This article reports results of a comprehensive genomic profiling to determine the prevalence of CDK12 loss‐of‐function genomic alterations in multiple tumor types and associations with focal tandem duplications, potentially leading to a genomic biomarker of immunotherapy benefit.
Keywords: CDK12, Tandem duplication, Immunotherapy, Genomics
Abstract
Background.
CDK12 loss‐of‐function (LOF) genomic alterations are associated with focal tandem duplications (FTDs) in ovarian and prostate cancers. Because these FTDs may produce fusion‐induced neoantigens (FINAs), CDK12 alteration is a candidate biomarker for immune checkpoint inhibitor sensitivity. Here we determine the prevalence of CDK12‐LOF alterations and their association with FTDs across diverse tumor types.
Materials and Methods.
A total of 142,133 tumor samples comprising 379 cancer types were sequenced (August 2014 to April 2018) by hybrid capture‐based comprehensive genomic profiling (Foundation Medicine, Cambridge, MA) as part of routine clinical care. Results were analyzed for base substitutions, short insertions/deletions, rearrangements, and copy number alterations. CDK12‐LOF genomic alterations were assessed for zygosity status and association with FTDs/focal copy number gain.
Results.
CDK12 genomic alterations were detected in 1.1% of all cases, most frequently in prostate cancer (5.6%), but were also observed at >1% frequency in 11 cancer types. Across multiple cancer types, including prostate, gastric/esophageal, ovarian, breast, and endometrial cancer, the number of FTDs was significantly increased in CDK12‐LOF versus CDK12 wild‐type cases. Notably, CDK12‐LOF was not consistently associated with a homologous recombination deficiency genomic signature. Quantitative assessment of CDK12‐associated FTDs by measurement of single copy number gains identified novel likely deleterious CDK12 kinase‐domain mutations in prostate and ovarian cancers.
Conclusion.
Detection of CDK12‐LOF genomic alterations and their association with FTDs in a diverse spectrum of malignancies suggests that immunotherapy approaches targeting FINAs derived from CDK12‐associated FTDs may be a broadly applicable strategy that could be explored across cancer types in a tumor‐agnostic manner.
Implications for Practice.
CDK12 inactivation in ovarian and prostate cancer results in the generation of focal tandem duplications, which can cause fusion‐induced neoantigens. In prostate cancer, CDK12 alterations have demonstrated promise as a potential predictive biomarker for response to immune checkpoint blockade. This study evaluated genomic profiling data from >142,000 tumors to determine the prevalence of CDK12 loss‐of‐function genomic alterations across tumor types and demonstrated that CDK12 alterations are associated with the tandem‐duplicator phenotype in cancer types other than ovarian and prostate cancer. The association of CDK12 alterations with focal tandem duplications across broad cancer types suggests that CDK12 inactivation warrants further investigation as a pan‐cancer biomarker for immunotherapy benefit.
Introduction
Recurrent loss‐of‐function (LOF) alterations in the CDK12 tumor suppressor gene, which encodes cyclin‐dependent kinase 12, have been described in prostate and ovarian cancers [1], [2], [3], [4]. The CDK12‐cyclin K heterodimer regulates RNA polymerase II [5] and is implicated in transcriptional control of homologous recombination repair genes [6]. Therefore, CDK12 genomic alteration (GA) has emerged as a potential attractive candidate biomarker for synthetic lethal targeting by poly (ADP‐ribose) polymerase (PARP) inhibitors [7]. However, preliminary clinical evidence suggests that PARP inhibitor therapy may be largely ineffective in treatment of patients with advanced prostate cancer harboring CDK12 GAs [8].
In ovarian and prostate cancers, CDK12 loss‐of‐function (CDK12‐LOF) alterations are not associated with a homologous recombination deficiency (HRD) phenotype but are instead associated with the tandem‐duplicator phenotype (TDP), a genomic signature characterized by focal tandem duplications (FTDs) with a bimodal size distribution with modes of either 0.2–0.4 Mb or 1.7–3.0 Mb [1], [3], [9], [10]. CDK12‐associated FTDs can result in expressed gene fusions and fusion‐induced neoantigens (FINAs), raising the possibility of CDK12‐LOF alteration as a predictive biomarker for immune checkpoint inhibitor (ICI) sensitivity [1]. In prostate cancer, CDK12‐LOF alterations are associated with increased immune infiltrates, and some patients with CDK12‐altered metastatic castration‐resistant prostate cancer have derived clinical benefit from ICI therapy [1].
