Abstract
In humans, chronic glucocorticoid use is associated with side effects like muscle wasting, obesity, and metabolic syndrome. Intermittent steroid dosing has been proposed in Duchenne Muscular Dystrophy patients to mitigate the side effects seen with daily steroid intake. We evaluated biomarkers from Duchenne Muscular Dystrophy patients, finding that, compared with chronic daily steroid use, weekend steroid use was associated with reduced serum insulin, free fatty acids, and branched chain amino acids, as well as reduction in fat mass despite having similar BMIs. We reasoned that intermittent prednisone administration in dystrophic mice would alter muscle epigenomic signatures, and we identified the coordinated action of the glucocorticoid receptor, KLF15 and MEF2C as mediators of a gene expression program driving metabolic reprogramming and enhanced nutrient utilization. Muscle lacking Klf15 failed to respond to intermittent steroids. Furthermore, coadministration of the histone acetyltransferase inhibitor anacardic acid with steroids in mdx mice eliminated steroid-specific epigenetic marks and abrogated the steroid response. Together, these findings indicate that intermittent, repeated exposure to glucocorticoids promotes performance in dystrophic muscle through an epigenetic program that enhances nutrient utilization.
Keywords: Metabolism, Muscle Biology
Keywords: Muscle, Neuromuscular disease
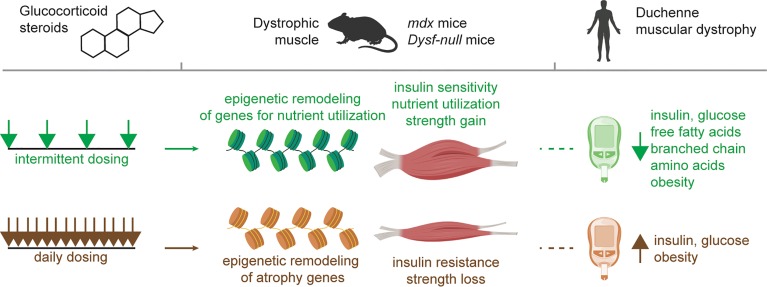
Repeated pulses of glucocorticoids reprogram dystrophic muscle by activating a cascade an epigenomic changes that promote nutrient uptake and enhance muscle performance.
Introduction
Tightly regulated control of muscle metabolism is essential to performance and the well being of the whole organism (1–3). With exercise, muscle coordinately upregulates the metabolism of glucose, fatty acids, and amino acids for energy production, and this response varies with time of day (4). In Duchenne Muscular Dystrophy (DMD), loss of dystrophin produces a fragile muscle membrane, leading to muscle degeneration. In addition to the primary loss of dystrophin, muscle mitochondrial function and ATP synthetic capacity are secondarily impaired (5, 6). Glucocorticoid (GC) steroids are used in DMD, where their use prolongs ambulation (7). However, the mechanisms by which steroids exert benefit in DMD is not completely understood, and chronic daily intake of GCs has adverse consequences, including metabolic dysfunction and obesity (8, 9). GC steroids have not been recommended for other genetic forms of muscular dystrophy. In dysferlin-deficient muscular dystrophy, a randomized study of deflazacort in 25 patients for 6 months showed a trend toward worsening muscle function and steroid-induced side effects compared with placebo (10). Alternative GC dosing strategies have been suggested to limit side effects (11, 12), but the clinical benefit of these strategies are debated. Recently, we compared daily GC steroids to an intermittent once-weekly (pulsatile) GC steroid protocol for the ability to promote muscle repair in dystrophic mice (13, 14). Although we found that both pulsatile and daily steroids could improve muscle repair, we made the unexpected observation that pulsatile dosing enhanced muscle performance, while daily dosing elicited muscle wasting.
Because GC steroids act through the GC receptor (GR) with cofactors to regulate gene expression (15), we hypothesized that once-weekly GC steroids would elicit distinct epigenomic profiles in order to have a sustained effect on gene expression. Here, we demonstrate that pulsatile GC steroids induced a distinct epigenomic program in dystrophic muscle, supported by the coordinated action of GR and the additional transcription factors KLF15 and MEF2C. In the mdx model of DMD, pulsatile steroids enhanced insulin sensitivity and muscle uptake of glucose, fatty acids, and branched chain amino acids (BCAA); this was associated with increased mitochondrial respiration. We further tested this concept in dysferlin-deficient mice and found similar metabolic reprogramming. In DMD patients, we found that weekend GC steroids, compared with daily GC steroids, were associated with reduced obesity and biomarkers of insulin resistance. Together, these findings define favorable metabolic reprogramming from GC treatments in mice and humans with muscular dystrophies.
Results
Weekend steroid exposure blunts metabolic stress in GC-treated DMD patients.
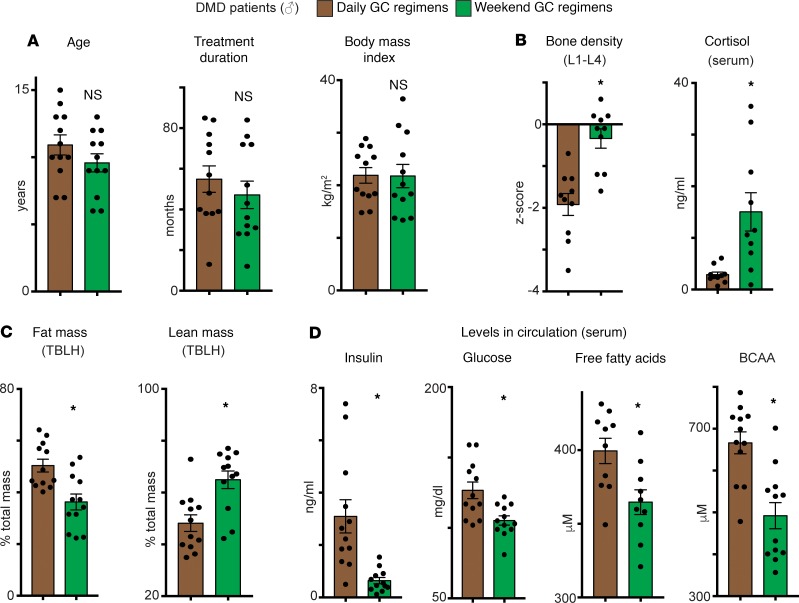
Although chronic daily steroid use improves outcomes in DMD patients (7), prolonged use of steroids also induces adverse outcomes like obesity, insulin resistance, and endogenous corticosteroid suppression (9). Insulin resistance is characterized by increased circulating insulin, glucose, free fatty acids (FFAs), and BCAA (16, 17). In DMD, most patients receive daily steroids, but weekend high-dose treatment (1–4 mg/kg given 2 consecutive days per week) has been proposed as an alternative to limit side effects (11). We compared clinical data and serum biomarkers from DMD boys receiving daily (1–2.5 mg/kg) or weekend (1–4 mg/kg) steroids (n = 12 patients/group, 5 prednisone/7 deflazacort in the daily group, and 7 prednisone/5 deflazacort in the weekend group). Steroid doses averaged 1.01 ± 0.22 mg/kg for daily prednisone, 0.77 ± 0.06 mg/kg for daily deflazacort, 2.44 ± 3.9 mg/kg for weekend prednisone, and 3.79 ± 0.15 mg/kg for weekend deflazacort (mean ± SEM). These doses calculated to the following cumulative doses in mg/week: 235.2 ± 30.9 daily prednisone, 200.6 ± 30.3 daily deflazacort, 218.5 ± 22.3 weekend prednisone, and 253.2 ± 39.3 for weekend deflazacort. Importantly, the groups were matched for BMI, age, and steroid treatment duration (48–55 months) (Figure 1A and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.132402DS1). As shown by dual-energy X-ray absorptiometry (DEXA) scans and serum analyses, weekend steroid use correlated with improved bone mineral density and increased serum cortisol, as compared with daily steroids, consistent with less osteoporosis and adrenal suppression (Figure 1B). DMD patients receiving weekend steroids had decreased fat mass and increased lean mass compared with those receiving daily steroids (Figure 1C). Weekend dosing associated with lower levels of glucose, insulin, FFAs, and BCAA in circulation (Figure 1D and Supplemental Table 1). Weekend steroid treatment was associated with comparable effects on ambulation, serum creatinine kinase levels, and cardiac function compared with daily steroid treatment (Supplemental Table 1). Although the total weekly GC dose in weekend-treated DMD patients was higher than the total GC dose in the once-weekly regimen previously tested in mice, the decrease in biomarkers of obesity and insulin resistance seen in the weekend-treated DMD patients supports that pulsatile dosing maintains clinical benefits and highlights a significant reduction in the metabolic stress generally induced by long-term GC regimens.
Figure 1. Duchenne Muscular Dystrophy (DMD) patients treated with weekend glucocorticoid steroids have reduced markers of obesity and insulin resistance compared with those receiving daily glucocorticoids.
(A) Age, treatment duration, and BMI did not differ between daily (1–2.5 mg/kg/dose × 7 days/week) and high-dose weekend (1–4 mg/kg/dose × 2 consecutive days/week) cohorts. (B) Weekend dosing correlated with normalization of bone density score (DEXA scans at L1–L4 vertebrae) and less suppression of endogenous cortisol levels. (C) Compared with daily steroid treated patients, weekend steroid dosing correlated with decreased fat mass gain and increased lean mass preservation (TBLH, total body less head; DEXA scans). (D) Weekend steroid dosing, as compared with daily steroid dosing, associated with reduced serum insulin, glucose, branched chain amino acids (BCAA), and free fatty acids, consistent with reduced metabolic stress. Histograms depict single values and mean ± SEM; n = 12 patients/cohort; *P < 0.05 vs daily; Welch’s unpaired t test (2-tailed).
Epigenetic programs induced by weekly versus daily steroids in dystrophic mouse muscles.
