Abstract
Eukaryotic genome is packaged in a nucleus in the form of chromatin. The fundamental structural unit of the chromatin is the protein-DNA complex, nucleosome, where DNA of about 150 bp is wrapped around a histone core almost twice. In cellular processes such as gene expression, DNA repair and duplication, the nucleosomal DNA has to be unwrapped. Histone proteins have their variants, indicating there are a variety of constitutions of nucleosomes. These different constitutions are observed in different cellular processes. To investigate differences among nucleosomes, we calculated free energy profiles for unwrapping the outer superhelical turn of CENP-A nucleosome and compared them with those of the canonical nucleosome. The free energy profiles for CENP-A nucleosome suggest that CENP-A nucleosome is the most stable when 16 to 22 bps are unwrapped in total whereas the canonical nucleosome is the most stable when it is fully wrapped. This indicates that the flexible conformation of CENP-A nucleosome is ready to provide binding sites for the structural integrity of the centromere.
Keywords: CENP-A, nucleosome, unwrapping, free energy, molecular dynamics
Significance.
Because histone proteins have their variants and are subject to posttranslational modification, there are a variety of constitutions for nucleosomes which are the fundamental structural unit of chromatin. To investigate differences between the canonical H3 nucleosome and a H3 variant, CENP-A nucleosome, free energy profile for DNA unwrapping of CENP-A nucleosome was calculated. The profile shows that 16 to 22 bp unwrapped states are the most stable for CENP-A nucleosome, indicating that the CENP-A nucleosome is poised to provide scaffolds for kinetochore proteins
Nucleosomes are a fundamental structural unit of chromatin which is composed of histone proteins and DNA. The crystal structure of the nucleosome provides the atomic details [1] and deepens our understanding of how DNA is stored in a nucleus. In the structure, DNA of about 150 bp is wrapped around a histone core about 1.7 times. However, this nucleosomal DNA must be unwrapped and become accessible to regulatory proteins for achieving gene function such as transcription, duplication and repairs. To study the dynamics of nucleosomes including DNA unwrapping, all atom molecular dynamics simulations were carried out [2–17], however, they mostly suffered from short of conformational samplings due to the large size of the simulation systems and the inclusion of intrinsically disordered regions in the systems.
Histone proteins have their variants. Crystal structural analyses for nucleosomes containing a different histone constitution from the canonical histones show that the structures are basically similar to the canonical nucleosome except a histone variant of H3, CENP-A nucleosome. The crystal structure of CENP-A containing nucleosome core particle (CENP-A NCP) has invisible regions at both ends of DNA, indicating the large fluctuation of the regions because of the short length of the αN helix of CENP-A compared with that of H3 [18]. Replacement of the αN helix of CENP-A with that of H3 leads to H1 recruitment, causing mitotic and cytokinesis defects [19]. We previously carried out molecular dynamics simulations on the canonical and CENP-A nucleosome core particles, which showed that two Arg residues, R49 and R52, in the helix are responsible for the stability of DNA at both ends of DNA in the canonical nucleosome [7]. In the CENP-A NCP, Lys corresponds to these residues and is likely to lose a hydrogen bond with DNA more easily than Arg. Furthermore, we showed a free energy profile for unwrapping DNA from the canonical nucleosome. The profile suggested that a free energy for unwrapping the outer superhelical turn is about 11.5 kcal/mol which agrees well with values obtained in single molecule experiments [20]. The detailed analysis showed that asymmetric unwrapping of DNA is dominant, i.e., either end of DNA firstly unwraps from the histone core by up to 10 bps, then the other end of DNA starts to unwrap.
In this study, we report free energy profiles for unwrapping the outer superhelical turn of CENP-A nucleosome which were calculated using a massive MD simulation with K computer. The free energy profile clearly showed that the CENP-A NCP is the most stable when 8 to 11 bps are unwrapped at both sides of DNA. The gentle shape of free energy curve of CENP-A NCP around the minimum indicates that DNA can be highly flexible between open and closed conformations under a physiological condition.