In this study, we assess comprehensive genomic profiling results from 142,133 cancer specimens to determine the prevalence of CDK12‐LOF alterations and their association with FTDs in multiple tumor types beyond ovarian and prostate cancers. We hypothesized that CDK12 inactivation would be observed at a reasonable frequency in many cancer types, potentially leading to a tumor‐agnostic genomic biomarker of immunotherapy benefit.
Materials and Methods
Approval for this study, including a waiver of informed consent and Health Insurance Portability and Accountability Act waiver of authorization, was obtained from the Western Institutional Review Board (Protocol 20152817). Tumor tissue specimens (n = 142,133; 44% metastatic site specimens) underwent comprehensive genomic profiling (CGP) using a validated hybrid capture‐based next‐generation sequencing (NGS) assay (FoundationOne) [11] in a Clinical Laboratory Improvement Amendments‐certified, College of American Pathologists‐accredited, New York State‐approved laboratory (Foundation Medicine, Cambridge, MA). CGP was performed on hybridization‐captured, adaptor ligation‐based libraries to a median coverage depth of 764× for exons of 395 cancer‐related genes plus select introns from 31 genes frequently rearranged in cancer (supplemental online Table 1). Results were analyzed for base substitutions, short insertions/deletions (indels), rearrangements, and copy number alterations (amplification and homozygous deletion). Custom filtering was applied to remove benign germline variants as described [12]. CDK12 GAs included protein‐truncating mutations, homozygous deletions, genomic rearrangements, and likely pathogenic kinase‐domain missense mutations (supplemental online Table 2); all other alterations were classified as variants of unknown significance (VUS). Zygosity status for mutations was determined as previously described [13]. Bi‐allelic alterations were defined as (a) mutations with loss of heterozygosity (LOH) at the wild‐type allele, as determined by zygosity status [13], (b) copy number loss (homozygous deletion), or (c) ≥2 CDK12 GAs in a given sample. Percent genome‐wide loss of heterozygosity (gLOH) was used as a surrogate marker of HRD status and was calculated as previously described, with a gLOH score of ≥16% being designated as gLOH‐High [14]. FTD burden was assessed by counting the number of detected duplicating rearrangements per case. FTD‐derived gene fusions were defined as FTDs that resulted in an in‐strand juxtaposition of two coding sequences. For prostate cancer, the TMPRSS2‐ERG duplication was excluded from the FTD burden score because this driver event has been shown to be independent of CDK12 inactivation [1]. Single copy number gain (CN + 1) score was derived by evaluating the number of segments (between 10 kb and 10 Mb) with single copy gains relative to the median ploidy of the sample. CN + 1 score was assessable for cases that met copy number metrics based on the intersegment signal‐to‐noise ratios of normalized and GC corrected log‐2‐ratio data; samples with low signal‐to‐noise ratio were excluded from analysis.
