Abstract
In Nature, DNA molecules carry the hereditary information. But DNA has physical and chemical properties that make it attractive for uses beyond heredity. In this Review, we discuss the potential of DNA for creating machines that are both encoded by and built from DNA molecules. We review the main methods of DNA nanostructure assembly, describe recent advances in building increasingly complex molecular structures, and discuss strategies for creating machine-like nanostructures that can be actuated and move. We highlight opportunities for applications of custom DNA nanostructures as scientific tools to address challenges across biology, chemistry and engineering.
Introduction
The cellular machinery is composed of a multitude of molecules working in tandem to assure the viability of cells and functionality of different mechanisms. In a broad sense, many of those biomolecules can be referred to as biomolecular machines, because they perform a specific task in response to a particular stimulus using their moving parts. Natural biomolecular machines have a vast scope of functionalities and include but are not limited to motor proteins, enzymes and sensory proteins, as uncovered through decades of research in molecular biology. DNA polymerases, RNA polymerases, the ribosome and ATPases are some familiar examples of biomolecular machines that play pivotal roles in DNA replication, gene expression, translation and cell energy production, respectively. By contrast, human-made biomolecular machines, which mimic or aim to eventually surpass the functions of their natural counterparts, remain in their infancy. Although substantial progress has been made in investigating and describing molecular-scale phenomena in detail, our abilities to design and build on such a fine scale are still comparably limited. The ultimate goal of designing artificial biomolecular machines is to achieve sophisticated tasks in a controllable modular manner. This, in turn, would enable the engineering of molecular interactions and motions to execute a list of functions or even create artificial cells or life-like entities.
One of the striking features of nature's molecular machinery is its structural sophistication. Typically, thousands of atoms come together in intricate 3D molecular complexes. The structural complexity is presumably an important feature to achieve robust and regulated functionality within the cellular context. Artificial biomolecular machines should be similarly robust, and users must also be able to regulate their function. It is thus likely that to satisfy these criteria, they will resemble natural molecular assemblies in overall dimensions, and structural complexity and intricacy. Building artificial molecular structures that include thousands or millions of atoms presents a formidable challenge to the traditional methods of chemical synthesis. But nature presents a path to meeting this challenge. Nature uses biopolymers made of amino acids or nucleic acids, each featuring a defined alphabet of chemical building blocks. The sequences of building blocks in such a biopolymer encode the structures of natural molecular machines, which form in a self-assembly [G] process called folding. One possible route to creating complex artificial molecular structures consists in investigating how both the materials and the principles that nature uses can be adapted to build synthetic molecular structures. This is the strategy followed by biomolecular designers in the fields of de novo protein design1, RNA nanotechnology and DNA nanotechnology [G], which are all driven by the idea of encoding structures in sequences.
Glossary.
Sticky-ended DNA
A DNA partial duplex with a single-stranded overhang which can hybridize to another complementary single-stranded overhang, thus “sticking” the two partial duplexes together.
DNA crossover
The point at which a DNA single strand exits its hybridization axis and enters an adjacent helix to continue its hybridization in the second helical axis.
DNA tile
A motif self-assembled from multiple single-stranded DNA oligomers to form a unit for further assembly of a nanostructure. There are usually one or more crossovers in each tile rendering it more rigid.
DNA origami
A DNA nanostructure formed by folding a long single-stranded DNA scaffold via hybridization of many short DNA complements known as “staple strands”.
Origami scaffold
The long single-stranded viral genome running through a whole DNA origami structure in a raster pattern.
Staple strands
The short DNA oligomers (usually 20-60 nt long) used to staple different segment of the scaffold together and form a pre-determined geometry.
Honeycomb packing
The spatial arrangement of helices in which each helix forms crossovers with its three neighbouring helices at a 120 ° exit angle.
Square packing
The spatial arrangement of helices in which each helix forms crossovers with its four neighbouring helices at a 90 ° exit angle.
Segment length
The distance between two consecutive crossovers which is a multiple of 7 base pairs in the honeycomb packing and a multiple of 8 base pairs for the square packing.
Base insertion/ deletion
Lengthening or shortening a segment to create a twist along the helical axis is called a base pair insertion or deletion, respectively. This terminology could be confusing in the sense that the sequence of scaffold is fixed and no actual insertion or deletion of a nucleotide is possible. Shortening or lengthening a segment will shift the crossover position upstream or downstream imposing a strain at the crossover point.
ssDNA brick assembly
A version of DNA tile assembly in which each DNA tile is a single-stranded DNA brick with a unique sequence and multiple segments (or domains) of a certain length.
Strand displacement reaction (SDR)
A hybridization scheme in which a longer complement (fuel strand) displaces a shorter complement (output strand) via branch migration to form the more stable duplex.
DNA toehold
The unpaired segment of a partial duplex which can act as a seeding region to start a branch migration and an SDR.
DNA walker
A small DNA oligomer that can move on a track.
Click contacts
The topological surface features of a DNA nanostructure in the forms of protrusions and recessions capable of forming base stacking interactions between two shape-complementary features and thus, binding them.
Exit/ branch angle periodicity
The smallest distance (in base pairs) possible between two consecutive staple crossovers shared by the same two helices.
Self-assembly
A process in which the disordered components of a system spontaneously organize themselves into a defined ordered state in the absence of any external force controlling them. The whole process is guided by minimization of the free energy of the system. Protein folding and lipid bilayer formation are two examples of molecular self-assembly.
DNA nanotechnology
The design and self-assembly of DNA into pre-defined patterns in order to control the shapes and functions of the assembled nanostructures.
Rotaxanes
A class of mechanically interlocked molecules consisting of a ring entrapped between the two bulky ends of a dumbbell-shaped molecule.
Catenanes
A class of mechanically interlocked molecules comprised of two or more interchained macrocyclic rings.
Brownian motors
A molecule or a molecular system converting the random Brownian motion to the directional motion at the nanoscale by doing work on the environment.
DNA switch
A molecular switch made of DNA that transitions between at least two distinct states using a trigger such as pH, metal ions, etc.
Persistence length
A mechanical property indicating the stiffness of a polymer such as DNA and is defined as the length over which the molecule behaves like a rigid rod.
Wireframe tessellation
Approximating a geometrical shape at its edges through tiling its surfaces by non-overlapping polygons without leaving a gap.
DNAzyme
Also known as deoxyribozyme, DNA enzyme, or catalytic DNA, is a DNA oligonucleotide with a specific sequence performing a chemical reaction similar to enzymes.
DNA walker
A DNA oligonucleotide moving on a molecular track by a series of hybridization-dehybridization cycles.
Tensegrity
Originally an architectural concept, tensegrity is particular type of structure maintaining its integrity through pervasive tensional forces. In a tensegrity, each individual structural element is under stress but the overall structure is perfectly stable.
Aptamers
Aptamers are oligonucleotides or small peptides binding specifically to a target molecule.
Ratchet mechanisms
The mechanisms by which molecular motors utilize the random thermal noise to produce directional motion.
ToC blurb
This Review discusses the potential of DNA for creating machines that are both encoded by and built from DNA molecules. Alongside an overview of DNA nanostructure assembly, the authors describe recent advances and remaining challenges, highlighting applications of custom DNA nanostructures as scientific tools.
The synthesis and powering of small artificial molecular machines (AMMs) have progressed extensively in the past three decades, as recognized by the 2016 Nobel prize in chemistry. These advances have led to a much improved understanding of the requirements for building simple molecular machines, particularly from the design standpoint, and to the successful driving of motion using chemical fuels, light or electrochemical reactions2,3. Mechanically interlocked molecules such as a rotaxanes [G] and catenanes [G] were at the centre of developing AMMs4–6. These molecules were carefully designed and studied in an attempt to control the movements of their parts and their directionality at the molecular level. Consequently, theoretical frameworks were developed for a number of approaches to drive the random Brownian motion [G] of molecules or their parts in a defined direction using AMMs7–9 as well as the physicochemical analyses of Brownian motors [G] 10–12 (Box 1).
Box 1. Energetic concepts in designing molecular machines.
One key step towards formulating the energetics of artificial molecular machines (AMMs) was clarifying the definitions based on the thermodynamics of the systems under study. Such clarification led to the distinction between molecular machines and switches159. A molecular machine works on the environment by responding to a stimulus using the motions of its sub-molecular parts (for instance, in F1-ATPase) or the entire molecule159,160 (for example, biomolecular motors such as kinesin and myosin). Maintaining the system away from its equilibrium and keeping it in a kinetic steady state demands constant intake of energy and its conversion into work on the environment (resulting in, for example, molecular motion). By contrast, a molecular switch might transition between different states by consuming energy, but the overall work it does on the environment is zero, meaning the work done to go from an initial state to a final one is fully reversed when the system goes back to its initial state160,161.
The dominant players at the molecular scale and in solution are thermal noise and viscosity, whereas inertia and gravity can be neglected160. Molecules in solution diffuse randomly in all directions, as described by the Brownian motion. Imparting directionality to the molecular motion is challenged by the presence of thermal noise, delivering almost eight orders of magnitude more power to a molecule than the power provided for example by hydrolysis of adenosine triphosphate (ATP) as a chemical source of energy162,163. The behaviour of molecules can be described using an underlying free energy landscape, where each state or configuration of a molecule is associated with a particular point in this energy landscape. As such, gaining control over the molecular behaviour is inevitably intertwined with understanding how to determine and sculpt the energy barriers and minima in the free energy landscape.
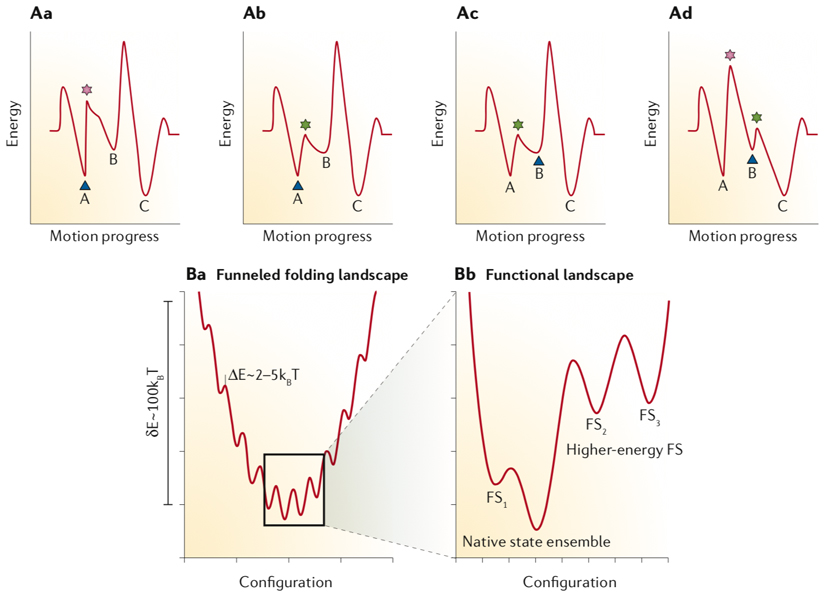
Timing the proper sequence of events in a motion cycle demands creating appropriate energy barriers between two consecutive steps. The challenge is driving the system uphill (also known as ratcheting) at the end of each step to roll it over the energy barrier, initiating the next step. This can be done in two manners using either an ‘energy ratchet’ concept or the ‘information ratchet’ 11,161. In the ‘energy ratchet’ type of control, the energy pumped into the system is used to either populate the higher energy states of the system without changing the energy landscape or to modify the minima and maxima of the energy landscape to navigate the reaction coordinate. The input energy brings the system to the point that the existing thermal noise in the microenvironment is enough to bias the Brownian motion of the whole molecule or a sub-molecular domain of it toward the desired directional motion162. The ‘information ratchet’ system also relies on changing the amplitudes of energy barriers but as a function of the system’s state. As a result, there is an information feedback loop between the current state of the system and the next energy barrier that should be modified (see the figure, part A). In the figure, the blue triangle indicates the current state of the system, the pink star shows a large energy barrier, and the green star marks a lowered energy barrier circumventable by thermal noise. The minima labelled with 1, 2, and 3 are the three steps in a directional motion cycle. As shown in Aa, the current state of system lies in 1 and for the motion to progress from 1 to 2 there is a large energy barrier. In Ab, a trigger decreases the marked saddle point between 1 and 2. In Ac, the motion updates the current state of the system to trapped in minimum 2. In the last snapshot (Ad), the next relevant saddle point between 2 and 3 is modified. Here, the reaction progresses by stepwise modification of the energy landscape to decide which step would be the next one based on which energy barrier is lowered the most to provide the path for an entire event to unfold. The ‘information ratchet’ mode of navigating a path through an energy landscape is frequently used in nature in a variety of molecular motions ranging from protein folding to stepwise movement of motor proteins on their tracks. For instance, the folding energy map of proteins is funnel-shaped, containing some kinetic traps (see the figure, part B). The zoom-in of the global energy minimum illustrates that there might be several functional states (FS) with their own energy barriers, transitioned to accomplish a function150.