Methods
Atomic models
Crystal structure of CENP-A NCP was derived from the Protein Data Bank: PDB code: 3AN2. The coordinates for both DNA ends (13 bp at each end) were missing from the CENP-A NCP. They were modeled based on the DNA structure of the canonical H3 NCP (PDB code: 1KX5). After structurally aligning the phosphate atoms of backbones (residue numbers, from −60 to 60) of 3AN2 with those of H3 NCP, we modelled the missing DNA of the CENP-A NCP using the same conformational parameters as those of the corresponding DNA of the H3 NCP. The conformational parameters were calculated using X3DNA [21]. The missing histone tails of CENP-A NCP were not modelled because the simulation system was set as similar as possible to our previous simulation of H3 NCP [7]. The effect of the histone tails, likely to be of significance, will be the subject of a future investigation. Finally, the simulation system had about 31,500 atoms in a box of 147 Å×147 Å×147 Å. All the simulations were carried out under NVT condition.
Adaptively biased MD (ABMD) combined with multiple walker method
We followed the simulation protocol of our previous H3-NCP unwrapping calculation [7]. Therefore, we briefly describe the method. To obtain various, unwrapped nucleosome conformations along the reaction coordinates shown in Figure 1, we used the adaptively biased molecular dynamics (ABMD) method [22] combined with multiple walker method [23] implemented in an in-house molecular dynamics simulation software, SCUBA [16,24–28] which was initiated to develop by Prof. Go and was named after his hobby. The reaction coordinate d was defined as a distance between two phosphorus atoms of T73 in chains I and J at both DNA ends. Force fields used were AMBER ff99SB [29] for proteins, ff99bsc0 [30] for DNA and ff99ions08 [31] for ions, and the TIP3P [32] for water. The system was solvated in the 120 mM solution of NaCl, and the excess negative charges of the nucleosome complex were neutralized by the excess number of sodium ions. In the ABMD with the multiple walker method, we ran 100 independent MD simulations with a sharing biased potential. We carried out ABMD until the biased potential became almost flat for the entire range on the reaction coordinate we desired to sample.
Figure 1.
CENP-A nucleosome structure and the reaction coordinate. The reaction coordinate is a distance between two phosphorus atoms at the ends of DNA (T73 in chains I and J).
The range of the reaction coordinate was set 20 to 200 Å and a wall potential with a harmonic shape and a constant of 10.0 kcal/mol was applied at d=25 and 195 Å when a walker goes out of the predefined range above. The resolution of the reaction coordinate, Δd was set at 1.0 Å. The relaxation time in the ABMD, τF was set at 100 ps for maintaining dsDNA conformation based on our previous calculation [20], and the ABMD biasing potential was updated every step.
Umbrella sampling and Free-energy profile using WHAM
Umbrella sampling was followed for enhancing equilibrium sampling. In the umbrella sampling, the reaction coordinate was divided into 76 windows with a width of 2 Å which covers 40 to 190 Å. The sampled conformations in ABMD at less than 40 Å and more than 190 Å on the reaction coordinate were discarded because their DNA structures were corrupted or highly distorted. The umbrella potential for each windows is a harmonic function with a force constant of 0.2 kcal/(mol Å2).
The weighted histogram analysis method (WHAM) [33] was used to refine the free-energy landscape from the sampled trajectories in the umbrella sampling simulations. The conformation of the nucleosome was stored ever 1 ps from 15 ns long umbrella sampling. The free energy profiles were calculated based on the end to end distance and the number of unwrapped base pairs.
Results
All adaptively biased MD and umbrella sampling simulations were carried out using K computer. For each of walkers (replicas), we assigned 48 nodes (384 cores). We used about 2.3 million node×hours in total.
Adaptively biased MD calculations
To obtain different CENP-A NCP conformations as naturally as possible, we carried out adaptively biased MD (ABMD) simulation using 100 walkers. The 100 walkers sitting on similar positions at the beginning were distributed along the reaction coordinate in 5 ns and started to fluctuate (Supplementary Fig. S1). Because the dropped energies for the duration of every 1 ns along the reaction coordinate nearly converged at 14 ns (Supplementary Fig. S2), we stopped the ABMD simulations at 20 ns and switched to umbrella sampling simulations for further refining conformation sampling.
