Astroblastoma is a rare central nervous system tumor. This article reports on the common clinical and molecular features of astroblastoma in a cohort of 14 adults and one adolescent.
Keywords: Astroblastoma, Next‐generation sequencing, MN1‐BEND2, BRAF mutation
Abstract
Background.
Astroblastoma (ABM) is a rare glial brain tumor. Recurrent meningioma 1 (MN1) alterations have been recently identified in most pediatric cases. Adolescent and adult cases, however, remain molecularly poorly defined.
Materials and Methods.
We performed clinical and molecular characterization of a retrospective cohort of 14 adult and 1 adolescent ABM.
Results.
Strikingly, we found that MN1 fusions are a rare event in this age group (1/15). Using methylation profiling and targeted sequencing, most cases were reclassified as either pleomorphic xanthoastrocytomas (PXA)‐like or high‐grade glioma (HGG)‐like. PXA‐like ABM show BRAF mutation (6/7 with V600E mutation and 1/7 with G466E mutation) and CD34 expression. Conversely, HGG‐like ABM harbored specific alterations of diffuse midline glioma (2/5) or glioblastoma (GBM; 3/5). These latter patients showed an unfavorable clinical course with significantly shorter overall survival (p = .021). Mitogen‐activated protein kinase pathway alterations (including FGFR fusion, BRAF and NF1 mutations) were present in 10 of 15 patients and overrepresented in the HGG‐like group (3/5) compared with previously reported prevalence of these alterations in GBM and diffuse midline glioma.
Conclusion.
We suggest that gliomas with astroblastic features include a variety of molecularly sharply defined entities. Adult ABM harboring molecular features of PXA and HGG should be reclassified. Central nervous system high‐grade neuroepithelial tumors with MN1 alterations and histology of ABM appear to be uncommon in adults. Astroblastic morphology in adults should thus prompt thorough molecular investigation aiming at a clear histomolecular diagnosis and identifying actionable drug targets, especially in the mitogen‐activated protein kinase pathway.
Implications for Practice.
Astroblastoma (ABM) remains a poorly defined and controversial entity. Although meningioma 1 alterations seem to define a large subset of pediatric cases, adult cases remain molecularly poorly defined. This comprehensive molecular characterization of 1 adolescent and 14 adult ABM revealed that adult ABM histology comprises several molecularly defined entities, which explains clinical diversity and identifies actionable targets. Namely, pleomorphic xanthoastrocytoma‐like ABM cases show a favorable prognosis whereas high‐grade glioma (glioblastoma and diffuse midline gliome)‐like ABM show significantly worse clinical courses. These results call for in‐depth molecular analysis of adult gliomas with astroblastic features for diagnostic and therapeutic purposes.
Introduction
Astroblastoma (ABM) is a rare neuroepithelial tumor and has been recognized by the World Health Organization as a distinct tumor entity [1]. It was first described in 1926 by Bailey and Cushing [2] and further characterized by Bailey and Bucy [3] in 1930. ABM is generally regarded as an entity occurring in children and young adults [4], [5], [6]. However, in a recent epidemiological survey, 56% of cases were diagnosed after 30 years of age [7]. The diagnosis of ABM is based on a typical histomorphology with perivascular astroblastic pseudorosettes and vascular hyalinization [2], [3], [8]. However, perivascular arrangement of tumor cells is not specific for ABM and may also be seen in other central nervous system tumors such as glioblastoma (GBM), primitive neuroectodermal tumors, angiocentric glioma, and ependymomas [9], [10]. The lack of specific neuropathological features explains why the validity of ABM as a distinct entity is still a matter of debate [11].
Recently, efforts have been made to identify specific molecular alterations in ABM. In 2016, Sturm et al. [12] described a new molecularly defined tumor entity characterized by meningioma 1 (MN1) and BEN domain containing 2 (BEND2) fusion genes that histologically frequently corresponds to ABM. Several reports have confirmed the existence of recurrent MN1 alterations in pediatric as well as adult ABM [13], [14], [15]. In contrast, Lehman and colleagues [16] showed that tumors with the histological diagnosis of ABM frequently harbor BRAF V600E mutations.
Here, we report a comprehensive clinical, radiological, histological, and molecular characterization of a retrospective series of adolescent and adult ABM and show that they share common clinical and molecular features with other glial tumor entities.
Materials and Methods
Patient Cohort and Histological Review
Patients were retrospectively identified using systematic archival review for the term “astroblastoma” between 1990 and 2017 at the Pitié Salpêtrière University Hospital (i.e., cases with a final diagnosis of astroblastoma or astroblastoma being mentioned in the pathology report as a differential diagnosis). Tumors samples were stored with signed consent form in the tumor tissue bank OncoNeuroTek. All identified cases were centrally reviewed by experienced neuropathologists (K.M. and/or F.B.). Required histologic features for inclusion in the study were (a) the presence of perivascular astroblastic pseudorosettes; (b) the presence of hyalinized vessels; (c) cuboidal, columnar, or tapered perivascular cellular processes, occasionally ending in broad endfeet; and (d) absence of definitive criteria for other central nervous system (CNS) tumors. Hematoxylin and eosin staining as well as immunohistochemistry for Ki67, GFAP, IDH1 R132H, ATRX, EMA, OLIG2, vimentin, FGFR3, H3K27M, p53, and NFkB (commonly found in supratentorial ependymomas [17]) were performed. Additional immunohistochemical assessment of CD34 was done post hoc on all cases with a confirmed diagnosis of ABM.