Part B was adapted from reference150 with permission from Elsevier.
Box 1 figure.
Here, we focus on recent developments in structural DNA nanotechnology and its potential for creating biomolecular machines that are both encoded by and built from DNA sequences. In this context, ‘encode’ refers to using the DNA sequence as a way of programming the self-assembly process rather than the involvement of a translational step. We first discuss how DNA is employed as a molecular construction material and explore how motion can be rendered to assembled DNA nanostructures. In the last section, we point out those applications of DNA nanotechnology that, upon integration with moving DNA parts, hold great promise to bring about the required functional complexity in DNA machines. Most of these applications arise from the spatial addressability of DNA nanostructures — any point of interest in a DNA nanostructure can be ‘addressed’ by modifying the sequence of DNA of that particular location of the structure — resulting in their capability to act as a template to position molecules of interest on a precise point in the nanostructure. DNA switches [G], which can be used as parts of supramolecular DNA nanomachines, are not discussed for the sake of brevity; recent reviews on this topic can be found elsewhere13–16.
Encoding structures in DNA sequences
Why DNA?
Why consider building molecular structures or machines from DNA, given that DNA assumes a rather passive role as an information carrier in nature? The answer lies in the attractive physical and chemical properties of DNA. Custom DNA sequences are easily available via solid-phase chemical synthesis, gene synthesis as well as via biotechnological methods. DNA molecules show remarkable chemical stability compared to RNA and proteins, and the mechanics of DNA double-helices and single strands are well understood17,18. The deoxyribose-phosphate backbone of DNA has multiple rotational degrees of freedom per base. DNA single-strands are thus flexible polymers, whereas double-helical DNA domains are fairly rigid, with persistence lengths [G] on the scale of ~150 base pairs (bp) (approximately 45–50 nm at sodium concentrations above 10 mM)19,20. Hence, a wide range of local stiffness can be attained by combining flexible and rigid elements to meet the structural and functional requirements of any particular design. Also, effective persistence lengths much beyond those of individual DNA helices can be achieved in helical bundles. For example, bundling 6-8 DNA helices yields persistence lengths on the order of 2.0–3.5 μm21, which is comparable to the persistence lengths found for proteinaceous filaments22. The thermodynamics of double-helical DNA domain formation are well established, and the duplex stability can be accurately calculated using the nearest-neighbour model23, which increases the predictability of the self-assembly process outcome. Furthermore, the propensities of DNA single strands to form secondary structures can be reliably computed24. The Watson–Crick base pairing between DNA strands with complementary sequences offers a strong interaction mode to build up secondary structures, which can then be hierarchically arranged into tertiary and quaternary structures. By contrast, the limited chemical diversity of naturally occurring DNA may narrow the scope of functionalities achievable with structures built from canonical DNA bases. Fortunately, a large number of chemical modifications, conjugation methods25,26, non-canonical bases, and other modes of hybridization, such as triplex-forming nucleic acids27, exist that can be introduced into DNA nanostructures in a site-selective manner to expand chemical diversity. Moreover, even in their standard chemical compositions, DNA molecules can be catalytically active, as discovered by in vitro evolution experiments28,29. These results point to a hidden functional potential of DNA, and catalysis by DNA could potentially be exploited to power molecular motion or add more functional features to the assembled structures.
Progression of structural complexity in designed DNA objects
Nadrian Seeman, the founding father of DNA nanotechnology, laid out a key concept for the possible construction of nucleic acid junctions and lattices in a theoretical paper in in 198230. He suggested that a two-dimensional DNA structure could be achieved by fixing the branch migration in a Holliday junction. He later reported the first self-assembly of a 3D DNA cube31 in 1990, a 2D DNA crystal in 199832 and a 3D lattice in 200433. Seeman’s design strategy started with small DNA motifs or ‘tiles’, formed by self-assembly of a few short complementary DNA single-strands in the first step, that were attached to each other using sticky-ended overhangs in a second ‘tile assembly’ step. From the beginning, the main problem with the assembly and characterization of DNA nanostructures in three dimensions was the floppiness of the final products. Consequently, Seeman resorted to designing more rigid DNA tiles by weaving together strands using more crossovers34. An increasing number of other research groups joined the effort in the late 1990s, reporting new DNA nanostructures mostly as a support to immobilize molecules or to grow inorganic structures, such as nanowires35.
A new DNA assembly approach was reported in 2004, whereby a 1.7 kb single-stranded DNA strand was hybridized with five short oligomers as the supporting strands to fold into an octahedron36, in contrast to the traditional tiles made of DNA oligonucleotides of almost the same length. The long DNA strand was designed to form many internal loops interspaced by short intra-strand helical regions. Five edges of the octahedron were formed through hybridization of the five supporting strands, whereas the other seven struts were made by strand exchange among the loops of two different half-edges. In 2006, Paul Rothemund reported the one-pot assembly of single-layered DNA nanostructures, called scaffolded DNA origamis, using a single-stranded 7.3-kb-long M13 phage genome stapled together at many points using 100–200bases short DNA oligomers37. Scaffolded DNA origami and DNA tile assembly form the bases of many current design methodologies for the construction of nanostructures of defined size and shape, which may consist of hundreds of heavily intertwined DNA single strands and thousands of base pairs.
Principles of DNA origami assembly
A DNA origami is a nanostructure typically formed from one DNA single-strand of a few thousand bases long that serves as a template (the ‘scaffold’) and a multitude of much shorter DNA single-strands with lengths below 100 bases (the ‘staples’). DNA origami may be conceptualized as tying together DNA duplexes similar to the way a raft is built by putting together logs or reeds. Each DNA double-helix can be considered as a rigid rod, and the attachment points between two neighbouring double-helices are called crossovers (Fig. 1A). The scaffold strands raster back and forth to provide one of the strands of the helices. The second strands of the helices, the short complementary DNA staple strands, are often synthesized chemically. One common design approach starts by approximating the desired shape of a DNA nanostructure with parallel rods of different lengths. At each crossover point, one of the two strands of a duplex (scaffold or staple) swaps a complementary segment with a neighbouring duplex, thus, completing its hybridization by running through a different helix. To avoid generating and propagating strains, and as a result, twists, only certain crossover exit or branching angles are allowed depending on the packing type.
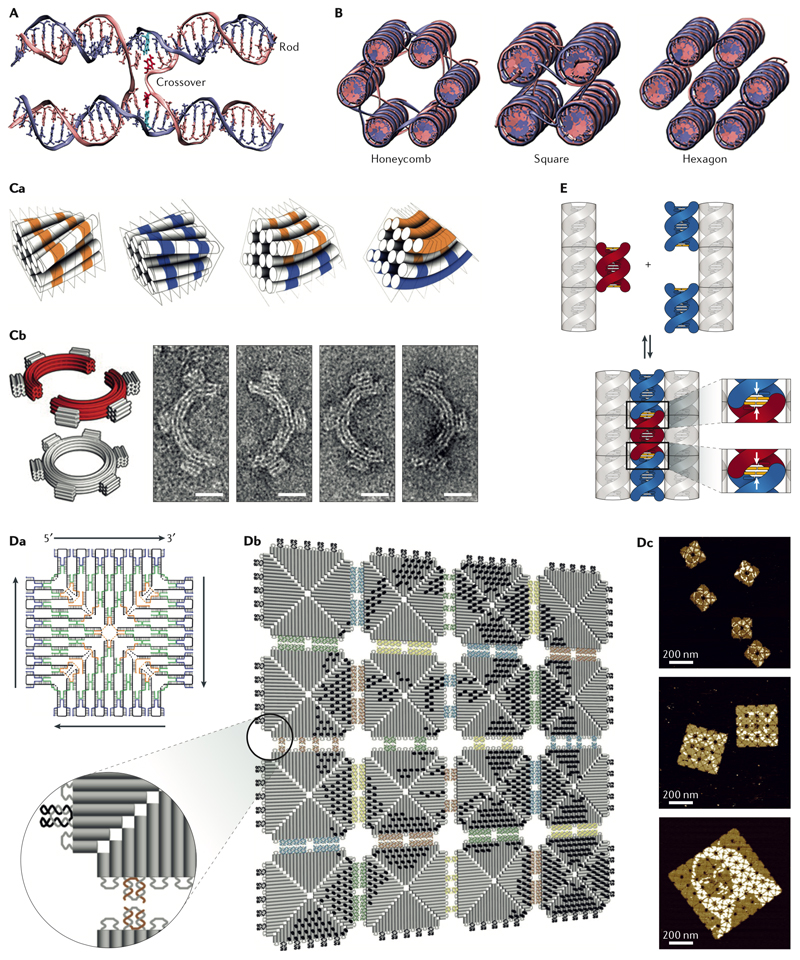
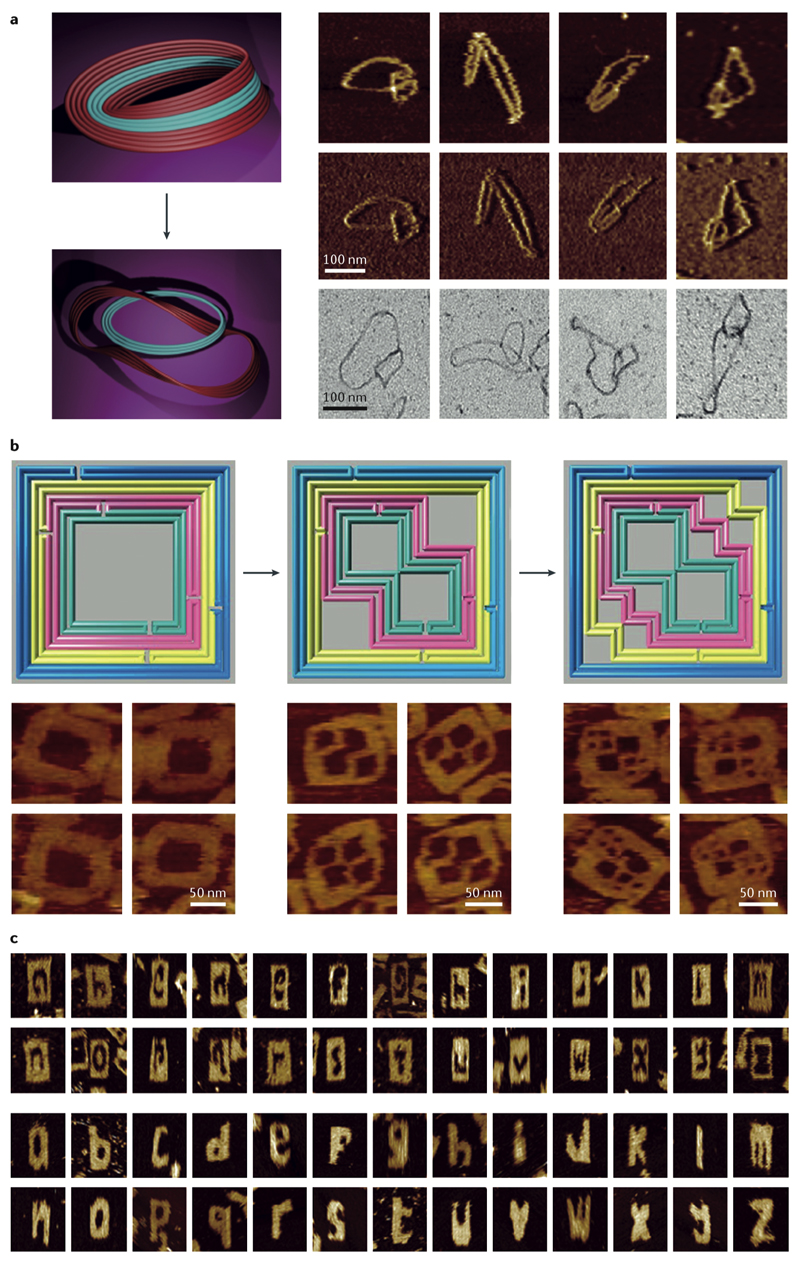
Fig. 1. Design principles of DNA origami structures and higher-order self-assembly using origami tiles.