Convergence of free energy calculations
First, to examine if free energy calculation converges, we obtained free energy profiles for different simulation times of the umbrella sampling against the reaction coordinate of the DNA end to end distance (Fig. 1). We observed changes in the profile curves from 5 ns to 10 ns, but not from 10 to 15 ns (Fig. 2), indicating that the profiles converged at 10 ns. Hereafter we show the results based on 15 ns long simulation data which converged well.
Figure 2.
Convergence of free energy profile. Each of the profiles was calculated as a function of DNA end to end distance (see Fig. 1 legend) using an ensemble obtained by the umbrella sampling of 5, 10, 12, 14 or 15 ns. The minimum of the free energy profiles were aligned to 0 kcal/mol.
Free energy as a function of DNA end to end distance
Figure 2 shows that the free energy profile curve is nearly flat between 85 and 140 Å (the difference is less than 0.5 kcal/mol). This indicates that the unwrapped states are the most stable for CENP-A NCP. In a crystal structure of the canonical NCP (H3 NCP; PDB ID 1KX5) in which DNA is fully wrapped, the end to end distance is 63 Å. The profile shows that it requires about 1.5 kcal/mol to adopt the fully wrapped state in CENP-A NCP.
Free energy as a function of the number of unwrapped base pairs
To interpret the unwrapping process from a structural point of view, we plotted the free energy as a function of the number of unwrapped base pairs (Fig. 3a). We defined unwrapped base pairs as those in which the center of the base pair deviated by more than 4 Å deviation from the center of the histone core in the reference structure. As the reference structure, we used the initial structure for ABMD subject to the energy minimization and 1 ns long relaxation run without any constraint. The profile curve is nearly flat when a total of between 15 and 25 bps are unwrapped although a small minimum is observed at 22 bp.
Figure 3.
Free energy profiles of CENP-A NCP unwrapping. (a) Free energy profile for unwrapping DNA as a function of the total number of unwrapped bps. (b) Free energy profile for unwrapping DNA as a function of the total number of unwrapped bps and DNA end to end distance. Ensemble obtained by an umbrella sampling of 15 ns was used.
We also calculated the 2-dimensional free energy profile as a function of the number of the total unwrapped bp and the end to end distance (Fig. 3b). Interestingly, the 2D profile shows that there are a few local minima along the end to end distance at between 15 and 25 bps in total unwrapped bp, indicating CENP-A NCP adopts different conformations under the same number of total unwrapped bp. In fact, CENP-A NCP has a variety of unwrapped states, as shown in Figure 4. In this figure, difference in unwrapped bp between the two ends is plotted against the total number of unwrapped bp. From the figure, we can see symmetric or asymmetric unwrapping of the nucleosome. For instance, when 15 bp of DNA are unwrapped in total, the most dominant conformation is one where 11 bp unwrapped from one end of DNA and 4 bp from the other end. However, other combinations of unwrapping states were also observed. The existence of these multi-conformations demonstrates that both DNA ends are highly flexible, which gives small changes in free energy. This differs from the canonical NCP where asymmetric unwrapping conformations were dominantly observed (see Fig. 6 in a reference [20]).
Figure 4.
Differences in unwrapped bps between two DNA ends. The differences in number of unwrapped bp in the two DNA ends (shown as bp diff., the ordinate) are plotted as a function of the total number of unwrapped bps (the abscissa). The bp differences are normalized to the number of conformations with the same total number of unwrapped bps. For instance, when one of the ends is unwrapped by 10 bp and the other end is fully wrapped around the histone octamer as shown by insets, we have a plot at a position of (total unwrapped bp, bp diff) = (10, 10).