Clinical and Radiological Data
For all patients, age at diagnosis, sex, clinical presentation, treatments, and clinical outcome were retrospectively collected. From initial diagnosis, progression‐free survival and overall survival were compared using a log‐rank test and plotted according to the Kaplan‐Meier method.
When available, brain magnetic resonance imaging (MRI) data (T1w, T2w, and contrast‐enhanced T1w sequences) were reviewed and annotated for supra‐ versus infratentorial location, perilesional edema, contrast enhancement, cystic component, and type of tumor boundary (well defined or diffuse).
Panel DNA Sequencing
All exons of genes frequently mutated in brain tumors were sequenced using a custom next‐generation sequencing panel (details in supplemental online Table 1). Briefly, tumor DNA was extracted from formalin‐fixed paraffin‐embedded or fresh‐frozen tumor specimens using the QIAamp DNA mini kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. DNA was quantified using a QuantiFluor dsDNA assay (Promega, Madison, WI). Target regions were captured from fragmented genomic DNA samples using a custom SeqCap EZ choice kit (Roche, Basel, Switzerland), and paired‐end 75‐bp massively parallel sequencing was carried out on a NextSeq500 sequencer (Illumina, San Diego, CA) according to the manufacturer's protocols.
Mutation and Copy Number Profiling
Quality control of the reads was performed with FastQC on demultiplexed data. Trimmomatic [18] was used to remove low‐quality segments (phred base quality <20) of the reads at 3′ and 5′ ends. Reads smaller than 40 base pairs after trimming were discarded. Reads were aligned against the hg19 assembly of the human genome using BWA‐MEM (version 0.7.15) [19]. We then applied Genome Analysis Toolkit (GATK) [20] for base quality score recalibration and indel realignment. PatternCNV [21] was used to estimate copy number variation based on read depth. Mutations and indels were called using GATK4 MuTect2 (beta version). Single nucleotide variants were annotated using Variant Effect Predictor [22]. Putative somatic variants were selected by filtering out all Single nucleotide polymorphisms in the gnomAD release 2.0.1 with an overall population allele frequency > 0.01 [23]. Variants were filtered for missense and nonsense mutations, and a minimum variant allele frequency > 0.1 was required. Disease‐causing variants were annotated using known cancer hotspots [24] and ClinVar [25].
RT‐PCR
First strand complementary DNA (cDNA) was generated from 500 ng total RNA using Maxima First Strand Synthesis Kit (Thermo Fisher Scientific, Illkirch, France) and diluted 1:10 in molecular biology‐grade water. Reverse transcription polymerase chain reaction (RT‐PCR) with primers specific for MN1‐BEND2 fusions was performed using 0.02 U/μL Q5 polymerase (New England Biolabs, Ipswich, MA), 200 μM dNTPs, 500 nM forward and reverse primers, Q5 reaction buffer with High GC Enhancer, and 5 μL template cDNA in total reaction volume of 20 μL. Thermal cycling was performed as follows: 98°C initial denaturation for 2 minutes, followed by 30 cycles of denaturation at 98°C for 10 seconds, annealing at 65°C for 20 seconds, and extension at 72°C for 90 seconds, as well as a final extension at 72°C for 2 minutes. Amplicons were analyzed using a Caliper LabChip GX DNA 5K assay (Perkin Elmer, Waltham, MA).
qPCR
Quantitative real‐time polymerase chain reaction (qPCR) of BEND2 expression was performed in 20 μL reactions using 10 μL 2X LightCycler 480 SYBR Green master mix (Roche, Basel, Switzerland), 1 μL of a 10 μM forward and reverse primer mix, 4 μL H2O, and 5 μL of 1:10 prediluted cDNA. The following PCR program was used: 10 minutes of preincubation at 95°C, 45 cycles of 10 seconds of denaturation at 95°C, 10 seconds of annealing at 60°C, and 10 seconds of extension at 72°C. All reactions were run in duplicates and normalized to a housekeeping gene (PPIA) using the ddCt method.
RNA Sequencing and Fusion Gene Discovery
Five hundred nanograms of total RNA was used for library preparation using NextSeq High Output Kit v2.5 kits (Illumina) and subjected to 2 × 75 bp paired‐end sequencing on a NextSeq 500 device (Illumina). Quality control of raw sequencing data (e.g., for potential ribosomal contamination) and insert size estimation were performed using FastQC, Picard tools, samtools, and rseqc [26]. Reads were mapped using STAR v2.4.0 [27] to the hg19 human genome assembly. Gene expression analysis was performed as described previously [28]. Briefly, for each gene present in the Human FAST DB v2016_1 annotation, reads aligning on constitutive regions (that are not prone to alternative splicing) were counted. Based on these read counts, normalization was performed using DESeq2 [29] and R (v.3.2.5).
Fusion detection was performed using five different tools: Defuse v0.6.0 [30], FusionCatcher v0.99.5a [31] without BLAT, JAFFA v1.06 [32], SoapFuse v1.27 [33], TopHat fusion v2.06.13 [34]. Comparison of results between algorithms was done by FuMa v2 [35] with FASTD DB v2016_1 annotations. High‐confidence fusion candidates identified by >3 tools were reviewed manually.
Methylation Profiling
Genome‐wide methylation profiling was performed as previously reported [36]. Briefly, 500 ng of DNA was subjected to bisulfite conversion and hybridized to Infinium MethylationEPIC BeadChip microarrays (Illumina). Raw IDAT files were used as input for online methylation‐based random forest classification (www.molecularneuropathology.org) using classifier version v11b4.
Data Availability
All raw sequencing data have been deposited at the European Genome‐phenome Archive (accession no. EGAS00001003604). Microarray‐based methylome data have been made available at ArrayExpress (accession no. E‐MTAB‐7490).
