Abstract
Distinct subsets of Tregs reside in nonlymphoid tissues where they mediate unique functions. To interrogate the biology of tissue Tregs in human health and disease, we phenotypically and functionally compared healthy skin Tregs with those in peripheral blood, inflamed psoriatic skin, and metastatic melanoma. The mitochondrial enzyme, arginase 2 (ARG2), was preferentially expressed in Tregs in healthy skin, increased in Tregs in metastatic melanoma, and reduced in Tregs from psoriatic skin. ARG2 enhanced Treg suppressive capacity in vitro and conferred a selective advantage for accumulation in inflamed tissues in vivo. CRISPR-mediated deletion of this gene in primary human Tregs was sufficient to skew away from a tissue Treg transcriptional signature. Notably, the inhibition of ARG2 increased mTOR signaling, whereas the overexpression of this enzyme suppressed it. Taken together, our results suggest that Tregs express ARG2 in human tissues to both regulate inflammation and enhance their metabolic fitness.
Keywords: Immunology, Metabolism
Keywords: Adaptive immunity
Regulatory T cells in human tissues express Arg2 to enhance both their suppressive capacity and metabolic fitness.
Introduction
Tregs play an indispensable role in establishing and maintaining immune tolerance. Over the life of an individual, subsets of these cells accumulate in nonlymphoid organs, with a predilection to stably reside in barrier tissues, such as skin, lung, and the gastrointestinal tract (1–4). In addition, populations of Tregs can be found in visceral adipose tissue, skeletal muscle, and heart (5–7). A growing body of research suggests that tissue Tregs have unique functions that are largely dependent on the specific organs in which they reside. Treg production of amphiregulin within skeletal muscle, lung, and brain facilitates normal tissue regeneration after injury (6–8), whereas adipose tissue Tregs are crucial in attenuating inflammatory processes in fat and maintaining insulin sensitivity (5, 9). IgA selection is specifically regulated by a subset of Tregs in the colon (10), whereas skin Tregs facilitate full-thickness wound healing, epidermal repair, and regulate hair follicle cycling (11–13).
Implicit in the observation that Tregs have different functions in different tissues is the idea that these cells take cues from and adapt to their local environment. Perhaps the cellular process that is most influenced by the local tissue environment is metabolism. Oxygen and nutrient availability vary greatly between healthy tissues and change profoundly during disease. Tregs in the gastrointestinal tract are influenced by specific bacterial-derived metabolites (14–17), whereas prolyl hydroxylase protein expression in the lungs induces Tregs in response to changing oxygen tension, a process exacerbated by lung cancer metastasis (18). Adipose tissue Tregs both sense and metabolize lipids through a distinct transcriptional program, including LDLR, Dgat, and Pgat1 (9). Thus, tissue Tregs respond to signals from their local environment and acquire specific metabolic programs to both survive and optimally mediate their functions in the face of dynamically changing nutrient availability and oxygen tension.
One major sensor integrating multiple environmental cues is mTOR, a key regulator of cell growth and driver of metabolic activity (19). Whereas in many immunological contexts, mTOR promotes proliferation and differentiation, Tregs use mTOR signaling, mediated by 2 separate complexes (mTORC1 and mTORC2), as a rheostat to modulate and balance growth and suppressive function (20). Signaling mediated by mTORC1 inhibits Treg expansion and differentiation, as the prototypical mTORC1 inhibitor rapamycin restricts these processes in vitro and in vivo (21–23). However, absolute loss of mTORC1 in Tregs leads to widespread autoimmunity, whereas mTORC2 signaling appears to be dispensable for Treg function (24, 25). Thus, Tregs residing in peripheral tissues must dynamically regulate signaling through the mTORC1 pathway, requiring some degree of mTORC1 signaling for survival but reducing excessive signaling to allow for proliferation (20). Indeed, tissue Treg expression of cMaf has recently been described as a means by which Tregs residing in the colon intrinsically suppress mTOR activity (26).
Our group and others have observed that Tregs in lesional skin of psoriasis patients are dysfunctional, in that they are unable to effectively control inflammation despite an expansion in numbers (27–30). Here, we comprehensively interrogate these cells and identify a candidate enzyme, arginase 2 (ARG2), that is specifically and preferentially expressed by Tregs in healthy skin and reduced in these cells in psoriatic skin. Whereas ARG2 is not expressed in peripheral blood Tregs, its expression can be driven by T cell receptor (TCR) stimulation. We have found that ARG2 increases Treg suppressive capacity and accumulation in tissues and increases the intrinsic capacity of Tregs for suppression of mTOR signaling. Expression of ARG2 is a mechanism by which Tregs have adapted to optimally function in tissues, and this pathway is altered in human autoimmunity and cancer.
Results
Tregs in human skin preferentially express ARG2.
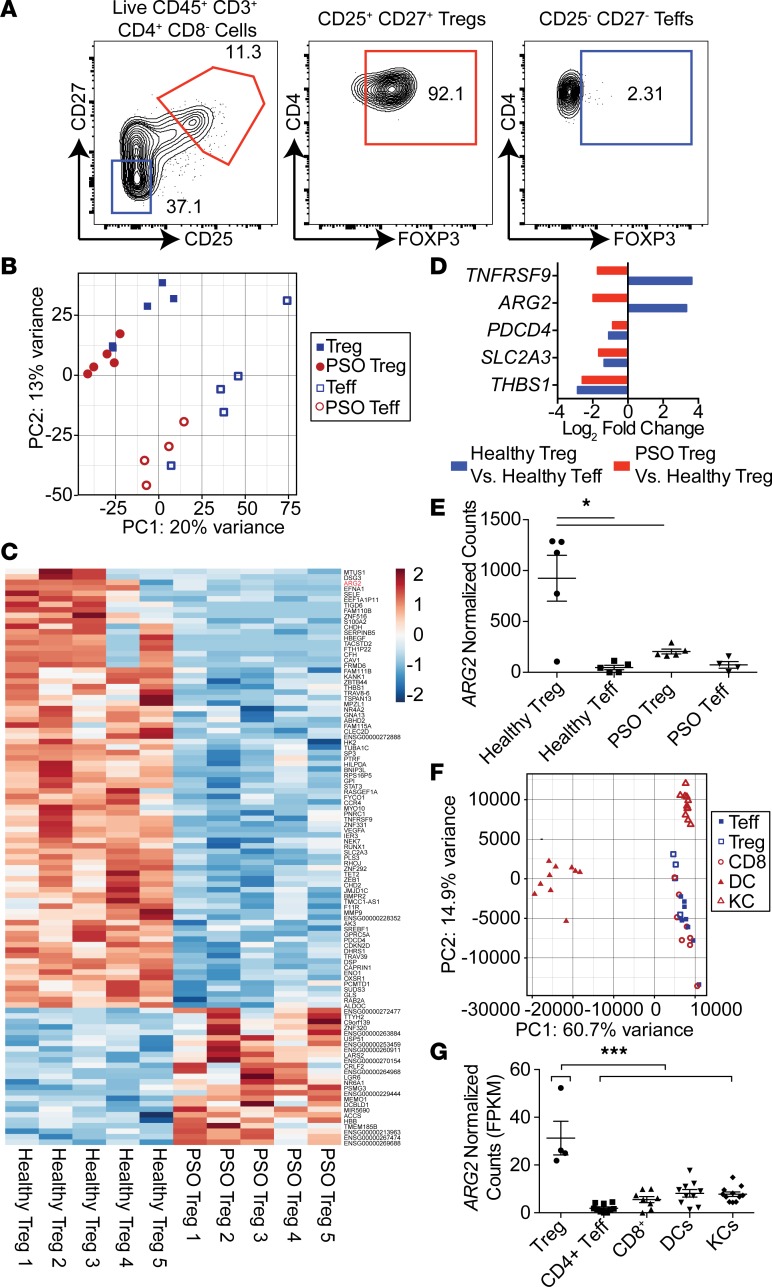
Our group and others have shown that Tregs are dysfunctional in psoriatic skin, despite being increased in numbers (27, 28, 30). To identify pathways that are altered in Tregs in psoriatic human skin, we sort-purified Tregs and CD4+ effector T cells (Teffs) from normal human skin from 5 healthy donors and from inflamed skin from 5 psoriatic patients and performed whole-transcriptome RNA-Seq analysis (Figure 1A, Supplemental Figure 1, and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.129756DS1). As validation of our sort-purification strategy, we found that skin Tregs highly expressed “core” Treg signature genes (31, 32), including FOXP3, CTLA4, IL2RA (CD25), and IKZF2 (Helios) (Supplemental Table 2). Principal component analysis (PCA) revealed that psoriatic Tregs more closely resembled healthy Tregs when compared with psoriatic and healthy Teffs; however, psoriatic Tregs could be clearly distinguished from healthy Tregs (Figure 1B), with 105 genes differentially expressed between these cells (Figure 1C). To identify genes most important in healthy tissue Treg function, we asked which of these genes were highly and differentially expressed between healthy tissue Tregs and psoriatic Tregs (Figure 1D). Of this subset, 2 genes, TNFRSF9 and ARG2, were present in healthy tissue Tregs and lost in psoriatic Tregs. Whereas TNFRSF9 (also known as CD137 or 4-1BB) has been previously characterized as a molecule marking stable, highly suppressive Tregs (33, 34), ARG2 has not been reported to be expressed by Tregs or play a role in Treg biology.
Figure 1. Arginase 2 is highly expressed in Tregs in healthy human skin and reduced in Tregs in psoriatic skin.
(A) Flow cytometric plot depicting FOXP3 expression in human skin CD25+CD27+ sort-purified Tregs (1) and CD25–CD27– sort-purified Teffs (2). (B) Principal component analysis (PCA) of RNA-Seq data from sort-purified Tregs and CD4+ effector T cells (Teffs) from 5 healthy skin samples and 5 psoriasis (PSO) skin lesions. (C) Heat map of significantly different gene counts (normalized by row) comparing healthy human skin Tregs with Tregs from PSO lesions (P < 0.05, Wald test). (D) Log2 fold change of genes differentially expressed in both healthy Tregs, compared with healthy skin Teffs, and PSO skin Tregs, compared with healthy skin Tregs (P < 0.05, minimum Treg expression > 200 normalized counts). (E) Normalized RNA-Seq counts of samples depicted in A (*P < 0.05, Wald test). (F) PCA analysis of RNA-Seq data from Tregs, CD4+ Teffs, CD8+ T cells, dendritic cells (DC), and keratinocytes (KC) isolated from healthy human skin (10 donors). (G) Normalized RNA-Seq counts of cells depicted in F (***P < 0.001, ordinary 1-way ANOVA, Dunnett’s multiple comparisons test).