Results
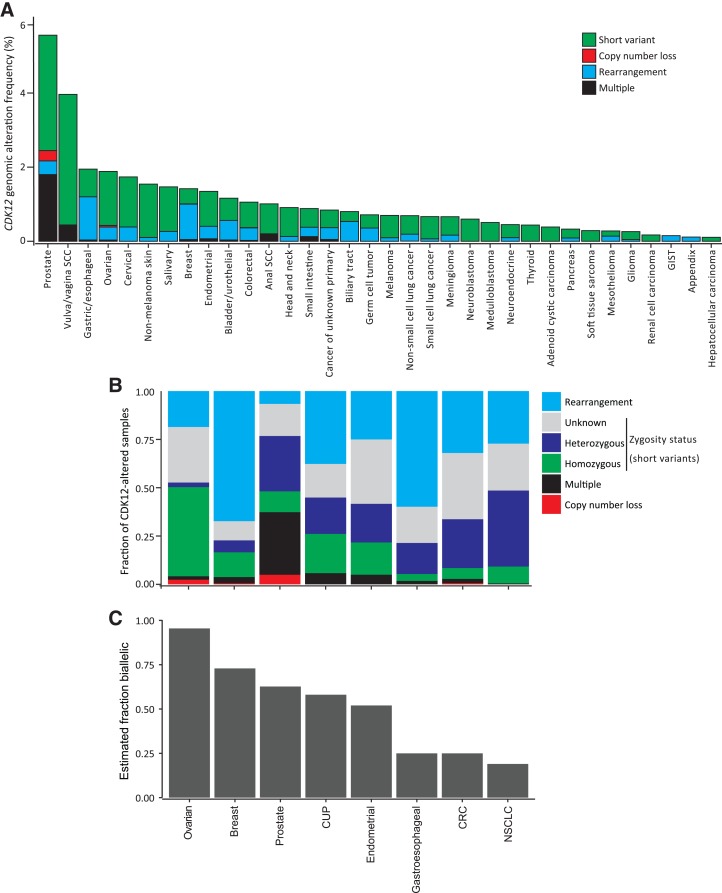
To assess the prevalence of at least mono‐allelic CDK12 GAs across tumor types, we assessed CGP results from 142,133 tumors that were sequenced in the context of routine clinical care (Fig. 1A; supplemental online Fig. 1; supplemental online Table 3). Overall, CDK12 GAs were observed in 1.1% of cases. As expected, CDK12 GAs were most prevalent in prostate cancer (5.6%). Across other cancer types, CDK12 GAs were observed at a 0.9% frequency overall, most commonly in vulvar/vaginal squamous cell carcinoma (4.0%), gastric/esophageal cancer (1.9%), ovarian cancer (1.9%), cervical cancer (1.7%), nonmelanoma skin cancer (1.7%), salivary gland cancer (1.5%), breast cancer (1.4%), endometrial cancer (1.3%), bladder/urothelial cancer (1.1%), and colorectal cancer (1.1%). In most other diseases, CDK12 GAs were detected below a 1% prevalence.
Figure 1.
Pan‐cancer analysis of CDK12 alterations. (A): Frequency of CDK12 loss‐of‐function (LOF) alterations across multiple tumor types. Cancer types included are those with at least 100 sequenced samples and with ≥1 case with CDK12‐LOF alteration. Full details of cancer‐type prevalence are included in supplemental online Table 3. “Multiple” indicates samples with ≥2 concurrent CDK12 genomic alterations (GAs). (B): Relative fraction of cases with each class of CDK12 alteration. Cancer types with ≥50 CDK12 GA cases were individually assessed in this analysis. Short variant alterations were evaluated for zygosity. (C): Bi‐allelic fraction was determined for cases where mono‐allelic versus bi‐allelic status could be determined. Bi‐allelic alterations included copy number loss, mutation with wild‐type copy under loss of heterozygosity, or multiple detected alterations in a given case. Mono‐allelic mutations were those determined as heterozygous.
Abbreviations: CRC, colorectal cancer; CUP, cancer of unknown primary; GIST, gastrointestinal stromal tumor; NSCLC, non‐small cell lung cancer; SCC, squamous cell carcinoma.
To estimate the fraction of CDK12 GAs likely to result in bi‐allelic inactivation, we examined the relative fraction of CDK12‐altered cases harboring each different class of GA (Fig. 1B); to ensure sufficient power to evaluate correlation of CDK12 genotype with phenotype, cancer types with a raw count of ≥50 cases with CDK12 GAs were chosen for in‐depth analysis. As demonstrated, bi‐allelic CDK12 inactivation by copy number loss was most often observed in prostate (0.3% overall, 5.0% of CDK12‐altered specimens) and ovarian cancers (0.05% overall, 2.4% of CDK12‐altered specimens) but was not generally observed in other disease types (Fig. 1A, 1B). Short variant mutations were assessed for zygosity status to determine whether CDK12 GAs were bi‐allelic (pathogenic mutation, with LOH of the wild‐type allele) or mono‐allelic (heterozygous mutation); the ratio of bi‐allelic to mono‐allelic short variant mutations was highest in ovarian and breast cancers (Fig. 1B). Multiple CDK12 GAs in a given tumor were presumed bi‐allelic based on the association of such alterations with FTDs in prostate cancer [3]: 32.4% of prostate cancer cases with CDK12 GAs harbored multiple alterations, but multiple alterations were also observed in CDK12‐altered cancer of unknown primary (5.8%) and endometrial cancers (5.0%). For the CDK12‐altered cases where bi‐allelic/mono‐allelic status could be reliably determined, we estimated the fraction of cases with bi‐allelic alteration (Fig. 1C): the majority of ovarian, breast, prostate, unknown primary, and endometrial cancer cases harbored bi‐allelic CDK12 GAs. Interestingly, in contrast to other cancer types, CDK12 GAs in breast and gastroesophageal cancers were primarily rearrangements (Fig. 1B; 67.3% and 59.8% of CDK12‐altered specimens, respectively); however, bi‐allelic/mono‐allelic status could not generally be determined for such rearrangements.