Results
Patient and Tumor Characteristics
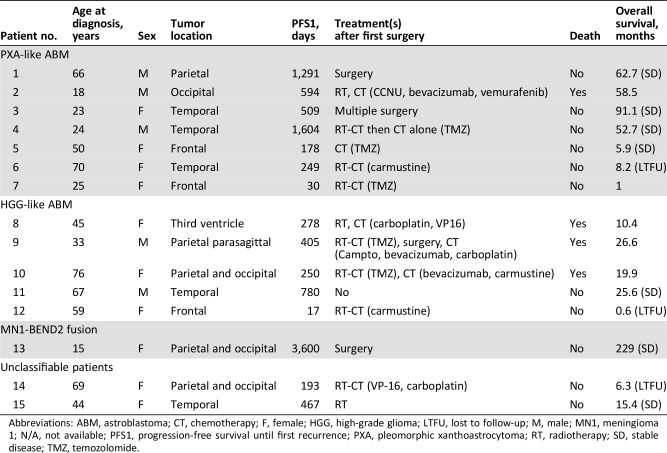
We initially identified 25 patients with an archival diagnosis of ABM. After centralized neuropathological review, a histological diagnosis of ABM was confirmed in 15 cases: 14 adults and 1 adolescent. Ten cases were excluded because of a morphological diagnosis of glioblastoma in seven cases, no availability of tumor DNA and RNA in two cases, and co‐occurrence of astroblastoma and IDH‐mutant glioblastoma within the same tumor in one patient. Typical ABM histology with astroblastic pseudorosettes, perivascular pattern, vascular hyalinization, and immunoreactivity for GFAP, OLIG2, and vimentin were seen in all included cases. Positive immunostaining for EMA and p53 was found in only three cases and four cases, respectively. Of 15 ABMs, 10 were classified as “high grade” based on hypercellular zones with increased mitotic index, vascular proliferation, and necrosis.
The 15 cases included in the current study were investigated further (10 females and 5 males, sex ratio 2:1). Median age at diagnosis was 44.5 years (range: 24‐66) with a bimodal age distribution: four patients were <25 years of age and seven were >50 years of age. The most common initial symptom was neurological deficit (8/15 patients), followed by headaches (6/15) and seizure (4/15).
All patients underwent brain computed tomography or MRI scan. All cases were supratentorial, including one intraventricular tumor (third ventricle). Location was temporal, frontal, parietal, occipital, and parieto‐occipital in six, three, two, one, and three cases, respectively. Brain MRI was available for review in 11/15 patients. All tumors appeared well circumscribed and demarcated from normal brain on MRI. Contrast enhancement was detected in all patients, and a cystic component was observed in 6/11 patients. All patients underwent surgical resection (13 patients with gross total resection, 2 with partial resection).
After surgery, 11 of 15 patients received adjuvant treatment (6 with combined radio‐ and chemotherapy, 4 with radiotherapy alone, and 1 with chemotherapy alone) because of high‐grade morphology (10 cases) or partial resection (1 case).
After first‐line treatment, eight patients did not experience tumor relapse. In contrast, two, four, and one patients experienced one, two, and three or more tumors relapses, respectively. Four patients died during the follow‐up period (26.7%). The median progression‐free survival and overall survival were 1.6 and 4.9 years, respectively (Table 1).
Table 1. Patient characteristics.
Abbreviations: ABM, astroblastoma; CT, chemotherapy; F, female; HGG, high‐grade glioma; LTFU, lost to follow‐up; M, male; MN1, meningioma 1; N/A, not available; PFS1, progression‐free survival until first recurrence; PXA, pleomorphic xanthoastrocytoma; RT, radiotherapy; SD, stable disease; TMZ, temozolomide.
Molecular Profiling
Depending on DNA and RNA availability, quantity, and quality, molecular profiling was performed. Targeted next‐generation panel sequencing was performed in all 15 patients, transcriptome sequencing in 4 samples, and methylation profiling in 6 cases.
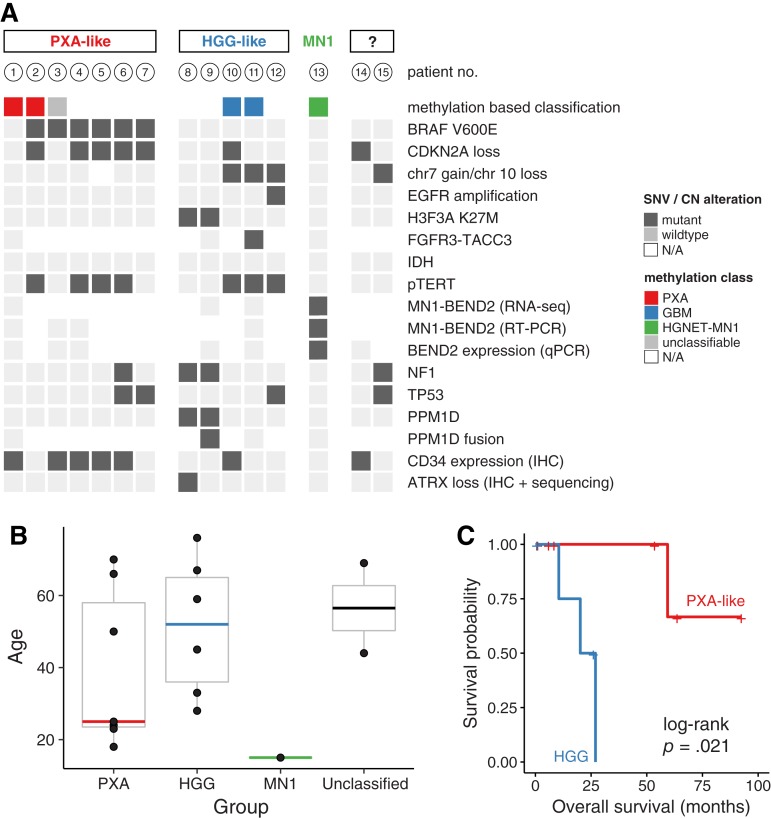
Together with immunohistochemistry, molecular profiling allowed the reclassification of cases into three main groups (Fig. 1A).