Lost interactions between histones and DNA
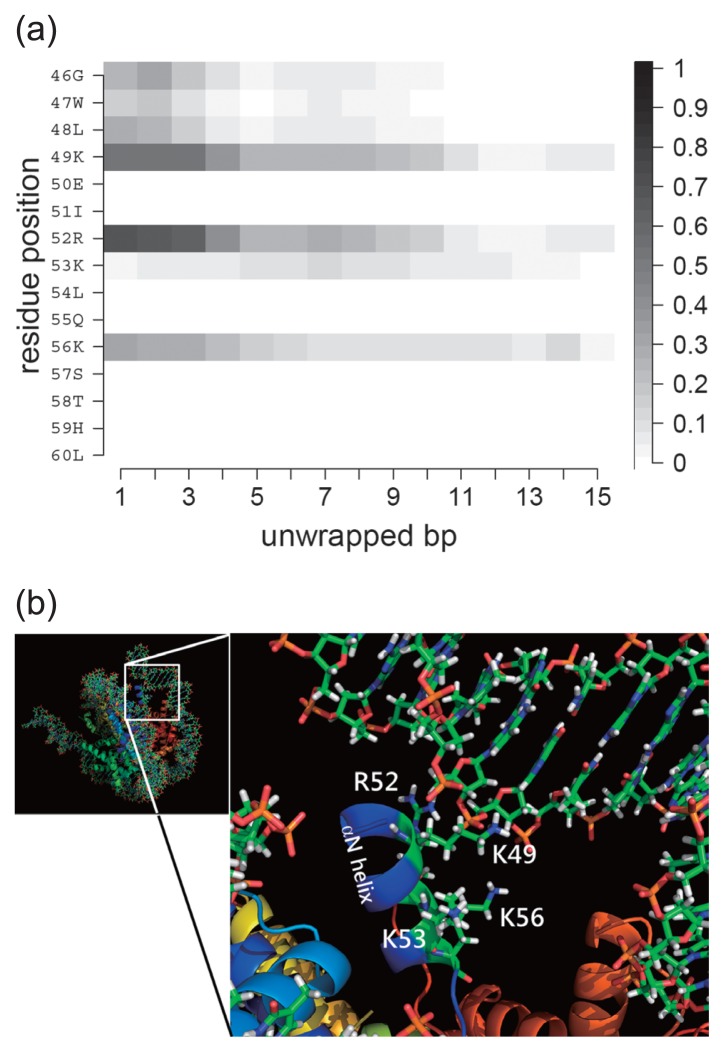
To investigate the interaction between histones and DNA, we calculated residue-wise contact probabilities between protein and DNA as a function of the total number of unwrapped base pairs (Fig. 5). Three residues, K49, R52 and K56 formed hydrogen bonds with DNA in the fully wrapped state. We also observed contacts in G46, W47 and L48 with DNA through their mainchain atoms. Surprisingly, these contacts were somehow maintained up to 10 bp unwrapped states, indicating that these residues were dragged by unwrapped DNA. In contrast, the contacts of most of the residues were lost at 4 bp unwrapped state in the canonical NCP [20], probably because the αN helix in which these residues are involved is longer than that of CENP-A NCP and has a stronger packing to the histone H4.
Figure 5.
Histone-DNA contacts changing as a function of the number of unwrapped bps at one DNA end. (a) CENP-A-DNA end contacts. Plotted are the contact probabilities of each residue in the conformational ensemble. Contact is counted if at least one pair of atoms in the histone and DNA is within 4 Å. (b) Close-up view of the CENP-A-DNA end interface. Residues 46 to 60 of CENP-A are located between the outer DNA and the inner DNA. Residues K49, R52, K53 and K56 and the α-N helix are labeled.
Discussion
Nucleosomes are the fundamental structural unit of chromatin; however, there exist many different types of nucleosomes each of which is constituted with histone variants or histones with posttranslational modification although their 3D structures are essentially similar to each other. The question to be addressed is what the differences are among different types of nucleosomes and how they contribute to different roles of individual nucleosomes. Here, we compare the CENP-A NCP with the canonical NCP in terms of free energy profile.