In prior literature, CDK12 alterations have been associated with two distinct genomic signatures (TDP and HRD) [1], [3], [7], [9]; therefore, we assessed the association between CDK12 GAs with these phenotypes by quantifying FTD burden (the number of detected duplicating rearrangements per sample) and percentage gLOH, respectively. Because heterozygous CDK12 short variant mutations potentially represent passenger events, we focused this analysis on CDK12 bi‐allelic alterations or rearrangements only (referred to as CDK12‐LOF alterations). Consistent with previous studies, for ovarian and prostate cancers, FTD burden was significantly increased in samples with CDK12‐LOF compared with CDK12 wild‐type (CDK12‐WT) samples (Fig. 2A).
Figure 2.
CDK12 alteration and the tandem‐duplicator phenotype (TDP). (A): The TDP was quantified by evaluating the number of FTDs detected per case. The distribution of FTD burden was compared for CDK12‐WT versus CDK12‐LOF for the overall data set (“all samples”), eight individual tumor types with ≥50 CDK12 genomic alteration samples, and all other disease types that were grouped together and analyzed as a single group (“all other”). (B): Histogram showing the relative size distribution of FTDs identified in CDK12‐WT and CDK12‐LOF cases.
Abbreviations: CRC, colorectal cancer; CUP, cancer of unknown primary; FTD, focal tandem duplication; LOS, loss‐of‐function; NSCLC, non‐small cell lung cancer; WT, wild‐type.
CDK12‐LOF was also significantly associated with increased FTD burden across all other cancer types examined (Fig. 2A). The effect size was greatest in prostate cancer, where the mean number of FTDs per tumor was 1.05 for CDK12‐LOF versus 0.13 for CDK12‐WT samples (odds ratio [OR] = 10.9, p = 9.2 × 10−37), but it was also high in cancer of unknown primary, endometrial, ovarian, gastroesophageal, and colorectal cancers (Fig. 2A; supplemental online Table 4). When considering all CDK12 GAs including heterozygous mutations, there were also more FTDs observed relative to CDK12‐WT; however, the magnitude of the association in each cancer type was reduced (supplemental online Table 4). Consistent with prior studies [1], [3], [9], [10], FTDs in cases harboring CDK12‐LOF alterations had a bimodal size distribution with peaks around 0.4 Mb and 2.5 Mb (Fig. 2B). The fraction of FTDs giving rise to potential gene fusions was similar irrespective of CDK12 genotype (11% [44/410] for CDK12‐LOF tumors vs. 12% [1,621/13,847] for CDK12 wild‐type tumors); therefore, the enrichment of FTDs in CDK12‐LOF cases is also predicted to result in an enrichment in gene fusions that when expressed potentially result in fusion‐associated neoantigens [1], [4].
When assessed across all samples, CDK12‐LOF was associated with HRD (increased gLOH); however, in contrast to the pan‐cancer association of CDK12‐LOF alterations with FTDs, the association of CDK12‐LOF with HRD was specific to certain cancer types: CDK12‐LOF was associated with increased gLOH in non‐small cell lung cancer and cancer of unknown primary but not for the other six cancer types assessed (supplemental online Table 5; supplemental online Fig. 2); for all other cancer types without sufficient CDK12‐LOF samples to be assessed individually, there was an overall association between CDK12‐LOF and HRD. These results suggest that the role of CDK12 in homologous recombination repair may be tumor type context‐dependent.
