Fifty patients with high-grade gliomas from the authors’ hospital and 128 patients with high-grade gliomas from The Cancer Genome Atlas were included in this study. For each patient, the authors calculated 348 hand-crafted radiomics features and 8192 deep features generated by a pretrained convolutional neural network. They then applied feature selection and Elastic Net-Cox modeling to differentiate patients into long- and short-term survivors. In the 50 patients with high-grade gliomas from their institution, the combined feature analysis framework classified the patients into long- and short-term survivor groups with a log-rank test P value <.001. In the 128 patients from The Cancer Genome Atlas, the framework classified patients into long- and short-term survivors with a log-rank test P value of .014. In conclusion, the authors report successful production and initial validation of a deep transfer learning model combining radiomics and deep features to predict overall survival of patients with glioblastoma from postcontrast T1-weighed brain MR imaging.
Abstract
BACKGROUND AND PURPOSE:
Patient survival in high-grade glioma remains poor, despite the recent developments in cancer treatment. As new chemo-, targeted molecular, and immune therapies emerge and show promising results in clinical trials, image-based methods for early prediction of treatment response are needed. Deep learning models that incorporate radiomics features promise to extract information from brain MR imaging that correlates with response and prognosis. We report initial production of a combined deep learning and radiomics model to predict overall survival in a clinically heterogeneous cohort of patients with high-grade gliomas.
MATERIALS AND METHODS:
Fifty patients with high-grade gliomas from our hospital and 128 patients with high-grade glioma from The Cancer Genome Atlas were included. For each patient, we calculated 348 hand-crafted radiomics features and 8192 deep features generated by a pretrained convolutional neural network. We then applied feature selection and Elastic Net-Cox modeling to differentiate patients into long- and short-term survivors.
RESULTS:
In the 50 patients with high-grade gliomas from our institution, the combined feature analysis framework classified the patients into long- and short-term survivor groups with a log-rank test P value < .001. In the 128 patients from The Cancer Genome Atlas, the framework classified patients into long- and short-term survivors with a log-rank test P value of .014. For the mixed cohort of 50 patients from our institution and 58 patients from The Cancer Genome Atlas, it yielded a log-rank test P value of .035.
CONCLUSIONS:
A deep learning model combining deep and radiomics features can dichotomize patients with high-grade gliomas into long- and short-term survivors.
Glioblastoma multiforme (GBM), the largest diagnostic subcategory of high-grade glioma (HGG) and the most common malignant adult brain tumor, afflicts 12,000–13,000 new patients annually in the United States. GBM, comprising a genetically and phenotypically heterogeneous category of tumors, has a very poor prognosis and a low rate of treatment response. The standard combined treatment of surgery, temozolomide, and chemoradiation has improved the median overall survival (OS) of GBM to roughly 2 years. Presently, there is no method to reliably predict the OS of patients with GBM as a response to treatment. The absence of such reliable prediction is a barrier in designing clinical trials and selecting optimal treatments for patients.
In the existing literature, MR imaging features of a brain tumor, including its volume, intensity, shape, and texture of contrast enhancement and evidence of tumor necrosis, diffusivity, infiltration, and blood volume, have been demonstrated to correlate with the OS of HGG.1-6 A large number of these features were included for radiomics, techniques that leverage the wealth of information in images by extracting semiquantitative or quantitative predefined image features to derive a relationship between the features and clinical outcomes of interest.
Radiomics feature analysis has been shown to correlate with molecular and histologic tissue types and outcomes, such as response and OS of HGG, but the correlation remains imperfect. A major weakness that likely constrains the performance of radiomics is that predefined features are low-order features selected on the basis of heuristic knowledge about oncologic imaging. Therefore, extracting higher level and more complicated features and integrating these additional features into the framework of radiomics may result in improved predictive power.
Recently, a deep convolutional neural network (CNN) achieved outstanding performance in many areas of medical image analysis, such as segmentation,7,8 classification,9,10 prediction of tumor grade,11 and patient survival.12,13 A typical CNN structure is a feed-forward network that includes an input layer, multiple hidden layers, and an output layer. Because the convolutional filters and other parameters are adjusted automatically during the training process, the parameters of CNN are learned in a way that optimizes the use of information contained in the input images. In this process, a CNN may create and select a large number of features at its hidden layers. These features, termed “deep features” in this work, may be exploited for predicting the OS of patients with HGG in an unbiased fashion that does not require any prior definition and may contain extensive abstract information from the hidden layers. In training a CNN, when the available dataset is of limited size, one can apply a pretrained CNN for the task on hand. This practice, called “transfer learning,” has been shown to be an effective way of using deep learning in many cases.12,14,15 In this study, we integrated deep transfer learning and traditional radiomics techniques to explore a very large number of features in brain MR imaging of patients with HGGs. We then classified patients into longer term and shorter term survivors by training a machine learning model to predict OS on the basis of these image features.