Figure 1.
Summary of genetic alterations (A), age distribution (B), and overall survival (C) by subgroups.
Abbreviations: CN, copy number; EGFR, epidermal growth factor receptor; GBM, glioblastoma; HGG, high‐grade glioma; HGNET, high grade neuroepithelial tumor; IHC, immunohistochemistry; MN1, meningioma 1; N/A, not available; PXA, pleomorphic xanthoastrocytoma; qPCR, quantitative real‐time polymerase chain reaction; RT‐PCR, reverse transcription polymerase chain reaction; SNV, single nucleotid variant.
ABM with Molecular Features of PXA.
Of 15 tumors, 7 (47%) were either classified as pleomorphic xanthoastrocytoma (PXA) using methylation‐based classification or showed BRAF mutation with concomitant CDKN2A loss and/or immunohistochemical CD34 expression. Six tumors harbored a BRAFV600E mutation, which was associated with CDKN2A homozygous deletion in five cases and TERT C228T promoter mutations in four cases.
One patient's tumor harbored a BRAF G466E alteration, a class III mutation resulting in a kinase‐dead form of BRAF [37]. This alteration was accompanied by a truncating NF1 mutation, a combination that is frequently observed in melanoma and known to result in a mechanistically different, yet functionally similar, ERK pathway activation [38].
None of these cases harbored typical histological features of PXA (i.e., cellular pleomorphism with spindle cells, mononucleated and multinucleated giant cells, granular bodies, or positive reticulin staining) [39]. Using immunohistochemistry, these cases harbored nuclear OLIG2 expression in all cases and cytoplasmic CD34 expression in 5/7 patients (whereas only 2/8 cases without BRAF mutation expressed CD34). Mean age at diagnosis was 25 years (Fig. 1B). In this subgroup, one patient out of seven died during follow‐up (Fig. 1C). The median progression‐free survival was 2.6 years.
ABM with Molecular Patterns of HGG.
Of 15 patients, 5 were classified as high‐grade glioma because of the presence of H3.3 K27M mutations or glioblastoma molecular features (methylation class GBM or any two of the following criteria: combined chr7 gain/chr10 loss, epidermal growth factor receptor (EGFR) amplification, and/or TERT promoter mutation) [40]. Three patients had TERT C228T promoter mutations, one of which harbored a FGFR3:TACC3 fusion. Interestingly, the two cases with molecular pattern of diffuse midline glioma (i.e., combination of H3F3A, PPM1D, and NF1 mutations) did not arise within classic midline location (i.e., thalamus, pons, and spinal cord [41]): one case was located in the third ventricle and the other one in frontoparietal parasagittal localization. Of note, none of the H3F3A‐wildtype cases in this group showed loss of histone H3 lysine 27 trimethylation.
Mean age at diagnosis in this group was 52 years (Fig. 1B). Strikingly, this subgroup was associated with poor clinical outcome compared with PXA‐like cases: three out of five patients (60%) died during the follow‐up. The median progression‐free survival and overall survival were 0.9 and 1.9 years, respectively (Fig. 1C).
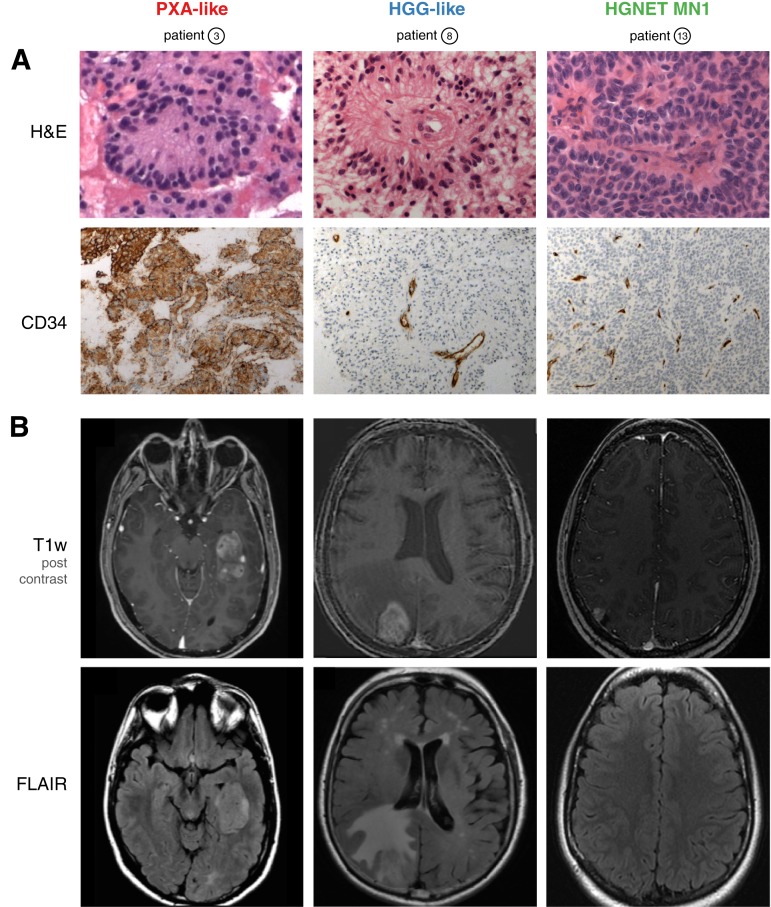
In contrast to PXA‐like cases, MRI of high‐grade glioma (HGG)‐like cases showed moderate‐to‐diffuse peripheral edema with a median volume of 23.5 mL (18.8–90) versus 2.7 mL (0.7–129) in PXA‐like subgroup (p < .001; Fig. 2).