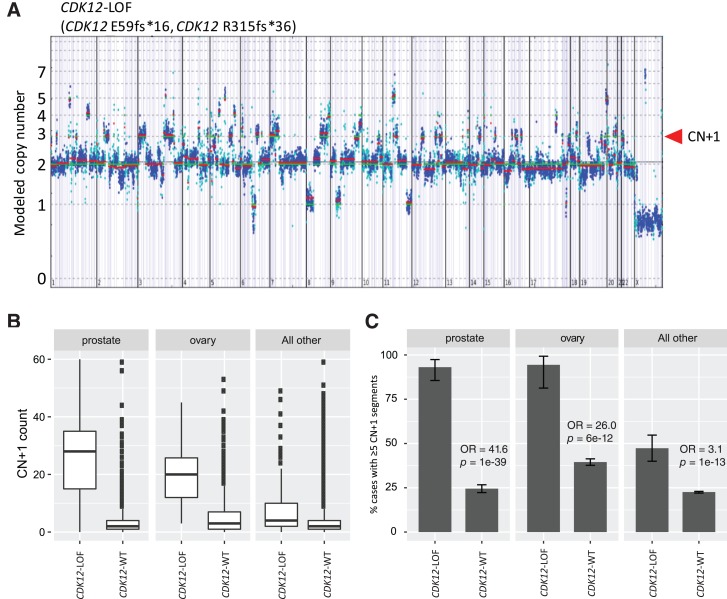
In ovarian and prostate cancers, CDK12‐associated FTDs result in a distinct copy number profile with single copy gains dispersed throughout the genome (Fig. 3A) [1], [3], [9]. In contrast to FTDs that are detected at precise rearrangement breakpoints, copy gains are measurable over the entire duplicated segment and therefore provide a further opportunity to quantify the TDP. As a surrogate measure of FTDs, we generated a CN + 1 score by evaluating the number of segments (between 10 kb and 10 Mb) with single copy gains greater than the ploidy of the sample. We assessed the association of CDK12‐LOF genotype with CN + 1 scores in ovarian and prostate cancers. CDK12‐LOF cases had significantly higher CN + 1 scores compared with CDK12‐WT samples (Fig. 3B; supplemental online Table 6). The fraction of cases with ≥5 CN + 1 segments was increased in CDK12‐LOF tumors for prostate cancer (OR = 41.6, p = 10−39) and ovarian cancer (OR = 26.0, p = 10−12) and also significantly increased in CDK12‐LOF cases for all other non‐prostate/ovarian cancer types, albeit with reduced magnitude (OR = 3.1, p = 10−13; Fig. 3C; supplemental online Table 6).
Figure 3.
CDK12 alterations are associated with copy number gains. (A): Example of copy number plot from a CDK12‐LOF prostate tumor harboring multiple CDK12 truncating mutations. The red arrow indicates short chromosomal segments at copy number 3 (CN + 1), indicative of focal tandem duplications. x‐axis indicates copy number, y‐axis indicates genomic position ordered by chromosome number. (B): The tandem‐duplicator phenotype was quantified by CN + 1 score for CDK12‐WT versus CDK12‐LOF cases in ovarian, prostate, and all other cancer types. Box‐and‐whisker plots: box spans first and third quartiles, the median is denoted by the horizontal line in the box, whiskers indicate maximum and minimum values within 1.5× the interquartile range, black dots indicate outlier events. (C): The percentage of cases harboring ≥5 CN + 1 segments was compared for CDK12‐WT and CDK12‐LOF cases for prostate, ovarian, and all other cancer types. Error bars indicate 95% confidence interval.
Abbreviations: CN + 1, copy number gain; LOF, loss‐of‐function; OR, odds ratio; WT, wild‐type.