MATERIALS AND METHODS
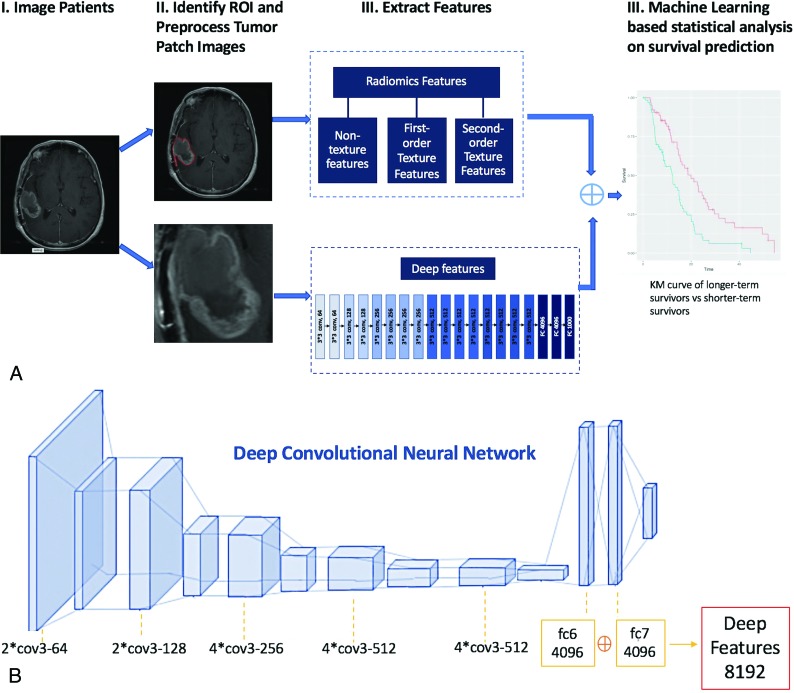
Our general workflow is depicted in Fig 1A, consisting of ROI-marking, image-preprocessing, feature extraction by traditional radiomics and deep learning, and statistical analysis.
Fig 1.
A, Flowchart shows our survival prediction system. B, The framework of VGG-19 model for deep-feature extraction.
Datasets
This study was approved by our institutional review board. We retrospectively retrieved 2 patient cohorts for this study. The first cohort is 50 patients with World Health Organization IV GBM, with known OS information, who had brain MRIs at the Brigham and Women’s Hospital between 2006 and 2011. Brain MR imaging after maximal surgical resection and before radiation therapy was retrieved for each patient. Only the gadolinium-enhanced T1-weighted spin-echo sequences acquired on 1.5T and 3T scanners from GE Healthcare (Milwaukee, Wisconsin) were used. The parameters for these axial contrast-enhanced T1-weighted images were as follows: TR = 416–566 ms, TE = 8–22 ms, FOV = 200–240 mm, matrix size = 256 × 256 or 512 × 512, section thickness = 5–6 mm. The second cohort set is 128 patients with World Health Organization IV GBM brain MR imaging retrieved from gadolinium-based contrast-enhanced T1-weighted images from The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov), publicly available, and The Cancer Image Archive (http://cancerimagingarchive.net/) with known OS information. The MRIs were acquired on 1.5T and 3T scanners from GE Healthcare (Milwaukee, Wisconsin), Siemens (Erlangen, Germany), Philips Healthcare (Best, the Netherlands) between 1995 and 2008. For the acquisition of the axial contrast-enhanced T1-weighted images, there were 58 2D spin-echo (SE), 49 3D gradient recalled-echo, and 21 other sequences. Scan parameters for SE sequences were as followings: TR = 409–809 ms, TE = 8–20 ms, FOV = 200–280 mm, matrix size = 488–683 × 421–683, section thickness = 1–6 mm. For gradient recalled-echo sequences, 28 were acquired using echo-spoiled gradient echo with TR = 25–250 ms, TE = 2.48–13.8 ms, flip angle = 25°–70°, FOV = 200–260 ms, matrix = 536–634 × 421–634, section thickness = 1–4 mm. Others were acquired using an MPRAGE sequence, with TR = 1160–2160 ms, TE = 2.75–4.24 ms, TI = 600–1100 ms, flip angle = 9°–15°, matrix = 561–624 × 421–624, section thickness = 0.9–5 mm.
From the 2 cohorts, we constituted 3 data groups. The first group is the 50 patients from Brigham and Women’s Hospital. Scans for this group were all acquired using the SE sequence. The second group is the 128 patients from TCGA, and scans were acquired using SE or gradient recalled-echo sequences. The third group comprised the 50 SE scans from Brigham and Women’s Hospital and the 58 selected SE scans from TCGA.
Tumor Segmentation and Image Preprocessing
For both patient cohorts, ROIs were manually traced by a radiologist on the section with the largest tumor area.
Before the extraction of quantitative features, several preprocessing techniques were applied to improve texture discriminations. First, intensity normalization was performed in a nonlinear way to convert MR images into standardized intensity ranges for all subjects.16 Second, to improve the computational performance and the signal-to-noise ratio of the texture outcome, we used gray-level quantization, which maps the full intensity range of the tumor region to different levels of gray.17 Two gray-level quantization algorithms (equal-probability quantization, uniform quantization) and 2 numbers of gray levels (16 and 32) were adopted. Finally, all images were resampled to an isotropic pixel size using bilinear interpolation. Scale values of 1 mm (pixel size = 1 × 1 mm3) and initial in-plane resolution were both tested.
For deep features, we cropped the MR images by finding a rectangular ROI that enclosed the outlined tumor. Then we resized the tumor patch to a 224 × 224 square to fulfill the requirement for the input size of the pretrained CNN model that we used. Also considering that the CNN model that we used was pretrained on natural images with a color range of 0–255, we normalized the intensity of tumor patch images to the same color range.