A| The schematic of an antiparallel Holliday junction forming a double crossover between two adjacent helices in a honeycomb DNA origami six-helix bundle. Note, how the nucleobases at the crossover position (highlighted in red and cyan in the left crossover) still maintain their natural angle for duplex formation. B| The honeycomb (left), square (middle), and hexagonal (right) helical packings are the most common helical arrangements. The crossovers were deleted in the hexagonal packing due to multiple possible segment lengths and scaffold routings. C| Curvature design through base pair deletion or insertion. Ca| The base deletion (in orange) with a segment size smaller than 7 bp in the honeycomb arrangement causes a left-handed twist whereas a base insertion (in blue) results in a right-handed twist. Calculated combination of these two types of twists can be used to create curvature without causing marked global deformation. Ca| Negative-stain transmission electron microscopy (TEM) micrographs of two curved half-circles origami monomers and their six-tooth gear dimer (scale bar 20 nm)43. D| Fractal assembly. Da| The strand map of the origami tile used for fractal assembly. The staples are shown in blue, green, and yellow and the scaffold in black. Db| Fractal assembly of DNA origami tiles using edge loops and 2-nucleotide staple hybridization. The Mona Lisa’s pattern could be printed by adding double-stranded extensions to the selected staples on the surfaces of the tiles55. Dc| Atomic force microscopy confirmed the correct assembly of the tiles. E| Click contacts. Association of shape-complementary surface features can be driven by base stacking57.
Panel D adapted from REF. 55, with permission from Springer Nature.
The two commonly used helix packing patterns are the honeycomb and square packings (Fig. 1B), which require crossover exit angles of 120° and 90°, respectively. This is because each internal helix in the honeycomb form has three adjacent neighbouring helices or in case of the square pattern four immediately adjacent helices38,39. Assuming the commonly accepted value of 34.3° rotation per base pair (10.5 bp per pitch or turn) in the abundant B-DNA conformers, a natural base rotation angle of +240.1° or -119.9° would occur once every 7 bp along a helical axis. Hence, in the honeycomb helical arrangement, branching is possible in segments of 7 bp, and the exit angles have a periodicity of 21 bp, which is the smallest distance possible between two consecutive staple crossovers shared by the same two helices. Similarly, the approximate natural exit angle in the square packing occurs once every 8 bp along the helical axis, which results in a +274.4° (-85.6°) base rotation and an exit angle periodicity of 32 bp. Since the natural exit angle in the honeycomb packing is much closer to the geometrically desired angle compared to the square packing, the twist density is much less pronounced in the honeycomb arrangement of helices. By contrast, the square packing offers a higher packing density.
The helical packing density plays an important role in the rigidity of DNA nanostructures. The highest packing density reported was obtained in the hexagonal helical arrangement, in which the geometrically ideal exit angle is 60°, and each internal helix has six neighbours39 (Fig 1B). This type of packing was reported using crossover spacings (segment lengths) of both 9 bp and 13 bp long. The scaffold routing from one helix to the next is possible in multiple ways to keep the antiparallel relationship between the two neighbouring helices. It is, therefore, not mandatory for each helix to form staple crossovers with all of its six neighbours, but depending on its location, sharing antiparallel crossovers with three to five immediately adjacent helices is enough. Hexagonal packing is not frequently used, partly because it is not directly supported in the DNA origami design software packages. The other complication with the hexagonal helical arrangement is the substantial global twist it induces, as it assumes the values of 10.8 bp and 11.1 bp per turn in its 9 bp and 13 bp long segments, respectively. The lowest helical densities were obtained in the lattice-free origami wireframe nanostructures, as these nanostructures are made of single or double helical edges connecting the vertices40,41. The advantage of using a single helix or a two-helix bundle to build a shape is the possibility to approximate the envelope of a target object by wireframe tessellation [G]40,42; however, this comes at the cost of the rigidity of the assembled structures.
The square lattice packing requires underwinding of the right-handed helices from the canonical ~10.50 bp per turn to 10.67 bp per turn. The underwinding creates right-handed restoring torques at all junctions, which collectively lead to global twist deformation43 of the square lattice object, which may be undesirable and hard to predict quantitatively. Counter torques can be engineered into the structures by site-directed base-pair deletions to straighten out the square lattice objects43 (Fig. 1Ca). The twist along the helical axis can also be exploited as a design tool to generate curvature in a structural motif by imposing an unnatural crossover exit angle if the crossover position is shifted a base pair down or up43 (Fig. 1Cb). Honeycomb lattice packing structures were long thought to be practically twist-free due to the fact that realizing the internal crossover packing seemingly does not require helical deformations, but it was found that honeycomb structures also exhibit non-linear global deformations44.
There are many possible ways of breaking down the complement sequence for the scaffold into 100–200 short staple sequences. The main considerations are branching at the natural exit angles — which defines the crossover positions — and their distances, the lengths of the complementary segments along each helix, and the plausible accessibility of the three or four neighbouring helices in the honeycomb and square packings, respectively. The popular origami design software package caDNAno45 specifies the possible crossover positions based on the afore-mentioned design requirements. Tools such as computer-aided engineering for DNA origami (CanDo)46,47, oxDNA48 and ENERG-MD49 can be used to compute the shape of a DNA origami designed by caDNAno based on mechanical models with different levels of details. The mixture of staple strands and the scaffold is typically self-assembled by temperature annealing, although isothermal or small-temperature interval assembly of the structures is also possible and was shown to produce superior yields50. The folded DNA origami structures can be purified by agarose gel electrophoresis, ultrafiltration devices, polyethylene glycol (PEG) precipitation51, ultracentrifugation, and size exclusion chromatography52. DNA origami structures were also purified after functionalization with antibody-DNA or Ferritin–DNA conjugates53.
Higher-order assemblies are possible when a supramolecular structure is assembled stepwise from building blocks that are DNA origami themselves. The conventional duplex formation in the process of staple-strand exchange between two DNA origami scaffolds can be used to oligomerize tiles, as demonstrated by hetero-trimerization of an icosahedron54. Another example of such oligomerization is called ‘fractal assembly’, in which a limited number of designed interactions between staples and scaffold loops at the edges of the 2D origami tiles create a complex array of origami tiles55 (Fig. 1D). Furthermore, the 2D origami array can act as a nanometer-scale canvas to print any arbitrary patterns by simply modifying specific staples on each tile55.
Besides DNA hybridization, blunt-end stacking forces have proved to be a powerful tool in connecting DNA origami building blocks56 (Fig. 1E). Proper end-to-end alignment of the DNA helices is essential to form strong base stackings. To achieve such alignment, topological features are designed at the edges of a 2D origami tile56 or on the surface of a 3D DNA origami monomer57. The topological features (click contacts) are designed based on the key-and-lock concept and, therefore, their shapes are complementary57. The additional benefit of using the shape complementary docking features is that they work based on simple geometrical considerations, avoiding the need for sequence design. The shape complementarity also ensures specificity of the interacting sites. The shape-complementary base stacking association of DNA origami monomers is robust and can hold together a DNA super-assembly of up to 1.2 gigadalton in size44. Other recent origami design approaches involve the elimination of staple strands to fold a nanostructure from a single-stranded DNA or RNA scaffold58,59 and using DNA-binding proteins to guide the folding of a double-stranded DNA scaffold60.
Tile assembly of DNA nanostructures
Tile assembly relies on the self-assembly of DNA tiles and connecting those tiles, mostly via sticky ends. In order to produce stiff DNA tiles or motifs, the sequences must be unique, the complementary stretches at the branching points should be excluded from the sequences, and cross-hybridization should be minimized34. Before the introduction of DNA origami in 2006, only a limited number of DNA tiles had been reported that addressed the geometrical needs of most shapes34 (Fig. 2A). Traditionally, DNA tiles consisted of a few synthetic oligomers and suffered from low sequence diversity because of the costs associated with the solid-phase synthesis of DNA oligomers in the early days of DNA nanotechnology. More recent approaches to assembling DNA nanostructures using a modular design take advantage of DNA tiles (or so-called single-stranded DNA ‘bricks’) made of DNA single-strands of 32 nucleotides (nt)61, 42 nt62,63 or 52 nt64 in length with unique sequences (Fig. 2B).
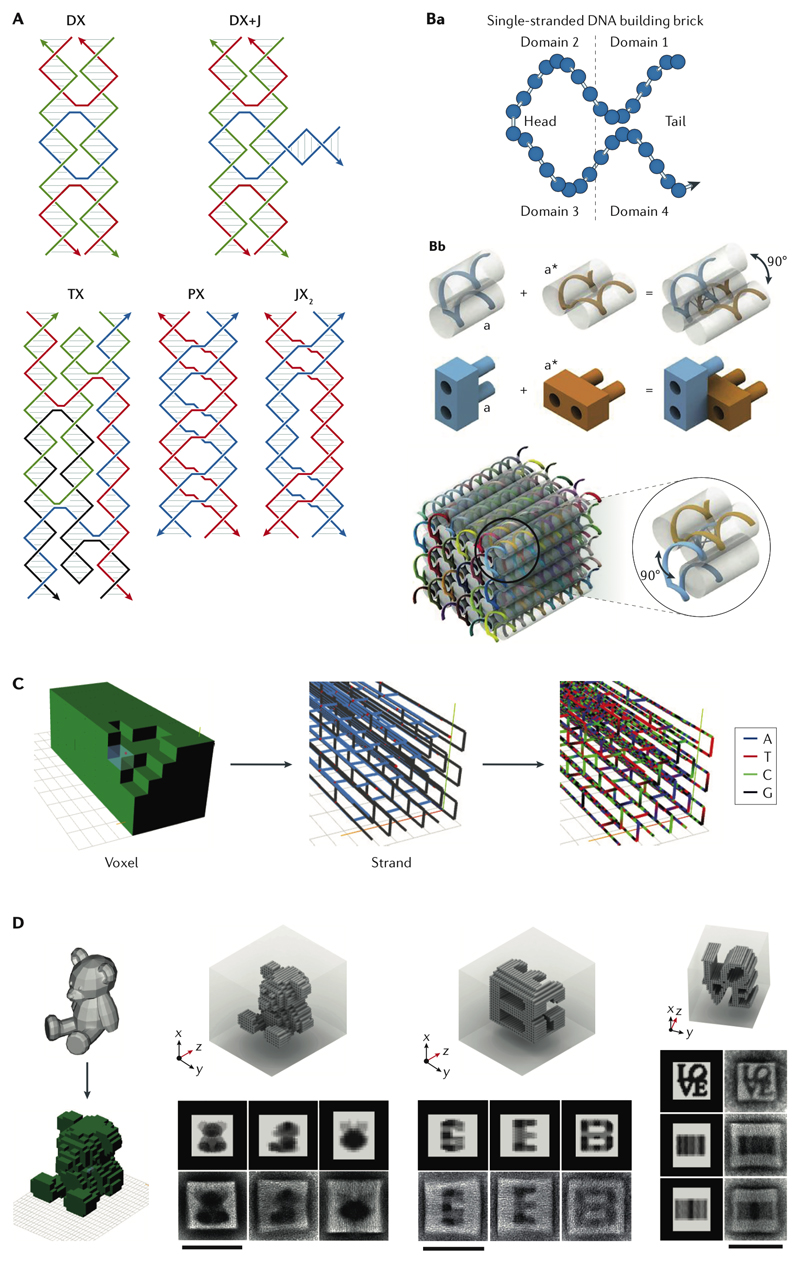
Fig. 2. Tile assembly of DNA nanostructures.
A| Different Seeman tiles with diverse strand polarities34. B| Principles of ssDNA brick assembly: each brick is a DNA single strand with four domains. When each domain is 8 nucleotides long, its hybridization with an adjacent brick produces a dihedral angle of 90 °. Each brick could be thought as a LEGO binding to four other complementary LEGOs to form a cube61. C| The design workflow starts with removing 3D pixels (voxels) from a 3D canvas followed by generation of the corresponding strand diagram and sequences64. D| Transmission electron microscopy (TEM) characterization of designed arbitrary shapes and geometries (scale bars 100 nm)64. Panels A-C were adapted from REFs. 61, 64, 64, respectively, with permissions from AAAS and Springer Nature. DX, double-crossover; JX, juxtaposed crossover; PX, paranemic crossover; TX, triple-crossover,
Similar to DNA origami assembly, this is a one-pot reaction, and by changing the length of ssDNA bricks, different packings are possible. For instance, the 32 nt brick is composed of 4 segments of 8 nt, providing the branching exit angle of roughly -90°, exactly in the same way as the square helix arrangement in DNA origami (Fig. 2B). A big cube assembled with the ssDNA bricks is then considered to be a 3D molecular canvas; by eliminating specific bricks in the folding reaction, any desired shapes can be sculpted out. A software package starts with the input of the geometrical shape, which is converted to a strand diagram, and finally generates the sequences needed (Fig. 2C). Complex shapes can be generated by carving out the right set of bricks (Fig. 2D). The advantage here over DNA origami is the lack of a long scaffold with a fixed sequence, which in principle enables making DNA nanostructures with arbitrary sequences and dimensions. The latter point was highlighted recently with DNA-tile based nanostructures that were 100 times larger than single-scaffold DNA origami64.