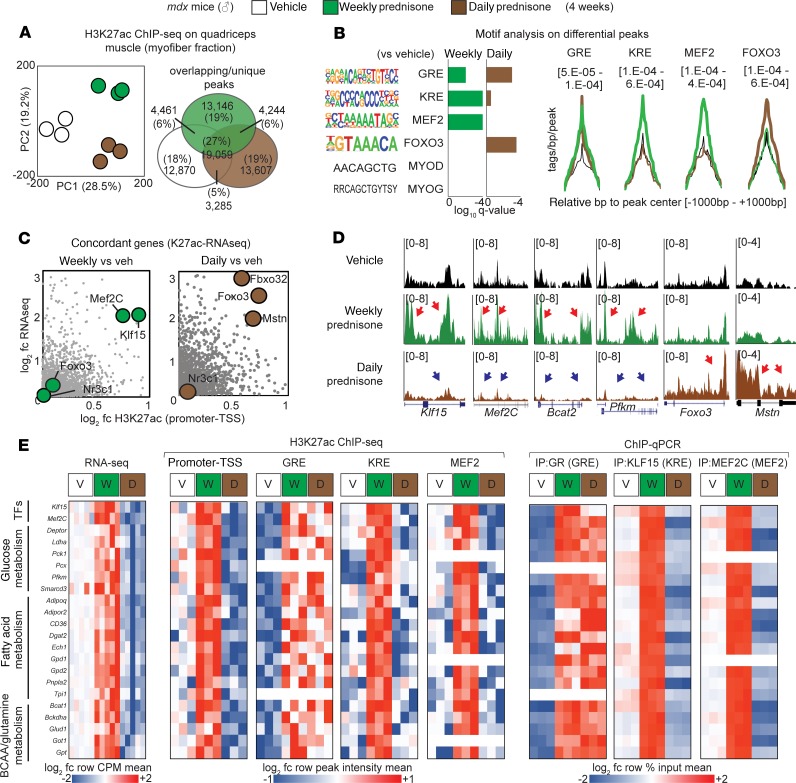
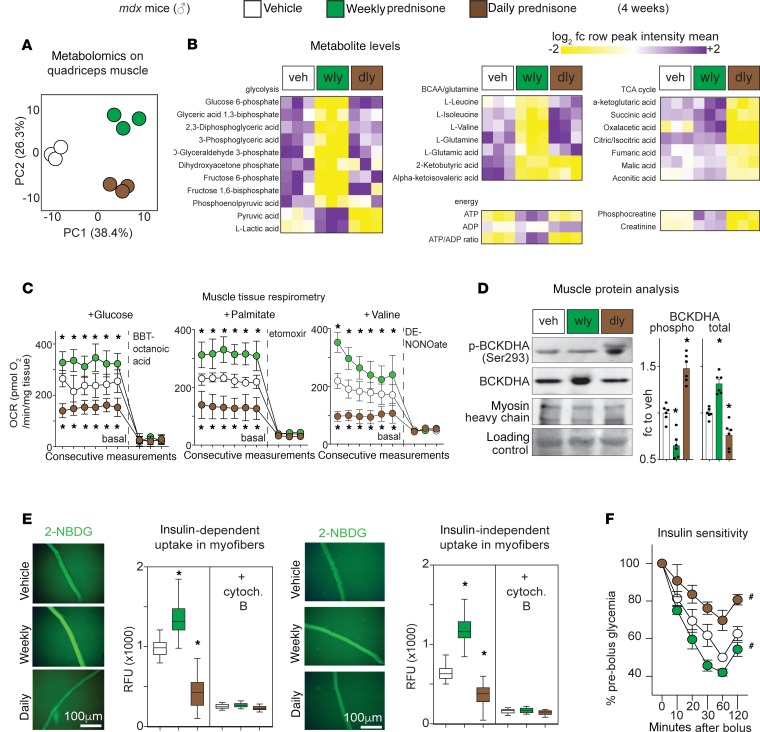
We previously enhanced muscle repair and gene expression changes in response to once-weekly steroids (14). We reasoned that once-weekly steroids would generate distinct epigenomic profiles to sustain these gene expression changes. To define the epigenomic programs elicited by steroid treatments of dystrophic muscle, we analyzed the genome-wide distribution of histone 3 lysine 27 acetylation (H3K27ac), a marker of transcriptional activation at enhancers and promoters (18), in once-weekly versus daily steroid–treated mdx mice receiving steroids for 4 weeks. H3K27ac analysis of the myofiber fraction of mdx muscle (n = 3 mice/group) was integrated with the muscle-matched RNA sequencing (RNA-seq) transcriptome (NCBI’s Gene Expression Omnibus [GEO] database; GSE137992 and GSE95682; n = 5 mice/group); both data sets were derived from samples collected 48 hours after the last drug administration. Principal component analysis (PCA) of global H3K27ac data readily clustered the profiles, distinguishing weekly from daily administration, from vehicle controls (Figure 2A). Unbiased analysis for transcription factor binding motifs highly represented in the differential H3K27ac peaks revealed significant enrichment for the GR-binding element (GRE) after both weekly and daily steroid use. However, weekly prednisone induced higher H3K27ac signal in KLF-responsive elements (KRE) and MEF2C-binding sites (MEF2), while FOXO3-binding sites were preferentially enriched in H3K27ac signal in the daily prednisone group (Figure 2B). KLF15 is a direct GR target that has been implicated in muscle metabolism and nutrient utilization (19), and MEF2C is a master regulator of muscle growth (20), while FOXO3 is a critical driver of muscle atrophy (21). Overlaying H3K27ac and RNA-seq data sets was used to identify concordant subsets of genes (i.e., genes with concordant up- or downregulation of both mRNA levels and H3K27ac signal in the promoter). After weekly prednisone, both Klf15 and Mef2C were upregulated and enriched in H3K27ac signal. In contrast, daily prednisone induced upregulation and H3K27ac signal on Foxo3 and other atrophy agonists like Fbxo32 and Mstn, and induced concordant downregulation of Klf15 and Mef2C (Figure 2C and Supplemental Figure 1A). Notably, the GR gene Nr3c1 was not significantly changed in H3K27ac enrichment or expression, indicating alternative regulators of GR activity or downstream cascades as mediators of this differential response.
Figure 2. Epigenomic programs in steroid-treated dystrophic mouse muscles.
(A) PCA of H3K27ac profiles derived from quadriceps myofibers. Weekly vs. daily prednisone regimens cluster from each other and from vehicle-treated controls. (B) Motif enrichment analysis highlights GRE, KRE, and MEF2 as acetylation-enriched motifs after weekly prednisone, while FOXO3 motifs were enriched after daily prednisone. (C) Concordant genes enriched for both H3K27ac and expression included Klf15 and Mef2C after weekly prednisone, while daily prednisone promoted acetylation and expression of Foxo3 and other atrophy genes. (D) Representative H3K27ac markings across gene loci of interest demonstrated divergent acetylation enrichment with respect to weekly or daily prednisone (signal is tags/bp normalized to 1e7 reads; red arrows, gain; blue arrow, loss of H3K27ac signal). (E) Pathway analysis showed that pulsatile weekly prednisone increased transcription of genes regulating glucose, fatty acid, and BCAA metabolism. H3K27ac ChIP-seq showed GR enrichment after both weekly and daily steroids, but it showed increased enrichment of KRE and MEF2 sites only after weekly prednisone. ChIP-qPCR confirmed enriched occupancy for GR, KLF15, and MEF2C on the same sites (n = 3 mice/group for K27ac ChIP-seq, n = 5 mice/group for RNA-seq; q value, Benjamini-Hochberg test).
Gene ontology (GO) analysis on all concordant genes revealed that GO terms for nutrient metabolism and muscle function were significantly enriched after weekly prednisone. Conversely, GO terms for muscle atrophy were enriched after daily prednisone (Supplemental Figure 1B). H3K27ac enrichment profiles on Klf15, Mef2c, Bcat2, and Pkm were visually distinct from the profile on atrophy genes like Foxo3 and Mstn (Figure 2D). Pathway analysis of concordant genes and their annotated regulatory regions showed that weekly prednisone induced H3K27ac enrichment on GRE, KRE, and MEF2 sites of upregulated genes involved in glucose, fatty acid, and BCAA-glutamine metabolism (Figure 2E, left). Those same regulatory regions were enriched for GR, KLF15, or MEF2C occupancy after pulsatile dosing, as shown by ChIP–quantitative PCR (ChIP-qPCR) assays (Figure 2E, right; and Supplemental Table 2). In contrast, after daily prednisone, H3K27ac and transcription factor occupancy was enriched for GRE sites, but not KRE or MEF2 sites, in these genes. These data highlight the importance of KLF15 and MEF2C in discriminating between GR-induced programs of favorable versus atrophic remodeling in muscle (Figure 2E). Daily prednisone induced H3K27ac enrichment at GRE and FOXO3 sites of atrogenes such as Fbxo32, Trim63, Mstn, Atf4, Gadd45a, and Cdkn1a (p21) (Supplemental Figure 1, C and D), consistent with a muscle wasting profile (22, 23). Thus, pulsatile GCs induce an epigenomic program in dystrophic muscle highly distinct from the epigenomic program induced by daily steroids.
Pulsatile GCs act through a coordinated program of GR, KLF15, and MEF2C activation and require epigenetic remodeling.
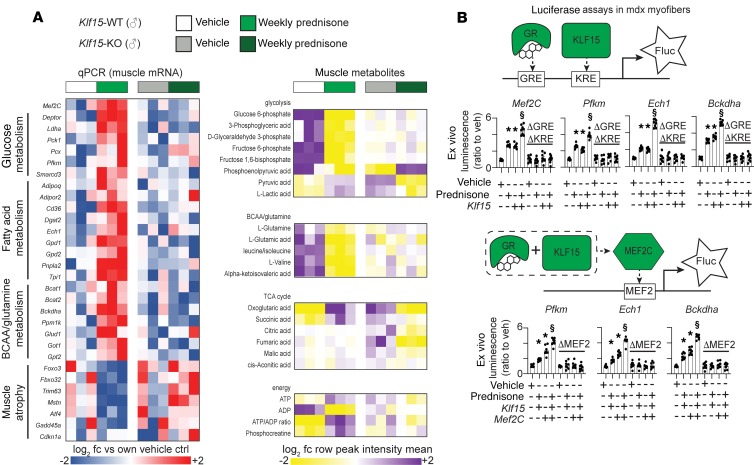
Because Klf15 was previously shown to be a GR target in dystrophic muscle (19), we asked whether the transcriptional program enabled by pulsatile prednisone was altered in Klf15-null mice (24). Homozygous Klf15-null mice versus littermate WT controls (Klf15-KO vs. Klf15-WT; n = 3 mice/group) were treated with vehicle or weekly prednisone for 4 weeks, and quadriceps muscle was isolated. In Klf15-WT mice, genes implicated in glucose, fatty acid, and BCAA metabolism were increased, but this upregulation was absent in Klf15-KO muscles, demonstrating a KLF15-dependent effect of intermittent steroid dosing (Figure 3A, left). After steroid dosing, downregulation of atrophy genes was also seen in Klf15-WT muscle, with trends toward upregulation of atrophy genes in Klf15-KO muscle.
Figure 3. Pulsatile weekly prednisone acts through a GR-, KLF15-, and MEF2C-mediated transcriptional program and requires epigenetic remodeling.
(A) Genetic loss of Klf15 disrupts the transcriptional and metabolic changes induced by weekly prednisone in muscle, including Mef2C upregulation. (B) Luciferase reporter plasmids were electroporated into mdx myofibers, carrying regulatory regions from Mef2c, Pfkm, Ech1, and Bckdha loci. Prednisone pulse and Klf15 overexpression had additive effects in increasing GRE-KRE activation. Prednisone pulse, Klf15, and Mef2C overexpression had additive effects on MEF2 activation. Changes were lost after specific deletion of target sites (Δ). Histograms depict single values and mean ± SEM; n = 3 mice/group (A), n = 8 muscles/group (B). *P < 0.05 vs. own vehicle control, $P < 0.05 vs single-factor treatment, 1-way ANOVA with Tukey’s multiple comparison.