Figure 2.
Representative histological (A) and magnetic resonance imaging (MRI) (B) features of subgroups. (A): H&E staining shows large astroblastic pseudorosettes in all cases (upper panel). Unlike HGG and HGNET‐MN1, PXA‐like astroblastoma (ABM) demonstrate an intense cytoplasmic and pericellular expression of CD34 (lower panel). (B): Representative MRI features of subgroups. Axial post‐contrast T1‐weighted magnetic resonance images demonstrate a solid component in all cases (upper panel). Unlike the other subgroups, HGG‐like ABM show moderate‐to‐extensive perifocal edema on FLAIR sequences (lower panel).
Abbreviations: FLAIR, T2‐weighted fluid‐attenuated inversion recovery; H&E, hematoxylin and eosin; HGG, high‐grade glioma; HGNET, high grade neuroepithelial tumor; MN1, meningioma 1; PXA, pleomorphic xanthoastrocytoma; T1w, T1‐weighted.
ABM with MN1‐BEND2 Fusion.
Only one young patient (15 years of age) was epigenetically classified as high‐grade neuroepithelial tumor with MN1 alteration (CNS HGNET‐MN1) [12]. RNA sequencing identified an MN1:BEND2 fusion, which could be confirmed by RT‐PCR and BEND2 expression by real‐time PCR, whereas all others cases did not express BEND2 (supplemental online Fig. 1). The patient has had stable disease since tumor resection over 19 years of follow‐up.
Finally, two patients could not be assigned to either subgroup given the available molecular data. One patient (lost during follow‐up) harbored an NF1 mutation and combined chr7 gain/chr10 loss. However, because this patient did not harbor EGFR amplification, this case may correspond to PXA or GBM‐like ABM [42]. The other patient harbored CDKN2A homozygous deletion and was positive for CD34 expression immunohistochemically.
Discussion
In this study, we retrospectively characterized a series of 14 adults and 1 adolescent with centrally reviewed ABM. The present cohort shares characteristics with previous published studies [4], [5], [6], [9], [43], [44], [45], [46] with ABM being a supratentorial tumor with a clear female predominance and heterogeneous clinical courses. Comprehensive interrogation of genetic alterations in these 15 patients (including methylation profiling, targeted DNA sequencing, RNA sequencing, RT‐PCR, and qPCR) allowed reclassification of cases in three subgroups (PXA‐like ABM, HGG‐like ABM, and ABM with MN1‐BEND2 fusion) with distinct radiological and histological features as well as clinical outcomes.
ABM has been controversial in recent years, as some literature supports it as true entity with frequent MN1 alterations [12], [13], [15] whereas other studies suggest it does not exist as an established and unique tumor entity but could overlap with other well‐known tumors [11], [14], [16], [47].
Especially, in accordance with our study, gliomas with astroblastic features may harbor molecular signatures of PXA [14], [15], [16] and HGG [11], [14], [47]. Moreover, reclassification of ABM into a more specific molecularly defined entity could explain the clinical unpredictability and difficulty in grading these tumors.
The largest group was ABM with a molecular signature of PXA. Previously, methylation‐based reclassification of an ABM case into PXA [14], [15] and high prevalence of BRAF mutations [15], [16] have been reported. BRAF mutations are also frequently observed in ganglioglioma (GG) and pilocytic astrocytoma (PA). However, combined BRAF V600E mutation with (a) CDKN2A loss, usually only seen in PXA [48], (b) TERT promoter mutations, common in anaplastic PXA but virtually never seen in PA and rare in GG [49], [50], and (c) positive CD34 staining demonstrates compelling evidence that indeed many ABM share the molecular identity of PXA.
The second group includes ABM with molecular features of HGGs (i.e., GBM and diffuse midline glioma, K27M‐mutant). As expected, these patients experienced an unfavorable clinical course. These tumors were accompanied by larger perifocal edema, a typical feature of high‐grade glioma. It is interesting to note that mitogen‐activated protein kinase (MAPK) pathway alterations including BRAF mutation, NF1 mutation, or FGFR fusion were present in 67% of cases (10/15) in the entire cohort. Even though MAPK pathway alterations are frequent in GBM (mostly affecting EGFR), they rarely involve BRAF, NF1, and FGFR [51], [52]. Rather, the mutational spectrum resembles that of pilocytic astrocytomas [53]. Strikingly, these alterations are actionable drug targets, potentially subject to BRAF inhibition (e.g., vemurafenib), MEK inhibition (e.g., trametinib), or FGFR inhibition (e.g., NCT02052778). In addition to BRAF mutations, a NRAS mutation has previously been reported in a case of ABM [14], extending the spectrum of observed MAPK/RAS pathway mutations. Because all of these alterations were identified retrospectively, none of the reported patients received targeted therapy.