Feature Extraction
Two types of features were extracted. The first type is hand-crafted features that were manually extracted from an ROI. Hand-crafted features were divided into 3 groups: 1) nontexture features, including volume, size, and intensity features (such as solidity, eccentricity); 2) first-order histogram-based texture features, including skewness, kurtosis, variance, and others; and 3) second-order texture features, including features from the gray-level co-occurrence matrix, gray-level run length matrix, gray-level size zone matrix, and neighborhood gray-tone difference matrix. In total, we calculated 348 radiomics features for each ROI.
The second type of features were deep features. We chose the VGG-19,18 which was pretrained on the natural image dataset ImageNet (http://www.image-net.org/),19 which contains >1.2 million images as our CNN. VGG-19 has 19 layers with weights, formed by 16 convolutional layers and 3 fully connected layers. All the convolutional layers are built with a fixed kernel size of 3 × 3, and the stride and padding are fixed at 1. The network has 5 max-pooling layers with a window size of 2 × 2 and uses rectified linear units as the nonlinear activation function. The first 2 fully connected layers have 4096 features each, while the last FC layer has 1000 features with SoftMax activation (https://www.moleculardevices.com/products/microplate-readers/acquisition-and-analysis-software/softmax-pro-software) (Fig 1B). After we ran the front propagation of the VGG-19 model with pretrained weights as the initialization, a total number of 8192 deep features were extracted from the first 2 fully connected layers. All features were normalized by transforming the data into z scores with a mean of 0 and an SD of 1.
Feature Reduction
Feature reduction is a critical step because with 8192 deep features and 348 radiomics features, the number of features may result in overfitting in OS prediction. In addition, some features may have zero variance, have high correlations with other features, or have little relevance to the goal of OS prediction. Thus, the number of features needs to be reduced. We adopted 3 steps for feature reduction, namely, median absolute deviation, concordance indices (C-indices), and the Pearson coefficient correlation, to improve the generalizability and interpretability of our model.
Statistical Analysis
For censored survival data, we used the Elastic Net-Cox proportional hazards model20 to analyze the OS of patients with HGG. We denoted the sample size as . To maximize the use of the limited data, we applied leave-one-out cross validation times as the outer loop to split the data into training and test sets. Each pair of training and test sets was examined independently. The model was fit on the basis of the training set by maximizing the penalized partial log-likelihood function for the Cox model with a penalty term. In this model, the penalty parameter was optimized within the 10-fold cross-validation loop.; , which was used to determine the influence of the L1 penalty on the L2 penalty was set to be 0.1. After optimization, the model output a survival score for each patient in the training and test sets. We used the median of survival scores in each training set as the threshold to classify each patient in the test set into either a longer term or a shorter term survivor cohort. Finally, Kaplan-Meier analysis was used to estimate survival probabilities of each cohort,21 and a log-rank test was implemented to test the hypothesis that the survival curves differed statistically significantly between the 2 cohorts. A P value representing the statistical significance of the curve separation was used as an index to model predictive performance. For all analyses, a P value < .05 was indicative of statistical significance. All testing was 2-tailed.
Hardware and Software
The ROI was manually labeled using Hermes on DICOM images (https://hermes-router.github.io/). Radiomics features were computed using the Radiomics Matlab package (https://www.mathworks.com/matlabcentral/fileexchange/51948-radiomics).3 Pretrained CNN models were run using Keras with Tensorflow backend (https://keras.io/applications/#vgg19) on servers equipped with Xeon CPU and Tesla K80 GPU. Elastic Net-Cox model was built using R package ‘glmnet’ (https://cran.r-project.org/web/packages/glmnet/index.html).
RESULTS
Clinical Characteristics of Patients
The demographic and clinical characteristics of patients in all 3 datasets are shown on Table 1. The median and mean OS were 503 days and 690 days for the patients in group one, 352 days and 449 days for patients in group 2, and 490 days and 642 days for group 3.
Table 1:
Demographic and clinical characteristics of patients in all 3 datasets
| Demographics | DATA 1 (n = 50) | DATA 2 (n = 128) | DATA 3 (n = 108) |
|---|---|---|---|
| Sex | |||
| Female (No.) (%) | 23 (46%) | 46 (35.9%) | 43 (39.8%) |
| Male (No.) (%) | 27 (54%) | 82 (64.1%) | 65 (60.2%) |
| Mean | 0.46 ± 0.50 | 0.35 ± 0.48 | 0.39 ± 0.49 |
| Age (yr) | |||
| Range | 23–87 | 17–86 | 23–87 |
| Mean | 57 ± 13 | 58 ± 14 | 56 ± 12 |
| OS (days) | |||
| Range | 149–3156 | 7–1638 | 7–3156 |
| Mean | 690.34 ± 625.31 | 449.30 ± 352.93 | 642.1 ± 519.30 |
Feature Extraction
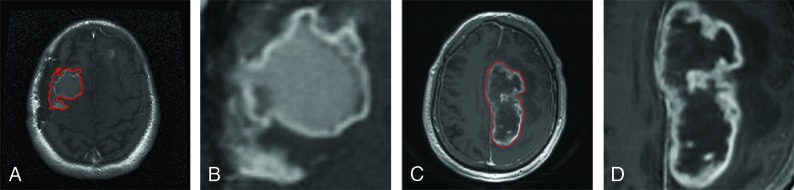
An example of a contrast-enhanced T1-weighted MR image of a longer term survivor and that of a shorter term survivor are shown in Fig 2A, -C. With different quantization algorithms, different numbers of gray-levels, and different scales for isotropic pixel resampling, we collected 348 hand-crafted quantitative radiomics features: 4 nontexture features, 24 first-order histogram-based texture features, 72 second-order texture features from the gray-level co-occurrence matrix, 104 second-order texture features from the gray-level run length matrix, 104 second-order texture features from the gray-level size zone matrix, and 40 second-order texture features from neighborhood gray-tone difference matrix. Table 2 summarizes the radiomics features described in Chmelik et al.9 Meanwhile, we generated and preprocessed the tumor patch images (Fig 2B, -D) as the input for the deep CNN architecture. Then we extracted 8192 features from the first 2 fully connected layers of the pretrained CNN model. Finally, a set of 8540 features was generated for each ROI.