In vitro and in vivo production of DNA or RNA nanostructures
Phagemid-mediated cloning of the ssDNA is a viable option to the M13 phage genome for producing DNA nanostructures65 and can be employed to produce a variety of scaffolds66 with fully customizable sequences67. The short oligonucleotide staple strands are chemically synthesized, and due to their inferior purity relative to scaffold, they are added in 4–10 times molar excess to the folding solution. Alternatively, biotechnological methods based on rolling-circle amplification of a set of staples interspaced by hairpin cleavage sites for restriction enzymes68, circle-to-circle rolling amplifications69, and primer exchange reaction (PER) cascades70 were also reported. These methods are more likely to yield higher-quality oligos compared with chemical synthesis, making it possible to use a staple:scaffold stoichiometry closer to 1:1. This was proved by coding both the scaffold and staple sequences in a phagemid with two DNAzyme, flanking the sequences of the scaffold and each staple; the procedure enabled mass production of both components71. The DNAzyme was a small ssDNA enzyme with a fixed sequence attached to its DNA substrate of known sequence, forming two catalytic hairpins72. The DNAzyme cut its substrate only in the presence of zinc ions. Upon purification of the cloned DNA, zinc was added to produce the scaffold and the required set of staples, which were then folded into the predesigned DNA origami71.
In situ production and folding of nucleic acid nanostructures inside cells (that is, in vivo) were also demonstrated for RNA and DNA assemblies. In vitro transcription and folding of a ssRNA into 2D hexagonal or rectilinear networks were demonstrated by designing two new RNA crossover motifs based on the A-form RNA–RNA duplex59. The first half of the RNA scaffold folds into helices as it is being synthesized, and the second half forms kissing loops with the first one upon transcription and folding. Recently, a similar methodology was used to produce RNA nanostructures inside cells73. Different RNA motifs at the level of secondary and tertiary structures, such as hairpins, kissing loops, kinks or 3-way loops, were designed and arranged rationally to define a sequential and hierarchical folding pathway. In vitro transcription or plasmid transformation plus in vivo transcription of the rationally designed DNA template generates a nascent RNA that folds into the secondary motifs. The unpaired regions between the motifs can bridge the more distant domains of the ssRNA under transcription and bring about the final 3D geometry of interest, analogous to the way protein chains fold in the course of translation.
DNA nanostructures were produced in vivo by adopting a similar approach, although the generated RNA had to be retro-transcribed to the DNA strand folding into the desired nanostructures74. The DNA nanostructure sequence was inserted into the genome of a particular strain of Escherichia coli that is incapable of degrading ssDNA. Two reverse transcriptases, from HIV (HIVRT) and murine leukaemia virus (MLRT), were co-expressed to convert the transcribed RNA of the DNA nanostructure’s gene back to ssDNA. A eukaryotic version of the HIVRT recruiter t-RNA (t-RNALYS) was designed and added to the end of the gene sequence encoding the DNA nanostructure, so that the resulting RNA can recruit HIVRT. The t-RNALYS loop also acts as a transcription terminator. MLRT was needed to improve the speed of transcription, but as it is a DNA-dependent polymerase it could not replace HIVRT. The ssDNA folded into the designed shapes, and its production was induced by isopropyl-β-D-thiogalactoside (IPTG). The nanostructures included zinc finger-binding sites to bring the two halves of a yellow fluorescent protein fused to the zinc finger domains together as proof of proper assembly74.
Encoding machines in DNA sequences
Dynamic DNA devices based on strand displacement reaction
DNA has a much larger persistence length when it forms a duplex (2.275–4.676 nm for the ssDNA compared to 45–50 nm for the DNA duplex19,20). This increase in rigidity and hybridization-associated changes in length can be exploited to generate moving parts within DNA assemblies when combined with a DNA strand displacement reaction (SDR). In an SDR, two complements compete for the same template strand, and the complement with the larger stretch of base pairs (fuel strand) will replace the weaker-binding complement (output strand)77 (Fig. 3a). The template must first form a partial duplex with the output strand and in doing so, a single-stranded overhang on the template (referred to as the toehold) is exposed, which is complementary to the fuel strand. Adding the fuel strand to the solution will then start a branch migration (similar to homologous recombination) that eventually leads to the release of the output strand. The SDR can be used to isothermally control the hybridization or dehybridization of DNA duplexes connecting two different parts of an origami, as demonstrated by the opening of the lid of an origami ‘box’ upon addition of fuels78.
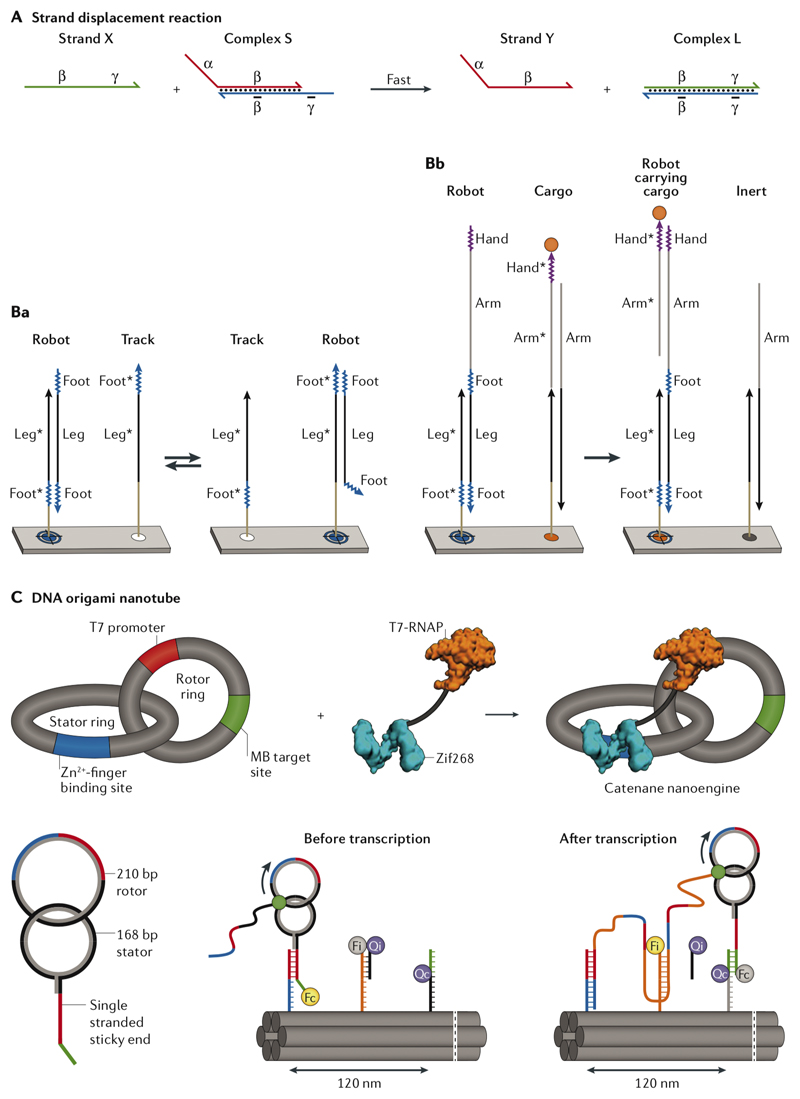
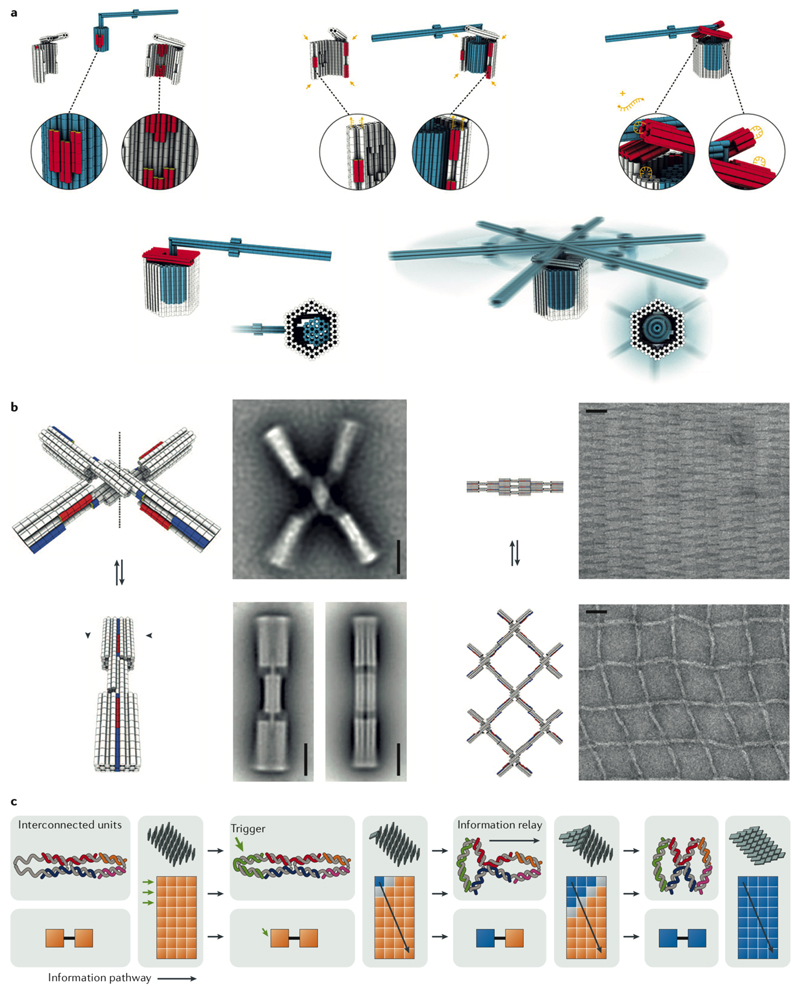
Fig. 3. Strand displacement reaction (SDR) and SDR-based walkers.
A| Strand X is the fuel and strand Y is the output77. The final complex (L) is thermodynamically more stable than the initial duplex (S), driving the equilibrium toward formation of more complex L77. Ba| In the single-stranded DNA (ssDNA) walker, walking is performed through two toeholds of equal sizes (Foot) flanking a template region (Leg) complementary to the ‘track strands’ immobilized on the surface of a DNA origami pegboard in equal distances80. All track strands share the template complement region (Leg*), but each has only one of the two complements (Foot*) to the walker’s feet in an alternating pattern. The walker starts binding to one of the track strands via its template and the first toehold, and its second toehold is exposed, which in turn can bind a neighbouring track sequence containing the complement to the second toehold. The track strand at the destination docking site has the complements to both toeholds of the walker (feet) and forms the most stable duplex so that no further movement is possible. Bb| A cargo could be picked up by adding a second template region (arm) and a new toehold (hand) to the walker. Adding a second toehold to the arm (not shown in this scheme) and its complement at a destination site will deliver the cargo to the designated site80. C| The assembly of a zinc finger-fused T7 RNA polymerase (RNAP) rotation mechanism. As the RNA polymerase transcribes its template, the rotor ring rotates against the stator ring kept at a fixed distance using the fused zinc finger domain binding. The molecular beacon (MB) target site helps with monitoring the transcription progress82. The ring rotation of the DNA catenane system is coupled to an SDR-based linear walk on a six-helix DNA origami nanotube. Panels A-C were adapted from REFs. 77, 80, 82, respectively, with permissions from American Chemical Society, AAAS, and Springer Nature.
DNA origami was used as a molecular track to control the motion of small DNA molecules such as DNAzymes and DNA walkers [G]79. In this context, the motion involves displacements of the entire small DNA molecule with respect to its position on the DNA origami support. A well-established type of DNA walker is composed of a single strand of DNA80 or a tensegrity triangle [G]81 with multiple toeholds for random walking, destination point docking and cargo pick-up (Fig. 3b). The same strategy is applicable for sorting and carrying a molecular cargo conjugated to another set of template–toehold sequences80. SDR-based walkers use a passive mode of motion driven by thermodynamics of the reactions involved. As shown by fluorescence and atomic force microscopy, the directional redistribution from the loading to destination sites occurs because the cargo strand can form more base pairs at delivery sites80.