ARG2 is a mitochondrial enzyme homologous to arginase 1 (ARG1), which is expressed in the cytoplasm (35). Both enzymes metabolize the free arginine required by proliferating cells, and expression of ARG1 by myeloid cells is thought to be a major mechanism by which these cells control local inflammation (35). More recently, ARG1 has been shown to intrinsically regulate innate lymphoid cell metabolic processes (36). ARG2 has been shown to play an immunoregulatory role in fetal dendritic cells, CD71+ erythroid cells, and CD8 T cells (37–39).
We hypothesized that the ARG2 pathway is a means by which Tregs regulate inflammation in tissues. In support of this hypothesis, we found that Tregs in human skin express multiple arginine transporters (40) and thus should be fully capable of taking up and metabolizing extracellular arginine through ARG2 (Supplemental Figure 2). When comparing healthy Tregs with psoriatic Tregs, healthy Tregs expressed almost 4-fold more ARG2, whereas Teffs in healthy and psoriatic skin expressed minimal levels of this transcript (Figure 1E). To determine if other cell subsets express ARG2 in normal human skin, we recruited a cohort of 10 healthy volunteers and performed whole-transcriptome RNA-Seq on Tregs, CD4+ Teffs, CD8+ T cells, dendritic cells, and keratinocytes sort purified from skin (Supplemental Figures 3 and 4). PCA revealed tight cell, lineage-specific segregation (Figure 1F). Interestingly, Tregs were the only cell subset that expressed appreciable levels of ARG2 in healthy human skin (Figure 1G). This enzyme was not expressed in Tregs in murine skin or murine lymphoid organs at the RNA or protein level (Supplemental Figure 5) and thus appears to be specific for human Tregs.
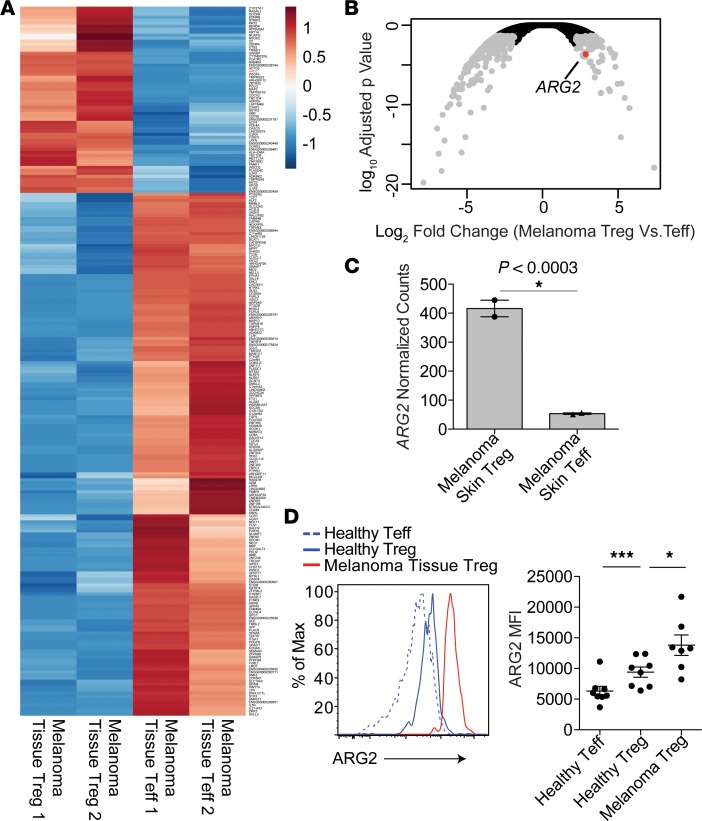
Multiple groups have reported that Tregs infiltrating tumors are highly suppressive (41–43). Thus, we hypothesized that Tregs infiltrating skin tumors would express high levels of ARG2. To test this, we performed whole-transcriptome RNA-Seq of Tregs isolated from human melanoma tumors that were metastatic to skin (Figure 2A). When compared with tumor-infiltrating CD4+ Teffs, Tregs expressed high levels of ARG2 (Figure 2, B and C). To validate these results at the protein level and to directly compare expression of ARG2 between Tregs in healthy skin and metastatic melanoma, we quantified ARG2 protein expression by flow cytometry in Tregs purified from metastatic tumors isolated from 8 separate melanoma patients. Consistent with our RNA-Seq data, Tregs in metastatic melanoma lesions express high levels of ARG2 protein, and these levels were higher in tumor-infiltrating Tregs compared with Tregs isolated from healthy skin (Figure 2D). Taken together, these results suggest that ARG2 is expressed in Tregs in healthy human skin and that expression of this enzyme is reduced in chronically inflamed psoriatic skin and increased in Tregs infiltrating skin tumors.
Figure 2. Tregs infiltrating human metastatic melanoma express high levels of arginase 2.
(A) Heat map of significantly different gene counts (normalized by row) comparing cutaneous melanoma tissue Tregs with cutaneous melanoma tissue Teffs (P < 0.05, Wald test). (B) Volcano plot of statistical significance (Wald test) vs. fold-change comparing skin Treg RNA-Seq profiles from cutaneous melanoma tissue Tregs with cutaneous melanoma tissue Teffs. ARG2 is indicated in red. (C) Normalized RNA-Seq counts of samples depicted in A. (*P < 0.0003, Wald test). (D) Flow cytometric quantification of ARG2 protein expression in Tregs isolated from healthy skin or cutaneous metastatic melanoma lesions from 7 to 8 independent donors (*P < 0.05, unpaired t test; ***P < 0.001, paired t test).
Tregs use the ARG2 pathway to suppress Teff proliferation.
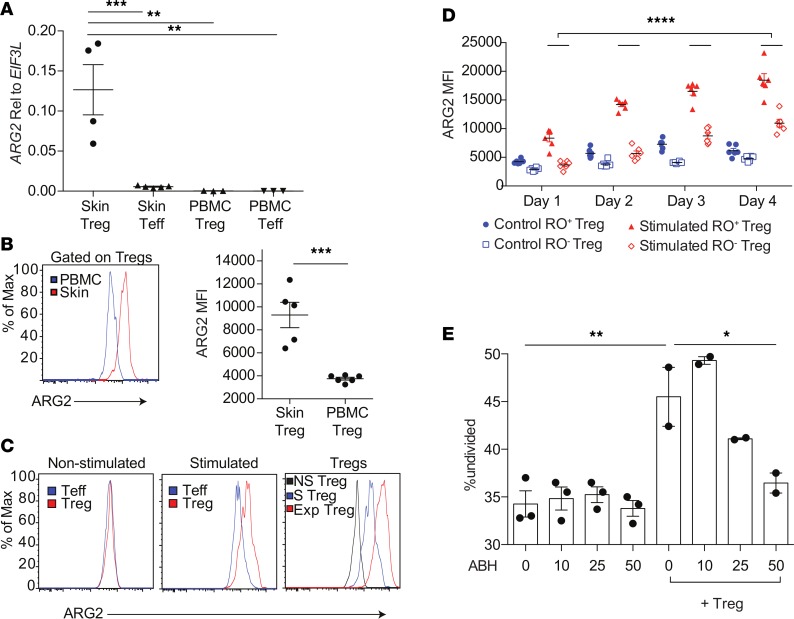
To determine if ARG2 is expressed in Tregs in human peripheral blood, we first quantified expression of this enzyme in Tregs and Teffs isolated from peripheral blood and skin by qRT-qPCR. Relative to Tregs isolated from skin, peripheral blood Tregs and Teffs expressed very little ARG2 mRNA (Figure 3A). Consistent with these results, flow cytometric quantification of ARG2 protein expression revealed significantly higher levels in skin Tregs compared with peripheral blood Tregs (Figure 3B). We next sought to determine whether ARG2 expression in peripheral blood Tregs could be induced upon activation ex vivo. TCR stimulation with anti-CD3 and costimulation with anti-CD28 antibodies preferentially induced ARG2 expression in peripheral blood Tregs compared with peripheral blood CD4+ Teffs (Figure 3C). In addition, ARG2 expression on sort-purified peripheral blood Tregs increased with time after activation, as Tregs activated for 7 days had higher levels of expression compared with Tregs stimulated for 4 days (Figure 3C). Tregs in peripheral blood that express CD45RO have an activated “effector” phenotype, with more potent immunosuppressive capacity (44). Furthermore, almost all Tregs in healthy human skin express CD45RO and have an effector memory phenotype (28). Thus, we asked whether CD45RO+ effector Tregs and CD45RO–“resting” Tregs differentially express ARG2 upon activation ex vivo. Interestingly, CD45RO+ effector Tregs in human peripheral blood preferentially increased ARG2 with time after activation (Figure 3D). Taken together, these results suggest that ARG2 is selectively expressed by Tregs in human skin and can be preferentially induced in effector Tregs in human peripheral blood.
Figure 3. Arginase 2 is preferentially induced on “effector memory” Tregs in human peripheral blood and plays a role in Treg-mediated suppression.