Comparison with the canonical NCP
The free energy comparison with the canonical NCP clearly indicates that canonical NCP is the most stable when DNA is fully wrapped around the histone core [20] whereas the most of CENP-A NCP populations adopt various conformations in which both ends of DNA are partially unwrapped. In the canonical NCP, once the first 5 bp of DNA were unwrapped from either end of DNA, the unwrapping proceeded spontaneously up to 10 bp in the same end. It went further up to 20 bp with an increase in free energy of less than 1 kcal/mol. Except for the first 5 bp unwrapping, the unwrapping did not require any work up to 23 bp in CENP-A NCP. The contacts of the first 5 bp in the DNA end with the histone are mainly from the αN helix and the precedent loop. Thus, the difference in stability is produced from the structural difference of this helix. Our previous MD simulations demonstrated that this strong interaction is likely to come from Arg residues located in the αN helix of H3 histone [7]. The corresponding residues in CENP-A NCP are Lys which can easily lose hydrogen bonds with DNA compared with Arg. In CENP-A NCP, the entropy from fluctuation of DNA and residues which lost interaction with the DNA will over-whelm the gain of enthalpy by the hydrogen bonds. The unwrapping states are completely different between CENP-A NCP and the canonical NCP. In CENP-A NCP, conformations with 15 to 22 bp unwrapped in total are stable.
The unwrapped bp difference map indicates that either end of DNA always has 11 bp unwrapped in the stable conformations and one of the ends repeats unwrapping and wrapping. In contrast, the canonical NCP starts to unwrap one of the ends and once the unwrapping reaches 10 bp, another end starts unwrapping. Therefore, conformations with 3 to 4 bp unwrapped from both ends were rarely observed in the canonical NCP while such conformations did exist in the CENP-A NCP (Fig. 4). This characteristic of CENP-A may be an advantage for providing the scaffold for proteins which are required to form the kinetochore.
Comparison with crystal and cryo-EM structures
In this study, the simulations were carried out based on the crystal structure of CENP-A NCP [18] in which 13 bp from each end of DNA are invisible. Our free energy profile has a minimum at 22 bp (Fig. 3a) where the most dominant conformation is 12 bp uwrapped from one end and 10 bp from the other end (Fig. 4). This agrees well with the invisible region in the crystal structure. Recently, another structure of CENP-A NCP has been determined by cryo-EM analysis [19]. In this structure, both ends of DNA are visible, but the ends are shifted outward by about 6 Å compared with the canonical NCP, consistent with the result in our simulation that CENP-A NCP favors more open conformations.
A linker histone H1 is known to stabilize nucleosomes by binding at the dyad region and bridging two linker DNAs. Roulland, Y., et al. showed that the flexibility of DNA ends of CENP-A NCP prevent H1 recruitment and that the flexibility can be suppressed by replacing the αN helix of CENP-A with that of H3 [34]. This indicates that the flexibility is facilitated by the short αN helix of CENP-A. Our simulation strongly supports that CENP-A NCP favors 6 bp or more unwrapped conformations from either or both ends.
Comparison with DNA unzipping studies
Dechassa, M. L., et al. reconstituted budding yeast centromeric nucleosomes to probe the structure of the centromeric NCPs containing the human analog of Cse4 by SAXS, MNase digestion and single-molecule DNA unzipping [35]. Their SAXS and MNase digestion data showed that Cse4-containing NCPs have fewer interactions with the outer turn DNA than the canonical one does (110 to 120 bp of DNA wrapped around the histone core) and adopt more extended conformation. Furthermore, the single molecule unzipping data showed that the peak forces for unzipping in super helical location (SHL) 3.5 to 6.5 regions of the Cse4-nucleosomes were clearly lower than those of yeast H3-nucleosomes. This interpretation may be unconvincing because in SHL 3.5 to 6 regions, both nucleosomes have the same H2A-H2B dimer-DNA interactions although they have different αN helix-DNA interactions in SHL 6 to 7 regions. In fact, our simulations showed that a similar, sharp increase of the free energy was observed in both H3 and CENP-A profiles (after 25 to 27 bp unwrapped). It should be noted that there is some ambiguity in mapping of DNA position in which unwrapping occurs from the unzipping experimental data because the experiment measures the shift of coverslip to which one end of DNA attached under a constant force [36]. As we shown in Figure 3b, the DNA end to end distance has a relatively wide distribution against the total number of unwrapped bp. For example, at 15 bp unwrapped state in total, the distance can be 90 to 110 Å, which leads to ambiguity in mapping the DNA position where the change in the force occurs.