The other approach for creating motion takes advantage of DNA-binding enzymes such as DNA or RNA polymerases. As an example, rotational and linear molecular motions were combined on a DNA origami nanotube82 (Fig. 3c). Two interlocked DNA rings were made, one of which (the rotor ring) was used as a template for rolling circle transcription using an engineered T7 RNA polymerase fused to a zinc finger DNA-binding domain82. The zinc finger binding sequence was embedded in the stator ring, whereas the rotor ring contained the promoter sequence for T7 RNA polymerase. The stator ring also comprised a DNA anchor sticky end for attachment to the track strands on the origami support. The rotation was powered through hydrolysis of nucleotide triphosphate (NTP) by T7 RNA polymerase in an in vitro rolling circle transcription, whereas the linear movement was based on a passive SDR-mediated linear walk on the origami nanotube. As the RNA polymerase moves along its double helical template, transcribing RNA, the second ring starts to rotate against the fixed stator ring. As the second ring rotates, the growing RNA amplicon successively ‘walks’ on the DNA origami support using the previously discussed SDR mechanism, thus pushing the rings forward (Fig. 3c). The walking was confirmed using fluorescence and atomic force microscopy82.
Structural reshaping using strand displacement reaction
Strand displacement reactions can be used to reconfigure DNA origami structures in a stepwise manner. First, the staples corresponding to a particular domain of the structure may be removed via their toehold extensions after adding the fuel strands. Then, a new subset of staples needs to be added to reshape the stripped scaffold domain. The first step of this procedure was demonstrated by cutting a DNA origami Möbius strip along its edges, resulting in a new strip double its original length or two interchained strips depending on the position of the cut line across the width of the strip (Fig. 4a)83. The second step, leading to reshaping of the domain of interest, was done both in DNA origami84 (Fig. 4b) and in DNA structures assembled from ssDNA bricks (Fig. 4c)85. Based on a similar SDR-mediated actuation strategy, a 2D DNA mesh made of vertical and horizontal two-helix DNA beams was formed by excluding the DNA strands responsible for fixing the beam angles from the folding mixture86. Addition and SDR-assisted removal of those strands caused expansion or contraction of the mesh, or the nanotubes made of it, owing to the change in beam angles86. This actuation mechanism was also employed to switch between the compact and expanded states in an origami Bennett linkage87or to release a moving DNA origami ring fixed to its dumbbell-shaped origami track by hybridization88.
Fig. 4. Reshaping nanostructures using strand displacement reaction (SDR).
a| A DNA origami Möbius was cut along its strips using SDR to produce two interlocked rings (left). The atomic force microscopy (AFM) height (right, top row), amplitude (right, middle row), and transmission electron microscopy (TEM) (right, lower row) images of the products83. b| The left origami frame was transformed to the middle and then the right frame using staple subset removal by SDR and addition of a new subset of staples as confirmed by the AFM (bottom row)84. c| AFM images of the SDR-mediated carved letters of alphabet off a 2D molecular canvas assembled using single-stranded DNA brick assembly technique85. Panels A-C were adapted from REFs. 83, 84, 85, respectively, with permissions from Springer Nature, American Chemical Society, Wiley-VCH Verlag.
Free and interlocked molecular motion in DNA nanostructures
Certain 3D features can be incorporated in a design to allow for free movement of a specific part of the structure and yet limit the motion to a desired degree of freedom. 3D DNA components have been designed to exhibit rigid domains and to create molecular analogues of macroscopic mechanisms such as pivots, hinges, crank sliders87 (to couple rotational with linear motion) or Bennet linkages87. In these and other mechanisms, DNA strand linkages or particular design features, such as mechanically interlocked but not directly connected parts, limit the range of movements88. Analysis of a rotary device illustrates how design features can be used to produce free motion in a specified plane of rotation89 (Fig. 5a). The apparatus was designed to tightly constrain the motion of a rotor module to one rotational degree of freedom, without actually restricting the angular range of rotation. The rotary device was constructed from three different multilayer DNA origami components: a rotor unit, and two clamp elements that form an axle bearing. At lower magnesium concentrations, the rotor underwent random rotary motions and the rotation was visualized using single-molecule fluorescence microscopy of the apparatus, with the crank-lever fluorescently labelled at both ends.
Fig. 5. Modes of molecular motion in DNA nanostructures.
A| The design and assembly scheme of a DNA rotary device that limits the motion of its lever through spatial confinements laid out in the design89. The rotary apparatus has a rotor unit, and two clamp elements that form an axle bearing (top left). The rotor unit also features an axial protrusion shown in red. The clamp units also feature shape-complementary recessions. The clamp elements have a cross section resembling a bisected hexagon. For assembly, the rotor is first docked onto one clamp unit with the help of shape-complementary surface features (top middle). To close the axle bearing, the second half-clamp is clicked into the edge contacts of the first half-clamp already hosting the rotor (top middle). Hybridization of the DNA single-strands along the mutual binding interfaces of the two half-clamps glues them together tighter. Two brackets (coloured red in top right) were then capped the top opening of the bearing through DNA hybridization. The brackets rigidify the bearing and trap the rotor in the cavity. The lever rotates at the lower magnesium concentrations (bottom). B| A DNA actuator with shape complementary surface features switches open and close at magnesium chloride concentrations of 5 mM and 25 mM, respectively (scale bar 20 nm)57. A switchable 2D grid made of this actuator also responds similarly to different ionic stregnths (scale bar 50 nm)57. C| Propagation of conformation flipping across a DNA nanostructure. Ca| Two interconnected Holliday junctions have two minimum conformations (symbolized with a pair of orange or blue boxes at the bottom) and one higher energy transition state (the orange–blue box at the bottom). Addition of a trigger strand to a system of two connected Holliday junctions flips the conformation of the first junction, which is relayed to the second junction because the stacking interactions must be maximized again among the base pairs located at the junctions91. Cb| Depending on the start point, the interlocked connectivity of the Holliday junctions relays the initial flipping of the conformation across the whole array in a domino effect91. Panels A-C were adapted from REFs. 89, 57, 91, respectively, with permissions from AAAS and Wiley-VCH Verlag.
Ionic strength or temperature can be exploited to bring about conformational changes in DNA nanostructures. Shape-complementary topological features (surface recessions and protrusions) on DNA origami offer not only specificity for the higher-order assembly of superstructures but also weak interactions for reversibility, because of short-range nucleobase stacking forces57. Taking advantage of this latter aspect, various reconfigurable devices were produced, among them an actuator, a 2D DNA grid, and a foldable gear57. The actuator is a cross-shaped DNA origami with shape-complementary recessions and protrusions on its arms. The arms click together in the closed state at high magnesium concentrations, when there is effective charge screening, or at low temperatures, owing to much smaller thermal fluctuations. When the actuator was polymerized into a 2D grid, the whole grid could be switched between the compact and expanded conformations by changing the magnesium concentration (Fig. 5b). Temperature-jump experiments on the actuator highlighted the robust reversibility of the shape recognition scheme and the absence of structural or functional degradation. More than 1,000 state transitions, occurring on the time-scale of seconds, in the two-state switch could be realized through temperature cycling (between 25°C and 50°C) without signs of wear-out or structural degradation 59. In a different work, direct thermal actuation was used to switch between open and closed conformations of a DNA origami hinge that had been functionalized with a thermoresponsive polymer90.
The modular design of a nanostructure with cooperative interconnectivity of its reconfigurable building blocks ensures propagation of the reconfiguration across the whole structure. Based on this concept, a DNA array was developed that could be reconfigured using isomerization cascades of Holliday junctions91 (Fig. 5c). Due to the coordinated connectivity of the junctions, the conformational switching at a given point in the structure can grow all over the structure in a domino effect. Thereby, 2D helix networks and nanotubes can be switched by a DNA trigger strand from one minimum energy configuration to another (Box 1), where they pass through a transition state featuring many small barriers, each representing the transition state for isomerization of a single Holliday junction.
External magnetic or electric fields and light have also been explored for generating motion in DNA nanostructures. A rod-like DNA arm mounted on a flat DNA origami plate was switched back and forth between discrete sites using alternating voltage in a four-electrode microfluidic chamber92. In analogy to the run and tumble propulsion of bacteria, helical DNA filaments were attached to magnetic nanoparticles to create microswimmers93, which could then be actively propelled in the solution by rotating external magnetic fields. The cis–trans photoisomerization of azobenzene-functionalized DNA strands controls their hybridization or melting behaviour (Fig. 5d), providing a new actuation mechanism94. Loop formation and melting were used to switch a DNA shaft mounted on a DNA origami plate between two perpendicular states95. The hybridization of two loops at the ends of the shaft to the two anchor sites on the origami was initiated by UV irradiation. This led to loop melting, and the two sticky ends were freed up to hybridize, halting the rotor at a fixed rotational state while the visible light could release the rotor. The rotation was monitored by high-speed atomic force microscopy97.
Exemplary applications of DNA nanostructures
One of the early applications of periodic DNA assemblies was positioning molecules with nanometer precision. Positional accuracy is an essential prerequisite for many advanced functions such as cargo pick-up and release, molecular recognition, active-site binding, conformational change and catalysis. The structural addressability of DNA nanostructures will play a central role in controlling the interaction and coordinating the motion of future DNA nanomachines. 2D DNA arrays can serve as molecular pegboards, for instance in solid support applications, whereas 3D DNA structures are useful to encapsulate or display other molecules, for example, in the context of co-crystallization and drug delivery. Extensive control over spatial functionalization of DNA nano-assemblies has been demonstrated by building enzyme networks, in super-resolution microscopy, and in organizing plasmonic elements into predesigned arrangements. The application of DNA nanostructures as positioning devices in the fields of plasmonics96,97 and photonics98,99 is growing rapidly and is beyond the scope of this Review. In this section, we discuss a number of the current applications of DNA nanotechnology that hold great promise to expand the functional spectrum of the future DNA nanomachines.
DNA crystals
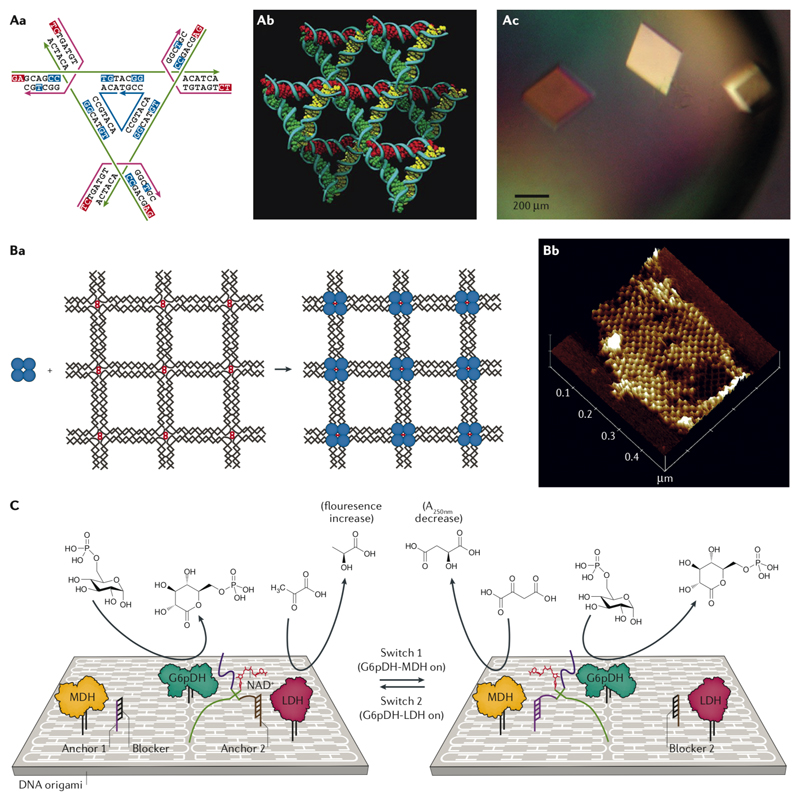
The co-crystallization of proteins inside a crystalline DNA lattice was the initial driving force for DNA nanotechnology. The first 3D DNA crystal lattice, made of a 13-mer DNA oligomer and reported in 2004, was held together by parallel and antiparallel DNA base pairing plus homopurine stacking interactions33. In 2009, the first instance of a rationally designed 3D DNA lattice formed exclusively by canonical Watson–Crick base pairing was published100. The crystal unit cell was composed of 7 short DNA strands forming a tensegrity triangle, which could then associate with neighbouring units using a 2-nt sticky end (Fig. 6a)100. The solvent channels within the lattice formed cavities, and the crystal was resolved by X-ray diffraction at a resolution of 4 Å100. The more recent crystals (diffracted at 3.1 Å), designed rationally based on the same principles, enable tuning of the cavity size and shape as well as chirality of the DNA-made crystal cells101,102. Further studies on these DNA crystals proved the robustness of the crystallization process, whereby introducing different defects, such as single-base mismatches, did not prevent lattice formation103. It was also shown that post-crystallization entrapment of the guest molecules inside the solvent channels and cavities of the crystal is possible104. Efforts to crystalize DNA origami unit cells have not been successful so far, although an ordered micrometer-scale 3D array of tensegrity-triangle-like DNA origami monomers was recently assembled105.