(A) qRT-PCR of sort-purified healthy human skin Tregs, human skin CD4+ Teffs, healthy peripheral blood Tregs, and healthy peripheral blood CD4+ Teffs, quantifying ARG2 relative to housekeeping gene EIF3L (**P < 0.01, ***P < 0.005, ordinary 1-way ANOVA). (B) Flow cytometric quantification of ARG2 protein expression on peripheral blood Tregs compared with Tregs in healthy human skin (***P < 0.004, Student’s t test). (C) Representative flow cytometric plots of ARG2 protein expression on peripheral blood Tregs and CD4+ Teffs that were not stimulated (left) or stimulated for 4 days with soluble anti-CD3 and anti-CD28 in the presence of IL-2 (middle) or Tregs expanded over 7 days in vitro (Exp) compared with nonstimulated (NS) Tregs and Tregs after 4 days of anti-CD3/anti-CD28 stimulation (S) (right). Data are representative of at least 3 independent experiments. (D) Flow cytometric quantification of ARG2 protein expression in peripheral blood CD45RO+ “effector memory” or CD45RO– “resting” Tregs from 6 donors over 1, 2, 3, or 4 days of stimulation (****P < 0.001, 2-way repeated measures ANOVA). (E) In vitro Treg suppression assay showing the percentage of CFSE-labeled CD4+ Teffs remaining undivided after 3 days of stimulation with anti-CD3/anti-CD28, with or without preactivated peripheral blood Tregs (1:4 ratio) and/or increasing concentrations of the arginase inhibitor (ABH). Experiment performed with 3 replicates and is representative of 2 independent experiments (*P < 0.05, **P < 0.01, ordinary 1-way ANOVA).
We set out to determine if Tregs use ARG2 to regulate the proliferation of Teffs via degradation of extracellular arginine. Peripheral blood mononuclear cells (PBMCs) from healthy volunteers were stimulated to induce ARG2 expression, and CD45RO+Tregs were sort purified and used in standard in vitro Treg suppression assays, with or without increasing concentrations of 2 different pan-arginase inhibitors. As previously reported (39), Teffs from peripheral blood do not express appreciable levels of ARG1 or ARG2 (Supplemental Figure 6). In addition, Tregs from peripheral blood do not express appreciable levels of ARG1 in the steady state or after activation (Supplemental Figure 6). Thus, the only significant cell source of arginase that is expressed in these assays was Treg expression of ARG2. Consistent with this, arginase inhibition had no effect on Teff proliferation in the absence of Tregs (Figure 3E). In contrast, Tregs effectively suppressed Teff proliferation and this suppression was attenuated in a dose-dependent fashion in the presence of either the arginase inhibitor, 2(S)-amino-6-boronohexanoic acid (ABH; Figure 3E), or the arginase inhibitor, S-(2-boronoethyl)-L-cysteine (BEC; Supplemental Figures 7 and 8). The effect of arginase inhibition was dependent upon arginine concentrations in the media, as arginase inhibition of Treg-mediated suppression was ameliorated when these experiments were carried out in arginine low media (Supplemental Figure 8). These results suggest that Tregs can use ARG2 to metabolize extracellular arginine and suppress Teff proliferation.
ARG2 expression facilitates Treg accumulation in tissues.
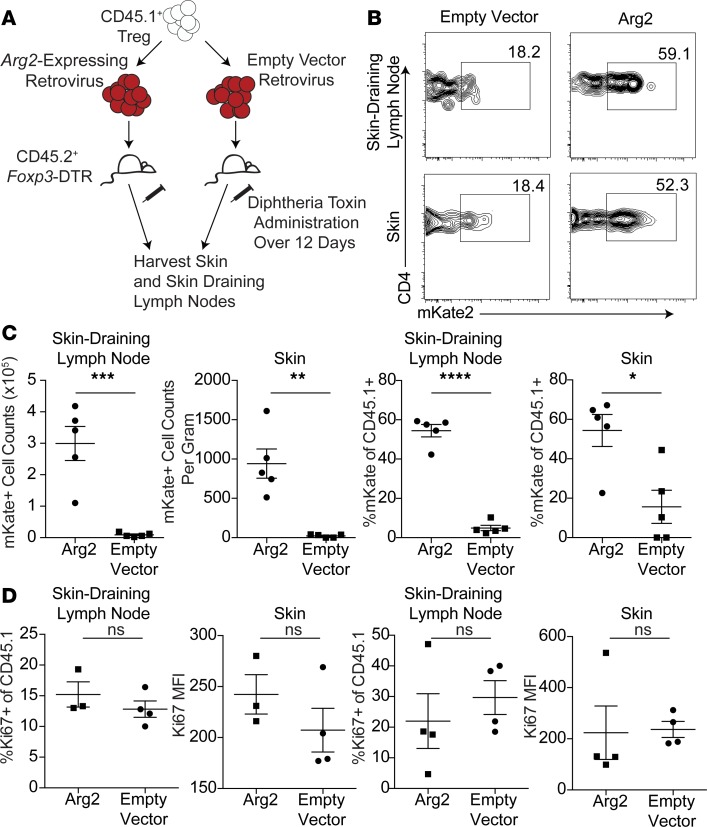
Given that ARG2 expression is relatively absent in Tregs in peripheral blood and highly expressed in Tregs residing in skin (Figure 3), we next queried whether the arginase metabolic pathway could play a role in establishing and maintaining Tregs in tissues. To test this, we expressed Arg2 in murine Tregs and adoptively transferred these cells to mice in which endogenous Tregs had been acutely ablated, providing a niche for transferred cells (45). ARG2-containing retrovirus or empty vector control retrovirus was transduced into ex vivo expanded, congenically labeled, murine Tregs. Cells were then transferred into Foxp3–diphtheria toxin receptor (Foxp3-DTR) hosts (46) that were given diphtheria toxin (DT) to deplete endogenous Tregs (Figure 4A). At all time points examined, we observed that Arg2-expressing Tregs accumulated to a greater degree in skin and skin-draining lymph nodes when compared with empty vector–transduced controls (Figure 4, B and C). There were no significant differences in Treg proliferation, as measured by Ki67 expression (Figure 4D). These results suggest that Arg2 expression provides Tregs with a selective advantage to accumulate and persist in tissues in vivo without markedly influencing the proliferative capacity of these cells.
Figure 4. Treg expression of arginase 2 maintains Tregs in tissues.
(A) Schematic of adoptive transfer of Arg2 overexpressing Tregs or control Tregs into FOXP3-DTR mice. (B) Representative flow plot of viral expression (mKate+) gate in skin and skin-draining lymph node cells isolated after adoptive transfer and host Treg depletion (gated on transferred CD45.1+ Tregs). (C) Cell counts and percentage of skin and skin-draining lymph node transferred Tregs expressing control vector or Arg2 vector, normalized to input (gated on transferred CD45.1+ events). Experiment is representative of 3 independent studies (*P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001, unpaired t test). (D) %Ki67+ and Ki67 MFI of CD45.1+ transferred events expressing control vector or Arg2 vector in the skin-draining lymph node and skin. Experiment is representative of 3 independent studies.
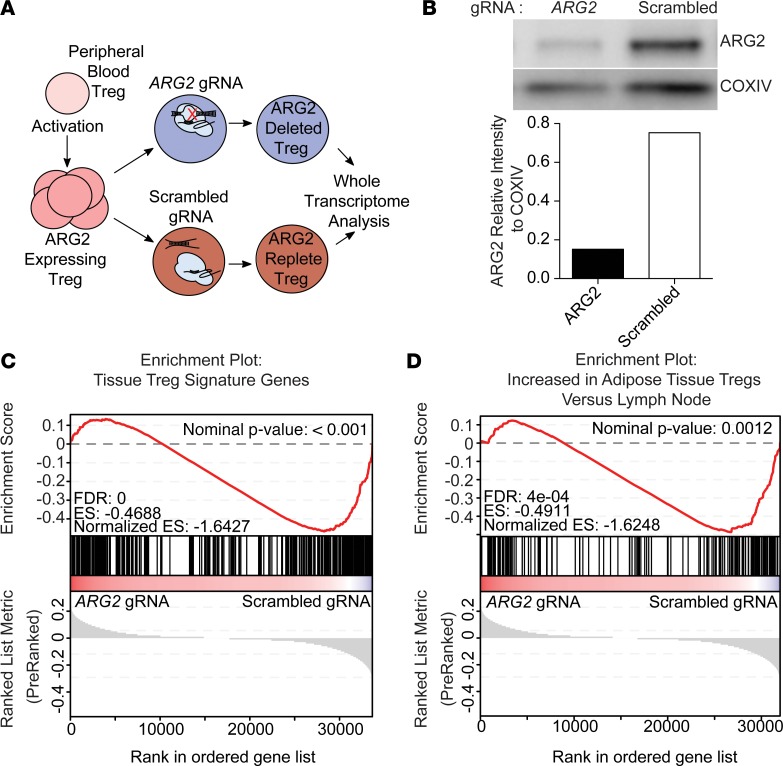
ARG2 expression is required to maintain a healthy tissue Treg signature.
In addition to the well-defined role of arginine metabolism in immune regulation (reviewed in ref. 35), expression of ARG1 and ARG2 can influence metabolism and other cellular processes in a cell-intrinsic fashion (36, 39). Given that ARG2-expressing Tregs had a selective advantage to accumulate in skin, we asked how ARG2 expression could intrinsically affect Treg transcriptional programming. To this end, we engineered an approach to delete ARG2 in primary human Tregs using CRISPR/Cas9 ribonucleoprotein (RNP) technology (47). Tregs were purified from peripheral blood of healthy human donors and activated in vitro to induce ARG2 expression. After activation/expansion, we treated these cells with recombinant Cas9 protein complexed with either a pool of guide RNAs specific for the first exon of ARG2 or a control scrambled guide RNA. After 3 more days of expansion, we isolated total RNA and performed whole-transcriptome RNA-Seq to interrogate the transcriptional signature of ARG2-edited human Tregs (Figure 5A). Tracking Indels by DEcomposition (TIDE) analysis (48) of the targeted ARG2 genome region after CRISPR-mediated editing revealed site-specific DNA disruption (Supplemental Figure 9). Western blot analysis revealed ARG2 protein expression to be substantially reduced in ARG2-edited Tregs compared with controls; however, ARG2 protein was still detectable in these cells (Figure 5B). In an attempt to elucidate how ARG2 expression influences the Treg transcriptome, we compared Tregs targeted with ARG2 Cas9 RNPs with those treated with control scrambled guide RNPs. Fold changes in gene expression were compared with known Treg gene sets via gene set enrichment analysis (49, 50). In addition to these publicly available gene sets, we developed a human tissue Treg gene signature using our comparison of Tregs to Teffs isolated from healthy human skin and a published gene set defining the transcriptome of peripheral blood effector memory Tregs (Supplemental Figure 10, Supplemental Table 3, and ref. 51). Strikingly, ARG2-edited Tregs had significantly (P < 0.001) reduced expression of the “healthy tissue” Treg gene set when compared with control Tregs (Figure 5C). Furthermore, ARG2-targeted Tregs had significantly reduced expression of the gene set preferentially expressed by adipose tissue-resident Tregs (Figure 5D and ref. 9). Taken together, these results suggest that Treg expression of ARG2 helps to maintain a transcriptional signature associated with Tregs found in peripheral tissues.