Fig 2.
Example of contrast-enhanced T1-weighted MR images of longer term survivors (A and B) with an overall survival of 1405 days and shorter term survivors (C and D) with an overall survival of 447 days. A and C, Contrast-enhanced T1-weighted MR images with tumor contours in red. B and D, Tumor patches segmented from A and C, respectively.
Table 2:
List of radiomics features in this study
| Method | Quant. algo. | Ng | NS | Features | NF |
|---|---|---|---|---|---|
| Non-textures | Volume, size, solidity, eccentricity | 4 | |||
| Histogram | Equal probability, uniform | 16 32 |
2 | Variance, skewness, and kurtosis | 24 |
| GLCM | Equal probability, uniform | 16 32 |
2 | Energy, Contrast, Correlation, Homogeneity, Variance, Sum Average, Entropy, Dissimilarity and Autocorrelation | 72 |
| GLRLM | Equal probability, uniform | 16 32 |
2 | Short Run Emphasis (SRE), Long Run Emphasis (LRE), Gray-level Non-uniformity (GLN), Run-Length Non-uniformity (RLN), Run Percentage (RP), Low Gray-level Run Emphasis (LGRE), High Gray-level Run Emphasis (HGRE), Short Run Low Gray-level Emphasis (SRLGE), Short Run High Gray-level Emphasis (SRHGE), Long Run Low Gray-level Emphasis (LRLGE), Long Run High Gray-level Emphasis (LRHGE), Gray-level Variance (GLV) and Run-Length Variance (RLV) | 104 |
| GLSZM | Equal probability, uniform | 16 32 |
2 | Small Zone Emphasis (SZE), Large Zone Emphasis (LZE), Gray-level Non-uniformity (GLN), Zone-Size Non-uniformity (ZSN), Zone Percentage (ZP), Low Gray-level Zone Emphasis (LGZE), High Gray-level Zone Emphasis (HGZE), Small Zone Low Gray-level Emphasis (SZLGE), Small Zone High Gray-level Emphasis (SZHGE), Large Zone Low Gray-level Emphasis (LZLGE), Large Zone High Gray-level Emphasis (LZHGE), Gray-level Variance (GLV) and Zone-Size Variance (ZSV) | 104 |
| NGTDM | Equal probability, uniform | 16 32 |
2 | Coarseness, Contrast, Busyness, Complexity and Strength | 40 |
Note:—Quant. algo. indicates quantization algorithm; Ng, number of gray levels; NS, number of scales; NF, number of features; GLCM, gray-level co-occurrence matrix; GLRLM, gray-level run length matrix; GLSZM, gray-level size zone matrix; NGTDM, neighborhood gray-tone difference matrix.
Feature Reduction and Multivariate Statistical Analysis
Feature reduction was performed on the training set of each leave-one-out 10-fold cross-validation loop. We noticed that the deep feature matrix is relatively sparse and there are many uninformative deep features with zero variance. We set zero as the threshold of median absolute deviation to reduce about 60% of the total features, which were all from deep features. The remaining deep features are less noisy, and some of them have high C-indices. The ranges of C-indices among the 3 datasets vary, so the threshold of C-indices was slightly different. Because the distribution of C-indices for groups 2 and 3 was similar, the threshold of C-indices for these 2 datasets were the same at 0.66. The C-indices in the group 1 dataset were relatively higher at 0.685. The threshold for the Pearson coefficient correlation was set at 0.85 for all 3 datasets. Because the feature reduction was performed within each cross-validation loop and was based only on the subselected training set for the loop, the number of surviving features and the names of those features vary among the different training loops. In all loops, the number of surviving features was less than 100 and most of the surviving features were deep features.