We then asked whether there were any distinctive features associated with CDK12‐WT cases harboring high CN + 1 scores. We evaluated prostate and ovarian tumors where the association with CN + 1 score and CDK12 status was strongest, and in doing so, we found that CDK12 VUS were significantly enriched in CDK12‐WT cases with high CN + 1 scores versus those with low CN + 1 scores (Fig. 4A, 4B). The majority of the CDK12 VUS mutations in high CN + 1 cases were located in the kinase domain (31/48, 65%); in contrast, fewer CDK12 kinase domain VUS were found in low CN + 1 cases (8/37, 22%; Fig. 4A, 4C). Although these CDK12 VUS alterations that were associated with high CN + 1 scores have not been previously been functionally characterized as deleterious mutations or observed as somatic mutations in cancer, mutated residues included those involved in kinase activation (H857Y/R, F878S, T893I) or interactions between CDK12 and its heterodimeric cyclin K partner (G807R), as well as recurrently mutated residues, suggesting functional importance (Fig. 4) [15].
Figure 4.
CDK12 variants of unknown significance (VUS). (A): Details of CDK12 VUS alterations identified in CDK12‐wild‐type (WT) prostate and ovarian cancer cases and the number of CN + 1 segments identified in each case. (B): CDK12‐WT cases at different CN + 1 thresholds were evaluated for the frequency at which CDK12 VUS alterations were identified. (C): CDK12 VUS alterations identified in CDK12‐WT cases with ≥5 CN + 1 segments and their location relative to the kinase domain.
Abbreviations: CN + 1, copy number gain; OR, odds ratio; VUS, variants of unknown significance.
Discussion
The recent tumor‐agnostic approval of ICI agents targeting the programmed cell death 1 (PD‐1) checkpoint for patients with mismatch repair deficiency and/or microsatellite instability has demonstrated the clinical utility of biomarker strategies based on high predicted neoantigen burden to select patients likely to derive benefit from ICI. CDK12‐LOF is associated with expressed fusion‐induced neoantigens derived from FTDs and has therefore emerged as a potential candidate predictive biomarker for ICI sensitivity in prostate cancer [1], [4]. However, CDK12 status and its association with FTDs has not previously been studied beyond ovarian and prostate cancer.
To assess the pan‐cancer landscape of CDK12‐LOF genomic alterations and to evaluate whether the association with FTD extends broadly across tumor types, we assessed our database of 142,133 consecutive tumor samples that were sequenced using a clinical‐grade CGP assay. Although CDK12 alterations have been reported in The Cancer Genome Atlas (TCGA) [16], the specimens evaluated in this study were from routine clinical testing enriched in patients with advanced disease [17], [18]; these results therefore represent the landscape of CDK12 alteration in the real‐world setting that may be distinct from unselected primary tumors. We identify CDK12 GA in 1.1% of all cancers; although the overall prevalence of CDK12 GAs is relatively low, consistent with an analysis of CDK12 alteration prevalence in the TCGA [16], specific cancer types including gastroesophageal, breast, and endometrial harbored CDK12 GAs at a > 1% frequency. Notably, NGS‐based screening strategies have enabled successful completion of basket trials targeting NTRK alterations that also occur at low frequencies [19], recently leading to the approval of larotrectinib for NTRK fusion‐positive cancers of any histology.
Our understanding of the functional role of CDK12 is evolving. It was originally thought that CDK12 may be a mediator of homologous recombination DNA repair, and that CDK12‐LOF mutations would result in homologous recombination deficiency [7]. However, consistent with more recent data from human ovarian and prostate cancers [1], [9], we found no association between CDK12‐LOF alterations and the HRD phenotype (as measured by gLOH) in prostate and ovarian cancers, and only marginal associations in other cancer types evaluated. This finding is in keeping with emerging clinical data suggesting that PARP inhibitors are largely ineffective in prostate cancers with CDK12 deficiency [8]. Therefore, the role of CDK12 in homologous recombination repair and the role of CDK12 alteration as a biomarker for PARP inhibitors might be limited to specific tumor types. To this end, our results demonstrating the association between CDK12‐LOF and the TDP across tumor types suggests immune checkpoint blockade may be a more rational approach that may be broadly applicable for the treatment of patients with CDK12‐altered cancers, and such basket trials evaluating CDK12 as a biomarker for ICI are currently underway (NCT03570619).