Fig. 6. Applications of DNA nanostructures.
A| The strand design of the unit cell of the tensegrity triangle crystal (Aa), the stereoscopic image of the triangles in the lattice (Ab), and the microscope image of the crystals (Ac) 100. B| Atomic force microscopy (AFM) image of streptavidin (blue circles) placed on a 2D DNA grid (Ba) and the AFM proof of assembly (Bb)35. C| Switching between two different enzyme pathways on a DNA origami support is possible by controlling the location of the cofactor (NADH)112. Panels A-C were adapted from REFs. 100, 35, 112, respectively, with permissions from Springer Nature, AAAS, and Wiley-VCH Verlag.
DNA molecular pegboards and nanoplatforms
One of the first applications of 2D DNA grids was the generation of periodic streptavidin arrays and silver nanowires (Fig. 6b)35. The control over spacing of different molecular entities on a DNA pegboard enabled the systematic investigation of multivalent binding among two aptamers with different binding sites and thrombin: under optimal distances, the two aptamers showed 10 times higher avidity toward thrombin106. The 2D DNA origami support was recently shown to be able to control the polymerization of polydopamine into defined nanopatterns107. A single-layer DNA origami support has a spatial resolution of 6 nm for positioning molecules, but it is assembled in solution and there is no control over its orientation when deposited on a surface108. A lithographically patterned surface could control the orientation of individual origami assemblies deposited on the surface, thus extending control over molecular pattering in the range of 6 to a few hundred nanometers108,109. The use of a 3D multilayer DNA origami hinge as a positioning device could increase the precision of positioning to the sub-nanometer scale and was exploited to control the efficiency of a crosslinking chemical reaction110.
Enzyme networks on DNA origami
Multienzyme cascades can be assembled on DNA origami supports to study the role of substrate diffusion and kinetics111. A biological circuit was made on a DNA origami support by putting three dehydrogenase enzymes, glucose-6-phosphate dehydrogenase (G6PDH), malate dehydrogenase (MDH) and lactate dehydrogenase (LDH), in a trigonal arrangement, in which accessibility to the NADH cofactor could be programmed (Fig. 6c)112. The DNA–NADH conjugate was attached to one arm of a fixed Holliday junction, while the other two arms had two different switch strands and the last arm had a pivot strand latching the whole Holliday junction onto the surface of a DNA origami platform. Two anchor sites between G6PDH and the other two enzymes were devised to hybridize to one of the switch strands and control the location of the cofactor. The NADH molecules produced by G6PDH could then be used by LDH or MDH depending on where the cofactor was anchored. Binary enzyme pathway activation is possible through SDRs between the switch strands and the anchor sites112.
Many studies on an enzyme cascade composed of glucose oxidase (GOx) and horseradish peroxidase (HRP) assumed that the close proximity of the enzymes immobilized on the DNA supports facilitated diffusion of the substrates, resulting in an enhanced enzymatic activity113. However, according to a more recent study113, no increased activity was observed when the two enzymes were conjugated together but not immobilized on a DNA origami. It was therefore proposed that the lower pH close to the surface of the origami might be responsible for the observed enhancement, not the local concentrations of substrates114. Encapsulation of the GOx and HRP pair plus four other single enzymes in a DNA origami cage also proved that the contribution of co-localization is small, and the increase in turnover numbers is the main reason for increased activity114.
Super-resolution microscopy: DNA-PAINT and STORM
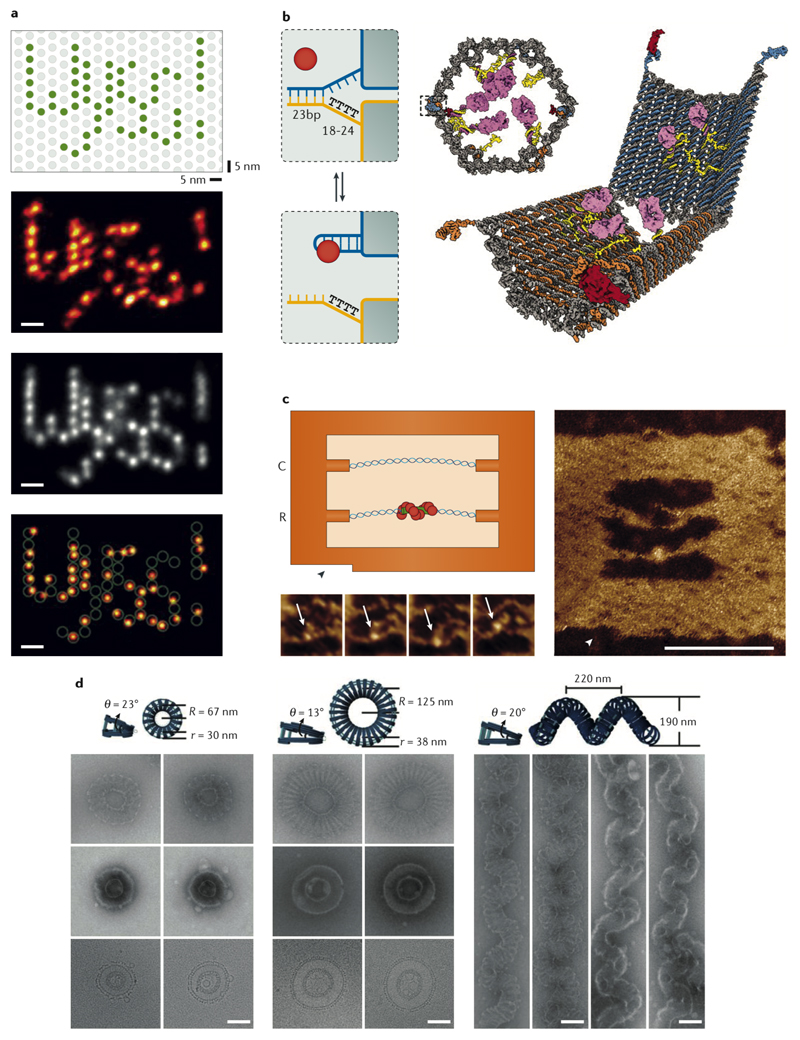
PAINT (points accumulation for imaging in nanoscale topography) is a sub-diffraction limit imaging technique based on the transient binding of fluorescently labelled probes to a binding site on the object115. The adaptation of this method for DNA (called DNA-PAINT) utilizes binding of short DNA probes to a DNA origami for validation and calibration116. By optimizing the length and binding parameters of the labelled DNA probes, and using DNA origami as a molecular canvas, DNA-PAINT achieved resolutions below 5 nm (Fig. 7a)117 and was developed into a quantitative technique (DNA-qPAINT) capable of measuring the number of bound probes118. The positional accuracy provided by a DNA origami chassis was also used to quantitively measure the number of bound molecules to the binding sites in a super-resolution microscopy technique called STORM (stochastical optical reconstruction microscopy)119.
Fig. 7. Applications of DNA nanostructures.
A| High-resolution DNA-PAINT: A single-molecule resolution could be obtained by activating particular target binding sites on the origami (top scheme), single image processing (second row), and averaging (third row) to resolve fluorophores only 5 nm apart. The overlay of the designed pattern and the single image is shown at the very bottom117. B| Schemes of the antibody-loaded DNA origami barrel. The locks are two aptamers that will release the two half-barrels upon binding to their targets and expose their payloads120. C| High-speed AFM observation of the Rec-A protein bound to a region of sequence homology (scale bar 40 nm)130. D| DNA origami-guided self-assembly of the membrane tubules with different internal diameters (left) and membranes with complex curvatures (right) (scale bars 100 nm)139. Panels A-D were adapted from REFs. 117, 120, 130, 139, respectively, with permissions from Springer Nature, AAAS, and Wiley-VCH Verlag.
DNA nanocarriers for targeted drug delivery
With the high level of control over designing structural details and shapes, functionalization of DNA nanostructures for encapsulation and targeting drugs to their site of action has been accomplished. The most prominent prototype of such a drug carrier was demonstrated by a hexagonal origami barrel carrying antibody fragments against human leukocyte antigens (HLAs)120. The barrel was composed of two halves locked together using two aptamers, one on each side. Upon binding to the aptamer target, in this case cell receptors, the barrel opened and exposed its antibody payload for binding to a different set of receptors, as proved by labelling the cells of interest or activating cell receptor-mediated signalling pathways (Fig. 7b)120. In vitro studies show that cellular uptake of DNA origami structures depends on their shapes; more compact DNA origamis with lower aspect ratios have a higher efficiency of cellular uptake121,122.
Two major concerns regarding the use of DNA nanostructures under physiological conditions arise from their potential instability at very low salt concentrations and their degradation by endonucleases in serum (after 4–8 hours in 10% fetal bovine serum122). Different strategies have been presented to enhance the physicochemical and enzymatic degradation stabilities of DNA nanostructures, such as chemical crosslinking by alkylating agents123, UV photocrosslinking124 via cyclobutane pyrimidine dimer bonds, DNA triplex formation125, encapsulation in PEGylated lipid bilayers126, coating with proteins such as bovine serum albumin (BSA)127, and oligolysine–PEG copolymers128,129. The incubation of DNA origami with oligolysine–PEG copolymer to replace divalent cations and form a protective layer around the nanostructures proved to be a powerful stabilization method129. The coating protected different DNA origami assemblies even in the absence of any salt in the buffer and stabilized them against nuclease degradation by three orders of magnitude compared to untreated DNA nanostructures129. Moreover, in mice, the in vivo stability of the multilayer origami improved 5-fold over that of the uncoated origami129. UV photo crosslinking is an alternative stabilization method for DNA origami. The so-called UV ‘point welding’ method enhanced the stability so remarkably that such crosslinked multilayer DNA origami structures withstood exposure to temperatures up to 90°C and maintained their integrities in essentially salt-free distilled water124. In a different approach, the octahedral DNA origami functionalized with lipid–DNA staples on the surface was encapsulated in liposomes126. The encapsulated octahedrons showed >100 times less immune response in an in vitro assay with spleen cells, 17 times more bioavailability, and 9 times longer elimination half-life in mice (the elimination half-life of the non-encapsulated octahedron in mice was about 50 minutes in mice, compared with 38 minutes for a control oligonucleotide). These studies highlight the importance of considering the physicochemical and enzymatic protection of DNA nanocapsules as an essential part of any DNA-made drug delivery system and provide solutions to achieve such protection.
DNA nanotechnology for functional studies
DNA origami is a useful tool for performing high-resolution measurements to quantify biomolecular interactions and elucidate their mechanisms. For instance, the mechanisms of homology search by the RecA protein and the DNA methylation regulation by EcoRI methyltransferase were revealed by observing how these proteins move along their DNA double-stranded tracks, tied at both ends to a DNA origami breadboard (Fig. 7c)130,131. In a similar approach, a DNA origami force clamp was designed for measuring the bending energy of the TATA-binding protein when it binds its recognition site132. The force clamp has a partial duplex hooked at both ends to the origami and labelled with a Förster resonance energy transfer (FRET) pair at both ends of the duplex. To increase the force, the length of the single-stranded regions flanking the duplex was shortened in six different origami variants and the FRET efficiencies under different forces were measured. The protein binding causes a 90° bend in the duplex, changing the FRET efficiency, which could be converted to the bending energy. Another DNA origami force stage was built from a V-shaped origami hinge with a single nucleosome complex immobilized on each arm and a FRET pair prefixed on the arms133. The energy landscape of nucleosome interactions could thus be inferred from the angular distribution profile of the force stage under transmission electron microscopy (TEM) or FRET efficiencies under different binding conditions133.
The rigidity of DNA origami can be combined with the single-molecule optical tweezer technique to improve the sensitivity of such measurements, especially in the lower force regime21. This experimental setup has been used to determine the unfolding energy of DNA loops and the stacking energy of DNA blunt ends134. Synthetic nanopores were constructed from DNA origami units in different shapes with cholesterol anchors for insertion into lipid membranes135,136. DNA origami rings were also used to organize the proteins involved in nuclear pore complex (NPC) formation into nanopores to study the mechanism of NPC assembly137,138. Other curved DNA origami shapes were able to guide formation and membrane dynamics of liposomes139,140 as well as controlling their diameter141 and lipid vesicles curvature142, mimicking the functions of membrane-shaping proteins such as BAR (Bin/amphiphysin/Rvs protein)142, and membrane remodelling machineries such as dynamins and ESCRT (endosomal sorting complexes required for transport)140 (Fig. 7d). Small DNA structures were shown to promote lipid exchange in bilayers143. Membrane-inserted DNA origami plates were employed to programme cell–cell adhesion via DNA hybridization144.