Metabolomic profiling of steroid-treated Klf15-WT muscle showed signatures consistent with increased utilization of glucose, with a gain in pyruvate and lactate and a reduction in glycolytic precursors. Increased BCAA/glutamine consumption was accompanied by a gain in α-ketoglutarate and succinate, as well as increased ATP and phosphocreatine levels in WT muscles after weekly prednisone. In Klf15-KO muscle, these prednisone-induced trends were lost, together with a general decline in TCA cycle intermediate levels (Figure 3A, right). These findings demonstrate that KLF15 regulates transcriptional and metabolic changes induced by pulsatile GCs in dystrophic muscle.
The motif enrichment analysis from prednisone-treated mdx muscles identified GRE, KRE and MEF sites enriched in H3K27ac signal after weekly dosing (Figure 2B). This observation suggested functional cooperation of activated GR, KLF15, and MEF2C in driving metabolic gene upregulation in dystrophic muscle after repetitive pulsed exposure to GCs (Figure 3B, diagrams). We tested this hypothesis in myofibers by expressing reporter constructs carrying GRE-KRE and MEF2 sites from genes important for glycolysis (Pfkm; ref. 25), fatty acid oxidation (Ech1; ref. 26), and BCAA catabolism (Bckdha; ref. 27). We also tested the regulatory regions of MEF2C itself (Supplemental Table 3). We monitored gene expression by measuring firefly luciferase (Fluc) reporter activity in mdx muscles (n = 8 muscles/group) in the presence of either a prednisone pulse (1 mg/kg), Klf15 overexpression, or the combination thereof. Experiments were controlled with similar vectors specifically deleted for the GRE and KRE sequences (ΔGRE-KRE). Prednisone pulse plus Klf15 overexpression had an additive effect on reporter activity, whereas reporter activity was lost when the GRE or KRE sites were deleted (Figure 3B, top). Prednisone pulse, Klf15 overexpression, and Mef2C overexpression had additive effects on reporter activation, while reporter activity remained unchanged when the MEF2 sites were deleted (Figure 3B, bottom). Therefore, GR and KLF15 act together to enhance MEF2C’s transcriptional response and then, together, enhance transcriptional upregulation of metabolic target genes.
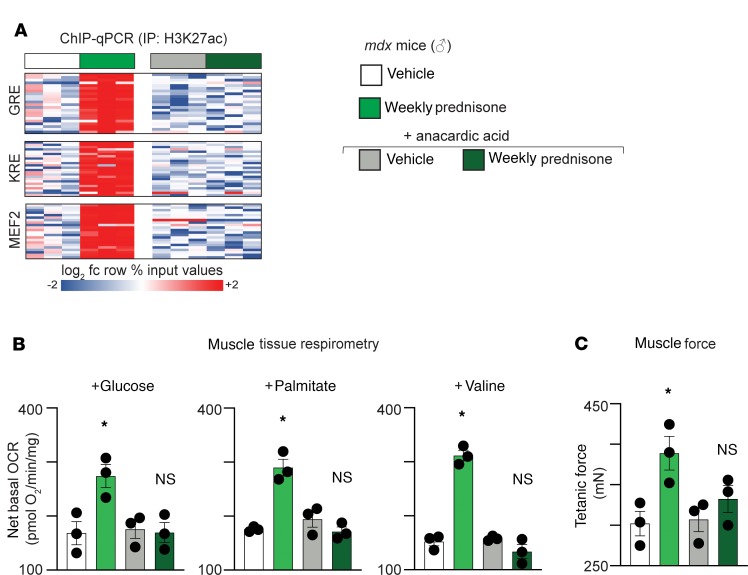
Because the H3K27ac patterns were strikingly different between pulsed prednisone– and daily prednisone–treated dystrophic muscle, we queried the role of histone acetylation in this response. The mdx mice were cotreated with the histone acetyltransferase (HAT) inhibitor anacardic acid (28). The mdx mice were treated for 4 weeks with anacardic acid (daily 5 mg/kg i.p. administration), together with vehicle or weekly prednisone regimens. HAT activity was reduced in anacardic acid–treated muscle, consistent with expected activity of the compound (Supplemental Figure 2A). After 4 weeks of anacardic acid exposure, the gain in H3K27ac marks on the GRE, KRE, and MEF2 sites previously seen with weekly prednisone was blunted, and this correlated with loss of upregulation of Klf15, Mef2C, and the metabolic pathway genes (Figure 4A and Supplemental Figure 2B). Furthermore, we measured muscle oxygen consumption rates (OCR) and monitored muscle force production after weekly prednisone with HAT inhibition. Anacardic acid coadministration blunted the gain in mitochondrial respiration produced by weekly prednisone, and this correlated with loss of prednisone-induced force improvement (Figure 4, B and C). In aggregate, these data demonstrate that pulsatile exposure to GCs elicits specific epigenomic remodeling, which — when blocked — abrogates the metabolic and functional benefit to dystrophic muscle.
Figure 4. Pulsatile weekly prednisone requires epigenetic remodeling.
(A) Coadministration of anacardic acid, a histone acetyl-transferase inhibitor, blunted gain of H3K27ac marking in identified GRE, KRE, and MEF2 sites in muscle observed after weekly prednisone. (B) Coadministration of prednisone and anacardic acid blunted gains in oxygen consumption rates with glucose, palmitate, or valine. (C) Coadministration of prednisone and anacardic acid blunted gains in muscle force (Tibialis anterior). Histograms depict single values and mean ± SEM; n = 3 mice/group (C). *P < 0.05 vs. vehicle, 1-way ANOVA with Tukey’s multiple comparison.
Pulsatile GCs enhanced glucose uptake and insulin sensitivity in mdx mice.
Based on the epigenetic/transcriptional changes induced by pulsatile GCs, we evaluated how weekly prednisone altered mdx muscle metabolism. We performed unbiased metabolomics on mdx muscles (n = 3 mice/group, 4 weeks exposure) after treatment with either weekly or daily prednisone, evaluating 171 hydrophilic metabolites. Metabolite profiles clustered according to steroid regimen, and daily and weekly steroids were distinct from vehicle treatment (Figure 5A). Weekly prednisone–treated muscle had reduced glycolytic precursors and intermediates and increased pyruvate and lactate, which is consistent with enhanced glycolysis. Weekly prednisone also promoted oxidative catabolism of BCAA and glutamine; this was seen as gain in α-ketoglutarate and succinate over anaplerotic precursors and intermediates. These findings correlated with a gain in ATP and phosphocreatine, consistent with improved energy production (Figure 5B). Conversely, daily prednisone induced the opposite patterns, with substantial impairment of BCAA and glucose catabolism seen as loss of downstream metabolites, accumulation of precursors, loss of TCA cycle intermediates, and lower levels of ATP and phosphocreatine (Figure 5B). The changes in ATP were confirmed by HPLC analyses of quadriceps and diaphragm muscles (Supplemental Figure 3A). Furthermore, free amino acid levels were decreased in muscles after weekly steroids, suggesting that pulsed prednisone induces a general shift toward higher utilization of amino acids for energy or protein production (Supplemental Figure 3B).
Figure 5. Pulsatile glucocorticoids enhanced nutrient utilization for mitochondrial respiration in dystrophic muscle.
(A) PCA of 171 metabolites showed treatment-specific clustering of muscle tissues. (B) Heatmaps of metabolite levels showed that pulsatile prednisone activated consumption of glucose, BCAA, and glutamine, increasing ATP and phosphocreatine levels. (C) Muscle respirometry showed that weekly prednisone led to higher basal oxygen consumption in the presence of either glucose, palmitate, or valine. (D) Weekly prednisone increased BCKDHA levels and reduced its phosphorylation in muscle. (E) Weekly prednisone increased glucose uptake in muscle, as shown by 2-NBDG uptake in live dystrophic myofibers. (F) Weekly prednisone increased whole-body insulin sensitivity, while daily regimen induced insulin resistance. Curves depict mean ± SEM; histograms depict single values and mean ± SEM; box plots, Tukey distribution; n = 3 mice/group (A and B), n = 6 mice/group (C–F). *P < 0.05 vs. vehicle, 2-way ANOVA with Tukey’s multiple comparison (C), 1-way ANOVA with Tukey’s multiple comparison (D and E). #P < 0.05 vs. vehicle, 2-way ANOVA.
To assess the effect of GCs on oxygen consumption rates, we performed respirometry on prednisone-treated dystrophic muscles in the presence of glucose, palmitate, or valine. Weekly prednisone increased the basal mitochondrial respiration levels with glucose, palmitate (fatty acid), or valine (BCAA) in quadriceps muscle tissue (n = 6 mice/group), while daily prednisone produced a reduction in the basal respiration in these conditions relative to vehicle-treated controls (Figure 5C). We next examined BCAA catabolic enzymes, given their known importance in muscle metabolism (27). We found that weekly prednisone increased the total amount and reduced phosphorylation of branched chain keto acid dehydrogenase (BCKDHA), which commits BCAA to oxidative metabolism and is inhibited by phosphorylation (Figure 5D). Thus, pulsatile GCs promote mitochondrial respiration and energy production in dystrophic muscle.
Considering the changes in nutrient utilization, we investigated how prednisone regimens impacted nutrient uptake into muscle. In contrast to daily prednisone, weekly prednisone enhanced muscle glucose uptake, as quantitated through 2-NBDG (fluorescent glucose analog) in isolated myofibers (Figure 5E). This myofiber-specific result also correlated with effects on whole-body insulin sensitivity. Weekly prednisone–treated mice showed higher insulin sensitivity (Figure 5F and Supplemental Table 4), offsetting GC-driven gluconeogenesis and normalizing glycemia (Supplemental Figure 3C). Conversely, daily prednisone promoted hyperglycemia, adipocyte hypertrophy, and insulin resistance (Figure 5F, Supplemental Figure 3C, and Supplemental Table 4). In aggregate, these data indicate that pulsatile prednisone promotes nutrient utilization and insulin sensitivity in dystrophic muscle, improving glucose homeostasis.
Isotope tracing validates activated rates of nutrient catabolism to the TCA cycle.