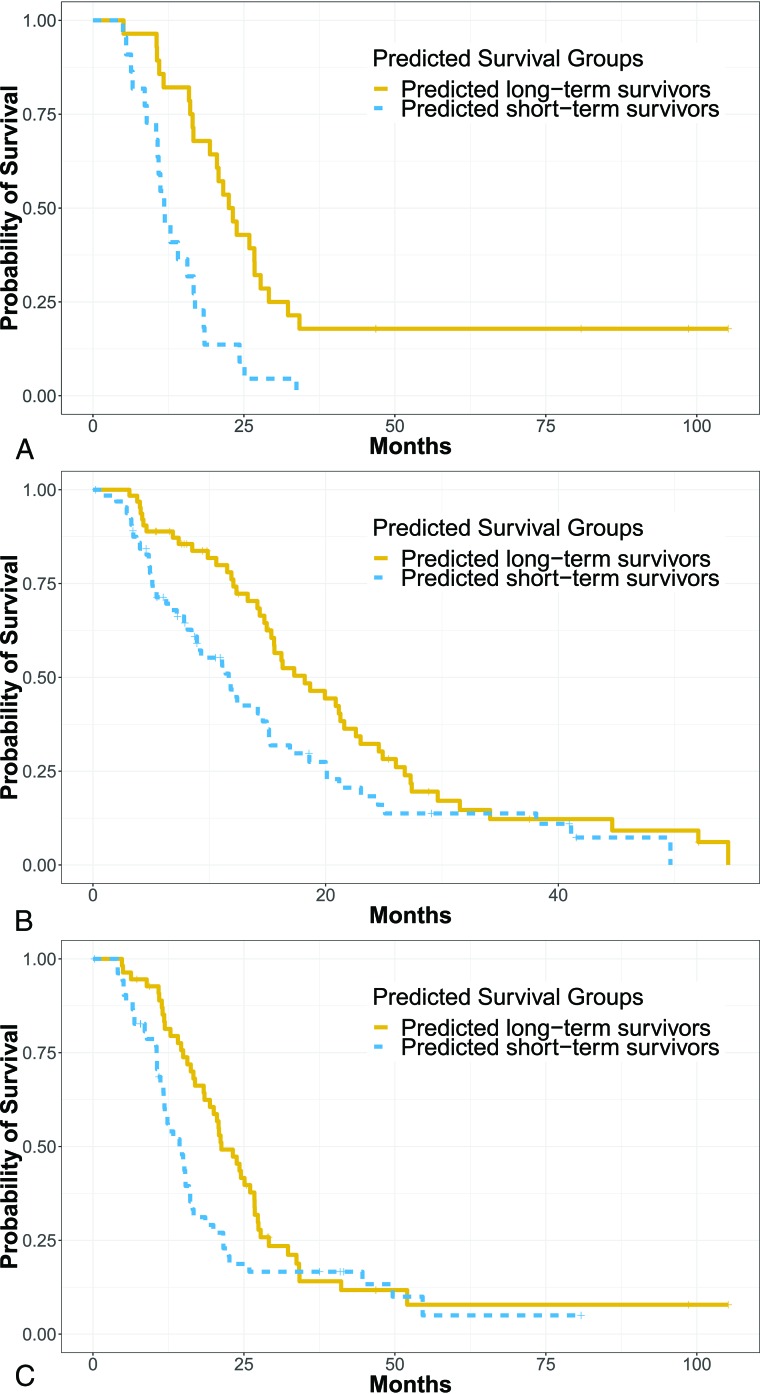
In this study, individuals who were lost to follow-up or were still alive at the end of the study were right-censored. The ratio of noncensored-to-censored data in the 3 datasets was 45:5 for DATA 1, 97:31 for DATA 2, and 90:18 for DATA 3. The Elastic Net-Cox proportional hazards model was used as the multivariate statistical model to generate survival scores for each patient. On the basis of the survival scores, we were able to dichotomize patients into 2 groups: predicted long-term and short-term survivors. In Fig 3, we demonstrate the overall performance of our method in the group 1 dataset (Fig 3A) with a log-rank test P value < .001 (hazard ratio = 3.26; 95% CI, 1.7–6.0), group 2 dataset (Fig 3B) with a log-rank test P value = .014 (hazard ratio = 1.65; 95% CI, 1.1–2.4), and the group 3 dataset (Fig 3C) with a log-rank test P value = .035 (hazard ratio = 1.71; 95% CI, 1.0–2.3). The ratio of shorter term-to-longer term survivors and the percentage of shorter term survivors was 22:28 (44%) for DATA 1, 63:65 (49%) for DATA 2, and 57:51 (53%) for DATA 3.
Fig 3.
Kaplan-Meier curve of predicted longer term and shorter term survival in a dataset (A) with 50 patients with HGG from Brigham and Women’s Hospital, a dataset (B) with 128 patients with HGG from the TCGA, and a dataset (C) with 108 patients with HGG from Brigham and Women’s Hospital and the TCGA.
DISCUSSION
In this study, we demonstrated that machine learning–based statistical analysis of an image feature set comprising both radiomics and deep learning features extracted from gadolinium-enhanced brain MR imaging of patients with GBM can be used to distinguish longer term from shorter term survivors. The model was proved efficient to analyze anonymized brain MR imaging data from both publicly available sources and our hospital.
Previous literature on quantitative image-based prediction of OS has suggested that deep features play a complementary role to radiomics features.12,14,22,23 Our study demonstrates that this remains true when using deep features extracted by VGG-19, an advanced model with documented excellent performance in image classification. Deep features are not limited to previously identified image attributes or even to those understandable by humans. This is an advantage because it leads to the possibility of discovering information in medical images that is not observable to human readers, which, in turn, raises the rational hope of adding diagnostic value beyond simple quantification of information already accessible in MR images. The abstract “features” represented in the weights of the deep CNN have a number of limitations. The meaning of individual features is not easy for humans to clearly understand. Also, it remains uncertain how reproducible the deep feature output is from the current CNN operating with available dataset sizes and processing power. As such, radiomics features were integrated to the pipeline, which were well-defined and selected a priori to comprise image attributes known or rationally expected by human experts to contain predictive information. At least in the near-term, we believe that the combination of radiomics and deep features may be rationally expected to provide greater value than deep feature–based analyses alone.