Despite thorough multimodal screening for MN1 alterations, only one adolescent patient was assigned the diagnosis of CNS high‐grade neuroepithelial tumor with MN1 alteration. This result differs from three previous studies in adult ABM [13], [14], [15] in which MN1 rearrangements were frequent. Of note, we cannot entirely rule out MN1 alterations with fusion partners other than BEND2 in tumors that only were screened by RT‐PCR and for BEND2 expression and did not undergo methylation profiling (six cases). However, all of these cases harbored glioblastoma molecular features or a BRAF or H3.3 K27M mutation and could thus reliably be assigned to one of the other subgroups. This finding is in line with evidence of previous studies, in which MN1 alterations, BRAF mutation, and HGG molecular features were mutually exclusive [12], [13], [14], [15], [47].
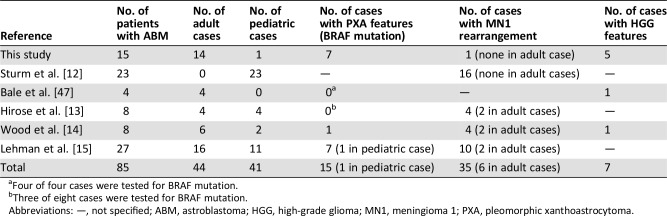
Our results differ partly from those of five other retrospective studies [12], [13], [14], [15], [47]. Comparing our results directly with these studies is not possible as we encounter major differences in study population (majority of adult patients in our study vs. a significant proportion of pediatric cases in others) [12], [13], [14], [15], required histologic features, number of patients, and molecular analyses performed. However, analysis of available data (summarized in Table 2) suggests that ABM with MN1 alterations appears to be frequent in children (found in 29/41 [70.7%] pediatric ABM cases) and uncommon in adults (found in 6/44 [13,6%]). In contrast, ABM with molecular features of PXA (i.e., BRAF mutation, CD34 expression, corresponding methylation class) appears to be recurrent in adolescents and adults (found in 14/44 [31.8%]) and rare in children (1/41 [2.4%]). Finally, most of these studies did not specifically investigate molecular features of high‐grade glioma (chr7 gain/chr 10 loss, histone mutation, methylation‐based classification) and could explain why the majority of these cases (5/7) are found in our study. Additional studies with a focus on this issue would be necessary to further explore this entity.
Table 2. Review of literature.
Four of four cases were tested for BRAF mutation.
Three of eight cases were tested for BRAF mutation.
Abbreviations: —, not specified; ABM, astroblastoma; HGG, high‐grade glioma; MN1, meningioma 1; PXA, pleomorphic xanthoastrocytoma.
Our study has several limitations inherent to its retrospective observational design. The diagnosis of ABM is based on fairly exclusive histologic features. This could lead to differences in patient selection and also explain differences in results between studies. Survival data should be interpreted with caution considering the three patients lost to follow‐up and the study inclusion period (from 1990 to 2017): we cannot exclude the possibility that the evolution of imaging and histopathology techniques along with treatments of patients (including surgery techniques and chemotherapy and radiotherapy protocols) during the study period may have influenced our results. Finally, because of lack of material, methylation profiling could not be performed in all cases. Thus, we cannot prove that all BRAF‐mutant, CD34‐positive cases are indeed PXAs. Ganglioglioma, for example, remains a differential diagnosis.
Conclusion
Our data indicate that adult ABM comprises several molecularly defined entities. CNS high‐grade neuroepithelial tumor with MN1 alteration appears to be uncommon in adults. Adults’ ABM frequently harbor molecular features of PXA and HGG. Astroblastic morphology in adults should prompt thorough molecular investigation aiming at a clear histomolecular diagnosis and identifying actionable drug targets, especially in MAPK pathway genes.
See http://www.TheOncologist.com for supplemental material available online.
Acknowledgments
We thank Amithys Rahimian‐Aghda, Marine Giry, Inès Detrait, and Yohann Schmitt for biobanking, sequencing studies, and expert technical assistance. We also thank Noémie Robil and Pierre de la Grange for fusion gene analysis, David Capper for methylation profiling, and Jean‐François Côté for providing human testis tissue. P.E. is a participant of the BIH‐Charité Clinician Scientist Program funded by the Charité ‐ Universitätsmedizin Berlin and the Berlin Institute of Health. P.E. has been supported by the Deutsche Forschungsgemeinschaft (EU 162/1). This research was supported by a grant of the Ligue Contre le Cancer and the Institut Universitaire de Cancérologie (RS17/75‐130 to A.I.). INCA‐DGOS‐Inserm_12560 SiRIC CURAMUS is financially supported by the French National Cancer Institute, the French Ministry of Solidarity and Health, and Inserm. The research leading to these results has received funding from the program Investissements d'avenir (ANR‐10‐IAIHU‐06).
Contributed equally.
Footnotes
For Further Reading: Priscilla K. Brastianos, Franziska Maria Ippen et al. Emerging Gene Fusion Drivers in Primary and Metastatic Central Nervous System Malignancies: A Review of Available Evidence for Systemic Targeted Therapies. The Oncologist 2018;23:1063–1075.
Implications for Practice: Driver gene fusions involving receptor tyrosine kinases have been identified across a wide range of tumor types, including primary central nervous system (CNS) tumors and extracranial solid tumors that are associated with high rates of metastasis to the CNS (e.g., lung, breast, melanoma). This review discusses the systemic therapies that target emerging gene fusions, with a focus on brain‐penetrant agents that will target the intracranial disease and, where present, also extracranial disease.