Future perspectives: learning from nature
DNA nanomachines: challenges and solutions
Examination of natural biomolecular machines provides a list of the essential ingredients of a sophisticated molecular machine and shines light upon design challenges and possible solutions. Dissecting F-type ATP synthase as a rotary motor and kinesin as a linear transport motor reveals that these biomolecular machines are made of multiple moving parts that interface together so that they can transmit forces. These motors are triggered by binding of ATP, and the resulting conformational change leads to not only motion but also the activation of a chemical reaction. ATP powers the system autonomously owing to the catalytic nature of its breakdown. The motors play the role of a catalyst that is regenerated at the end of each cycle. By contrast, a non-autonomous powering mode always involves frequent external manipulation of the system to proceed from the current step of the cycle to the next, as demonstrated, for example, by the electrochemical or photochemical operation of AMMs2,3,145,146.
One key hurdle in designing functional macromolecules and supramolecules is the lack of deep understanding of how the energy landscapes of those molecules can be accurately designed and modified (Box 1). This process starts with the very fundamental question of folding predictions and decoding structure–activity relationships. One paradigmatic difference between the approaches to studying the chemistries of small molecules versus larger biomolecules lies in the fact that synthetic chemists have taken a more empirical approach to making and breaking chemical bonds long before they could find access to a detailed picture of the physical nature of chemical bonds. While such an empirical approach to designing larger molecules has been the subject of many efforts in the fields of supramolecular chemistry and protein engineering, it cannot furnish us with an in-depth and coherent macromolecular folding theory. A combined computational–experimental approach, however, seems to be more productive, as demonstrated by the successful rational design of proteins based on known motif libraries1,147–149.
The design and self-assembly of DNA nanostructures has succeeded considerably in folding predesigned nanostructures in the megadalton and gigadalton size range, even without comprehensive insights into the kinetics and thermodynamics of the folding procedure. Progress was possible despite limited mechanistic insight because thermal annealing of DNA nanostructures overcomes most kinetic traps and energy barriers that researchers cannot predict at the moment. When thermal annealing does not produce the desired folding in high yields, researchers simply iteratively redesign the nanostructure in a fashion very much akin to the trial-and-error approach of synthetic chemists. The limited scope of interactions involved in DNA duplex formation, namely hydrogen bonding and base stacking of DNA nucleobases, lends itself to an unprecedented level of predictability and robustness of DNA nanostructure assembly. However, the reduced diversity of possible interactions may also turn out to be a limiting factor in creating complex functions. Diversified and more orthogonal interactions may be needed to interface different modules of a DNA nanomachine together and to the environment. More efforts directed at integrating chemically modified DNA strands and RNA structural motifs would thus be highly beneficial.
Reliable prediction of the energy map of a particular self-assembly process would make it possible to compute the final 3D shape of the molecules and even customize the self-assembly path to meet particular geometrical requirements for a specific task such as catalysis or molecular recognition. This strategy is inspired by computing the potential energy surface of the folding and self-assembly of proteins, which are usually funnel-shaped, with local energy barriers that appear like small rough features on the surface (Box 1)150. The short and long range non-covalent interactions among amino acid residues and solvent molecules, plus the rigidity of the protein backbone and its excluded volume, yield a folding pathway that transitions through local minima to reach the final global energy minimum. Yet, the presence of functional amino acid residues responsible for forming the active sites might lead to different degrees of ‘frustration’, since formation of functional active sites serves the purpose of accomplishing a task, not minimizing the folding energy of the system151. The final outcome of folding is an energy state that is still close to the global energy minimum but not necessarily at the global energy minimum (see the roughness at the bottom of the funnel labelled FS (functional state) in the Box 1 figure). In many proteins, transition from one functional state to another occurs by generating temporary local unfolding, whereas the overall structure remains folded, a phenomenon referred to as ‘cracking’150. Having precisely computed the energy map of the folding, it would be possible in the future to predict functions of a given protein or DNA nanostructure and the structure–activity relationship data based on its primary sequence151. The accurate computation of energy landscapes would even permit devising transitions between multiple functional states through sculpting the energy barriers or via mechanisms similar to cracking. The problem with modelling not only the self-assembly of molecular machines but also their motion cycles is the extended time scale of such simulations, which need to reconcile between folding events happening in the matter of microseconds and conformational changes occurring at the scale of milliseconds or even seconds152.
The main source of energy in the locomotive DNA systems discussed in Fig. 3 is the free energy of SDR due to the formation of a more stable duplex as the product of the reaction. In SDR-driven motions and reshaping instances mentioned in Fig. 4, the chemical energy was put into the system in a stepwise manner, making the process non-autonomous. The domains of the DNA nanostructures programmed to do SDR react with the fuel only once and become exhausted quickly. In the other examples presented where a change in the environment is the energy source of the motion (for example, temperature change, ionic strength, light, magnetic or electric fields), the need for external manipulation of the system makes the powering step non-autonomous in nature.
Autonomous powering of DNA nanomachines requires finding a reaction that is catalytic, simultaneously gives rise to a marked conformational change at the molecular scale, and, finally, integrating it as a designed structural feature. One avenue to pursue is adding catalytic nucleic acid molecules such as RNAzymes and DNAzymes to the moving domains of a DNA nanostructure such that conformational changes upon catalytic activity are coupled to mechanical motions. However, it is unclear how large the range of molecular motion of a DNAzyme can be, if any, when catalysing a reaction. The catalytic activity might be completely lost upon conjugation into the DNA nanostructures, or residual flexibility may prevent them from triggering motions in the immediately attached parts. An alternative route could be to design DNA–protein hybrid nanomachines similar to those described in Fig. 3 or 7. The robustness of DNA nanostructure assembly enables the design of asymmetric structural elements to include the desired degrees of freedom of motion. Conformational changes in proteins could be used to exploit ratchet effects [G] (Box 1) to rectify otherwise non-directional random motions, translate linear motion into rotary movements, or to control the directionality and amplitude of a conformational change. However, borrowing modules from nature as the driving elements or using the ones produced by in vitro selections, such as DNAzymes, compromises the goal of founding a rational design framework fully based on first principles.
Allosteric versus mechanical gating
One of the advanced features of natural biomolecular machines is ‘allosteric gating’, whereby binding of a trigger molecule such as ATP or ADP can favour a particular state among multiple states transitioned in a motion cycle153. Although the detailed mechanism of mechanochemical coupling in natural biomolecular motors is still not fully understood, careful examination of ATP synthase and kinesin reveals that they both use ATP binding and ADP/Pi release as the timing measures to modify the heights of energy barriers between different discrete motion steps in an information ratchet type of energy landscape navigation (Box 1). The kinesin dimer has two identical ATP-hydrolysing walker heads that bind to the microtubule tracks, one at a time (Fig. 8a). The track binding releases ADP from one head, preparing it for ATP binding, upon which a directional conformational change (called neck linker docking) puts forward the second head to bind to the track while ATP hydrolysis in the first head generates ADP and inorganic phosphate (Pi). The ADP-bound head in the back loses its affinity for the track binding and detaches to allow for the motion cycle to repeat itself154. Here, the head-track binding is strong in the ATP-bound and unoccupied states whereas the opposite holds for the ADP-bound state of kinesin155. The two identical heads are also halted in different stages of the catalytic reaction progress (ATP binding, hydrolysis, Pi release, ADP release) through kinetic traps so that the chemical coupling to the structural motion can take place. This aspect is also shared in the rotary F1-ATP synthase motor in which the hexagonal part of the stator is composed of six subunits, (αβ)3, around the γ subunit of the rotor shaft (Fig. 8b). Each αβ heterodimer has an ATP binding site but is trapped at a different conformational state along the ATP hydrolysis catalytic reaction coordinate; one is unoccupied, another bound to ATP, and the third one to ADP. As the catalytic reaction progresses towards ATP hydrolysis, the rotor shaft rotates counterclockwise (when viewed from the membrane side). Here, also the asymmetric structural elements in the rotor and stator create asymmetric energy barrier profiles to synchronize the rotation with the different stages of the catalysis and also making the rotation unidirectional156. The magnesium-mediated ATP binding to the active site initiates a conformational change that propagates throughout the subunits and between them at their interfaces, causing the rotor shaft to revolve in an 80° sub-step and release an ADP from a different site, while ATP hydrolysis and inorganic phosphate release drive the rotor another 40° in the same direction157 (Fig. 8b). The spectacular feature of the chemo-mechanical coupling in ATP synthase is that the direction of rotation of the shaft could be reversed by the proton channel activity of the FO segment of the rotor to synthesize ATP158. Thus, ‘mechanical gating’ drives the ATP synthesis forward while “allosteric gating” leads to the mechanical movement of the ATP synthase parts powered by the ATP hydrolysis. It was also shown that the local stiffness of different parts of ATP synthase varies over an order of magnitude, causing an elastic transmission of torque and drastically enhancing the turnover rate of the enzyme157. It is also believed that the carefully engineered local rigidities of different parts considerably reduces the kinetic limping of the enzyme156.
Fig. 8. Natural biomolecular motors.
A| Front-head gating model for kinesin164: the kinesin dimer has two motor heads (blue) attached to a coiled coil (dark blue helix) via two neck linkers (NL, in yellow). Kinesin moves unidirectionally (from the negative end to the positive) on a molecular track (shown in brown) made of microtubules (MT). 1) the rear head is ADP-bound and has a low affinity for binding to MT while the front head is waiting for ATP to bind. 2) Upon ATP biding the NL docking occurs pulling forward the loosely-bound rear head while it loses ADP. 3) The new front head is now nucleotide-free (in the apo state) and tightly binds the MT. In this state both heads are tightly bound to the MT and the rear head has its NL oriented forward while the front head’s NL is oriented backward, leading to the ATP binding site of the front head becoming unavailable. 4) The phosphate release from the rear head returns it to the ADP-bound state, resulting in the weak binding to MT, and allowing the NL of front head to bind ATP again. B| The bacterial F-type ATP synthase. Ba| Structural diagram of the bacterial F-type ATP synthase. The six αβ heterodimer form a barrel and the peripheral stalk is made of “ab” subunits attached to the barrel via δ subunit. The central shaft has a γ subunit and an ε crank protrusion. The “c” subunits form the proton channel across the membrane connected to “a” subunit158. Bb| Chemo-mechanical coupling cycle of the F1 rotary motor of ATP synthase165. Each circle indicates the chemical state of one of the three β subunits and the red arrow represents the orientation of the γ rotor. ATP* shows the pre- or post-hydrolysis state of ATP. Here, the ATP binding the orange circle at 0º is hydrolyzed to ADP and Pi at 200º and the ADP and Pi release occurs at 240º and 320º, respectively. The blue and green circles go through the same reaction cycle starting with a 120º and 240º delay, respectively. Panel B (left) was adapted with permission from Elsevier.
No dynamic DNA nanostructure has been able to reach the intricacy and complexity of ATP synthase or kinesin so far. Engineering mechanochemical coupling in a supramolecular structure through alternative allosteric and mechanical gating presumably requires fully predictable and almost defect-free self-assembly of monomeric parts. Interfacing monomers to guide the propagation of the mechanical forces resulting in the conformational change demands computation of the energy landscape of the monomers to modify the energy barriers or to remove kinetic traps. Initiating a conformational change upon binding through allosteric gating depends on devising a chemically active site and characterizing a robust and reversible reaction, leading to a large enough molecular motion. In order to autonomously power the motion, the reaction must also be catalytic.
Conclusions
Producing highly accurate discrete objects or regular 3D lattices (crystals, ideally) using either DNA oligomers or DNA origami cell units is a highly useful approach to honing our design skills. As described, nanometer-scale resolutions are easily accessible for the modification of DNA nanostructures through DNA hybridization or direct chemical alteration of the staple strands. The main advantage of chemically modifying DNA nanostructures is that there is almost equal opportunity for functionalization throughout the structure, unlike other biomolecules for which chemical modifications are typically restricted by the presence of a particular chemical moiety (for example, lysine or cysteine residues) at the desired sites. The enzyme–DNA hybrid nanostructures put forth an important path toward autonomous powering of DNA nanomachines through a catalytic reaction. Advances in super-resolution techniques such as DNA-PAINT technology allow for a more accurate characterization of DNA nanomachines. DNA nanostructures also hold great promise as smart drug delivery vehicles, especially when more complicated stimulus-responsive conformational changes could be exploited. Other already established applications of DNA nanotechnology in the areas of sensing and functional studies could be utilized to enhance the functional scope of DNA nanomachines in the future.