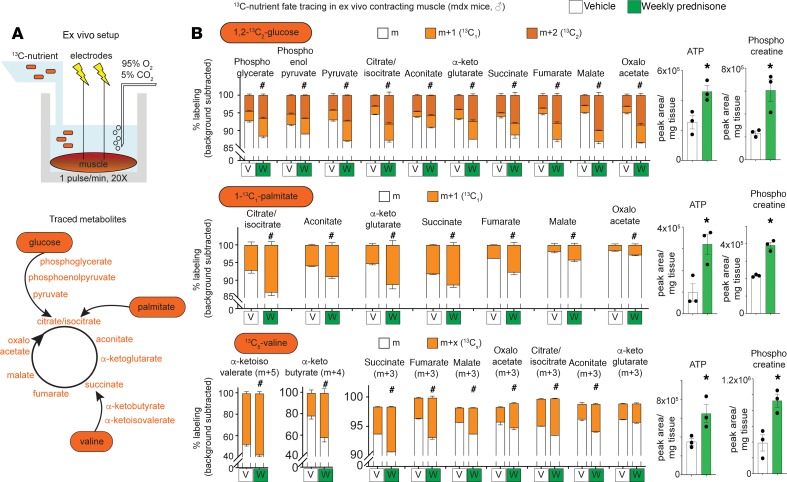
To confirm the effects of pulsatile prednisone on nutrient utilization in muscle, we performed 13C-isotope tracing in ex vivo contracting mdx muscle after a 4-week treatment with either weekly prednisone or vehicle. We adapted reported conditions (29) to isolated quadriceps muscles with repeated isometric contractions in the presence of 13C-labeled nutrients (i.e., 1,2-13C2-glucose, 1-13C1-palmitate, or 13C5-valine; Figure 6A, top). Each nutrient-specific catabolic cascade was analyzed quantitating the relative enrichment in appropriate 13C-labeled intermediates and TCA cycle metabolites (Figure 6A, bottom). Prednisone-treated muscles showed a higher 13C incorporation in target downstream metabolites of all 3 labeled nutrients, as compared with vehicle-treated muscles (Figure 6B). In the presence of labeled glucose, both 1- and 2-13C incorporation rates were generally increased, suggesting activated carbohydrate catabolism through glycolysis and pentose phosphate pathways (Figure 6B, top). Importantly, absolute levels of ATP and phosphocreatine levels were monitored in the same samples analyzed for mass resolution and were found increased in prednisone-treated muscles (Figure 6B, right). Thus, pulsatile GCs stimulate a coordinated increase in glucose, fatty acids, and BCAA catabolism to fuel the TCA cycle and energy production in dystrophic muscle.
Figure 6. Isotope tracing in ex vivo contracting muscle confirms higher nutrient flux to the TCA cycle after pulsatile weekly prednisone treatment.
(A) 13C incorporation into muscle metabolites was analyzed through mass resolution in an ex vivo contraction system. (B) Using either glucose, palmitate, or valine as source of 13C, weekly prednisone–treated muscles showed higher 13C incorporation than vehicle-treated muscles. In the same mass spectrometry analysis, ATP and phosphocreatine levels increased in prednisone-treated muscle. Histograms depict mean ± SEM. (stacked histograms), or single values and mean ± SEM; n = 3 mice/group. #P < 0.05 vs. vehicle, 2-way ANOVA; *P < 0.05 vs. vehicle, Welch’s t test (2-tailed).
Metabolic and functional effects of pulsatile GCs are maintained in long-term treatments of both mdx and Dysf-null mice.
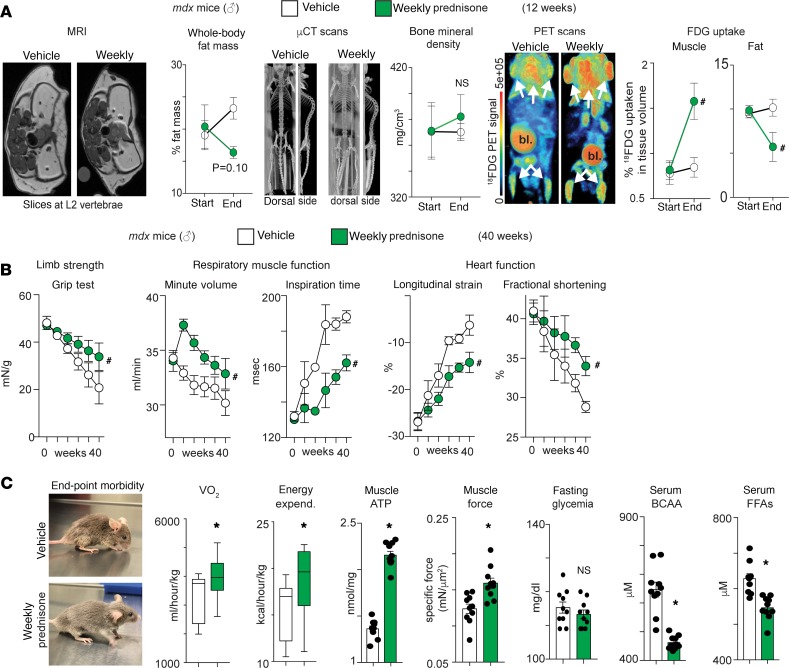
The animal studies above observed effects after 4 weeks of weekly prednisone. To test whether weekly steroids displayed this same effect over a longer term, mdx male mice were treated with vehicle or weekly prednisone for 12 weeks (n = 3 mice/group) and analyzed through live multimodal imaging at baseline and endpoint. Based on the known effects of long-term daily GCs on obesity and osteoporosis, whole-body fat mass was monitored through magnetic resonance imaging (MRI), bone mineral density was quantitated through μCT, and glucose uptake was monitored through PET of 18FDG, a glucose analog. All analyses were carried on the same animals. MRI showed a nonsignificant trend in fat mass reduction after weekly prednisone, while μCT showed no changes in bone mineral density (Figure 7A, left and center). We also found that weekly prednisone increased muscle uptake of 18FDG, while decreasing uptake in fat (Figure 7A, right). Thus, the combination of MRI, μCT, and PET imaging showed that pulsatile prednisone promoted muscle insulin sensitivity in mdx mice, without the induction of obesity and osteoporosis.
Figure 7. Long-term pulsatile prednisone improves insulin sensitivity, energy production, and muscle function in mdx mice.
(A) Multimodal imaging in live mice showed that weekly prednisone associated with a nonsignificant trend in fat mass reduction, no loss of bone mineral density, and significant changes in FDG uptake, upregulated in muscle and downregulated in fat (MRI-controled volumetric quantitation). (B) Treatment correlated with improved strength, respiratory function, and cardiac function over time, as compared with vehicle-treated mdx animals. (C) At endpoint, treated mice showed improved morbidity and increased VO2 and energy expenditure during the nocturnal activity phase. Treatment increased muscle ATP and force (Tibialis anterior), did not induce hyperglycemia, and reduced circulating levels of BCAA and free fatty acids. Curves, mean ± SEM; histograms depict single values and mean ± SEM; box plots, Tukey distribution; n = 3 mice/group (A), n = 10 mice/group (B and C). *P < 0.05 vs vehicle, Welch’s t test (2-tailed); #P < 0.05 vs vehicle, 2-way ANOVA.
We extended weekly prednisone treatment to a longer duration to comprehensively monitor functional and metabolic effects of pulsatile treatment. The mdx male mice were treated with either vehicle or weekly prednisone for 40 weeks beginning at 6 weeks of age (n = 10 mice/group). Over time, prednisone treatment significantly alleviated the functional decline in strength, respiratory muscle function, and left ventricle contractility as compared with vehicle treatment (Figure 7B). After 40 weeks, weekly prednisone improved morbidity and increased oxygen consumption (VO2) and energy expenditure during nocturnal activity (Figure 7C, left). Also, treatment-induced gains in muscle ATP and force were maintained in the long term (Figure 7C). Moreover, long-term treatment did not induce hyperglycemia, while favoring BCAA and FFA disposal in circulation and peripheral tissues (Figure 7C; Supplemental Table 5). Therefore, the beneficial reprogramming of muscle metabolism and function was durable in long-term weekly prednisone treatment of mdx mice.
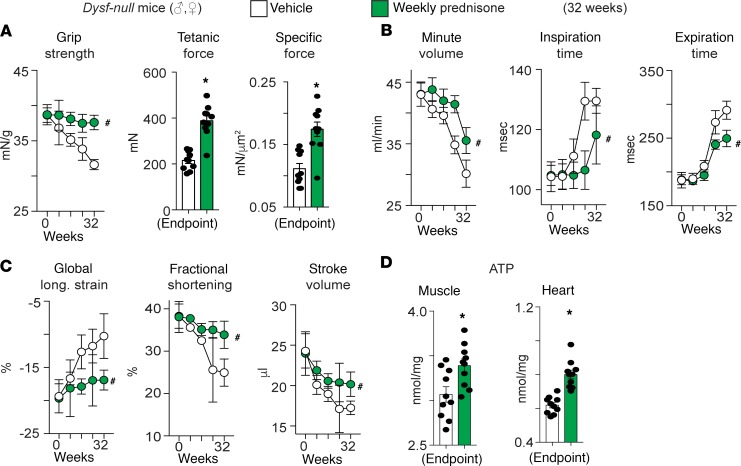
Finally, to explore whether pulsatile GCs may be useful in other forms of muscular dystrophy, we interrogated the effects of long-term weekly prednisone in a mouse model of limb girdle muscular dystrophy. We specifically selected a form of muscular dystrophy for which clinical data suggest deleterious effects from daily prednisone in patients (10). Dysferlin deficient (Dysf-null) mice, a genetic model of Limb Girdle Muscular Dystrophy type 2B, received either vehicle or weekly prednisone for 32 weeks from the age of disease onset (~9 months; n = 10 mice/group; randomized males/females). Pulsatile prednisone improved strength and muscle force (Figure 8A), as well as respiratory muscles and heart contractility, as compared with vehicle-treated mice (Figure 8, B and C). At endpoint, ATP levels in muscle and heart were increased by the treatment (Figure 8D). Consistent with observations in mdx mice and DMD patients, intermittent prednisone did not exacerbate insulin and glucose levels, and it decreased the levels of FFAs and BCAA in circulation and peripheral tissues (Supplemental Table 5). These data suggest that repetitive pulsed GCs may work in other forms of muscular dystrophy beyond DMD.
Figure 8. Pulsatile prednisone enhanced muscle metabolism and function in a distinct genetic model of muscular dystrophy, Dysf-null mice, a genetic model of Limb Girdle Muscular Dystrophy type 2B.
Mice lacking dysferlin (Dysf) serve as a model of Limb Girdle Muscular Dystrophy type 2B, and loss of dysferlin induces muscular dystrophy by impairing muscle repair. (A) Weekly prednisone exposure mitigated loss of grip strength over time seen in this model, and treatment increased muscle force at study endpoint (Tibialis anterior). (B and C) Weekly prednisone–treated mice had improved respiratory and cardiac muscle function over time, as compared with vehicle-treated controls. (D) ATP levels were increased in both muscle and heart tissues at endpoint. Curves, mean ± SEM; histograms depict single values and mean ± SEM; n = 10 mice/group. *P < 0.05 vs vehicle, Welch’s t-test (two-tailed); #P < 0.05 vs vehicle, 2-way ANOVA.
