a diverse number of mechanisms to inhibit S. aureus.
Recently, there has been a renewed interest in the use of bacterial-derived compounds as novel therapeutics to treat highly drug resistant infections. Indeed, these compounds are potentially even more valuable because of the dearth of new antibiotics that are entering the market for human use. Our studies identified several isolates that inhibited S. aureus growth independent of physical contact, presumably through the activity of a secreted and diffusible compound(s) (Figures 1, 4). These isolates exclusively belonged to the Corynebacterium and Staphylococcus genera. Given that Corynebacterium and Staphylococcus are the primary genera that have been found to inhibit S. aureus growth on the skin and within the nasal cavity, it is clear that there appears to be a selective pressure for members of these genera to compete with S. aureus. It is worth noting that a portion of the identified isolates mediated killing activity (Figure 5); S. epidermidis represented the majority of the isolates that mediated bactericidal activity. This finding is likely not unexpected given that S. epidermidis, a common member of the human microbiota, has been found to actively compete with S. aureus by a variety of mechanisms: production of S. aureus-specific anti-microbial peptides, production of anti-biofilm compounds, and rapid and efficient nutrient acquisition (Lina et al., 2003; Iwase et al., 2010; Nakatsuji et al., 2017).
While the finding that S. epidermidis inhibits S. aureus is not surprising, to our knowledge our results are the first to show that Staphylococcus saprophyticus has anti-S. aureus activity (Figures 4–6). Moreover, we observed that treatment with S. saprophyticus CCFM was able to rescue survival of S. aureus-infected G. mellonella caterpillars (Figure 7C). S. saprophyticus is the second most common cause of bacterial urinary tract infections (UTIs) and is not associated with the healthy urinary tract (Marrie et al., 1982). Thus, S. saprophyticus presumably must out-compete normal urinary tract flora during the process of colonization and ultimate disease causation. Given that S. aureus can also infrequently colonize the urinary tract and cause UTIs, it’s interesting to speculate that S. saprophyticus has evolved to kill S. aureus as a means to prevent competition for this niche.
We note that secreted bactericidal compound(s) from some of the characterized isolates may have the potential to be developed for use as novel therapeutics to treat or prevent S. aureus-mediated infection. This ascertain is supported by the fact that anti-S. aureus activity was retained in CCFM from the three tested isolates (Figure 6), suggesting that these species negatively impact S. aureus viability most likely through the secretion of a toxic compound(s). Thought the nature of these compound(s) are unclear, they may include compounds like lantibiotics (McAuliffe et al., 2001), which are peptide antibiotics that are produced by a broad range of Gram-positive bacteria, including Staphylococcus. Genes that code for lantibiotics are often located on plasmids and other mobile genetic elements, and have a wide range of target-species specificity. Lantibiotics from closely related Staphylococcal species, such as epidermin (Götz et al., 2014), have been found to have potent inhibitory activity against S. aureus, including MRSA. It is possible that the anti-S. aureus activity we observed from S. saprophyticus, and the other Staphylococcal tested species, is the result of a lantibiotic that maintains potent inhibitory properties. Combined, our results indicate that many Staphylococcal species have evolved strategies to compete with S. aureus.
While this work was designed as a proof of concept study to explore the extent of anti-S. aureus activity exhibited by various microbes, we acknowledge that there are limitations to the study. For example, while the patient population at WRMMC is fairly diverse, given that many of the patients are soldiers that may have incurred traumatic injuries during the course of their service, a substantial proportion of isolates were obtained from wounds; this undoubtedly affected the types of species of bacteria that we ultimately screened. In addition, while this study described the basic molecular mechanisms of these interactions, a more detailed study will be required to clearly identify specific compounds and/or mechanisms of action that are responsible for anti- S. aureus activity.
In summary, this proof of concept study indicates that multiple bacterial species possess strain-specific anti-S. aureus activity when co-cultured in a bacterial interaction assay. This study further highlights the multifarious nature of polymicrobial interactions, which remain poorly understood. Furthermore, this work expands upon the growing body of literature that supports that the study of ‘bacterial warfare’ and the toxic compounds created by microbes as a means to compete with one another may be a ‘next best option’ for the identification of novel therapeutics that will help in overcoming the significant increase in antimicrobial resistance that threatens the health and wellbeing of the population (Zipperer et al., 2016; Nakatsuji et al., 2017; Stubbendieck et al., 2019). As such, we hypothesize that several of the inhibitory isolates identified in this study may produce toxic compounds that have the potential to be used as novel therapeutics or intervention strategies. Our future work will pursue elucidation of the molecular mechanism by which both A. baumannii and S. saprophyticus inhibit S. aureus. Overall, our findings support the continued study of polymicrobial interactions as a means to identify novel therapeutics and/or molecular targets of S. aureus and other pathogens.