A limitation of this study is that we employed a targeted NGS assay (covering 1.1 Mb of sequenced DNA) rather than whole‐exome sequencing (WES) or whole‐genome sequencing (WGS); therefore; FTDs that may have occurred outside of the captured genomic regions would not be detected, resulting in limited sensitivity to robustly capture the CDK12‐associated FTD phenotype. Nevertheless, we were able to recapitulate the observation of CDK12‐associated FTD described in prior WGS or WES studies of ovarian and prostate cancers [1], [3], [9], and, using a much larger genomic data set of cancer samples, we further demonstrated that CDK12‐LOF is broadly associated with FTDs in many diverse tumor types; the association of CDK12 with FTDs in cancer types other than ovarian and prostate cancer has not been extensively evaluated in previous studies of CDK12 in the TCGA data sets [10], [16]. An additional limitation was our inability to interrogate genomic signatures in CDK12‐altered cancer types with fewer than 50 cases of CDK12 inactivation, therefore restricting cancer type‐specific analyses to only eight tumor types; however, CDK12‐LOF were also associated with FTDs in other tumor types overall (Fig. 2A). Finally, because we were unable to resolve the exact genomic coordinates of the FTD breakpoints, we were unable to assess actual or predicted gene‐fusion antigenicity or to estimate FINA burden.
CDK12 is emerging as a candidate genomic biomarker for ICI therapy, and potential biomarker strategies incorporating quantitative assessment of the CDK12‐associated TDP could provide additional information beyond CDK12 genotype alone. In contrast to WGS or WES that are usually employed in the research setting, targeted NGS is more routinely available as a tool in clinical practice; therefore, the ability to quantify CDK12‐associated TDP using targeted NGS may be required for clinical decision making. As an analogy, tumor mutational burden as determined by targeted NGS has been correlated with results from WES and is increasingly being used in the clinical arena [20]. In ovarian and prostate cancers in particular, quantitative measurement of the TDP using the CN + 1 score enabled further resolution of the FTD signature and also allowed post hoc identification of several recurrent kinase‐domain CDK12‐VUS alterations that are likely pathogenic. In other tumor types, CN + 1 score was not generally associated with CDK12 status, and further studies may be required to refine the CN + 1 score in non‐ovarian/prostate cancer histologies.
Conclusion
We report the first pan‐cancer analysis of CDK12 alterations across multiple tumor types, showing a modest prevalence (>1%) of this genetic alteration in at least 11 cancer types and association of CDK12‐LOF with increased FTD burden across all tumor types examined. We suggest that CDK12 warrants broad evaluation as a candidate genomic biomarker for ICI benefit, and that FTD signatures may further refine the predictive value of this potential marker.
See http://www.TheOncologist.com for supplemental material available online.
Contributed equally.
Contributor Information
Jon H. Chung, Email: jchung@foundationmedicine.com.
Emmanuel S. Antonarakis, Email: eantona1@jhmi.edu.
Author Contributions
Conception/design: Ethan S. Sokol, Jon H. Chung, Emmanuel S. Antonarakis
Provision of study material or patients: Ethan S. Sokol, Dean Pavlick, Garrett M. Frampton, Jeffrey S. Ross, Vincent A. Miller, Siraj M. Ali, Tamara L. Lotan, Drew M. Pardoll, Jon H. Chung, Emmanuel S. Antonarakis
Collection and/or assembly of data: Ethan S. Sokol, Dean Pavlick, Garrett M. Frampton, Jeffrey S. Ross, Vincent A. Miller, Siraj M. Ali, Tamara L. Lotan, Drew M. Pardoll, Jon H. Chung, Emmanuel S. Antonarakis
Data analysis and interpretation: Ethan S. Sokol, Dean Pavlick, Garrett M. Frampton, Jeffrey S. Ross, Vincent A. Miller, Siraj M. Ali, Tamara L. Lotan, Drew M. Pardoll, Jon H. Chung, Emmanuel S. Antonarakis
Manuscript writing: Ethan S. Sokol, Dean Pavlick, Garrett M. Frampton, Jeffrey S. Ross, Vincent A. Miller, Siraj M. Ali, Tamara L. Lotan, Drew M. Pardoll, Jon H. Chung, Emmanuel S. Antonarakis
Final approval of manuscript: Ethan S. Sokol, Dean Pavlick, Garrett M. Frampton, Jeffrey S. Ross, Vincent A. Miller, Siraj M. Ali, Tamara L. Lotan, Drew M. Pardoll, Jon H. Chung, Emmanuel S. Antonarakis
Disclosures