Discussion
GCs are among the most highly prescribed drugs worldwide and are part of the standard of care to prolong ambulatory age in DMD patients. However, GC administration has a significant side effect profile, particularly with long-term daily exposure (9). The use of GCs to treat a muscle disease has always been complicated by the observations that even short-term use of GCs can drive atrophic remodeling of muscle, which is especially prominent in mouse models (30). The striking atrophic response of mouse muscle, which we also observed in daily steroid–treated mice, may in part be driven by its skewed composition of fast fibers — specifically type IIb fibers, which have an especially high reliance on glycolysis (31). Fast myofibers are more susceptible than slow myofibers to FOXO3 activation and, therefore, to GC-driven atrophy (32). However, we discovered that repetitive pulsatile exposure to GCs enhanced the performance of dystrophic muscle and that this increased performance was supported by nutrient-fueled mitochondrial respiration and aerobic energy production. These favorable changes in muscle were directed by a distinct epigenomic program involving the transcription factors GR, KLF15, and MEF2C. KLF15 is a direct regulator of muscle gene programs that control amino acid metabolism and functional muscle remodeling downstream of GR activation (19, 33). KLF15 was also shown to play an important role in circadian control of metabolic physiology (34). MEF2C regulates muscle growth and is implicated in muscle metabolic response to exercise (35). The epigenomic analyses of GC effects in mouse models identified coordinate actions of the GR/KLF15 axis and MEF2C, as implicated in performance of dystrophic muscle.
Nutrient utilization in a large, energy-demanding tissue like muscle alters the body’s homeostasis and insulin sensitivity (36). The data presented here support that repetitive pulsatile GCs couple higher oxidative catabolism of glucose, fatty acids, and amino acids, with increased rates of nutrient consumption from the circulation and peripheral tissues, resulting in improved energy production and insulin sensitivity. This pulsatile regimen also associated with a general reduction of free amino acids in muscle and circulating BCAA. The gene expression and metabolic signatures of BCAA catabolism suggest that the BCAA disposal correlates with higher utilization for energy production in treated muscle. However, considering the beneficial effects of weekly prednisone on dystrophic muscle performance (13, 14), there may also be a concomitant effect on amino acid utilization for muscle protein synthesis. This pathway may be more broadly useful in conditions of chronic muscle degeneration, as the relative ratio of BCAA catabolism in striated muscle changes between insulin-sensitive and -resistant states (37).
Nonetheless, the epigenetically driven metabolic remodeling described here is impactful, considering that this muscle program was not elicited by interventions like exercise or calorie restrictions, but rather by a specific pharmacologic regimen. These data may also inform further refinement of clinical GC usage and development of biomarkers for monitoring their effects in muscle. It is important to note that the human DMD patients described in this report received a different GC steroid regimen compared with what was tested in mouse models. The weekend GC steroid–treated DMD patients received more GCs than the animal models, since clinical practice patterns for weekend GC treatment usually attempt to provide a similar cumulative weekly dose as to what would be given to DMD boys receiving daily steroids. Consistent with this concept, the average cumulative doses per week were similar between daily and weekend groups (217.9 vs. 235.9 mg/week, respectively). We specifically selected to compare groups with comparable BMIs in order to more critically contrast markers of insulin resistance, since obesity itself can alter these measures. The daily GC–treated DMD group showed increased markers of insulin resistance and fat mass without significant changes with BMIs, similar to the weekend GC–treated group. The similar ambulatory function in the weekend GC– and daily CG–treated groups hints that intermittent GC steroids provide comparable benefits but without the same metabolic stress, similar to other patient cohorts; however, this now provides data from longer treatment duration (11, 12, 38). Ultimately, optimizing GC regimens in dystrophic patients requires further human studies (7); the data presented herein provide some potential serum and body composition measures that could aid in tailoring steroid regimens to maximize benefit while reducing side effects. Whether less frequent or even lower doses would be sufficient to elicit the same favorable effects requires additional investigation. Given the well-established role of GCs in modulating immune cell function, future studies will also be required to elucidate how different GC regimens impact the cross-talk between immune and muscle cells.
Strikingly, metabolic programming by pulsatile GCs was not limited to dystrophin-linked muscular dystrophy but was also seen in a genetic model of limb-girdle muscular dystrophy, which is characterized by completely distinct genetic and cellular defects. There are currently no indications for GCs in muscular dystrophies beyond DMD, and efficacy has been questioned in small studies (10, 39). Intriguingly, it was recently reported that activation of the GC/KLF15/BCAA axis benefits a mouse model of spinal muscular atrophy, a genetic disorder with a significant neuronal component (40). It is, therefore, possible that favorable metabolic reprogramming by pulsed GC regimens is applicable in a broader range of neuromuscular diseases and target tissues — even those that feature prominent defects in nerve function. Together, our findings demonstrate that pulsatile GCs enable a distinctive epigenetic program in dystrophic muscle to favorably remodel nutrient utilization for energy production. Furthermore, these transcriptional and metabolic signatures may provide useful biomarkers for monitoring favorable responses to therapy in DMD or other forms of heritable or acquired myopathies.
Methods
Supplemental Methods are available online with this article.
Animal handling and steroid treatments.
Mice were housed in a pathogen-free dedicated vivarium. Euthanasia was performed through carbon dioxide inhalation, followed by cervical dislocation and heart removal. Mice were fed ad libitum with Mouse Breeder Sterilizable Diet (7904; Harlan Teklad) and maintained on a 14-hour/10-hour light/dark cycle. The mdx mice from the DBA/2J background were obtained from the Jackson Laboratory (stock no. 013141) and interbred. Male mice were used for reported experiments. Starting ages were about 6 months for short-term experiments and about 6 weeks for long-term experiments. Dysferlin-null (Dysf-null) mice from the 129T2/SvEmsJ background were previously characterized (41, 42). Starting ages were about 9 months for long-term experiments. For experiments with Dysf-null and WT mice, both females and males (~1:1 ratio) were randomized in treatment groups. KLF15-KO mice were previously generated and described (24). KLF15-KO and -WT male littermates were generated from heterozygous matings, and mice were injected for 4 weeks from the age of approximately 6 months. Prednisone (P6254; Sigma-Aldrich) was resuspended in DMSO (D2650; Sigma-Aldrich) to a stock concentration of 5 mg/mL. Dosing was based on weekly weight measurements (1 mg/kg body weight; ref. 43) in 100 μL total PBS volume per dose. Mice were injected daily via i.p. injection at 7 am. On injection days, stock solutions stored at –20°C were diluted into sterile Eppendorf tubes containing sterile PBS (14190; Thermo Fisher Scientific). Anacardic acid (catalog A7236; Sigma-Aldrich) was coadministered during the weekly prednisone regimen (4 weeks) as daily i.p. injection of 5 mg/kg in 50 μL volume (28). Inhibition of HATs by anacardic acid in tissues was monitored using the HAT activity colorimetric dosing assay (catalog P-4003; Epigentek). Sterile BD Micro-Fine IV Insulin Syringes (14-829-1A; Thermo Fisher Scientific) were used to inject the i.p. cavity of nonsedated animals. All animal analyses both during treatment and at endpoint were conducted blinded to treatment groups.
Human sample collection.
Subjects were recruited from Muscular Dystrophy Association Clinic at the Ann & Robert H. Lurie Children’s Hospital of Chicago with a confirmed genetic diagnosis of DMD. Blood samples were sterilely collected in a red-top tube at the end of each individual’s clinic appointment (generally late morning to early afternoon) on Thursdays. Samples were centrifuged at 2000 g for 10 minutes at 4°C. Serum was isolated, prealiquoted for downstream assays to avoid repeated freeze/thaw, and stored at –80°C. DEXA data were collected from regular measurements that individuals with DMD undergo annually as part of standard of care. All scans were performed on a GE Lunar iDXA during the same clinic visit as blood sample collection or at the most recent clinic visit, approximately 6 months prior. Z-scores were established based on age-standardized controls. For Brooke functional scoring, physical therapists assessed the Brooke functional scale score at each clinic visit and documented it as part of their clinic notes. The scale is scored on a 9-point scale: a score of 1 indicates the highest level of ambulation versus a score of 9, which indicates that the individual is confined to a wheelchair. Data were collected on day of blood collection. For 10-meter run tests, individuals diagnosed with DMD and who are ambulatory performed the 10-meter run test as part of their clinical assessment. A physical therapist timed individuals with a stopwatch. Individuals performed 10-meter run test as fast as safely permissible while barefoot. Data were collected on the day of blood collection. For ECG data, individuals with DMD underwent 12 lead ECGs on a GE MAC5500HD on standard ECG paper (10 mv, 25 mm/s, 150 Hz) as part of their clinical care. ECGs were collected at the same clinic visit as blood collection or at a prior clinic encounter, approximately 6 months prior. ECGs were read and confirmed by a pediatric cardiologist at our institution. For heart function measurements, individuals with DMD undergo routine echocardiogram assessment annually. Echocardiographic measurements used in this study were either performed at the same clinic visit as serum collection or during the most recent clinic encounter, approximately 6 months prior. Echocardiography was performed on a Philips iE33 Ultrasound machine and read routinely by pediatric cardiologists at our institution. All analyses related to serum samples were conducted blinded to treatment groups and to other clinical assessments.
Metabolic and endocrine biomarkers.
For measurement of whole-tissue ATP levels, approximately 25 mg of frozen-pulverized tissue was extracted in 10% perchloric acid and neutralized in 0.75 M K2CO3, as previously described (44). ATP was measured by high-pressure liquid chromatography (HPLC) with Shimadzu LC-20A pump (Shimadzu Scientific Instr Inc.) and UV-VIS detector, using a Supelco LC-18-T column (15 cm × 4.6 cm; 58970-U; MilliporeSigma). The HPLC was run at a flow rate of 1 mL/minute with 100% buffer A (0.5 M KH2PO4, 0.5 M K2HPO4) from 0–5 minutes, a linear gradient to 95% buffer A/5% buffer B (100% methanol) from 5–6 minutes, 95% buffer A/5% buffer B from 6–11 minutes, a linear gradient to 85% buffer A/15% buffer B from 11–13 minutes, 85% buffer A/15% buffer B from 13–23 minutes, and a linear gradient to 100% buffer A from 23–30 minutes. ATP eluted as a sharp peak at 3 minutes, and peak values were normalized to tissue weight of frozen tissue for calculating pmol/mg values. Corticosterone was measured in mouse serum, and cortisol was measured in human serum using dedicated ELISA kits (ADI-900-097, Enzo Life Sciences; K7430-100, BioVision) according to manufacturer instructions and internal standards to calculate ng/mL values. Insulin levels were quantitated in mouse and human serum with species-specific ELISA kits (10-1247-01, mouse-specific; 10-1113-01, human-specific; Mercodia), following manufacturer instructions and internal standards to calculate ng/mL values. FFAs were quantitated using Enzychrom Free Fatty Acid Assay kit (EFFA-100; BioAssay Systems), following the kit’s instructions and standards to calculate μM (serum) and nmol/mg (tissue) values. For BCAA dosing, BCAA levels (not discriminating individual amino acid concentrations) were assayed using a dedicated colorimetric kit (ab83374; Abcam), following manufacturer’s instructions and standards to calculate μM (serum) and nmol/mg extracted protein (tissue) values. All dosing assays relied on triplicates for each standard or sample; tests were run on either serum or approximately 25 mg of frozen-pulverized whole tissue (treated according to each kit’s procedure). Colorimetric reactions were quantitated using a Synergy HTX multimode plate reader (BioTek) and averaging 4 reads/sample at appropriate wavelengths. All dosing assays were conducted blinded to treatment groups.