Data Availability Statement
The datasets generated for this study can be found in the NCBI, GenBank, MN175920–MN175947.
Author Contributions
BH and DM designed the research study. EK and JB provided the clinical bacterial isolates utilized in all experiments. BH, GB, KH, AA, and SS performed the experiments. BH, GB, and DM analyzed the data. BH wrote the manuscript. All authors contributed substantially to revisions and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the members of Clinical Microbiology, Walter Reed National Military Medical Center for collecting the clinical isolates used in this study, the Uniformed Services University of the Health Sciences Infectious Disease Clinical Research Program for providing S. aureus strain 2014.N, and Emad Elassal and Michael Otto for providing S. aureus strains Mu50 and LAC, respectively. The contents of this manuscript are solely those of the authors and do not reflect the views of the Uniformed Services University of the Health Sciences, the Department of Defense, or the United States Government.
Funding. This study was supported by a U.S. Department of Defense Program project grant (HT9404-12-1-0019) and a Military Infectious Diseases Research Program award (#HU0001-15-2-0031).
References
- Bomar L., Brugger S. D., Yost B. H., Davies S. S., Lemon K. P. (2016). Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. mBio 7:e1725-15. 10.1128/mBio.01725-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler P. G., Duerden B. I., Armstrong D. G. (2001). Wound microbiology and associated approaches to wound management. Clin. Microbiol. Rev. 14 244–269. 10.1128/cmr.14.2.244-269.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugger S. D., Bomar L., Lemon K. P. (2016). Commensal–Pathogen interactions along the human nasal passages. PLoS Pathog. 12:e1005633. 10.1371/journal.ppat.1005633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie C. G., Bucci V., Stein R. R., McKenney P. T., Ling L., Gobourne A., et al. (2015). Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517 205–208. 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burian M., Bitschar K., Dylus B., Peschel A., Schittek B. (2017). The protective effect of microbiota on S. aureus skin colonization depends on the integrity of the epithelial barrier. J. Investig. Dermatol. 137 976–979. 10.1016/j.jid.2016.11.024 [DOI] [PubMed] [Google Scholar]
- Castellanos N., Nakanouchi J., Yüzen D. I., Fung S., Fernandez J. S., Barberis C., et al. (2019). A study on Acinetobacter baumannii and Staphylococcus aureus strains recovered from the same infection site of a diabetic patient. Curr. Microbiol. 76 842–847. 10.1007/s00284-019-01696-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulthurst S. (2019). The type VI secretion system: a versatile bacterial weapon. Microbiology 165 503–515. 10.1099/mic.0.000789 [DOI] [PubMed] [Google Scholar]
- Dalton T., Dowd S. E., Wolcott R. D., Sun Y., Watters C., Griswold J. A., et al. (2011). An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One 6:e27317. 10.1371/journal.pone.0027317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbois A. P., Coote P. J. (2011). Wax moth larva (Galleria mellonella): an in vivo model for assessing the efficacy of antistaphylococcal agents. J. Antimicrob. Chemother. 66 1785–1790. 10.1093/jac/dkr198 [DOI] [PubMed] [Google Scholar]
- Dowd S. E., Sun Y., Secor P. R., Rhoads D. D., Wolcott B. M., James G. A., et al. (2008). Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 8:43. 10.1186/1471-2180-8-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filkins L. M., Graber J. A., Olson D. G., Dolben E. L., Lynd L. R., Bhuju S., et al. (2015). Coculture of Staphylococcus aureus with Pseudomonas aeruginosa Drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J. Bacteriol. 197 2252–2264. 10.1128/JB.00059-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuno J. P., Hebden J. N., Standiford H. C., Perencevich E. N., Miller R. R., Moore A. C., et al. (2008). Prevalence of methicillin-resistant Staphylococcus aureus and Acinetobacter baumannii in a long-term acute care facility. Am. J. Infect. Control 36 468–471. 10.1016/j.ajic.2008.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacometti A., Cirioni O., Schimizzi A. M., Del Prete M. S., Barchiesi F., D’Errico M. M., et al. (2000). Epidemiology and microbiology of surgical wound infections. J. Clin. Microbiol. 38 918–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz F., Perconti S., Popella P., Werner R., Schlag M. (2014). Epidermin and gallidermin: staphylococcal lantibiotics. Int. J. Med. Microbiol. 304 63–71. 10.1016/j.ijmm.2013.08.012 [DOI] [PubMed] [Google Scholar]