Author Contributions
Conception/design: William Boisseau, Philipp Euskirchen, Karima Mokhtari, Ahmed Idbaih
Provision of study material or patients: Philipp Euskirchen, Karima Mokhtari, Caroline Dehais, Khê Hoang‐Xuan, Marc Sanson, Laurent Capelle, Aurélien Nouet, Carine Karachi, Franck Bielle, Justine Guégan, Yannick Marie, Nadine Martin‐Duverneuil, Luc Taillandier, Audrey Rousseau, Jean‐Yves Delattre, Ahmed Idbaih,
Collection and/or assembly of the data: William Boisseau, Philipp Euskirchen, Karima Mokhtari, Ahmed Idbaih
Data analysis and interpretation: William Boisseau, Philipp Euskirchen, Karima Mokhtari, Ahmed Idbaih
Final approval of manuscript: William Boisseau, Philipp Euskirchen, Karima Mokhtari, Caroline Dehais, Khê Hoang‐Xuan, Marc Sanson, Laurent Capelle, Aurélien Nouet, Carine Karachi, Franck Bielle, Justine Guégan, Yannick Marie, Nadine Martin‐Duverneuil, Luc Taillandier, Audrey Rousseau, Jean‐Yves Delattre, Ahmed Idbaih
Disclosures
Mehdi Touat: Taiho Oncology, Integragen (C/A), Agios Pharmaceuticals (SAB); Franck Bielle: Abbvie, Sanofi (RF [family member]), Celgene (E [family member]), Crossject (OI [family member]); Ahmed Idbaih: CarThera (travel), Transgene, Sanofi, Air Liquide, Gecko, Beta Innov (RF). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Louis DN, Perry A, Reifenberger G et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathologica 2016;131:803–820. [DOI] [PubMed] [Google Scholar]
- 2.Bailey P, Cushing H. Tumors of the Glioma Group. Philadelphia: Lippincott, 1924. [Google Scholar]
- 3.Bailey P, Bucy PC. Astroblastomas of the brain. Acta Psychiatr Neurol 1930;5:439–461. [Google Scholar]
- 4.Sughrue ME, Choi J, Rutkowski MJ et al. Clinical features and post‐surgical outcome of patients with astroblastoma. J Clin Neurosci 2011;18:750–754. [DOI] [PubMed] [Google Scholar]
- 5.Mallick S, Benson R, Venkatesulu B. Patterns of care and survival outcomes in patients with astroblastoma: An individual patient data analysis of 152 cases. Childs Nerv Syst 2017;33:1295–1302. [DOI] [PubMed] [Google Scholar]
- 6.Merfeld EC, Dahiya S, Perkins SM. Patterns of care and treatment outcomes of patients with astroblastoma: A National Cancer Database analysis. CNS Oncol 2018;7:CNS13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmed KA, Allen PK, Mahajan A et al. Astroblastomas: A Surveillance, Epidemiology, and End Results (SEER)‐based patterns of care analysis. World Neurosurg 2014;82:e291–e297. [DOI] [PubMed] [Google Scholar]
- 8.Aldape KD, Rosenblum MK, Brat DJ. Astroblastoma. In: Louis DN, Ohgaki H, Wiestler OD, eds. WHO Classification of Tumours of the Central Nervous System. Revised 4th ed Lyon, France: International Agency for Research on Cancer, 2016:121–122. [Google Scholar]
- 9.Brat DJ, Hirose Y, Cohen KJ et al. Astroblastoma: Clinicopathologic features and chromosomal abnormalities defined by comparative genomic hybridization. Brain Pathol 2000;10:342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burger PC, Scheithauer BW. Tumors of the Central Nervous System. Atlas of Tumor Pathology. Washington, DC: Armed Forces Institute of Pathology, 2007. [Google Scholar]
- 11.Mellai M, Piazzi A, Casalone C et al. Astroblastoma: Beside being a tumor entity, an occasional phenotype of astrocytic gliomas? Onco Targets Ther 2015;8:451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sturm D, Orr BA, Toprak UH et al. New brain tumor entities emerge from molecular classification of CNS‐PNETs. Cell 2016;164:1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirose T, Nobusawa S, Sugiyama K et al. Astroblastoma: A distinct tumor entity characterized by alterations of the X chromosome and MN1 rearrangement. Brain Pathol 2018;28:684–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood MD, Tihan T, Perry A et al. Multimodal molecular analysis of astroblastoma enables reclassification of most cases into more specific molecular entities. Brain Pathol 2018;28:192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehman NL, Usubalieva A, Lin T et al. Genomic analysis demonstrates that histologically‐defined astroblastomas are molecularly heterogeneous and that tumors with MN1 rearrangement exhibit the most favorable prognosis. Acta Neuropathol Commun 2019;7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehman NL, Hattab EM, Mobley BC et al. Morphological and molecular features of astroblastoma, including BRAFV600E mutations, suggest an ontological relationship to other cortical‐based gliomas of children and young adults. Neuro Oncol 2016;19:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker M, Mohankumar KM, Punchihewa C et al. C11orf95‐RELA fusions drive oncogenic NF‐κB signalling in ependymoma. Nature 2014;506:451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Durbin R. Fast and accurate short read alignment with Burrows‐Wheeler Transform. Bioinformatics 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna A, Hanna M, Banks E et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next‐generation DNA sequencing data. Genome Res 2010;20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C, Evans JM, Bhagwate AV et al. PatternCNV: A versatile tool for detecting copy number changes from exome sequencing data. Bioinformatics 2014;30:2678–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLaren W, Gil L, Hunt SE et al. The Ensembl Variant Effect Predictor. Genome Biol 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lek M, Karczewski KJ, Minikel EV et al. Analysis of protein‐coding genetic variation in 60,706 humans. Nature 2016;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang MT, Asthana S, Gao SP et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol 2016;34:155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landrum MJ, Lee JM, Benson M et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res 2016;44:D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Wang S, Wei L. RSeQC: Quality control of RNA‐seq experiments. Bioinformatics 2012;28:2184–2185. [DOI] [PubMed] [Google Scholar]