H3K27ac ChIP-seq on muscle myofibers.
Freshly isolated whole quadriceps muscles (both per mouse) were finely minced and digested in 10 mL/muscle of PBS supplemented with 1 mM CaCl2 and 100 U/mL collagenase II (catalog 17101, Invitrogen) at 37°C for 1 hour with shaking. The suspension was then filtered through a 40-μm strainer (catalog 22363547, Thermo Fisher Scientific), and the unfiltered fraction (enriched in myofibers) was kept for further steps. Separation of mononuclear fraction in the filtered fraction was confirmed at the microscope. Myofibers were fixed in 10 mL 1% PFA for 30 minutes at room temperature with gentle nutation. Fixation was quenched 1 mL of 1.375 M glycine (catalog BP381-5, Thermo Fisher Scientific) with gentle nutation for 5 minutes at room temperature. After centrifugation at 3000 g for 5 minutes, myofibers were lysed in 1.4 mL lysis buffer with about 250 μL 2.3-mm zirconia/silica beads (catalog 11079125z, BioSpec). Lysis buffer consisted of 10 mM HEPES (pH 7.3; catalog H3375), 10 mM KCl (catalog P9541), 5 mM MgCl2 (catalog M8266), 0.5 mM DTT (catalog 646563), 3 μg/mL cytochalasin B (C6762; all reagents from MilliporeSigma), and protease inhibitor cocktail (catalog 11852700, Roche). Myofibers were then homogenized by means of Mini-BeadBeater-16 (catalog 607, Biospec) for 30 seconds, followed by rotating at 4°C for 30 minutes. Samples were centrifuged at 3000 g for 5 minutes at 4°C, supernatant was removed, and the pellet was resuspended in cell lysis buffer as per reported conditions (45); the cell lysis buffer was supplemented with 3 μg/mL cytochalasin B and rotated for 10 minutes at 4°C. Nuclei were pelleted at 300 g for 10 minutes at 4°C and subsequently processed following reported protocol with the adjustment of adding 3 μg/mL cytochalasin B into all solutions for chromatin preparation and sonication, antibody incubation, and wash steps. Chromatin was then sonicated for 15 cycles (30 seconds, high power; 30 seconds pause; 200 μL volume) in a water bath sonicator set at 4°C (Bioruptor 300; Diagenode). After centrifuging at 10,000 g for 10 minutes at 4°C, sheared chromatin was checked on agarose gel for a shear band between ~150 and ~600 bp. A total of 2 μg of chromatin was kept for pooled input controls, whereas leftover chromatin (~50 μg) was used for each pull-down reaction: 5 μL H3K27ac primary antibody (catalog 39133, Active Motif) in 2 mL volume, rotating at 4°C overnight. Chromatin complexes were precipitated with 100 μL proteinA/G magnetic beads (catalog 88803; Thermo Fisher Scientific). After washes and elution, samples were treated with proteinase K (catalog 19131; Qiagen) at 55°C, and cross-linking was reversed through overnight incubation at 65°C. DNA was purified using the MinElute purification kit (catalog 28004; Qiagen) and quantitated using Qubit reader and reagents. Library preparation and sequencing were conducted at the NU Genomics Core using TrueSeq ChiP-seq library prep (with size exclusion) on 5 ng chromatin per ChIP sample or pooled input, along with HiSeq 50 bp single-read sequencing (~ 60 million read coverage per sample). Peak analysis was conducted using HOMER software (v4.10; ref. 46) and synthax (e.g., makeTagDirectory, makeUCSCfile, findPeaks, mergePeaks, annotatePeaks.pl, getDifferentialPeakReplicates.pl, findMotifsGenome.pl) after aligning fastq files to the mm10 mouse genome using bowtie2 (47). Homer motifs used for peak annotation after unsupervised motif analysis were gre.motif, klf3.motif, and mef2c.motif. PCA was conducted using ClustVis (48). GO pathway enrichment was conducted (cutoff, 1.5-fold transcriptional change) using the GO analysis tool (49).
Muscle metabolomics.
Total hydrophilic metabolite content was extracted from quadriceps muscle tissue at treatment endpoint through methanol/water (80:20) extraction, adapting conditions described previously (50). Briefly, total metabolite content from quadriceps muscle was obtained from about 100 mg (wet weight) quadriceps muscle tissue per animal. Frozen (–80°C) muscle was pulverized in liquid nitrogen and homogenized with approximately 250 μL 2.3-mm zirconia/silica beads (catalog 11079125z, BioSpec) in 1 mL methanol/water at a 80:20 (vol/vol) ratio by means of Mini-BeadBeater-16 (catalog 607, Biospec) for 1 minute. After centrifuging at 2350 g for 5 minutes, 200 μL of supernatant were transferred into a tube preadded with 800 μL of ice-cold methanol/water 80% (80:20 vol/vol). Samples were vortexed for 1 minute and then centrifuged at 20,160 g for 15 minutes at 4°C. Metabolite-containing extraction solution was then dried using SpeedVac (medium power). A total of 200 μL of 50% acetonitrile were added to the tube for reconstitution following by overtaxing for 1 minute. Sample solution was then centrifuged for 15 minutes at 20,000 g at 4°C. Supernatant was collected for liquid chromatography–mass spectrometry (LC-MS) analysis for hydrophilic metabolite profiling as follows. Samples were analyzed by high-performance LC and high-resolution MS and tandem MS (HPLC-MS/MS). Specifically, the system consisted of a Thermo Q-Exactive in line with an electrospray source and an Ultimate3000 (Thermo Fisher Scientific) series HPLC consisting of a binary pump, degasser, and autosampler outfitted with a Xbridge Amide column (Waters; dimensions of 4.6 mm × 100 mm and a 3.5-μm particle size). The mobile phase A contained 95% (vol/vol) water, 5% (vol/vol) acetonitrile, 20 mM ammonium hydroxide, 20 mM ammonium acetate, pH 9.0; B was 100% acetonitrile (all reagents from MilliporeSigma). The gradient was as following: 0 minutes, 15% A; 2.5 minutes, 30% A; 7 minutes, 43% A; 16 minutes, 62% A; 16.1–18 minutes, 75% A; 18–25 minutes, 15% A with a flow rate of 400 μL/min. The capillary of the ESI source was set to 275°C, with sheath gas at 45 arbitrary units, auxiliary gas at 5 arbitrary units, and the spray voltage at 4.0 kV. In positive/negative polarity switching mode, an m/z scan range from 70–850 was chosen, and MS1 data were collected at a resolution of 70,000. The automatic gain control (AGC) target was set at 1 × 106, and the maximum injection time was 200 ms. The top 5 precursor ions were subsequently fragmented, in a data-dependent manner, using the higher-energy collisional dissociation (HCD) cell set to 30% normalized collision energy in MS2 at a resolution power of 17,500. The sample volumes of 25 μL were injected. Data acquisition and analysis were carried out by Xcalibur 4.0 software and Tracefinder 2.1 software, respectively (both from Thermo Fisher Scientific). Metabolite levels were analyzed as peak area normalized to wet tissue weight (weight before cryopulverization). Metabolite analysis was performed blinded to treatment groups.
13C isotope tracing in ex vivo contracting muscle.
13C tracing from nutrients in muscle was performed adapting reported conditions (29) to our muscle stimulus settings used to probe muscle force (see below). Immediately after sacrifice, quadriceps muscles were dissected and immobilized on a Sylgard-coated well of a 12-multiwell plate by means of two 27-g needles at the muscle extremities. The well was prefilled with 1× Ringer’s solution (146 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES; pH, 7.4 [all reagents from MilliporeSigma]) containing 25 mU/mL insulin (catalog RP-10908; Thermo Fisher Scientific), and the appropriate 13C nutrient and kept at 37°C on a heated pad. 13C-labeled nutrients included 10 mM 1,2-13C2-glucose, 1 mM 1-13C-palmitate (BSA conjugated), and 5 mM 1,2,3,4,5-13C5-valine (catalogs 453188, 292125, and 758159, respectively; MilliporeSigma). The nutrient solution was constantly bubbled with a 95%O2/5%CO2 line (~2 psi). After 5-minute equilibration in solution, electrodes were inserted at the muscle extremities, securing them to the holder needles. Using a Whole Mouse Test System (catalog 1300A; Aurora Scientific) 20 contractions (1×/min) were induced with the following specifications: initial delay, 0.1 second; frequency, 200 Hz; pulse width, 0.5 ms; duration, 0.5 second; 100 mA stimulation. Muscles were then removed from the 13C nutrient solution, quickly rinsed in nutrient-free Ringer’s solution, dried, and immediately flash-frozen. Muscle metabolites were then extracted and analyzed as per metabolomics procedures (LC-MS, see above), and mass resolution was carried on predetermined metabolites, while control energetics (ATP, phosphocreatine) were analyzed from simultaneous quantitation from the LC-MS system. The metabolite labeling ratio was calculated on peak area/mg tissue values, subtracting the background 13C labeling ratio obtained from muscles exposed to unlabeled nutrients (the same reagents used for respirometry) and expressed as the percentage of total metabolites. Metabolite analysis was performed blinded to treatment groups.
Luciferase experiments in live myofibers.
Luciferase plasmids containing regulatory fragments were obtained cloning genomic sequences in the pGL4.23 backbone (E8411; Promega) using the KpnI-XhoI sites upstream of the minimal promoter site. Fragments were cloned conserving the genomic orientation with regard to transcriptional orientation, adding KpnI and XhoI tails to the appropriate extremities via Phusion PCR. WT fragments with responsive site ablation were cloned from WT C57BL/6J genomic DNA, while mutated fragments (Δ sites) were amplified from ad hoc synthetized DNA oligonucleotides, using genomic sequences from the C57BL/6J genomic background (see Supplemental Table 5 for a complete list of sequences). Flexor digitorum brevis (FDB) fibers were transfected by in vivo electroporation. Methods were described previously (51). Briefly, the hindlimb footpad was injected with 10 μL hyaluronidase (8 units) (catalog H4272, MilliporeSigma). After 2 hours, up to 40 μg in 20 μL of endotoxin-free plasmid (10 μL luciferase vector, 2 μL Renilla vector, 3 μL Klf15 vector [MR206548; Origene] or Mef2C vector [#32515; ref. 52]) was injected into the footpad. Electroporation was conducted by applying 20 pulses, each 20 ms in duration, at 1 Hz, at 100 V/cm. Animals were allowed to recover for a minimum of 7 days and not more than 10 days after electroporation to avoid examining injured muscle and to allow sufficient time for plasmid expression (53). GR activation was promoted with a pulse of 1 mg/kg i.p. prednisone 24 hours before luciferase analysis. Ex vivo luciferase assay was performed on whole, electroporated FDB muscles. Muscles were minced and homogenized in lysate buffer, and experiments were performed according to Dual Luciferase Assay Kit (catalog 1910; Promega) instructions. Luminescence was recorded at the Synergy HTX multimode 96-well plate reader (BioTek). Raw values were normalized to Renilla luciferase, then to protein content (MyHC), and finally to vehicle-treated muscles with the same plasmids. Results are expressed as fold change to average vehicle. All luciferase quantitation assays were conducted blinded to treatment groups.