Figure 5. Arginase 2 expression confers a healthy tissue Treg signature.
(A) Schematic of CRISPR-mediated ARG2 editing in Tregs expanded from human peripheral blood. (B) Western blot of expanded Tregs electroporated with CRISPR/Cas9 complex with either an ARG2-targeting guide RNA pool (left) or a scrambled guide RNA (right). ARG2 was quantified relative to the mitochondrial housekeeping protein COXIV. (C) Gene set enrichment analysis (GSEA) showing enrichment of human tissue Treg-specific genes within the ranked gene expression of control Tregs vs. ARG2-edited Tregs (n = 3). (D) GSEA showing enrichment of genes upregulated within adipose tissue Tregs vs. lymph node Tregs within the ranked gene expression of control Tregs vs. ARG2-edited Tregs (n = 3).
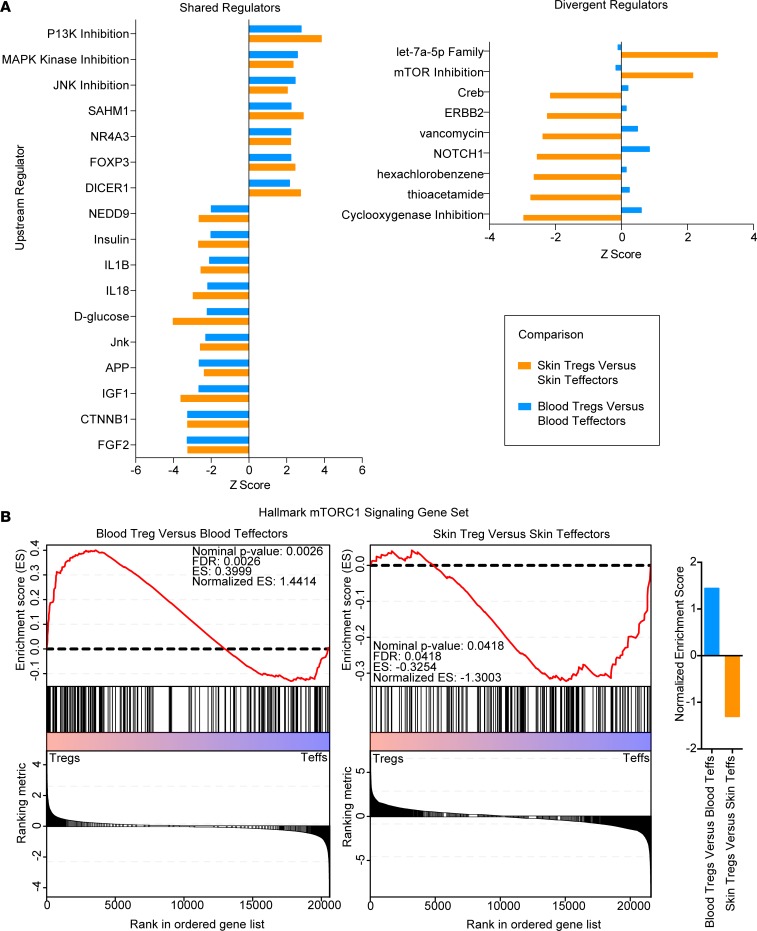
ARG2 attenuates mTOR activity in Tregs.
To identify candidate pathways that ARG2 might modulate in human tissue Tregs, we compared our RNA-Seq data with publically available data sets (52) of Tregs and Teffs purified from peripheral blood. Given that Tregs resident in healthy human skin have an “effector memory” phenotype (28), we used data from CD45RO+ peripheral blood Tregs as an ideal comparator population to skin Tregs. We used Ingenuity Pathway Analysis (IPA) to compare the transcriptome of blood Tregs versus blood Teffs with skin Tregs versus skin Teffs. Whereas many upstream regulators, including FOXP3, were shared between blood Tregs and skin Tregs (Figure 6A, left), others were differentially activated between these comparator groups (Figure 6A, right). Whereas broad inhibition of PI3 kinases was observed in both blood and tissue Tregs, an inhibition of mTORC1 activity was observed in skin Tregs but not in blood Tregs (Figure 6B). We confirmed this differential regulation in our transcriptomic data by performing gene set enrichment analysis (GSEA) using the Broad Institute data set of hallmark mTORC1 signaling genes. This analysis revealed mTORC1 signaling to be significantly enriched in blood Tregs and reduced in skin Tregs compared with Teffs (Figure 6B).
Figure 6. Tregs in human skin have a reduced mTOR signature.
(A) IPA displaying shared (left) and divergent (right) upstream regulators for blood Tregs vs. blood Teffs and skin Tregs vs. skin Teffs. (B) Gene set enrichment analysis for blood Tregs vs. blood Teffs (left) and skin Tregs vs. skin Teffs (right) reveals that blood Tregs have enriched mTORC1 signaling genes, whereas skin Tregs have reduced mTORC1 signaling.
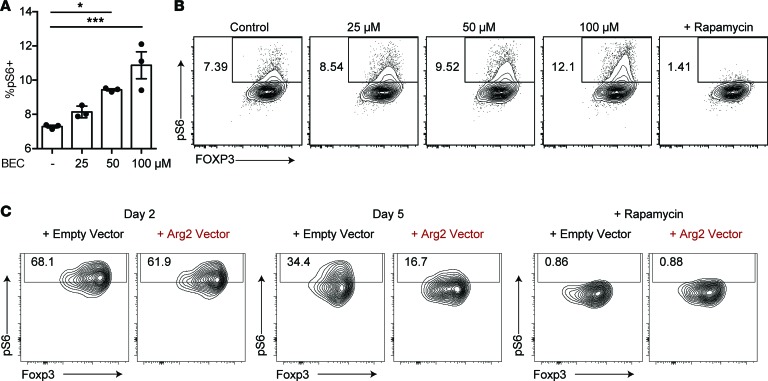
Intracellular arginine availability can be sensed by the mTOR complex, activating mTORC1 (53). Thus, we hypothesized that metabolism of arginine by ARG2 is a mechanism by which Tregs in tissues attenuate mTOR activity. To test this, we sort-purified Tregs from peripheral blood of healthy human donors, expanded these cells ex vivo, and treated them with increasing concentrations of the arginase inhibitor BEC. mTOR activation was quantified by flow cytometric measurement of S6 phosphorylation (phospho-S6), a well-accepted and robust indicator of mTOR signaling in leukocytes (54). Consistent with our hypothesis, the inhibition of ARG2 in Tregs during TCR-mediated activation resulted in a dose-dependent increase in phospho-S6 (Figure 7, A and B). To validate these findings, we took the opposite approach and overexpressed this enzyme in murine Tregs (that do not normally express Arg2; Supplemental Figure 5). When compared with control Tregs retrovirally transduced with empty vector, Tregs overexpressing Arg2 showed enhanced suppression of mTOR activity with time after Treg activation (Figure 7C). Taken together, these results indicate that ARG2 expression in Tregs plays a role in attenuating mTOR activity.
Figure 7. Treg expression of arginase 2 suppresses mTOR signaling.
(A and B) Flow cytometric analysis of pS6 expression on human Tregs expanded in vitro with increasing doses of the arginase inhibitor S-(2-boronoethyl)-L-cysteine (BEC). Rapamycin was used as a control to inhibit mTOR signaling (*P < 0.05, ***P < 0.005, ordinary 1-way ANOVA, data are representative of 3 independent experiments). (C) Flow cytometric analysis of pS6 expression on mouse Tregs expanded in vitro and transduced with a retrovirus expressing either Arg2 or an empty vector control. Data were collected on day 2 or day 5 after transduction. Rapamycin was used as a control to inhibit mTOR signaling. Data are representative of 2 independent experiments.
Discussion
Arginase metabolism has been characterized as a method of innate immune regulation by limiting the availability of this essential amino acid in the local tissue microenvironment (35). ARG2 has more recently been implicated in influencing immune cell function in a cell-intrinsic fashion, including CD71+ erythroid cells, fetal dendritic cells, and CD8 T cells (37–39). However, the molecular mechanisms underpinning how this enzyme influences immune cell function are poorly understood. Here, we demonstrate that Tregs resident in human skin preferentially express ARG2. Consistent with the role of this pathway in immune regulation, we found that ARG2 expression in Tregs attenuates Teff proliferation. In addition, we found that ARG2 plays a major role in maintaining a healthy tissue Treg identity and facilitates the accumulation of these cells in skin. A potential mechanism of how this is achieved is through the ability of this enzyme to attenuate mTOR activity, which plays a major role in enhancing many aspects of Treg biology (20, 23).
Immune cells in peripheral tissues have different metabolic demands and constraints than those found in secondary lymphoid organs. Furthermore, Tregs moving to tissues tend to be activated, effector memory-like cells, which can expand and suppress more effectively by dynamically regulating specific metabolic pathways, including mTOR (20, 24). Responses to mTORC1 signaling and to ARG2 expression clearly diverge between Tregs and Teffs. mTORC1 signals are essential for CD4 Teff differentiation and formation of CD8 T cell memory, whereas these signals restrain Treg responses (55). ARG2 in CD8+ T cells reduced their survival and proliferation (55). Our study shows that Treg expression of ARG2 enhances its ability to accumulate in tissues and suppress Teff responses. Given that a subset of tissue Tregs most likely constitutively receives signal through the TCR to a greater degree than both Tregs in the periphery as well as tissue Teffs (56), ARG2 induction should be enhanced within tissue Tregs. Taken together, preferential TCR signaling in tissue Tregs and differential mTORC1 signaling requirements could provide an elegant mechanism to promote Treg metabolic fitness in tissues by upregulating ARG2 and allowing for suppression of Treg-intrinsic mTORC1 signaling. Thus, ARG2 expression may allow Tregs to adapt to local cues to use mechanisms of suppression best suited for a given tissue microenvironment, and these mechanisms might be disrupted in specific disease states. It is interesting to speculate that Tregs that reside in skin have high levels of ARG2 to both regulate Teffs and intrinsically suppress mTORC1 activity, providing both a mechanism of immune regulation and a means to establish and maintain themselves in this tissue.