Besides deep features and radiomics features, we also investigated the effect of demographic features by adding sex and age information into the feature set of the TCGA dataset. This investigation did not change the performance substantially, likely because the areas under the curve of sex and age were both so low (area under the curve of sex = 0.53; area under the curve of age = 0.62) that these features were eliminated during feature reduction. We chose to include a deliberately heterogeneous cohort treated with a wide range of therapies to make the training set and model more readily generalizable to the heterogeneous patient mixture encountered in the clinic. Most of the patients received a standard treatment protocol of chemoradiation with temozolomide. Many received bevacizumab. Additional patients received a range of investigational therapies, including chemotherapies, targeted molecular therapies, and a few immunotherapies. Tumor genomics such as Isocitrate dehydrogenase 1 (IDH-1) mutation, MGMT methylation, and epidermal growth factor receptor application are powerful tumor markers that merit inclusion in future models. Because, unfortunately, tumor genomic data were unavailable for many patients in our cohort, we did not attempt to include this in our modeling. Twelve of 50 patients in our institutional dataset were IDH-1 wild-type, but IDH-1 status was not assessed in the remainder. Similarly, 11 of 50 were epidermal growth factor receptor–amplified, 20 of 50 were epidermal growth factor receptor not highly amplified, and 19 were unknown.
Like any statistical correlation approach, the CNN depends on the comprehensiveness of the training dataset, which is a statistically robust subset of the whole dataset. In general, the CNN model performance improves with increasing size of the dataset. Unfortunately, in the field of medical imaging and particularly in HGG brain MR imaging, annotating a large number of medical images remains challenging and time-consuming. While 50–150 fully annotated GBM datasets with known patient data represent a large amount of data to acquire, it is a small dataset from the perspective of the CNN training set and may result in overfitting. Under this circumstance, the transfer learning technique is introduced to apply CNN models from one field to another. For example, Paul et al14,22 generated deep features from a pretrained CNN model to improve survival prediction accuracy for patients with lung cancer. Lao et al12 adopted the pretrained CNN_S model for prediction of survival in GBM. Ahmed et al23 chose a pretrained CNN_F model in predicting the survival time of patients with brain tumor. Choi et al24 used a pretrained VGG-19 model to classify retinal images. On the other hand, transfer learning can reduce the number of training sets used by several orders of magnitude, but it introduces certain bias into the resulting CNN in the form of the pretrained parameters. It is likely that the success of the transferred model depends, in large part, on which CNN is selected, but to date, there is no systematic way to know which of the large number of available pretrained neural networks is best suited to a given task, which layers are optimal for choosing the features, or which pretrained dataset should be used. We chose VGG-19 to generate deep features for this study because it was trained on a large image data base and validated to provide excellent accuracy in the many applications; and it has been successfully applied to many medically related problems. In future work, it may be important to compare our results with the performance of other models such as VGG-16 (https://neurohive.io/en/popular-networks/vgg16/)18 or ResNet-50 (https://neurohive.io/en/popular-networks/resnet/).25
Transfer learning is not a fail-proof solution to the overfitting problem because overfitting may also occur in the retraining of the transferred CNN. Our initial output data matrix was very unbalanced and sparse, with only several dozen patients but 8540 output features. Our applied feature-reduction method eliminated roughly 99% of features and reduced the probability of divergence and computational cost. Reducing the number of features is essential to avoid overfitting in such small datasets, but feature reduction remains complicated and somewhat controversial.26,27 A number of methods exist. We chose not to use principal component analysis, a well-established method, because it complicates further the interpretability problems intrinsic to radiomics and deep learning models by generating new features in a new coordinate system, yet 1 more step removed from the original image.
In our experiments, we found that there were more downstream features left in DATA 1 than in DATA 2 and DATA 3. One reason could be the data homogeneity, ie, some predictors in the homogeneous dataset may lead to an overfitting problem because they may not be very predictive in a heterogeneous dataset. In fact, in our data, DATA 1 was the most homogeneous one of all 3 datasets because it included only SE MR images and the data were acquired from a single institution only. In addition, the hazard ratio in DATA 1 was found to be higher than in the second and third datasets, suggesting that data homogeneity may affect the performance of a machine learning model in some way.
On a large-scale and high-throughput data mining field, especially medical imaging analysis, machine learning–based statistical analysis techniques are widely used. For example, de Carvalho Filho et al28 used the support vector machine algorithm for lung nodule classification. Lao et al12 used the lasso Cox regression model to find a useful subset of reduced features, then constructed radiomics signatures to predict the OS of patients with GBM. Yin et al27 compared 3 feature-selection methods and 3 classification methods for differentiation of sacral chordoma and sacral giant-cell tumor. Yu et al29 used an Elastic Net-Cox hazard ratio model to predict survival of patients with squamous cell carcinoma and stage I adenocarcinoma. In our study, we did not model OS as a binomial classification problem because binary classifiers do not consider censored information and are highly dependent on the manually determined survival threshold. Instead we chose Cox regression, a time-to-event model that is better suited to handle censored data and model a continuous range of the survival probabilities. Because the traditional Cox model does not work well on high-dimensional data in which a number of covariates are much larger than the sample size, we selected the Elastic Net-Cox hazard ratio model for statistical analysis. This penalized Cox model has been proved able to handle high-dimensional data and obtain reliable survival prediction.29 It is possible that other statistical approaches may improve our ability to detect correlations in the data. This is an area that will benefit from future study.