Tissue respirometry.
Whole-tissue analysis of basal OCR was conducted adapting reported conditions for intact muscle tissue analysis (54) to the XF96 Extracellular Flux Analyzer platform (Agilent). Immediately after mouse sacrification, target muscle (quadriceps) tissues were quickly collected, rinsed in clean PBS buffer, and dissected into approximately 2 × 2 × 2 mm pieces. At least 3 biopsies were sampled for each tissue. Each biopsy was placed at the bottom of a dedicated 96-microplate well (101085; Agilent), covered with 225 μL of basal respirometry medium, and equilibrated at 37°C in a CO2-free incubator for 1 hour. Respirometry medium was based on XF Base Medium without phenol red (103335-100; Agilent) supplemented with either 10 mM glucose, 5 mM valine, or 1 mM palmitate-BSA. pH was adjusted to 7.4 for all media. Nutrients (G7021, V0500, P0500; MilliporeSigma) were diluted in XF Base Medium. During biopsy equilibration, a Seahorse XFe96 FluxPak cartridge (102601-100; Agilent), previously hydrated overnight with 300 μL/well XF calibrant (100840; Agilent) at 37°C in a CO2-free incubator, was loaded with 25 μL appropriate chemical compounds in designated ports and calibrated in the Analyzer. Respirometry analysis was then performed on equilibrated tissue biopsies using the following protocol for each basal or postinjection read: 3-minute mix, 5-minute delay, and 2-minute measure. Basal rate reads were collected for 6 consecutive courses; then, drugs were injected and control reads gathered for additional 3 consecutive courses. Drugs to validate basal respiration include the following, where catalog number, referenced inhibitory activity, and final concentration are reported after each compound (all compounds from MilliporeSigma): DE-NONOate (D184-50; inhibitor of methylmalonyl-CoA mutase [ref. 55]), 5 mM; BBT-octanoic acid (SML0404; inhibitor of PDH [ref. 56]), 1 mM; and etomoxir (E1905; inhibitor of CPT [ref. 57]), 1 mM. Compound concentrations were determined on literature and/or preliminary test assays on WT muscle biopsies, and the concentration of the compound when loaded in the cartridge port was ×10. OCR reads were averaged for the same tissue replicates and subtracted from background noise values (empty wells with only medium and appropriate compound). OCR reads were then normalized to biopsy dry weight and measured after overnight incubation of biopsy plate after respirometry analysis at 55°C, hence obtaining pmol O2/min/mg values for OCR. Net OCR values were calculated as basal OCR minus OCR after inhibitor supplementation. All respirometry analyses were conducted blinded to treatment groups.
2-NBDG uptake assay and glycemia/lactate monitoring.
2-NBDG uptake assay in live myofibers was conducted adapting previously reported conditions (58). FDB muscles were collected and carefully treated with collagenase type II and hand pipetting to liberate single myofibers, following reported procedures (51). Myofibers from 2 FDB muscles were collected in 1 mL Ringer’s solution (for 1 L, 7.2 g NaCl, 0.17 g CaCl2, 0.37 g KCl; pH, 7.4). A total of 200 μL of myofiber suspension was dispensed per well of chambered coverglass (155382; Thermo Fisher Scientific) and imaged as baseline condition for both transmitted light (1 ms integration) and green fluorescent channels (100 ms integration) at the Zeiss Axio Observer A1 microscope, using ×20 short-range objective and the ZEN 2 software (version 2011; Zeiss). Immediately after baseline imaging, myofibers were supplemented with 2 mM glucose (D8375-1g; Millipore Sigma) and 50 μM 2-NBDG (11046; Cayman Chemical). For insulin-dependent uptake, insulin (12585014; Thermo Fisher Scientific) was added to a final 85 μM concentration. To control Glut1-/Glut4-dependent uptake, negative control wells were further supplemented with 10 μM citochalasin B (C6762; Millipore Sigma). Myofibers were incubated for 30 minutes in a 37°C/10% CO2 incubator; they were then washed twice in Ringer’s solution and immediately imaged in fresh Ringer’s solution, using the same integration and objective settings used for preincubation pictures. 2-NBDG uptake was quantitated as relative fluorescent units, calculated as intramyofiber fluorescence after incubation subtracted of average baseline fluorescence. Fluorescence intensity was quantitated through serial analysis of acquired images (3 areas of ~85μm2 were analyzed for average fluorescence value per myofiber; >10 myofibers were analyzed per mouse) with ImageJ (NIH) software (59). All glucose uptake assays were conducted blinded to treatment groups.
Glucose was measured in blood (first drop from tail venipuncture) or serum (5 μL of 1:2 dilution) with an AimStrip Plus glucometer system (Germaine Laboratories) and expressed as mg/dL values. Lactate was measured in blood (second drop from tail venipuncture) or serum (5 μL of 1:2 dilution) with a Lactate Plus reader (Nova Biomedical) and expressed as mM values. Fasting glycemia was measured in mice after 4-hour fasting (7–11 am). Glucose, insulin, and pyruvate tolerance tests were conducted after 4-hour fasting in individual cages immediately after baseline fasting glucose monitoring. Mice were injected with either 1 g/kg glucose (D8375-1g; Millipore Sigma), 0.5 U/kg insulin (12585014; Thermo Fisher Scientific), or 2.5 g/kg pyruvate (P5280-25g; Millipore Sigma) in 200 μL i.p. injections, and glucose was then monitored by tail venipuncture at 10 minutes, 20 minutes, 30 minutes, 60 minutes, and 120 minutes after injection. All glucose- and pyruvate-tolerance tests were conducted blinded to treatment groups.
Data and materials availability.
Data sets for mdx muscle metabolomics, mdx myofiber H3K27ac ChIP-seq, and mdx muscle RNA sequencing are available as Data Track ID 1828 in NIH Metabolomics Workbench, GSE137992 and GSE95682 entries in GEO, respectively.
Statistics.
Statistical analyses were performed using Prism software v7.0a (Graphpad). Normality of data pools was tested with the Pearson-D’Agostino normality test. When comparing 2 groups, 2-tailed Student’s t test with Welch’s correction (unequal variances) was used. When comparing 3 groups of data for 1 variable, 1-way ANOVA with Tukey’s multicomparison was used. When comparing data groups for more than 1 related variable, 2-way ANOVA was used, and the Tukey’s multicomparison was additionally used when comparing more than 2 data groups. For ANOVA and t test analyses, a P value less than 0.05 was considered significant. Stacks of P values were analyzed with Benjamini-Hochberg test to calculate a q value (metabolomics, epigenomics). Data were presented as single values (dot plots, histograms) when the number of data points was fewer than 15. In analyses pooling larger data point sets per group (typically > 50 data points), Tukey distribution bars were used to emphasize data range distribution. Analyses pooling data points over time were presented as marked line plots. Tables, dot plots, histograms, and marked line plots depict mean ± SEM. Box plots depict the Tukey distribution of the data pool.
Study approval.
All methods using mice in this study were performed in ethical accordance with the American Veterinary Medical Association (AVMA) and under protocols fully approved by the IACUC of NU Feinberg School of Medicine (no. ISO00000761). Consistent with the ethical approvals, all efforts were made to minimize suffering.
Subject’s parents provided consent to participate (NCT03319030) under approval granted by the IRB of the Ann and Robert H. Lurie Children’s Hospital (no. 2017-1264). All protocols and consents were conducted in accordance with the Declaration of Helsinki and other international ethical guidelines.
Author contributions
MQ conceived and performed experiments, analyzed data, and drafted manuscript. ASZ and NLK provided and analyzed data and samples from patients. ZJ, CBP, ARD, GDB, SMH, and JB assisted with study conception, experiments, analyses, and manuscript drafting. EMM conceived the study, analyzed data, and edited the manuscript.
Supplementary Material
Acknowledgments
Metabolomics services were performed by the Metabolomics Core Facility at Robert H. Lurie Comprehensive Cancer Center of Northwestern University. Next-generation sequencing was performed at the NUSeq core at Center for Genetic Medicine of Northwestern University. Live animal multimodal imaging was performed at the Center for Advanced Molecular Imaging of Northwestern University. Metabolic cage testing was performed by the Northwestern Comprehensive Metabolic Core. We thank Christopher Newgard (Duke University, Durham, North Carolina, USA) for helpful suggestions.
This work was supported by NIH U54 AR052646, NIH RO1 NS047726, NU 320-8235910-30027355-01, the Parent Project for Muscular Dystrophy, and the American Heart Association. MQ is supported by NIH K01 DK121875 (NIDDK) and MDA-AANEM development grant no. 479350. SMH was supported by NIH R01 DK093821.
Version 1. 12/19/2019
Electronic publication
Footnotes
Conflict of interest: NU has filed a patent on intermittent glucocorticoid use based on this data (provisional number 62/876,238). SMH is an executive and shareholder of Amgen and holds equity stake in Tenaya Therapeutics. ZJ is an employee of Amgen as a member of the Amgen postdoctoral program.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(24):e132402.https://doi.org/10.1172/jci.insight.132402.
Contributor Information
Mattia Quattrocelli, Email: mattia.quattrocelli@northwestern.edu.
Aaron S. Zelikovich, Email: aszeliko@iu.edu.
Zhen Jiang, Email: zhen.jiang@gladstone.ucsf.edu.
Clara Bien Peek, Email: c-peek@northwestern.edu.
Alexis R. Demonbreun, Email: alexis.demonbreun@northwestern.edu.
Nancy L. Kuntz, Email: NKuntz@luriechildrens.org.
Grant D. Barish, Email: grant.barish@northwestern.edu.
Saptarsi M. Haldar, Email: saptarsi.haldar@gladstone.ucsf.edu.
Joseph Bass, Email: j-bass@northwestern.edu.
Elizabeth M. McNally, Email: elizabeth.mcnally@northwestern.edu.