Understanding the fundamental biology of immune cells resident in human tissues is of critical importance in our attempts to therapeutically manipulate these cells to treat human disease. Clinical augmentation of Tregs is currently being used to treat autoimmunity and inhibition of these cells is being applied to treat cancer (57, 58). Given that ARG2 is reduced within Tregs from inflamed skin, and even more highly expressed in Tregs infiltrating metastatic skin cancer, modulating ARG2 activity within Tregs is therefore a potential therapeutic target for both disease states. The means by which ARG2 becomes further upregulated in melanoma Tregs and reduced in psoriatic Tregs remain to be determined. Environmental hypoxia is known to affect ARG2 expression (59), and thus the differential oxygen tensions between a hypoxic tumor and a highly vascularized psoriatic lesion might result in altered ARG2 induction between disease states. The inflammatory cytokine milieu present within the tissue microenvironment might also alter ARG2 expression in Tregs, as TNF, IL-1B, and LPS can induce ARG2 within other cell types (60, 61). Determining which factors in psoriatic lesions and in melanoma tumors may influence ARG2 expression could provide targets for future therapeutics. Finally, it is interesting to note that peripheral blood arginine concentrations are increased in patients within severe psoriasis and decreased in the multiple cancer types (62, 63). Whereas it is likely that arginine concentrations within tissues are most relevant to tissue Treg homeostasis, arginine concentrations in the periphery could further exacerbate alterations in ARG2-mediated tissue Treg function. Thus, arginine metabolism through ARG2 is an immunoregulatory pathway enforced by Tregs in human tissues with relevance to both autoimmunity and cancer.
Methods
Human skin processing
Human skin from healthy donors resulting from surgical discards previously detailed (28), 6-mm punch biopsies from psoriatic lesions, or resected cutaneous melanoma tumor was minced and digested overnight with collagenase Type IV (0.8 mg/mL, Worthington, catalog LS004186) and DNAse (20 μg/mL, MilliporeSigma, catalog DN251G) in RPMI plus 10% FBS at 37°C. The resulting suspension was filtered (100 μm) and washed and lymphocytes were counted using a NucleoCounter NC-200 (ChemoMetec) or a hemocytometer.
Human peripheral blood processing
Blood from healthy human donors was obtained under IRB protocol (IRB 10-02830) or was purchased from Blood Systems Research Institute. PBMCs were isolated by Ficoll gradient prior to experiments; some PBMCs were cryopreserved before use.
Human Treg and Teff sort purification
Cells obtained from human skin were stained with Ghost 510 viability dye (Tonbo Biosciences), anti–CD45-FITC (HI30, BioLegend), anti–CD3-Alexa Fluor 700 (UCHT1, eBioscience), anti–CD4-PerCP-eFluor 710 (SK3, eBioscience), anti–CD8a-APC (OKT8, eBioscience), anti–CD25-PE-Cy7 (M-A251, BD Biosciences), and anti–CD27-APC-eFluor 780 (LG.7F9, eBioscience). An aliquot of cells were fixed and permeabilized with the eBioscience Foxp3/Transcription Factor Staining Buffer Kit and stained with anti–FOXP3-eFluor 450 (PCH101, eBioscience). Cells were sorted by Aria2 or Aria3u (BD Biosciences) and were gated on live, singlet, CD45+, CD3+, CD4+CD8– events. Tregs were selected as CD25+CD27+ events; gating strategy was verified with FOXP3 stained sample. Maximal Treg purity was targeted. Teffs were selected as CD25–CD27– events and were likewise validated by FOXP3 stained sample. Samples were sorted into RPMI plus 10% FBS with 40 U/mL RiboLock RNase inhibitor (Thermo Fisher Scientific), pelleted at 300 g for 10 minutes, and snap-frozen for RNA extraction.
Experimental animals
FOXP3-DTR mice (The Jackson Laboratory, B6.129(Cg)-Foxp3tm3(DTR/GFP)Ayr/J, catalog 016958) and CD45.1+ mice (The Jackson Laboratory, B6.SJL-Ptprca Pepcb/BoyJ, catalog 002014) were socially housed in specific pathogen–free conditions on a 12-hour light/dark cycle. All animals were used in scientific experiments for the first time.
Mouse skin processing
As described previously (13, 64), entire trunk skin was harvested, lightly defatted, and minced with scissors before resuspending in digestion media composed of 2 mg/ml collagenase XI (MilliporeSigma, catalog C9407), 0.5 mg/ml hyaluronidase (MilliporeSigma, catalog C9407), and 0.1 mg/ml DNase in RPMI with 1% HEPES, 1% penicillin/streptomycin (Gibco), and 10% fetal calf serum. The tissue was shaken at 220 rpm in an incubator (Excella E25, New Brunswick Scientific) at 37°C for 45 minutes. RPMI/HEPES/PS/FCS was added to quench the digestion, and the sample was shaken by hand and filtered in a 100-μm cell strainer followed by a 40-μm cell strainer. The sample was then pelleted and resuspended in PBS for counting and staining.
Mouse Treg and Teff sort purification
Mouse skin Tregs and Teffs were sort purified as described previously (13, 64). Briefly, FOXP3-DTR mice, which express GFP on all FOXP3+ cells, were stained with Ghost 510 viability dye (Tonbo Biosciences), anti–CD45-Alexa Fluor 700 (30-F11, eBioscience), anti–CD3-APC-eFluor 780 (145-2C11, eBioscience), anti–CD4-PE-Cy7 (RM4-5, eBioscience), and anti–CD25-APC (PC61.5, eBioscience). Live, CD3+, CD4+, CD25+, GFP+ cells were isolated as Tregs and live, CD3+, CD4+, CD25–, GFP– cells were isolated as Teffs. Cells were sorted into RPMI plus 10% FBS with 40 U/mL RiboLock RNase inhibitor (Thermo Fisher Scientific), pelleted at 300 g for 10 minutes, and snap-frozen for RNA extraction.
Generation of Arg2 retrovirus
DNA fragments containing Arg2 cDNA and mKate2 fluorescent marker were isolated by PCR from an Arg2 lentiviral vector (Applied Biological Materials Inc., catalog LV473328) and cloned into a pMIG-IRES-GFP backbone via a Gibson cloning strategy, with mKate2 replacing GFP. Plasmids were verified by restriction enzyme digestion and sequencing, and Arg2 and empty vector retrovirus were produced by the UCSF ViraCore.
Mouse Treg culture and infection
Lymph nodes and spleens from CD45.1+ mice were isolated into a single cell suspension. Red blood cells were lysed with ACK lysis buffer at 37°C for 4 minutes. CD4+ cells were enriched with the Mouse CD4 T Cell Isolation Kit (EasySep, catalog 19852) and stained with Ghost 510 viability dye (Tonbo Biosciences), anti–CD45-Alexa Fluor 700 (30-F11, eBioscience), anti–CD8-BV785 (53-6.7, BioLegend), anti–CD3-APC-eFluor 780 (145-2C11, eBioscience), anti–CD4-PerCP-Cy5.5 (RM4-5, Tonbo Biosciences), and anti–CD25-PE (PC61, BD Biosciences). Tregs were isolated as live, CD45+, CD3+, CD8–, CD4+, CD25hi. Tregs were cultured with a 3:1 ratio of the Dynabeads Mouse CD3/CD28 (Thermo Fisher Scientific, catalog 11456D) with 2000 IU/mL IL2 (UCSF Pharmacy) for 2 days prior to spinfection. Cells were counted and spinfected in 1.5-mL tubes (500,000 cells/tube) at 25°C for 90 minutes at 6000 g, with either Arg2-mKate2 or empty vector retrovirus (~1 × 106 particles/mL). Cells were plated with 2000 IU/mL and expanded for 3 more days prior to i.v. injection into FOXP3-DTR mice. Percentage of infection was assessed via flow cytometry on day 2–3 after infection.
Mouse Treg adoptive transfer and depletion
Expanded and infected Tregs were counted and approximately 150–200,000 cells were injected via tail vein into FOXP3-DTR mice. Equivalent numbers of cells were injected for Arg2 and control retrovirus groups for each experimental replicate. Three days after adoptive transfer, mice were intraperitoneally injected with diphtheria toxin (40 μg/kg) every other day for 12 days. Weight and activity levels were monitored every other day. On day 12, skin-draining lymph nodes and skin were harvested and counted. Cells were stained with Ghost 510 viability dye (Tonbo Biosciences), anti–CD45.1-Alexa Fluor 700 (A20, BioLegend), anti–CD8-BV785 (53-6.7, BioLegend), anti–CD3-APC-eFluor 780 (145-2C11, eBioscience), anti–CD4-PerCP-Cy5.5 (RM4-5, Tonbo Biosciences), and anti–CD25-BV650 (PC61, BioLegend).
RNA-Seq analysis of Tregs and Teffs
RNA isolation and sequencing were performed by expression analysis. Isolation was performed with QIAGEN RNeasy Spin Columns. RNA was quantified via a NanoDrop ND-8000 spectrophotometer (Thermo Fisher Scientific), and quality was checked by Agilent Bioanalyzer Pico Chip. One psoriatic Teff sample was excluded from sequencing for low RNA concentration. For primary cell analysis, cDNA was created from 220 pg of input RNA with the SMARTer Ultra Low Input Kit (50 bp, paired-end, unstranded reads) and was sequenced to a 25 M read depth. For CRISPR-electroporated Tregs, cDNA was created from 192 ng of input RNA with the Illumina TruSeq Stranded mRNA Preparation Kit (50 bp, paired-end, stranded reads) and was sequenced to a 25 M read depth. Reads were aligned to Ensembl hg19
GRCh37.75 reference genome with TopHat software (v. 2.0.12) (65). SAM files were generated with SAMtools from alignment results (66). Read counts were obtained with HTSeq-count (0.6.1p1) with the union option; for primary cells, the –stranded=no option was used, whereas for CRISPR-electroporated Tregs, the –stranded=reverse option was used (67). Differential expression was determined using the R/Bioconductor package DESeq2 (68).