Because postcontrast T1-weighted images are the critical mainstay of routine clinical brain tumor imaging,30 we chose these as the initial image type around which to build our pipeline. Addition of more sequences (precontrast T1, T2, FLAIR T2, SWI, DWI, PWI) with independent image contrast can be expected to improve model performance and is planned for future work.
Other factors that may affect the final model performance include interoperator variation in ROI selection. To investigate, we had our 2 radiologists label our largest TCGA dataset independently. The intraclass correlation coefficient on the downstream features yielded a median of 0.76. Most individual-feature intraclass correlation coefficients were >0.7. This high reproducibility implies that inter-operator variation in ROI selection had little effect on our dataset. We also performed a supplemental analysis to test the effect of normal tissue in the training set input. In the patients in the TCGA, we removed normal voxels from inside the bounding box and retrained the deep learning model. The result was similar to the performance with the normal tissue voxels retained in the bounding box, with a P value of .016. It remains unknown whether this result would be true using larger bounding boxes comprising more normal tissue. These analyses suggest that our model performance was robust to at least some variation in preprocessing. More investigation is needed with more heterogeneous and independent datasets to determine whether this is generally true.
CONCLUSIONS
We report successful production and initial validation of a deep transfer learning model combining radiomics and deep features to predict OS of patients with GBM from postcontrast T1-weighed brain MR images. Further optimization of these results will require systematic attention to each of the critical components of the pipeline, including choice and modification of the model, optimization of feature-reduction method, selection of statistical correlation strategy, incorporation of additional MR imaging data types, and inclusion of tumor genomics data. Necessary future steps before clinical translation will need to include interpretability analysis to determine the clinical significance of the surviving features in the model and testing in a dichotomized design that allows assessment or prediction performance on an individual patient basis.
ABBREVIATIONS:
- C-indices
concordance indices
- CNN
convolutional neural network
- GBM
glioblastoma multiforme
- HGG
high-grade glioma
- OS
overall survival
- SE
spin-echo
- TCGA
the Cancer Genome Atlas
Footnotes
W. Han and L. Qin are co-first authors.
This work was supported by National Institutes of Health award R01LM012434 and Harvard Data Science Fellowship.
Disclosures: Wei Han—RELATED: Grant: National Institutes of Health award R01LM012434.* Lei Qin—UNRELATED: Payment for Lectures Including Service on Speakers Bureaus: Purdue University, Biomedical Engineering Department, Comments: I gave a lecture to the Biomedical Engineering department about radiomics and machine learning. Kun-Hsing Yu—RELATED: Grant: Harvard Data Science Initiative, Comments: I received a fellowship from the Harvard Data Science Initiative*; UNRELATED: Patents (Planned, Pending or Issued): Harvard Medical School, Comments: Harvard Medical School submitted a nonprovisional patent on my work on digital pathology profiling (US Patent Application 16/179,101). Xiaoyin Xu—RELATED: Grant: National Institutes of Health, Comments: R01LM012434.* Geoffrey Young—RELATED: Grant: National Institutes of Health*; Support for Travel to Meetings for the Study or Other Purposes: National Institutes of Health*; UNRELATED: Expert Testimony: Risk Management Foundation; Grants/Grants Pending: Siemens, Comments: postprocessing software*; Payment for Lectures Including Service on Speakers Bureaus: Ray-Plus, Comments: honorarium for lecture; Travel/Accommodations/Meeting Expenses Unrelated to Activities Listed: Ray-Plus, Comments: travel expenses related to the lecture mentioned above; Other: collaborating institutions in China, Comments: support for visiting research fellows who participated in my lab as part of a research training program and participated in the work of the group. In the past 36 months, this has included fully externally supported MD fellows from the Henan Cancer Hospital, the Guangzhou First People’s Hospital, and the Peking Union Medical College Hospital.* *Money paid to institution.