References
- 1.Meng ZX, et al. Glucose Sensing by Skeletal Myocytes Couples Nutrient Signaling to Systemic Homeostasis. Mol Cell. 2017;66(3):332–344.e4. doi: 10.1016/j.molcel.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cokorinos EC, et al. Activation of Skeletal Muscle AMPK Promotes Glucose Disposal and Glucose Lowering in Non-human Primates and Mice. Cell Metab. 2017;25(5):1147–1159.e10. doi: 10.1016/j.cmet.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Ahn B, et al. MondoA coordinately regulates skeletal myocyte lipid homeostasis and insulin signaling. J Clin Invest. 2016;126(9):3567–3579. doi: 10.1172/JCI87382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato S, et al. Time of Exercise Specifies the Impact on Muscle Metabolic Pathways and Systemic Energy Homeostasis. Cell Metab. 2019;30(1):92–110.e4. doi: 10.1016/j.cmet.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Ryu D, et al. NAD+ repletion improves muscle function in muscular dystrophy and counters global PARylation. Sci Transl Med. 2016;8(361):361ra139. doi: 10.1126/scitranslmed.aaf5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Percival JM, Siegel MP, Knowels G, Marcinek DJ. Defects in mitochondrial localization and ATP synthesis in the mdx mouse model of Duchenne muscular dystrophy are not alleviated by PDE5 inhibition. Hum Mol Genet. 2013;22(1):153–167. doi: 10.1093/hmg/dds415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald CM, et al. Long-term effects of glucocorticoids on function, quality of life, and survival in patients with Duchenne muscular dystrophy: a prospective cohort study. Lancet. 2018;391(10119):451–461. doi: 10.1016/S0140-6736(17)32160-8. [DOI] [PubMed] [Google Scholar]
- 8.Nadal A, Quesada I, Tudurí E, Nogueiras R, Alonso-Magdalena P. Endocrine-disrupting chemicals and the regulation of energy balance. Nat Rev Endocrinol. 2017;13(9):536–546. doi: 10.1038/nrendo.2017.51. [DOI] [PubMed] [Google Scholar]
- 9.Matthews E, Brassington R, Kuntzer T, Jichi F, Manzur AY. Corticosteroids for the treatment of Duchenne muscular dystrophy. Cochrane Database Syst Rev. 2016;(5):CD003725. doi: 10.1002/14651858.CD003725.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walter MC, et al. Treatment of dysferlinopathy with deflazacort: a double-blind, placebo-controlled clinical trial. Orphanet J Rare Dis. 2013;8:26. doi: 10.1186/1750-1172-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connolly AM, Schierbecker J, Renna R, Florence J. High dose weekly oral prednisone improves strength in boys with Duchenne muscular dystrophy. Neuromuscul Disord. 2002;12(10):917–925. doi: 10.1016/S0960-8966(02)00180-3. [DOI] [PubMed] [Google Scholar]
- 12.Connolly AM, et al. Twice-weekly glucocorticosteroids in infants and young boys with Duchenne muscular dystrophy. Muscle Nerve. 2019;59(6):650–657. doi: 10.1002/mus.26441. [DOI] [PubMed] [Google Scholar]
- 13.Quattrocelli M, Salamone IM, Page PG, Warner JL, Demonbreun AR, McNally EM. Intermittent Glucocorticoid Dosing Improves Muscle Repair and Function in Mice with Limb-Girdle Muscular Dystrophy. Am J Pathol. 2017;187(11):2520–2535. doi: 10.1016/j.ajpath.2017.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quattrocelli M, et al. Intermittent glucocorticoid steroid dosing enhances muscle repair without eliciting muscle atrophy. J Clin Invest. 2017;127(6):2418–2432. doi: 10.1172/JCI91445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vockley CM, et al. Direct GR Binding Sites Potentiate Clusters of TF Binding across the Human Genome. Cell. 2016;166(5):1269–1281.e19. doi: 10.1016/j.cell.2016.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch CJ, Adams SH. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol. 2014;10(12):723–736. doi: 10.1038/nrendo.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Würtz P, et al. Metabolic signatures of insulin resistance in 7,098 young adults. Diabetes. 2012;61(6):1372–1380. doi: 10.2337/db11-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rivera CM, Ren B. Mapping human epigenomes. Cell. 2013;155(1):39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrison-Nozik A, et al. Glucocorticoids enhance muscle endurance and ameliorate Duchenne muscular dystrophy through a defined metabolic program. Proc Natl Acad Sci USA. 2015;112(49):E6780–E6789. doi: 10.1073/pnas.1512968112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276(5317):1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandri M, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117(3):399–412. doi: 10.1016/S0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bullard SA, et al. Gadd45a Protein Promotes Skeletal Muscle Atrophy by Forming a Complex with the Protein Kinase MEKK4. J Biol Chem. 2016;291(34):17496–17509. doi: 10.1074/jbc.M116.740308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bodine SC, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294(5547):1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 24.Gray S, et al. Regulation of gluconeogenesis by Krüppel-like factor 15. Cell Metab. 2007;5(4):305–312. doi: 10.1016/j.cmet.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.García M, et al. Phosphofructo-1-kinase deficiency leads to a severe cardiac and hematological disorder in addition to skeletal muscle glycogenosis. PLoS Genet. 2009;5(8):e1000615. doi: 10.1371/journal.pgen.1000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang D, Liu B, Huang K, Huang K. Enoyl coenzyme A hydratase 1 protects against high-fat-diet-induced hepatic steatosis and insulin resistance. Biochem Biophys Res Commun. 2018;499(3):403–409. doi: 10.1016/j.bbrc.2018.03.052. [DOI] [PubMed] [Google Scholar]
- 27.White PJ, et al. The BCKDH Kinase and Phosphatase Integrate BCAA and Lipid Metabolism via Regulation of ATP-Citrate Lyase. Cell Metab. 2018;27(6):1281–1293.e7. doi: 10.1016/j.cmet.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng C, et al. Inhibition of histone H3K9 acetylation by anacardic acid can correct the over-expression of Gata4 in the hearts of fetal mice exposed to alcohol during pregnancy. PLoS One. 2014;9(8):e104135. doi: 10.1371/journal.pone.0104135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerner J, Minkler PE, Lesnefsky EJ, Hoppel CL. Fatty acid chain elongation in palmitate-perfused working rat heart: mitochondrial acetyl-CoA is the source of two-carbon units for chain elongation. J Biol Chem. 2014;289(14):10223–10234. doi: 10.1074/jbc.M113.524314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schakman O, Gilson H, Kalista S, Thissen JP. Mechanisms of muscle atrophy induced by glucocorticoids. Horm Res. 2009;72 Suppl 1:36–41. doi: 10.1159/000229762. [DOI] [PubMed] [Google Scholar]
- 31.Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91(4):1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 32.Sandri M, et al. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci USA. 2006;103(44):16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun H, et al. Catabolic Defect of Branched-Chain Amino Acids Promotes Heart Failure. Circulation. 2016;133(21):2038–2049. doi: 10.1161/CIRCULATIONAHA.115.020226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeyaraj D, et al. Klf15 orchestrates circadian nitrogen homeostasis. Cell Metab. 2012;15(3):311–323. doi: 10.1016/j.cmet.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amoasii L, Sanchez-Ortiz E, Fujikawa T, Elmquist JK, Bassel-Duby R, Olson EN. NURR1 activation in skeletal muscle controls systemic energy homeostasis. Proc Natl Acad Sci USA. 2019;116(23):11299–11308. doi: 10.1073/pnas.1902490116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schenk S, Saberi M, Olefsky JM. Insulin sensitivity: modulation by nutrients and inflammation. J Clin Invest. 2008;118(9):2992–3002. doi: 10.1172/JCI34260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neinast MD, et al. Quantitative Analysis of the Whole-Body Metabolic Fate of Branched-Chain Amino Acids. Cell Metab. 2019;29(2):417–429.e4. doi: 10.1016/j.cmet.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Escolar DM, et al. Randomized, blinded trial of weekend vs daily prednisone in Duchenne muscular dystrophy. Neurology. 2011;77(5):444–452. doi: 10.1212/WNL.0b013e318227b164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Godfrey C, et al. Fukutin gene mutations in steroid-responsive limb girdle muscular dystrophy. Ann Neurol. 2006;60(5):603–610. doi: 10.1002/ana.21006. [DOI] [PubMed] [Google Scholar]
- 40.Walter LM, et al. Interventions Targeting Glucocorticoid-Krüppel-like Factor 15-Branched-Chain Amino Acid Signaling Improve Disease Phenotypes in Spinal Muscular Atrophy Mice. EBioMedicine. 2018;31:226–242. doi: 10.1016/j.ebiom.2018.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demonbreun AR, Fahrenbach JP, Deveaux K, Earley JU, Pytel P, McNally EM. Impaired muscle growth and response to insulin-like growth factor 1 in dysferlin-mediated muscular dystrophy. Hum Mol Genet. 2011;20(4):779–789. doi: 10.1093/hmg/ddq522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Demonbreun AR, et al. Dysferlin and myoferlin regulate transverse tubule formation and glycerol sensitivity. Am J Pathol. 2014;184(1):248–259. doi: 10.1016/j.ajpath.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sali A, et al. Glucocorticoid-treated mice are an inappropriate positive control for long-term preclinical studies in the mdx mouse. PLoS One. 2012;7(4):e34204. doi: 10.1371/journal.pone.0034204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324(5927):651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carey MF, Peterson CL, Smale ST. Chromatin immunoprecipitation (ChIP) Cold Spring Harb Protoc. 2009;2009(9):pdb.prot5279. doi: 10.1101/pdb.prot5279. [DOI] [PubMed] [Google Scholar]
- 46.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43(W1):W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bruno C, et al. The combination of four analytical methods to explore skeletal muscle metabolomics: Better coverage of metabolic pathways or a marketing argument? J Pharm Biomed Anal. 2018;148:273–279. doi: 10.1016/j.jpba.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 51.Demonbreun AR, McNally EM. DNA Electroporation, Isolation and Imaging of Myofibers. J Vis Exp. 2015;null(106):e53551. doi: 10.3791/53551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kozhemyakina E, Cohen T, Yao TP, Lassar AB. Parathyroid hormone-related peptide represses chondrocyte hypertrophy through a protein phosphatase 2A/histone deacetylase 4/MEF2 pathway. Mol Cell Biol. 2009;29(21):5751–5762. doi: 10.1128/MCB.00415-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kerr JP, et al. Dysferlin stabilizes stress-induced Ca2+ signaling in the transverse tubule membrane. Proc Natl Acad Sci USA. 2013;110(51):20831–20836. doi: 10.1073/pnas.1307960110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shintaku J, Guttridge DC. Analysis of Aerobic Respiration in Intact Skeletal Muscle Tissue by Microplate-Based Respirometry. Methods Mol Biol. 2016;1460:337–343. doi: 10.1007/978-1-4939-3810-0_23. [DOI] [PubMed] [Google Scholar]
- 55.Kambo A, Sharma VS, Casteel DE, Woods VL, Pilz RB, Boss GR. Nitric oxide inhibits mammalian methylmalonyl-CoA mutase. J Biol Chem. 2005;280(11):10073–10082. doi: 10.1074/jbc.M411842200. [DOI] [PubMed] [Google Scholar]
- 56.Liu X, et al. Acetate Production from Glucose and Coupling to Mitochondrial Metabolism in Mammals. Cell. 2018;175(2):502–513.e13. doi: 10.1016/j.cell.2018.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hossain F, et al. Inhibition of Fatty Acid Oxidation Modulates Immunosuppressive Functions of Myeloid-Derived Suppressor Cells and Enhances Cancer Therapies. Cancer Immunol Res. 2015;3(11):1236–1247. doi: 10.1158/2326-6066.CIR-15-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zou C, Wang Y, Shen Z. 2-NBDG as a fluorescent indicator for direct glucose uptake measurement. J Biochem Biophys Methods. 2005;64(3):207–215. doi: 10.1016/j.jbbm.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 59.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.