Supplementary Material
Acknowledgements
This paper is dedicated to the celebration of Professor Nobuhiro Go’s 80th birthday. We here appreciate his initial contribution for staring our research group and continuous encouragement. This research has been funded by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) Strategic Programs for Innovative Research, Computational Life Science and Application in Drug Discovery and Medical Development (hp170255, hp180191, hp190171) and by JSPS KAKENHI (No. 25116003 and JP18H05534) to HK and hp180027, the JSPS KAKENHI (18K06173) to HI. This work was also partly supported by the Platform Project for Supporting Drug Discovery and Life Science Research (BINDS) from AMED (JP19am0101106) to HK.
Footnotes
Conflict of Interest
Authors declare no conflicts of interest.
Author Contribution
H. K. designed the research. H. K. carried out all the simulations and H. I. and S. S. supported for carrying out simulations and a part of analysis. H. K. wrote the manuscript. H. I and S. S. checked, edited and agreed to the manuscript.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Ettig R, Kepper N, Stehr R, Wedemann G, Rippe K. Dissecting DNA-histone interactions in the nucleosome by molecular dynamics simulations of DNA unwrapping. Biophys J. 2011;101:1999–2008. doi: 10.1016/j.bpj.2011.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erler J, Zhang R, Petridis L, Cheng X, Smith JC, Langowski J. The Role of Histone Tails in the Nucleosome: A Computational Study. Biophys J. 2014;107:2911–2922. doi: 10.1016/j.bpj.2014.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop TC. Molecular dynamics simulations of a nucleosome and free DNA. J Biomol Struct Dyn. 2005;22:673–686. doi: 10.1080/07391102.2005.10507034. [DOI] [PubMed] [Google Scholar]
- 5.Biswas M, Langowski J, Bishop TC. Atomistic simulations of nucleosomes. Wiley Interdiscip Rev: Comput Mol Sci. 2013;3:378–392. [Google Scholar]
- 6.Biswas M, Voltz K, Smith JC, Langowski J. Role of histone tails in structural stability of the nucleosome. PLoS Comput Biol. 2011;7:e1002279. doi: 10.1371/journal.pcbi.1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kono H, Shirayama K, Arimura Y, Tachiwana H, Kurumizaka H. Two Arginine Residues Suppress the Flexibility of Nucleosomal DNA in the Canonical Nucleosome Core. PLoS One. 2015;10:e0120635. doi: 10.1371/journal.pone.0120635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, Kono H. Distinct Roles of Histone H3 and H2A Tails in Nucleosome Stability. Sci Rep. 2016;6:31437. doi: 10.1038/srep31437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaytan AK, Armeev GA, Goncearenco A, Zhurkin VB, Landsman D, Panchenko AR. Coupling between Histone Conformations and DNA Geometry in Nucleosomes on a Microsecond Timescale: Atomistic Insights into Nucleosome Functions. J Mol Biol. 2016;428:221–237. doi: 10.1016/j.jmb.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voltz K, Trylska J, Calimet N, Smith JC, Langowski J. Unwrapping of nucleosomal DNA ends: a multiscale molecular dynamics study. Biophys J. 2012;102:849–858. doi: 10.1016/j.bpj.2011.11.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winogradoff D, Echeverria I, Potoyan DA, Papoian GA. The Acetylation Landscape of the H4 Histone Tail: Disentangling the Interplay between the Specific and Cumulative Effects. J Am Chem Soc. 2015;137:6245–6253. doi: 10.1021/jacs.5b00235. [DOI] [PubMed] [Google Scholar]
- 12.Winogradoff D, Zhao H, Dalal Y, Papoian GA. Shearing of the CENP-A dimerization interface mediates plasticity in the octameric centromeric nucleosome. Sci Rep. 2015;5:17038. doi: 10.1038/srep17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saurabh S, Glaser MA, Lansac Y, Maiti PK. Atomistic Simulation of Stacked Nucleosome Core Particles: Tail Bridging, the H4 Tail, and Effect of Hydrophobic Forces. J Phys Chem B. 2016;120:3048–3060. doi: 10.1021/acs.jpcb.5b11863. [DOI] [PubMed] [Google Scholar]