Ethan S. Sokol: Foundation Medicine (E, OI); Dean Pavlick: Foundation Medicine (E); Garrett M. Frampton: Foundation Medicine (E, OI); Jeffrey S. Ross: Foundation Medicine (E, OI); Vincent A. Miller: Foundation Medicine (E), Revolution Medicines (C/A); Siraj M. Ali: Foundation Medicine (E, OI); Jon H. Chung: Foundation Medicine/Roche (E, OI). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Wu YM, Cieślik M, Lonigro RJ et al. Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell 2018;173:1770–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter SL, Cibulskis K, Helman E et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012;30:413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Viswanathan SR, Ha G, Hoff AM et al. Structural alterations driving castration‐resistant prostate cancer revealed by linked‐read genome sequencing. Cell 2018;174:433–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antonarakis ES. Cyclin‐dependent kinase 12, immunity, and prostate cancer. N Engl J Med 2018;379:1087–1089. [DOI] [PubMed] [Google Scholar]
- 5.Bartkowiak B, Liu P, Phatnani HP et al. CDK12 is a transcription elongation‐associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev 2010;24:2303–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blazek D, Kohoutek J, Bartholomeeusen K et al. The cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev 2011;25:2158–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bajrami I, Frankum JR, Konde A et al. Genome‐wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res 2014;74:287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abida W, Bryce AH, Vogelzang NJ et al. 793PDPreliminary results from TRITON2: A phase II study of rucaparib in patients (pts) with metastatic castration‐resistant prostate cancer (mCRPC) associated with homologous recombination repair (HRR) gene alterations. Ann Oncol 2018;29(suppl 8). [Google Scholar]
- 9.Popova T, Manié E, Boeva V et al. Ovarian cancers harboring inactivating mutations in CDK12 display a distinct genomic instability pattern characterized by large tandem duplications. Cancer Res 2016;76:1882–1891. [DOI] [PubMed] [Google Scholar]
- 10.Menghi F, Barthel FP, Yadav V et al. The tandem duplicator phenotype is a prevalent genome‐wide cancer configuration driven by distinct gene mutations. Cancer Cell 2018;34:197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frampton GM, Fichtenholtz A, Otto GA et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartmaier RJ, Albacker L, Chmielecki J et al. High‐throughput genomic profiling of adult solid tumors reveals novel insights into cancer pathogenesis. Cancer Res 2017;77:2464–2475. [DOI] [PubMed] [Google Scholar]
- 13.Sun JX, He Y, Sanford E et al. A computational approach to distinguish somatic vs. germline origin of genomic alterations from deep sequencing of cancer specimens without a matched normal. PLoS Comput Biol 2018;14:e1005965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swisher EM, Lin KK, Oza AM et al. Rucaparib in relapsed, platinum‐sensitive high‐grade ovarian carcinoma (ARIEL2 Part 1): An international, multicentre, open‐label, phase 2 trial. Lancet Oncol 2017;18:75–87. [DOI] [PubMed] [Google Scholar]
- 15.Dixon‐Clarke SE, Elkins JM, Cheng SW et al. Structures of the CDK12/CycK complex with AMP‐PNP reveal a flexible C‐terminal kinase extension important for ATP binding. Sci Rep 2015;5:17122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lui GYL, Grandori C, Kemp CJ. CDK12: An emerging therapeutic target for cancer. J Clin Pathol 2018;71:957–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadaps M, Funchain P, Mahdi H et al. Precision oncology in solid tumors: A longitudinal tertiary care center experience. JCO Precis Oncol 10.1200/PO.18.00186. [DOI] [PubMed] [Google Scholar]
- 18.Singal G, Miller PG, Agarwala V et al. Association of patient characteristics and tumor genomics with clinical outcomes among patients with non–small cell lung cancer using a clinicogenomic database. JAMA 2019;321:1391–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drilon A, Laetsch TW, Kummar S et al. Efficacy of larotrectinib in TRK fusion–positive cancers in adults and children. N Engl J Med 2018;378:731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan TA, Yarchoan M, Jaffee E et al. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann Oncol 2019;30:44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]