- 27.Dobin A, Davis CA, Schlesinger F et al. STAR: Ultrafast universal RNA‐seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noli L, Capalbo A, Ogilvie C et al. Discordant growth of monozygotic twins starts at the blastocyst stage: A case study. Stem Cell Reports 2015;5:946–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Bio 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McPherson A, Hormozdiari F, Zayed A et al. deFuse: An algorithm for gene fusion discovery in tumor RNA‐seq data. PLoS Comput Biol 2011;7:e1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicorici D, Satalan M, Edgren H et al. FusionCatcher ‐ A tool for finding somatic fusion genes in paired‐end RNA‐sequencing data. bioRxiv 2014;011650. [Google Scholar]
- 32.Davidson NM, Majewski IJ, Oshlack A. JAFFA: High sensitivity transcriptome‐focused fusion gene detection. Genome Med 2015;7:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia W, Qiu K, He M et al. SOAPfuse: An algorithm for identifying fusion transcripts from paired‐end RNA‐seq data. Genome Biol 2013;14:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim D, Salzberg SL. TopHat‐Fusion: An algorithm for discovery of novel fusion transcripts. Genome Biol 2011;12:R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoogstrate Y, Böttcher R, Hiltemann S et al. FuMa: Reporting overlap in RNA‐seq detected fusion genes. Bioinformatics 2016;8:1226–1228. [DOI] [PubMed] [Google Scholar]
- 36.Capper D, Jones DTW, Sill M et al. DNA methylation‐based classification of central nervous system tumours. Nature 2018;7697:469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao Z, Torres MN, Tao A et al. BRAF mutants evade ERK‐dependent feedback by different mechanisms that determine their sensitivity to pharmacologic inhibition. Cancer Cell 2015;28:370–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao Z, Yaeger R, Rodrik‐Outmezguine VS et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 2017;548:234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giannini C, Scheithauer BW, Burger PC et al. Pleomorphic xanthostrocytoma. What do we really know about it? Cancer 1999;85:2033–2045. [PubMed] [Google Scholar]
- 40.Stichel D, Ebrahimi A, Reuss D et al. Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and TERT promoter mutation in brain tumors and their potential for the reclassification of IDHwt astrocytoma to glioblastoma. Acta Neuropathol 2018;136:793–803. [DOI] [PubMed] [Google Scholar]
- 41.Solomon DA, Wood MD, Thian T et al. Diffuse midline gliomas with histone H3‐K27M mutation: A series of 47 cases assessing the spectrum of morphologic variation and associated genetic alterations. Brain Pathol 2016;5:569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brat DJ, Aldape K, Colman H et al. cIMPACT‐NOW update 3: Recommended diagnostic criteria for "Diffuse astrocytic glioma, IDH‐wildtype, with molecular features of glioblastoma, WHO grade IV." Acta Neuropathol 2018;136:805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asha U, Mahadevan A, Sathiyabama D et al. Lack of IDH1 mutation in astroblastomas suggests putative origin from ependymoglial cells? Neuropathology 2015;35:303–311. [DOI] [PubMed] [Google Scholar]
- 44.Salvati M, D'Elia A, Brogna C et al. Cerebral astroblastoma: Analysis of six cases and critical review of treatment options. J Neurooncol 2009;93:369–378. [DOI] [PubMed] [Google Scholar]
- 45.Port JD, Brat DJ, Burger PC et al. Astroblastoma: Radiologic‐pathologic correlation and distinction from ependymoma. AJNR Am J Neuroradiol 2002;23:243–247. [PMC free article] [PubMed] [Google Scholar]
- 46.Bell JW, Osborn AG, Salzman KL et al. Neuroradiologic characteristics of astroblastoma. Neuroradiology 2007;49:203–209. [DOI] [PubMed] [Google Scholar]
- 47.Bale TA, Abedalthagafi M, Bi WL et al. Genomic characterization of recurrent high‐grade astroblastoma. Cancer Genet 2016;209:321–330. [DOI] [PubMed] [Google Scholar]
- 48.Koelsche C, Sahm F, Wöhrer A et al. BRAF‐mutated pleomorphic xanthoastrocytoma is associated with temporal location, reticulin fiber deposition and CD34 expression. Brain Pathol 2014;24:221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koelsche C, Sahm F, Capper D et al. Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta Neuropathol 2013;126:907–915. [DOI] [PubMed] [Google Scholar]
- 50.Phillips JJ, Gong H, Chen K et al. The genetic landscape of anaplastic pleomorphic xanthoastrocytoma. Brain Pathol 2019;29:85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brennan CW, Verhaak RGW, McKenna A et al. The somatic genomic landscape of glioblastoma. Cell 2013;155:462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Stefano AL, Fucci A, Frattini V et al. Detection, characterization, and inhibition of FGFR‐TACC fusions in IDH wild‐type glioma. Clin Cancer Res 2015;21:3307–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones DT, Hutter B, Jäger N et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet 2013;45:927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All raw sequencing data have been deposited at the European Genome‐phenome Archive (accession no. EGAS00001003604). Microarray‐based methylome data have been made available at ArrayExpress (accession no. E‐MTAB‐7490).