- Hardy B. L., Dickey S. W., Plaut R. D., Riggins D. P., Stibitz S., Otto M., et al. (2019). Corynebacterium pseudodiphtheriticum exploits Staphylococcus aureus virulence components in a novel polymicrobial defense strategy. mBio 10:e2491-18. 10.1128/mBio.02491-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase T., Uehara Y., Shinji H., Tajima A., Seo H., Takada K., et al. (2010). Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465 346–349. 10.1038/nature09074 [DOI] [PubMed] [Google Scholar]
- Jarraud S., Mougel C., Thioulouse J., Lina G., Meugnier H., Forey F., et al. (2002). Relationships between Staphylococcus aureus genetic background, virulence factors, agr Groups (Alleles), and human disease. Infect. Immun. 70 631–641. 10.1128/iai.70.2.631-641.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Ellis M. W., Schlett C. D., Millar E. V., LaBreck P. T., Mor D., et al. (2016). Bacterial etiology and risk factors associated with cellulitis and purulent skin abscesses in military trainees. PLoS One. 11:e0165491. 10.1371/journal.pone.0165491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan F., Wu X., Matzkin G. L., Khan M. A., Sakai F., Vidal J. E. (2016). Streptococcus pneumoniae eradicates preformed Staphylococcus aureus biofilms through a mechanism requiring physical contact. Front. Cell. Infect. Microbiol. 6:104. 10.3389/fcimb.2016.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevens R., Morrison M. A., Nadle J., Petit S., Gershman K., Ray S., et al. (2007). Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298 1763–1771. [DOI] [PubMed] [Google Scholar]
- Kluytmans J. A. J. W., Wertheim H. F. L. (2005). Nasal carriage of Staphylococcus aureus and prevention of nosocomial infections. Infection 33 3–8. 10.1007/s15010-005-4012-9 [DOI] [PubMed] [Google Scholar]
- Korgaonkar A., Trivedi U., Rumbaugh K. P., Whiteley M. (2013). Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl. Acad. Sci. U.S.A. 110 1059–1064. 10.1073/pnas.1214550110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krismer B., Liebeke M., Janek D., Nega M., Rautenberg M., Hornig G., et al. (2014). Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathog. 10:e1003862. 10.1371/journal.ppat.1003862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M., Ohta T., Uchiyama I., Baba T., Yuzawa H., Kobayashi I., et al. (2001). Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357 1225–1240. 10.1016/s0140-6736(00)04403-2 [DOI] [PubMed] [Google Scholar]
- Laffineur K., Avesani V., Cornu G., Charlier J., Janssens M., Wauters G., et al. (2003). Bacteremia due to a novel Microbacterium species in a patient with leukemia and description of Microbacterium paraoxydans sp. nov. J. Clin. Microbiol. 41 2242–2246. 10.1128/jcm.41.5.2242-2246.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon K. P., Klepac-Ceraj V., Schiffer H. K., Brodie E. L., Lynch S. V., Kolter R. (2010). Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. mBio 1:e0129-10. 10.1128/mBio.00129-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina G., Boutite F., Tristan A., Bes M., Etienne J., Vandenesch F. (2003). Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl. Environ. Microbiol. 69 18–23. 10.1128/aem.69.1.18-23.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy F. D. (2003). Antimicrobial resistance: the example of Staphylococcus aureus. J. Clin. Investig. 111 1265–1273. 10.1172/jci200318535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrie T. J., Kwan C., Noble M. A., West A., Duffield L. (1982). Staphylococcus saprophyticus as a cause of urinary tract infections. J. Clin. Microbiol. 16 427–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuliffe O., Ross R. P., Hill C. (2001). Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 25 285–308. 10.1016/s0168-6445(00)00065-6 [DOI] [PubMed] [Google Scholar]