RNA-Seq analysis comparing keratinocytes, dendritic cells, CD8+ T cells, CD4+ Teffs, and Tregs from healthy skin
Cells were isolated and analyzed as described in Ahn et al. (69), with the addition of sequencing data from Tregs isolated concurrently as described above. Expression of ARG2 in the combined data set was analyzed by ANOVA.
Flow cytometry analysis of ARG2
Cells were stained with Ghost 510 viability dye (Tonbo Biosciences), anti–CD45-FITC (HI30, BioLegend), anti–CD3-Alexa Fluor 700 (UCHT1, eBioscience), anti–CD4-PerCP-eFluor 710 (SK3, eBioscience), and anti–CD8a-APC (OKT8, eBioscience) and then fixed and permeabilized with the eBioscience Foxp3/Transcription Factor Staining Buffer Kit. Before intracellular staining, cells were blocked with 10% goat serum. Cells were stained intracellularly with anti–FOXP3-eFluor 450 (PCH101, eBioscience) and an unconjugated anti-ARG2 (ab137069, Abcam) rabbit monoclonal followed with an anti–rabbit-PE secondary antibody (F(ab′)2-Donkey anti–Rabbit IgG Secondary Antibody, eBioscience). Events were collected on an LSRFortessa (BD Biosciences) flow cytometer, which was normalized between experiments using Sphero Ultra Rainbow Calibration particles. Samples were analyzed in FlowJo (v. 9.9.3).
RT-PCR
RNA was isolated with a PureLink RNA Minikit (Thermo Fisher Scientific). cDNA was transcribed with the iScript Advanced cDNA Synthesis Kit (Bio-Rad). Preamplification was performed for 10 cycles with the SSO Advanced PreAmp Supermix (Bio-Rad). ARG2 was measured with primers 5′-GAAGAAATCCGTCCACTCCGT-3′, 5′-TTAGGTGGCAGCCCAAACT-3′; ARG1 was measured with primers 5′-TGGACCCTGGGGAACACTAC-3′, 5′-CGAGCAAGTCCGAAACAAGC-3′, developed by NCBI Primer-BLAST (70); and EIF3L was measured with primers 5′-TGACCCCTACGCTTATCCCAG-3′, 5′-GTTTGCTGTTCATACTGACGTTC-3′ (PrimerBank ID 339275830c1) (71), with SsoAdvanced Universal SYBR Green master mix (Bio-Rad) on a StepOnePlus PCR System (Applied Biosystems).
Induction of ARG2 and Treg suppression assay
Cryopreserved PBMCs were thawed and rested before isolation of CD4 T cells using the EasySep Human CD4+ T Cell Enrichment Kit. Cells were stimulated with CD3 (clone HIT3a) (BD Pharmingen) and CD28 (clone CD28.2) (BD Pharmingen) in the presence of 4 × 10–10 M IL2 (Tonbo Biosciences) for 1–4 days and then stained with ARG2 panel as above. In some cases anti–CD45RO-FITC (UCHL1, eBioscience) was used in the place of anti–CD45-FITC. For Treg suppression assays, CD4 T cells isolated via the EasySep Kit were preactivated with anti-CD3 and anti-CD28 for 3 days and then stained with Ghost 510 viability dye (Tonbo Biosciences), anti–CD45RO-FITC (HI30, BioLegend), anti–CD3-Alexa Fluor 700 (UCHT1, eBioscience), anti–CD4-PerCP-eFluor 710 (SK3, eBioscience), anti–CD8a-eVolve 605 (RPA-T8, eBioscience), anti–CD25-PE-Cy7 (M-A251, BD Biosciences), and anti–CD127-PE (HIL-7R-M21, BD Biosciences), and Tregs were sorted as CD45RO+CD25+ CD127lo cells, which was validated by a separate FOXP3-stained control as above. Tregs were labeled with Cell Proliferation Dye eFluor 670 (eBioscience). Responding T cells were similarly stained and sorted as CD25–CD127hiCD4+ T cells and were labeled with carboxyfluorescein succinimidyl ester dye (Tonbo Biosciences). Tregs were added to responding T cells in a 1:5 ratio. Cells were stimulated with Human T Activator anti-CD3/anti-CD28 (Invitrogen) at a ratio of 1 bead/25 T responder cells for 3–4 days. Arginase inhibitors ABH or BEC (MilliporeSigma) were added to select wells. Some experiments were performed in arginine-depleted media (R1780, MilliporeSigma, with 10% FBS added). After proliferation, samples were stained with Ghost 510 viability dye (Tonbo Biosciences), anti–CD3-Alexa Fluor 700 (UCHT1, eBioscience), anti–CD4-PerCP-eFluor 710 (SK3, eBioscience), anti–CD8a-eVolve 605 (RPA-T8, eBioscience), then fixed and permeabilized with the eBioscience Foxp3/Transcription Factor Staining Buffer Kit, and stained intracellularly with anti–FOXP3-eFluor 450 (PCH101, eBioscience). Events were collected on an LSRFortessa (BD Biosciences) and analyzed in FlowJo (v.9.9.3).
Treg expansion
PBMCs were purified using a Ficoll gradient and were stained with Ghost viability dye (Tonbo Biosciences), anti–CD45-FITC (HI30, BioLegend), anti–CD3-Alexa Fluor 700 (UCHT1, eBioscience), anti–CD4-PerCP-eFluor 710 (SK3, eBioscience), anti–CD8a-APC (OKT8, eBioscience), anti–CD25-PE-Cy7 (M-A251, BD Biosciences), and anti–CD127-PE (HIL-7R-M21, BD Biosciences). An aliquot of cells was fixed and permeabilized and stained with anti–FOXP3-eFluor 450 (PCH101, eBioscience). Tregs were sorted as live, CD45+, CD3+, CD4+CD8–, CD25+, CD127lo, with gates drawn to maximize the percentage of FOXP3+ cells in the permeabilized sample within the CD25+ gate. Tregs were plated with Dynabeads Human T-Activator anti-CD3/anti-CD28 (Invitrogen) at a 1:1 ratio in XVIVO media supplemented with 10% FBS, sodium pyruvate, and nonessential amino acids. On day 2, 300 IU/mL human IL2 (UCSF Pharmacy) was added and was supplemented on days 5 and 7.
pS6 staining on human and mouse Tregs
pS6 staining was performed on human Tregs expanded for 7 days as described above with or without BEC in the culture medium. Rapamycin (20 nM) was added for 1 hour prior to stain to serve as a gating control by inhibiting mTOR signaling. Cells were stained extracellularly with Ghost viability dye (Tonbo Biosciences), anti–CD3-Alexa Fluor 700 (UCHT1, eBioscience), and anti–CD8a-BV650 (RPA-T8, BioLegend). Cells were fixed and permeabilized with the BD Biosciences Cytofix Buffer and Perm Buffer III per the kit protocol. Cells were then stained intracellularly with anti–CD4-PerCp-Cy5.5 (RPA-T4, BD Biosciences), anti–FOXP3-Alexa Fluor 647 (259D/C7, BD Biosciences), and anti–pS6-Ser240/244-PE (D68F8, Cell Signaling Technology). Mouse Tregs were stained for pS6 after retroviral infection as described above on day 2 or day 5 after infection. Rapamycin (20 nM) was added 1 hour prior to stain as a gating control. Cells were stained extracellularly with Ghost viability dye (Tonbo Biosciences), anti–CD3-FITC (17A2, BD Biosciences), anti–CD8a-BV785 (53-6.7, BioLegend), anti–CD4-BV650 (RM4-5, BD Biosciences), and anti–CD45-Alexa Fluor 700 (30-F11, eBioscience). Cells were fixed and permeabilized with the BD Biosciences Cytofix Buffer and Perm Buffer III per the kit protocol. Cells were then stained intracellularly with anti–FOXP3-eFluor 450 (FJK-16s, eBioscience), and anti–pS6-Ser240/244-PE (D68F8, Cell Signaling Technology). Events were collected on an LSRFortessa (BD Biosciences).
CRISPR/Cas9 editing of ARG2
Human Tregs on day 7 of expansion were electroporated using the Amaxa P3 Primary Cell Kit and 4D-Nucleofecter (Lonza) as previously described (47). Guide RNAs were prepared with chemically synthesized tracRNA and 3 pooled crRNAs (Dharmacon) targeting exon 1 of ARG2 (5′-ACGCGAGAGGCTGCCCCTTA-3′, 5′-CACCTGCCCTTGTGAGAACG-3′, 5′-GTGGACGGATTTCTTCAGGA-3′). A nontargeting scrambled crRNA (5′-GGTTCTTGACTACCGTAATT-3′) was used for comparator groups. tracRNA and crRNA were resuspended with 10 mM Tris-HCl pH 7.4 to generate 160 μM RNA stocks. The 3 crRNAs and tracrRNA were mixed 0.33:0.33:0.33:1 (or 1:1 for scrambled control) and incubated 30 minutes at 37°C to generate 80 μM crRNA:tracrRNA duplexes. An equal volume of 40 μM S. pyogenes Cas9 (Macrolab, Berkeley) was mixed with the crRNA:tracRNA and incubated for 15 minutes at 37°C to generate 20 μM Cas9-guideRNA RNP complexes. Tregs were resuspended at 10 × 106/mL in P3 buffer, and 20 μL of cells were mixed with 5 μl of 20 μM Cas9 RNP per well in a 96-well electroporation plate. We added 1 μL of a nonhomologous DNA ultramer (5′-TTAGCTCTGTTTACGTCCCAGCGGGCATGAGAGTAACAAGAGGGTGTGGTAATATTACGGTACCGAGCACTATCGATACAATATGTGTCATACGGACACG) resuspended at 100 μM in nuclease-free water to each well to enhance electroporation as previously described (72). Cells were nucleofected using program EH-115 and then rescued by adding 80 μl prewarmed XVIVO and incubating cells for 30 minutes at 37°C. Duplicate wells of the same treatment group were combined after electroporation, and cells were plated in XVIVO media with IL2 (300 IU/mL, UCSF Pharmacy) and Dynabeads Human T-Activator anti-CD3/anti-CD28 (Invitrogen) at a bead/cell ratio of 1:1, assuming 40% loss of cells during electroporation. Three days later cells were harvested for DNA analysis of editing, Western blot analysis, and RNA-Seq analysis.