References
- 1.Pope WB, Sayre J, Perlina A, et al. MR imaging correlates of survival in patients with high-grade gliomas. AJNR Am J Neuroradiol 2005;26:2466–74 [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou M, Scott J, Chaudhury B, et al. Radiomics in brain tumor: image assessment, quantitative feature descriptors, and machine-learning approaches. AJNR Am J Neuroradiol 2018;39:208–16 10.3174/ajnr.A5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vallieres M, Freeman CR, Skamene SR, et al. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol 2015;60:5471–96 10.1088/0031-9155/60/14/5471 [DOI] [PubMed] [Google Scholar]
- 4.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qu J, Qin L, Cheng S, et al. Residual low ADC and high FA at the resection margin correlate with poor chemoradiation response and overall survival in high-grade glioma patients. Eur J Radiol 2016;85:657–64 10.1016/j.ejrad.2015.12.026 [DOI] [PubMed] [Google Scholar]
- 6.Qin L, Li A, Qu J, et al. Normalization of ADC does not improve correlation with overall survival in patients with high-grade glioma (HGG). J Neurooncol 2018;137:313–19 10.1007/s11060-017-2719-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akkus Z, Galimzianova A, Hoogi A, et al. Deep learning for brain MRI segmentation: state of the art and future directions. J Digit Imaging 2017;30:449–59 10.1007/s10278-017-9983-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou X, Takayama R, Wang S, et al. Deep learning of the sectional appearances of 3D CT images for anatomical structure segmentation based on an FCN voting method. Med Phys 2017;44:5221–33 10.1002/mp.12480 [DOI] [PubMed] [Google Scholar]
- 9.Chmelik J, Jakubicek R, Walek P, et al. Deep convolutional neural network-based segmentation and classification of difficult to define metastatic spinal lesions in 3D CT data. Med Image Anal 2018;49:76–88 10.1016/j.media.2018.07.008 [DOI] [PubMed] [Google Scholar]
- 10.Acharya UR, Oh SL, Hagiwara Y, et al. A deep convolutional neural network model to classify heartbeats. Comput Biol Med 2017;89:389–96 10.1016/j.compbiomed.2017.08.022 [DOI] [PubMed] [Google Scholar]
- 11.Burlina P, Pacheco KD, Joshi N, et al. Comparing humans and deep learning performance for grading AMD: a study in using universal deep features and transfer learning for automated AMD analysis. Comput Biol Med 2017;82:80–86 10.1016/j.compbiomed.2017.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lao J, Chen Y, Li ZC, et al. A deep learning-based radiomics model for prediction of survival in glioblastoma multiforme. Sci Rep 2017;7:1035 10.1038/s41598-017-10649-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nie D, Zhang H, Adeli E, et al. 3D deep learning for multi-modal imaging-guided survival time prediction of brain tumor patients. Med Image Comput Comput Assist Interv 2016;9901:212–20 10.1007/978-3-319-46723-8_25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul R, Hawkins SH, Balagurunathan Y, et al. Deep feature transfer learning in combination with traditional features predicts survival among patients with lung adenocarcinoma. Tomography 2016;2:388–95 10.18383/j.tom.2016.00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhen X, Chen J, Zhong Z, et al. Deep convolutional neural network with transfer learning for rectum toxicity prediction in cervical cancer radiotherapy: a feasibility study. Phys Med Biol 2017;62:8246–63 10.1088/1361-6560/aa8d09 [DOI] [PubMed] [Google Scholar]
- 16.Collewet G, Strzelecki M, Mariette F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn Reson Imaging 2004;22:81–91 10.1016/j.mri.2003.09.001 [DOI] [PubMed] [Google Scholar]
- 17.Gibbs P, Turnbull LW. Textural analysis of contrast-enhanced MR images of the breast. Magn Reson Med 2003;50:92–98 10.1002/mrm.10496 [DOI] [PubMed] [Google Scholar]
- 18.Simonyan K, Zisseman A. Very deep convolutional networks for large-scale image recognition. Computer Vision and Pattern Recognition 2014;arXiv:1409.1556 [Google Scholar]
- 19.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. J Commun. ACM 2017;60:84–90 [Google Scholar]
- 20.Simon N, Friedman J, Hastie T, et al. Regularization paths for Cox’s proportional hazards model via coordinate descent. J Stat Softw 2011;39:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feinstein AR. Clinical Epidemiology: The Architecture of Clinical Research. Philadelphia: W.B. Saunders; 1985:xii [Google Scholar]
- 22.Paul R, Hawkins SH, Hall LO, et al. Combining deep neural network and traditional image features to improve survival prediction accuracy for lung cancer patients from diagnostic CT. In: Proceedings of the IEEE International Conference on Systems, Man, and Cybernetics, Budapest, Hungary October 9–12, 2016 10.1109/SMC.2016.7844626 [DOI] [Google Scholar]
- 23.Ahmed KB, Hall LO, Goldgof DB, et al. Fine-tuning convolutional deep features for MRI based brain tumor classification. In: Proceedings of Medical Imaging 2017: Computer-Aided Diagnosis, Orlando, Florida February 11–16, 2017 [Google Scholar]
- 24.Choi JY, Yoo TK, Seo JG, et al. Multi-categorical deep learning neural network to classify retinal images: a pilot study employing small database. PLoS One 2017;12:e0187336 10.1371/journal.pone.0187336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaiming H, Zhang X, Shaoqing R, Jian S. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, Navada June 26 to July 1, 2016 [Google Scholar]
- 26.Parmar C, Grossmann P, Bussink J, et al. Machine learning methods for quantitative radiomic biomarkers. Sci Rep 2015;5:13087 10.1038/srep13087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin P, Mao N, Zhao C, et al. Comparison of radiomics machine-learning classifiers and feature selection for differentiation of sacral chordoma and sacral giant cell tumour based on 3D computed tomography features. Eur Radiol 2019;29:1841–47 10.1007/s00330-018-5730-6 [DOI] [PubMed] [Google Scholar]
- 28.de Carvalho Filho AO, Silva AC, de Paiva AC, et al. Lung-nodule classification based on computed tomography using taxonomic diversity indexes and an SVM. J Signal Process Syst 2017;87:179–96 10.1007/s11265-016-1134-5 [DOI] [Google Scholar]
- 29.Yu KH, Zhang C, Berry GJ, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun 2016;7:12474 10.1038/ncomms12474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cha S. CNS tumors: monitoring therapeutic response and outcome prediction. Top Magn Reson Imaging 2006;17:63–68 10.1097/RMR.0b013e31802ee598 [DOI] [PubMed] [Google Scholar]