- Nakatsuji T., Chen T. H., Narala S., Chun K. A., Two A. M., Yun T., et al. (2017). Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 9:eaah4680. 10.1126/scitranslmed.aah4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2010). Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu. Rev. Microbiol. 64 143–162. 10.1146/annurev.micro.112408.134309 [DOI] [PubMed] [Google Scholar]
- Pastar I., Nusbaum A. G., Gil J., Patel S. B., Chen J., Valdes J., et al. (2013). Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS One 8:e56846. 10.1371/journal.pone.0056846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B. M., Jabra-Rizk M. A., O’May G. A., Costerton J. W., Shirtliff M. E. (2012). Polymicrobial interactions: impact on pathogenesis and human disease. Clin. Microbiol. Rev. 25 193–213. 10.1128/cmr.00013-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakr A., Brégeon F., Mège J.-L., Rolain J.-M., Blin O. (2018). Staphylococcus aureus nasal colonization: an update on mechanisms, epidemiology, risk factors, and subsequent infections. Front. Microbiol. 9:2419. 10.3389/fmicb.2018.02419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Rodriguez S. A., Cabanillas-Ramos J., Alcaraz U., Gomez-Gil B., Romalde J. L. (2013). Identification and virulence of Aeromonas dhakensis, Pseudomonas mosselii and Microbacterium paraoxydans isolated from Nile tilapia, Oreochromis niloticus, cultivated in Mexico. J. Appl. Microbiol. 115 654–662. 10.1111/jam.12280 [DOI] [PubMed] [Google Scholar]
- Stubbendieck R. M., May D. S., Chevrette M. G., Temkin M. I., Wendt-Pienkowski E., Cagnazzo J., et al. (2019). Competition among nasal bacteria suggests a role for siderophore-mediated interactions in shaping the human nasal microbiota. Appl. Environ. Microbiol. 85 e2406–e2418. 10.1128/AEM.02406-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay W. H., Chong K. K. L., Kline K. A. (2016). Polymicrobial–host interactions during infection. J. Mol. Biol. 428 3355–3371. 10.1016/j.jmb.2016.05.006 [DOI] [PubMed] [Google Scholar]
- Terra L., Dyson P. J., Hitchings M. D., Thomas L., Abdelhameed A., Banat I. M., et al. (2018). A novel alkaliphilic Streptomyces inhibits ESKAPE pathogens. Front. Microbiol. 9:2458. 10.3389/fmicb.2018.02458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. J.-Y., Loh J. M. S., Proft T. (2016). Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7 214–229. 10.1080/21505594.2015.1135289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Hamady M., Fraser-Liggett C. M., Knight R., Gordon J. I. (2007). The human microbiome project. Nature 449 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voyich J. M., Braughton K. R., Sturdevant D. E., Whitney A. R., Saïd-Salim B., Porcella S. F., et al. (2005). Insights into Mechanisms Used by Staphylococcus aureus to Avoid Destruction by Human Neutrophils. J. Immunol. 175 3907–3919. 10.4049/jimmunol.175.6.3907 [DOI] [PubMed] [Google Scholar]
- Wertheim H. F. L., Melles D. C., Vos M. C., van Leeuwen W., van Belkum A., Verbrugh H. A., et al. (2005). The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5 751–762. [DOI] [PubMed] [Google Scholar]
- Whitney J. C., Peterson S. B., Kim J., Pazos M., Verster A. J., Radey M. C., et al. (2017). A broadly distributed toxin family mediates contact-dependent antagonism between gram-positive bacteria. eLife 6:e26938. 10.7554/eLife.26938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, (2017). Global Priority List of Antibiotic-Resistant Bacteria To Guide Research, Discovery, And Development Of New Antibiotics. Available at: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf [Google Scholar]
- Wu X., Gordon O., Jiang W., Antezana B. S., Angulo-Zamudio U. A., del Rio C., et al. (2019). Interaction between Streptococcus pneumoniae and Staphylococcus aureus generates OH radicals that rapidly kill Staphylococcus aureus strains. J. Bacteriol. 201:e0474-19. 10.1128/JB.00474-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Pamp S. J., Fukuyama J., Hwang P. H., Cho D.-Y., Holmes S., et al. (2013). Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe 14 631–640. 10.1016/j.chom.2013.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipperer A., Konnerth M. C., Laux C., Berscheid A., Janek D., Weidenmaier C., et al. (2016). Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535 511–516. 10.1038/nature18634 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated for this study can be found in the NCBI, GenBank, MN175920–MN175947.