TIDE analysis
Genomic DNA was isolated from cells 3 days after electroporation using QuickExtract Buffer (EpiCentre) by incubating cells in buffer for 20 minutes at 65°C and 20 minutes at 95°C. DNA was amplified with primers flanking the targeted region (5′-ACGTCCCAGCCTTGCAGA-3′ and 5′-CTCCACTGGGGATTCTGACC-3′) with the Kapa HiFi HotStart ReadyMix Kit (Kapa Biosystems). Sanger sequencing of amplicons (Elim Biopharmaceuticals Inc.) was used for analysis of editing efficiency by sequence trace decomposition quantifying insertions and deletions (indels) using the TIDE algorithm (48).
Western blot
CRISPR experiment.
1 × 106 cells were washed in ice-cold PBS, resuspended in 1% NP-40 lysis buffer, and incubated at 4°C for 30 minutes. After pelleting, the supernatant was resuspended in 5% SDS buffer with 2% DTT, boiled for 5 minutes, and run on a Bis-Tris NuPAGE 4%–12% gradient gel (Thermo Fisher Scientific). After transfer, gel was probed with anti-ARG2 antibody (ab137069, Abcam), which was detected with anti–rabbit-HRP (4050-05, SouthernBiotech), and with anti–COX IV-HRP (ab62164, Abcam) as a loading control. Protein length was estimated with the Precision Plus Protein Dual Color Standards ladder (Bio-Rad). Quantification was performed with Image Lab 5.2.1 (Bio-Rad).
Comparison of human Tregs and mouse Tregs.
We collected 500,000 human and mouse Tregs after expansion for 14 days in vitro. Western blot was performed as described above, except a polyclonal anti-ARG2 antibody (ab81505, Abcam) was used for blotting.
IPA
A core analysis was performed through the use of IPA (QIAGEN Inc.; https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/) on genes significantly altered between healthy skin Tregs and healthy skin Teffs (adjusted P value < 0.05, log2 fold change ±1) and on genes significantly altered between peripheral blood Tregs and peripheral blood Teffs (adjusted P value < 0.05, log2 fold change ±1). Upstream regulators that were identified as shared were significantly (P < 0.05 for upstream regulator analysis) increased or decreased in both skin and blood (Z score ±2). Divergent regulators are depicted, which are significantly increased or decreased (P < 0.05 for upstream regulator analysis) in skin and blood, with a Z score ±2 in skin and a Z score in the opposite direction in blood. Some regulators were assigned biological activity (i.e., siroliumus was assigned as mTORC1 inhibition), and original regulator nomenclature is shown in Supplemental Figure 11.
GSEA
For CRISPR experiments, a preranked gene list ranking relative expression of genes of scrambled guide RNA control Tregs versus that of ARG2-edited Tregs was determined using DESeq2, and GSEAPreranked analysis was performed using the GSEA v2.2.0 tool with default settings (49, 50). For Figure 5C, the gene list tested was curated from genes differentially expressed in healthy human Tregs versus healthy human Teffs that were not increased in peripheral blood Tregs versus peripheral blood Teffs (ref. 51, Supplemental Figure 10, and Supplemental Table 3). For Figure 5D, all gene lists in the Broad Institute C7 collection: Immunological Signatures (73) were tested against the preranked gene set. For analysis of healthy skin Tregs versus healthy skin Teffs and blood Tregs versus blood Teffs (Figure 6B), a preranked gene list was generated for each comparison (healthy skin Tregs vs. healthy skin Teffs; blood Tregs vs. blood Teffs) with DESeq2, and GseaPreranked analysis was performed using the GSEA v2.2.0 tool with default settings on the Hallmark mTORC1 signaling gene set (74).
Data availability
RNA-Seq data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE139341, GSE139372, GSE115898, and GSE140031).
Statistics
Statistical analysis (apart from RNA-Seq analysis, described above) was performed with GraphPad Prism v.6.0f. Statistical tests were 2-sided unless noted otherwise, and 95% confidence intervals were used. Data are shown as mean ± SEM.
Study approval
Animal studies were approved by the UCSF IACUC under protocol AN176267. All experiments were performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011). Written informed consent for donation of patient biospecimens was obtained from all subjects prior to inclusion in the study or specimens received were deidentified and certified as Not Human Subjects Research. All human experimental protocols were approved by the UCSF IRB (IRB 10-02830/13-12246).
Author contributions
MR and ML conceived and designed the study. IB and SC contributed to retrovirus cloning and mouse Treg overexpression experiments. ML, SC, IB, KT, MP, KMM, and PS contributed to flow cytometry acquisition and analysis. ML, RA, WL, RS, and ZL performed bioinformatics analysis. DN, KS, and AM contributed to CRISPR-editing of ARG2. GK contributed to data analysis. SA, IN, HH, EK, US, MK, AD, and TS contributed to the drafting of the manuscript.
Supplementary Material
Acknowledgments
The authors thank the patient donors for providing tissue samples for this study. We thank James Mueller and Julie Zikherman for assistance with Western blotting. Flow cytometry data were generated in the UCSF Parnassus Flow Cytometry Core, which is supported by the Diabetes Research Center grant (NIH P30 DK063720). MML was supported in part by a research award sponsored by AbbVie Bioresearch Center and NIH T32 Immunology Training Grant (2T32AI007334-26/ 5T32AI007334-27). This work was primarily funded by MDR’s grants (NIH K08-AR062064, NIH DP2-AR068130, and Burroughs Wellcome Fund CAMS-1010934) and an AbbVie Bioresearch Center-sponsored research agreement. WL was funded in part by NIH grants R01AR065174, U01AI119125. AM holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund, is an investigator at the Chan Zuckerberg Biohub, is a member of the Parker Institute for Cancer Immunotherapy, and has received funding from the Innovative Genomics Institute.
Version 1. 12/19/2019
Electronic publication
Footnotes
Conflict of interest: This study was supported in part by funding from AbbVie Bioresearch Center. AM is a cofounder of Arsenal Biosciences, Spotlight Therapeutics, and Sonoma Biotherapeutics. AM serves as on the scientific advisory board of PACT Pharma and was a former advisor to Juno Therapeutics. The Marson Laboratory has received sponsored research support from Juno Therapeutics, Epinomics, and Sanofi and a gift from Gilead.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(24):e129756.https://doi.org/10.1172/jci.insight.129756.
Contributor Information
Sean Clancy, Email: Sean.Clancy@ucsf.edu.
Richard S. Ahn, Email: richardahn@ucla.edu.
Wilson Liao, Email: LiaoWi@derm.ucsf.edu.
David N. Nguyen, Email: David.Nguyen@ucsf.edu.
Kathrin Schumann, Email: kathrin.schumann@tum.de.
Alexander Marson, Email: alexander.marson@ucsf.edu.
Zheng Liu, Email: zheng.liu@boehringer-ingelheim.com.
Robert Sanchez Rodriguez, Email: reven@reven.org.
Mariela L. Pauli, Email: Mariela.Pauli@ucsfmedctr.org.
Keyon Taravati, Email: ktaravati1991@gmail.com.
Isaac M. Neuhaus, Email: NeuhausI@derm.ucsf.edu.
Hobart W. Harris, Email: hobart.harris@ucsf.edu.
Esther A. Kim, Email: Esther.Kim@ucsf.edu.
Uk Sok Shin, Email: uksok.shin@yahoo.com.
Adil Daud, Email: Adil.Daud@ucsf.edu.
Tiffany C. Scharschmidt, Email: tiffany.scharschmidt@ucsf.edu.
Michael D. Rosenblum, Email: rosenblummd@derm.ucsf.edu.