- 14.Bowerman S, Wereszczynski J. Effects of MacroH2A and H2A.Z on Nucleosome Dynamics as Elucidated by Molecular Dynamics Simulations. Biophys J. 2016;110:327–337. doi: 10.1016/j.bpj.2015.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winogradoff D, Aksimentiev A. Molecular Mechanism of Spontaneous Nucleosome Unraveling. J Mol Biol. 2019;431:323–335. doi: 10.1016/j.jmb.2018.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishida H, Kono H. H4 tails potentially produce the diversity in the orientation of two nucleosomes. Biophys J. 2017;113:978–990. doi: 10.1016/j.bpj.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Kono H. Investigating the Influence of Arginine Dimethylation on Nucleosome Dynamics Using All-Atom Simulations and Kinetic Analysis. J Phys Chem B. 2018;122:9625–9634. doi: 10.1021/acs.jpcb.8b05067. [DOI] [PubMed] [Google Scholar]
- 18.Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 19.Zhou BR, Yadav KNS, Borgnia M, Hong J, Cao B, Olins AL, et al. Atomic resolution cryo-EM structure of a native-like CENP-A nucleosome aided by an antibody fragment. Nat Commun. 2019;10:2301. doi: 10.1038/s41467-019-10247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kono H, Sakuraba S, Ishida H. Free Energy Profiles for Unwrapping the Outer Superhelical Turn of Nucleosomal DNA. PLoS Comput Biol. 2018;14:e1006024. doi: 10.1371/journal.pcbi.1006024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Babin V, Roland C, Sagui C. Adaptively biased molecular dynamics for free energy calculations. J Chem Phys. 2008;128:134101. doi: 10.1063/1.2844595. [DOI] [PubMed] [Google Scholar]
- 23.Raiteri P, Laio A, Gervasio FL, Micheletti C, Parrinello M. Efficient reconstruction of complex free energy landscapes by multiple walkers metadynamics. J Phys Chem B. 2006;110:3533–3539. doi: 10.1021/jp054359r. [DOI] [PubMed] [Google Scholar]
- 24.Ishida H. Branch migration of Holliday junction in RuvA tetramer complex studied by umbrella sampling simulation using a path-search algorithm. J Comput Chem. 2010;31:2317–2329. doi: 10.1002/jcc.21525. [DOI] [PubMed] [Google Scholar]
- 25.Ishida H. Essential function of the N-termini tails of the proteasome for the gating mechanism revealed by molecular dynamics simulations. Proteins. 2014;82:1985–1999. doi: 10.1002/prot.24553. [DOI] [PubMed] [Google Scholar]
- 26.Ishida H, Hayward S. Path of nascent polypeptide in exit tunnel revealed by molecular dynamics simulation of ribosome. Biophys J. 2008;95:5962–5973. doi: 10.1529/biophysj.108.134890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishida H, Matsumoto A. Free-energy landscape of reverse tRNA translocation through the ribosome analyzed by electron microscopy density maps and molecular dynamics simulations. PLoS One. 2014;9:e101951. doi: 10.1371/journal.pone.0101951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishida H, Matsumoto A. Mechanism for verification of mismatched and homoduplex DNAs by nucleotides-bound MutS analyzed by molecular dynamics simulations. Proteins. 2016;84:1287–1303. doi: 10.1002/prot.25077. [DOI] [PubMed] [Google Scholar]
- 29.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, SImmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez A, Marchan I, Svozil D, Sponer J, Cheatham TE, Laughton CA, et al. Refinement of the AMBER force field for nucleic acids: improving the description of α/γ conformers. Biophys J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joung IS, Cheatham TE., 3rd Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J Phys Chem B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 33.Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman PA. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 34.Roulland Y, Ouararhni K, Naidenov M, Ramos L, Shuaib M, Syed SH, et al. The Flexible Ends of CENP-A Nucleosome Are Required for Mitotic Fidelity. Mol Cell. 2016;63:674–685. doi: 10.1016/j.molcel.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 35.Dechassa ML, Wyns K, Li M, Hall MA, Wang MD, Luger K. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun. 2011;2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat Struct Mol Biol. 2009;16:124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.