References
- 1.Dudda JC, Perdue N, Bachtanian E, Campbell DJ. Foxp3+ regulatory T cells maintain immune homeostasis in the skin. J Exp Med. 2008;205(7):1559–1565. doi: 10.1084/jem.20072594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arpaia N, et al. A distinct function of regulatory T cells in tissue protection. Cell. 2015;162(5):1078–1089. doi: 10.1016/j.cell.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34(5):794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 5.Feuerer M, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. 2009;15(8):930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burzyn D, et al. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155(6):1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weirather J, et al. Foxp3+ CD4+ T cells improve healing after myocardial infarction by modulating monocyte/macrophage differentiation. Circ Res. 2014;115(1):55–67. doi: 10.1161/CIRCRESAHA.115.303895. [DOI] [PubMed] [Google Scholar]
- 8.Ito M, et al. Brain regulatory T cells suppress astrogliosis and potentiate neurological recovery. Nature. 2019;565(7738):246–250. doi: 10.1038/s41586-018-0824-5. [DOI] [PubMed] [Google Scholar]
- 9.Cipolletta D, et al. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486(7404):549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawamoto S, et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity. 2014;41(1):152–165. doi: 10.1016/j.immuni.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 11.Nosbaum A, et al. Cutting edge: regulatory T cells facilitate cutaneous wound healing. J Immunol. 2016;196(5):2010–2014. doi: 10.4049/jimmunol.1502139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ali N, et al. Regulatory T cells in skin facilitate epithelial stem cell differentiation. Cell. 2017;169(6):1119–1129.e11. doi: 10.1016/j.cell.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathur AN, et al. Treg-cell control of a CXCL5-IL-17 inflammatory axis promotes hair-follicle-stem-cell differentiation during skin-barrier repair. Immunity. 2019;50(3):655–667.e4. doi: 10.1016/j.immuni.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh N, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40(1):128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 17.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clever D, et al. Oxygen sensing by T cells establishes an immunologically tolerant metastatic niche. Cell. 2016;166(5):1117–1131.e14. doi: 10.1016/j.cell.2016.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18(16):1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 20.Chapman NM, Chi H. mTOR signaling, Tregs and immune modulation. Immunotherapy. 2014;6(12):1295–1311. doi: 10.2217/imt.14.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delgoffe GM, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30(6):832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Battaglia M, Stabilini A, Roncarolo MG. Rapamycin selectively expands CD4+CD25+FoxP3+ regulatory T cells. Blood. 2005;105(12):4743–4748. doi: 10.1182/blood-2004-10-3932. [DOI] [PubMed] [Google Scholar]
- 23.Battaglia M, Stabilini A, Migliavacca B, Horejs-Hoeck J, Kaupper T, Roncarolo MG. Rapamycin promotes expansion of functional CD4+CD25+FOXP3+ regulatory T cells of both healthy subjects and type 1 diabetic patients. J Immunol. 2006;177(12):8338–8347. doi: 10.4049/jimmunol.177.12.8338. [DOI] [PubMed] [Google Scholar]
- 24.Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499(7459):485–490. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chapman NM, et al. mTOR coordinates transcriptional programs and mitochondrial metabolism of activated Treg subsets to protect tissue homeostasis. Nat Commun. 2018;9(1):2095. doi: 10.1038/s41467-018-04392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neumann C, et al. c-Maf-dependent Treg cell control of intestinal TH17 cells and IgA establishes host-microbiota homeostasis. Nat Immunol. 2019;20(4):471–481. doi: 10.1038/s41590-019-0316-2. [DOI] [PubMed] [Google Scholar]
- 27.Sugiyama H, et al. Dysfunctional blood and target tissue CD4+CD25high regulatory T cells in psoriasis: mechanism underlying unrestrained pathogenic effector T cell proliferation. J Immunol. 2005;174(1):164–173. doi: 10.4049/jimmunol.174.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez Rodriguez R, et al. Memory regulatory T cells reside in human skin. J Clin Invest. 2014;124(3):1027–1036. doi: 10.1172/JCI72932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Remedios KA, et al. The TNFRSF members CD27 and OX40 coordinately limit TH17 differentiation in regulatory T cells. Sci Immunol. 2018;3(30):eaau2042. doi: 10.1126/sciimmunol.aau2042. [DOI] [PubMed] [Google Scholar]
- 30.Ahn R, et al. RNA-seq and flow-cytometry of conventional, scalp, and palmoplantar psoriasis reveal shared and distinct molecular pathways. Sci Rep. 2018;8(1):11368. doi: 10.1038/s41598-018-29472-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feuerer M, Hill JA, Mathis D, Benoist C. Foxp3+ regulatory T cells: differentiation, specification, subphenotypes. Nat Immunol. 2009;10(7):689–695. doi: 10.1038/ni.1760. [DOI] [PubMed] [Google Scholar]
- 32.Pfoertner S, et al. Signatures of human regulatory T cells: an encounter with old friends and new players. Genome Biol. 2006;7(7):R54. doi: 10.1186/gb-2006-7-7-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kachapati K, et al. The B10 Idd9.3 locus mediates accumulation of functionally superior CD137(+) regulatory T cells in the nonobese diabetic type 1 diabetes model. J Immunol. 2012;189(10):5001–5015. doi: 10.4049/jimmunol.1101013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nowak A, et al. CD137+CD154– expression as a regulatory T cell (Treg)-specific activation signature for identification and sorting of stable human tregs from in vitro expansion cultures. Front Immunol. 2018;9:199. doi: 10.3389/fimmu.2018.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray PJ. Amino acid auxotrophy as a system of immunological control nodes. Nat Immunol. 2016;17(2):132–139. doi: 10.1038/ni.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monticelli LA, et al. Arginase 1 is an innate lymphoid-cell-intrinsic metabolic checkpoint controlling type 2 inflammation. Nat Immunol. 2016;17(6):656–665. doi: 10.1038/ni.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGovern N, et al. Human fetal dendritic cells promote prenatal T-cell immune suppression through arginase-2. Nature. 2017;546(7660):662–666. doi: 10.1038/nature22795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elahi S, et al. Immunosuppressive CD71+ erythroid cells compromise neonatal host defence against infection. Nature. 2013;504(7478):158–162. doi: 10.1038/nature12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geiger R, et al. L-Arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell. 2016;167(3):829–842.e13. doi: 10.1016/j.cell.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Closs EI, Simon A, Vékony N, Rotmann A. Plasma membrane transporters for arginine. J Nutr. 2004;134(10 Suppl):2752S–2759S. doi: 10.1093/jn/134.10.2752S. [DOI] [PubMed] [Google Scholar]
- 41.Jie HB, et al. Intratumoral regulatory T cells upregulate immunosuppressive molecules in head and neck cancer patients. Br J Cancer. 2013;109(10):2629–2635. doi: 10.1038/bjc.2013.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Halim L, et al. An Atlas of Human Regulatory T Helper-like Cells Reveals Features of Th2-like Tregs that Support a Tumorigenic Environment. Cell Rep. 2017;20(3):757–770. doi: 10.1016/j.celrep.2017.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Syed Khaja AS, Toor SM, El Salhat H, Ali BR, Elkord E. Intratumoral FoxP3+Helios+ regulatory T cells upregulating immunosuppressive molecules are expanded in human colorectal cancer. Front Immunol. 2017;8:619. doi: 10.3389/fimmu.2017.00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyara M, et al. Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity. 2009;30(6):899–911. doi: 10.1016/j.immuni.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 45.van der Veeken J, et al. Memory of inflammation in regulatory T cells. Cell. 2016;166(4):977–990. doi: 10.1016/j.cell.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol. 2007;8(2):191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- 47.Schumann K, et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci U S A. 2015;112(33):10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42(22):e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mootha VK, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 50.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye L, et al. TCR usage, gene expression and function of two distinct FOXP3(+)Treg subsets within CD4(+)CD25(hi) T cells identified by expression of CD39 and CD45RO. Immunol Cell Biol. 2016;94(3):293–305. doi: 10.1038/icb.2015.90. [DOI] [PubMed] [Google Scholar]
- 52.Cuadrado E, et al. Proteomic analyses of human regulatory T cells reveal adaptations in signaling pathways that protect cellular identity. Immunity. 2018;48(5):1046–1059.e6. doi: 10.1016/j.immuni.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 53.Chantranupong L, et al. The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell. 2016;165(1):153–164. doi: 10.1016/j.cell.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perl AE, Kasner MT, Shank D, Luger SM, Carroll M. Single-cell pharmacodynamic monitoring of S6 ribosomal protein phosphorylation in AML blasts during a clinical trial combining the mTOR inhibitor sirolimus and intensive chemotherapy. Clin Cancer Res. 2012;18(6):1716–1725. doi: 10.1158/1078-0432.CCR-11-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones RG, Pearce EJ. MenTORing immunity: mTOR signaling in the development and function of tissue-resident immune cells. Immunity. 2017;46(5):730–742. doi: 10.1016/j.immuni.2017.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vasanthakumar A, et al. The transcriptional regulators IRF4, BATF and IL-33 orchestrate development and maintenance of adipose tissue-resident regulatory T cells. Nat Immunol. 2015;16(3):276–285. doi: 10.1038/ni.3085. [DOI] [PubMed] [Google Scholar]
- 57.Klatzmann D, Abbas AK. The promise of low-dose interleukin-2 therapy for autoimmune and inflammatory diseases. Nat Rev Immunol. 2015;15(5):283–294. doi: 10.1038/nri3823. [DOI] [PubMed] [Google Scholar]
- 58.Nishikawa H, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Curr Opin Immunol. 2014;27:1–7. doi: 10.1016/j.coi.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 59.Toby IT, Chicoine LG, Cui H, Chen B, Nelin LD. Hypoxia-induced proliferation of human pulmonary microvascular endothelial cells depends on epidermal growth factor receptor tyrosine kinase activation. Am J Physiol Lung Cell Mol Physiol. 2010;298(4):L600–L606. doi: 10.1152/ajplung.00122.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ming XF, et al. Arginase II promotes macrophage inflammatory responses through mitochondrial reactive oxygen species, contributing to insulin resistance and atherogenesis. J Am Heart Assoc. 2012;1(4):e000992. doi: 10.1161/JAHA.112.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fultang L, et al. Macrophage-derived IL1β and TNFα regulate arginine metabolism in neuroblastoma. Cancer Res. 2019;79(3):611–624. doi: 10.1158/0008-5472.CAN-18-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kamleh MA, et al. Increased levels of circulating fatty acids are associated with protective effects against future cardiovascular events in nondiabetics. J Proteome Res. 2018;17(2):870–878. doi: 10.1021/acs.jproteome.7b00671. [DOI] [PubMed] [Google Scholar]
- 63.Vissers YL, Dejong CH, Luiking YC, Fearon KC, von Meyenfeldt MF, Deutz NE. Plasma arginine concentrations are reduced in cancer patients: evidence for arginine deficiency? Am J Clin Nutr. 2005;81(5):1142–1146. doi: 10.1093/ajcn/81.5.1142. [DOI] [PubMed] [Google Scholar]
- 64.Scharschmidt TC, et al. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity. 2015;43(5):1011–1021. doi: 10.1016/j.immuni.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anders S, Pyl PT, Huber W. HTSeq — a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahn RS, et al. Transcriptional landscape of epithelial and immune cell populations revealed through FACS-seq of healthy human skin. Sci Rep. 2017;7(1):1343. doi: 10.1038/s41598-017-01468-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31(24):e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Richardson CD, Ray GJ, Bray NL, Corn JE. Non-homologous DNA increases gene disruption efficiency by altering DNA repair outcomes. Nat Commun. 2016;7:12463. doi: 10.1038/ncomms12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Godec J, et al. Compendium of immune signatures identifies conserved and species-specific biology in response to inflammation. Immunity. 2016;44(1):194–206. doi: 10.1016/j.immuni.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE139341, GSE139372, GSE115898, and GSE140031).