Creating autonomously running molecular motor units capable of driving directed transport of external loads remains a key goal of nanoscale science and technology. DNA nanotechnology could provide the means to fabricate the desired motors. One possible direction to realize such motors is the construction of mechanisms with well-defined rotary or translational degrees of freedom and having structural features that lead to asymmetric, periodic energy landscapes. Directional motion could then be powered by applying various kinds of deterministic or stochastic thermal, chemical or mechanical perturbations to drive the systems away from thermal equilibrium. DNA nanomachines would be complementary to synthetic AMMs due to their much larger size, water solubility and biocompatibility, which are properties that many current AMMs lack.
The realization of DNA biomolecular motors could help develop and experimentally test physical theories of transport phenomena occurring far from equilibrium. Both the process of building such functional motor units and the resultant units themselves could help uncover design principles underlying the function of natural macromolecular machines. Artificial biomolecular motors could help elucidate particularly how mechanical motions are coupled to chemical reactions. Finally, robustly functioning artificial motors could be of great practical use, for example, to guide chemical synthesis, to actively propel nanoscale drug delivery vehicles, to pump and separate molecules across barriers, or to package molecules into cargo components.
A plethora of coordinated and interconnected chemical processes maintain life as an ordered macro state far from thermal equilibrium, thus escaping the inevitable destruction by the second law of thermodynamics as long as energy is put into the system. The ability to design and customize energy landscapes will likely have far-reaching consequences well beyond the realm of molecular machines. It might not only allow forging transition pathways between different states of a single macromolecule but also enable design of handshakes, that is, coupling states of one system to those of others, thereby creating functional molecular networks, which is a key requisite for creating life-like entities.
In this article, we discussed the current efforts of biomolecular designers to build the theoretical and technological foundation for creating sophisticated molecular structures from DNA through rational design. There are a number of pivotal questions biomolecular designers will need to answer; what will be the molecular machines of the future and how can we build them? How can we build artificial biomolecular machines that can transport matter on the molecular scale, sense and process signals, and catalyse particular chemical reactions? How can we create miniscule robot-like assemblies that can make logic decisions whether or not to execute a task in certain environments? How can we imitate functions of the natural macromolecular machines in our synthetic objects and integrate such objects in a programmed way into autonomous systems with life-like properties? How can we construct artificial molecular clockwork to drive artificial cells? It will be interesting to see what the answers to these questions turn out to be and to what extent DNA nanotechnology will play a role in providing the corresponding technical solutions. So far, DNA has proven to be a versatile material for creating increasingly complex structures, laying the groundwork for building artificial DNA nanomachines.
Acknowledgements
The authors thank E. Feigl for making figure panels 2a and 2b. his work was financially supported by the Deutsche Forschungsgemeinschaft through the Gottfried-Wilhelm- Leibniz Program and by the European Commission through an ERC Consolidator Grant (#724261) to H.D.
Footnotes
Author contributions
The authors contributed equally to all aspects of the article.
Competing interests
The authors declare no competing interests.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Huang P-S, Boyken SE, Baker D. The coming of age of de novo protein design. Nature. 2016;537:320. doi: 10.1038/nature19946. [DOI] [PubMed] [Google Scholar]
- 2.Le Poul N, Colasson B. Electrochemically and Chemically Induced Redox Processes in Molecular Machines. ChemElectroChem. 2015;2:475–496. [Google Scholar]
- 3.Astumian RD. Optical vs. chemical driving for molecular machines. Faraday Discussions. 2016;195:583–597. doi: 10.1039/c6fd00140h. [DOI] [PubMed] [Google Scholar]
- 4.Sauvage J-P. From Chemical Topology to Molecular Machines (Nobel Lecture) Angewandte Chemie International Edition. 2017;56:11080–11093. doi: 10.1002/anie.201702992. [DOI] [PubMed] [Google Scholar]
- 5.Stoddart JF. Mechanically Interlocked Molecules (MIMs)—Molecular Shuttles, Switches, and Machines (Nobel Lecture) Angewandte Chemie International Edition. 2017;56:11094–11125. doi: 10.1002/anie.201703216. [DOI] [PubMed] [Google Scholar]
- 6.Cheng C, Stoddart JF. Wholly Synthetic Molecular Machines. ChemPhysChem. 2016;17:1780–1793. doi: 10.1002/cphc.201501155. [DOI] [PubMed] [Google Scholar]
- 7.Coskun A, Banaszak M, Astumian RD, Stoddart JF, Grzybowski BA. Great expectations: can artificial molecular machines deliver on their promise? Chemical Society Reviews. 2012;41:19–30. doi: 10.1039/c1cs15262a. [DOI] [PubMed] [Google Scholar]
- 8.Pezzato C, Cheng C, Stoddart JF, Astumian RD. Mastering the non-equilibrium assembly and operation of molecular machines. Chemical Society Reviews. 2017;46:5491–5507. doi: 10.1039/c7cs00068e. [DOI] [PubMed] [Google Scholar]
- 9.Astumian RD. Trajectory and Cycle-Based Thermodynamics and Kinetics of Molecular Machines: The Importance of Microscopic Reversibility. Accounts of Chemical Research. 2018;51:2653–2661. doi: 10.1021/acs.accounts.8b00253. [DOI] [PubMed] [Google Scholar]
- 10.Goychuk I. Molecular machines operating on the nanoscale: from classical to quantum. Beilstein journal of nanotechnology. 2016;7:328–350. doi: 10.3762/bjnano.7.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reimann P. Brownian motors: noisy transport far from equilibrium. Physics Reports. 2002;361:57–265. [Google Scholar]
- 12.Astumian RD, Mukherjee S, Warshel A. The Physics and Physical Chemistry of Molecular Machines. ChemPhysChem. 2016;17:1719–1741. doi: 10.1002/cphc.201600184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ranallo S, Porchetta A, Ricci F. DNA-Based Scaffolds for Sensing Applications. Analytical Chemistry. 2019;91:44–59. doi: 10.1021/acs.analchem.8b05009. [DOI] [PubMed] [Google Scholar]
- 14.Harroun SG, et al. Programmable DNA switches and their applications. Nanoscale. 2018;10:4607–4641. doi: 10.1039/c7nr07348h. [DOI] [PubMed] [Google Scholar]
- 15.Tang Y, Ge B, Sen D, Yu H-Z. Functional DNA switches: rational design and electrochemical signaling. Chemical Society Reviews. 2014;43:518–529. doi: 10.1039/c3cs60264h. [DOI] [PubMed] [Google Scholar]
- 16.Wang F, Liu X, Willner I. DNA Switches: From Principles to Applications. Angewandte Chemie International Edition. 2015;54:1098–1129. doi: 10.1002/anie.201404652. [DOI] [PubMed] [Google Scholar]
- 17.Gore J, et al. DNA overwinds when stretched. Nature. 2006;442:836–9. doi: 10.1038/nature04974. [DOI] [PubMed] [Google Scholar]
- 18.Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–9. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 19.Hagerman PJ. Flexibility of DNA. Annu Rev Biophys Biophys Chem. 1988;17:265–86. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- 20.Chuang HM, Reifenberger JG, Cao H, Dorfman KD. Sequence-Dependent Persistence Length of Long DNA. Phys Rev Lett. 2017;119 doi: 10.1103/PhysRevLett.119.227802. 227802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfitzner E, et al. Rigid DNA beams for high-resolution single-molecule mechanics. Angew Chem Int Ed Engl. 2013;52:7766–71. doi: 10.1002/anie.201302727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120:923–34. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.SantaLucia J, Jr, Hicks D. The thermodynamics of DNA structural motifs. Annu Rev Biophys Biomol Struct. 2004;33:415–40. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- 24.Zadeh JN, et al. NUPACK: Analysis and Design of Nucleic Acid Systems. Journal of Computational Chemistry. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 25.Trads JB, Torring T, Gothelf KV. Site-Selective Conjugation of Native Proteins with DNA. Acc Chem Res. 2017;50:1367–1374. doi: 10.1021/acs.accounts.6b00618. [DOI] [PubMed] [Google Scholar]
- 26.Singh Y, Murat P, Defrancq E. Recent developments in oligonucleotide conjugation. Chem Soc Rev. 2010;39:2054–70. doi: 10.1039/b911431a. [DOI] [PubMed] [Google Scholar]
- 27.Chandrasekaran AR, Rusling DA. Triplex-forming oligonucleotides: a third strand for DNA nanotechnology. Nucleic Acids Res. 2018;46:1021–1037. doi: 10.1093/nar/gkx1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hollenstein M. DNA Catalysis: The Chemical Repertoire of DNAzymes. Molecules. 2015;20:20777–804. doi: 10.3390/molecules201119730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silverman SK. Catalytic DNA: Scope, Applications, and Biochemistry of Deoxyribozymes. Trends Biochem Sci. 2016;41:595–609. doi: 10.1016/j.tibs.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seeman NC. Nucleic acid junctions and lattices. J Theor Biol. 1982;99:237–47. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 31.Chen JH, Seeman NC. Synthesis from DNA of a molecule with the connectivity of a cube. Nature. 1991;350:631–3. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 32.Winfree E, Liu F, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–44. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 33.Paukstelis PJ, Nowakowski J, Birktoft JJ, Seeman NC. Crystal structure of a continuous three-dimensional DNA lattice. Chem Biol. 2004;11:1119–26. doi: 10.1016/j.chembiol.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 34.Seeman NC. At the crossroads of chemistry, biology, and materials: structural DNA nanotechnology. Chem Biol. 2003;10:1151–9. doi: 10.1016/j.chembiol.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science. 2003;301:1882–4. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 36.Shih WM, Quispe JD, Joyce GF. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 2004;427:618–21. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 37.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 38.Ke Y, et al. Multilayer DNA origami packed on a square lattice. J Am Chem Soc. 2009;131:15903–8. doi: 10.1021/ja906381y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ke Y, Voigt NV, Gothelf KV, Shih WM. Multilayer DNA origami packed on hexagonal and hybrid lattices. J Am Chem Soc. 2012;134:1770–4. doi: 10.1021/ja209719k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benson E, et al. DNA rendering of polyhedral meshes at the nanoscale. Nature. 2015;523:441–4. doi: 10.1038/nature14586. [DOI] [PubMed] [Google Scholar]
- 41.Zhang F, et al. Complex wireframe DNA origami nanostructures with multi-arm junction vertices. Nat Nanotechnol. 2015;10:779–84. doi: 10.1038/nnano.2015.162. [DOI] [PubMed] [Google Scholar]
- 42.Veneziano R, et al. Designer nanoscale DNA assemblies programmed from the top down. Science. 2016;352:1534. doi: 10.1126/science.aaf4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dietz H, Douglas SM, Shih WM. Folding DNA into twisted and curved nanoscale shapes. Science. 2009;325:725–30. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagenbauer KF, Sigl C, Dietz H. Gigadalton-scale shape-programmable DNA assemblies. Nature. 2017;552:78–83. doi: 10.1038/nature24651. [DOI] [PubMed] [Google Scholar]
- 45.Douglas SM, et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009;37:5001–6. doi: 10.1093/nar/gkp436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim DN, Kilchherr F, Dietz H, Bathe M. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 2012;40:2862–8. doi: 10.1093/nar/gkr1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castro CE, et al. A primer to scaffolded DNA origami. Nat Methods. 2011;8:221–9. doi: 10.1038/nmeth.1570. [DOI] [PubMed] [Google Scholar]
- 48.Snodin BEK, et al. Introducing improved structural properties and salt dependence into a coarse-grained model of DNA. The Journal of Chemical Physics. 2015;142 doi: 10.1063/1.4921957. 234901. [DOI] [PubMed] [Google Scholar]
- 49.Maffeo C, Yoo J, Aksimentiev A. De novo reconstruction of DNA origami structures through atomistic molecular dynamics simulation. Nucleic Acids Res. 2016;44:3013–9. doi: 10.1093/nar/gkw155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sobczak JP, Martin TG, Gerling T, Dietz H. Rapid folding of DNA into nanoscale shapes at constant temperature. Science. 2012;338:1458–61. doi: 10.1126/science.1229919. [DOI] [PubMed] [Google Scholar]
- 51.Stahl E, Martin TG, Praetorius F, Dietz H. Facile and scalable preparation of pure and dense DNA origami solutions. Angew Chem Int Ed Engl. 2014;53:12735–40. doi: 10.1002/anie.201405991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagenbauer KF, et al. How We Make DNA Origami. Chembiochem. 2017;18:1873–1885. doi: 10.1002/cbic.201700377. [DOI] [PubMed] [Google Scholar]
- 53.Shaw A, Benson E, Hogberg B. Purification of functionalized DNA origami nanostructures. ACS Nano. 2015;9:4968–75. doi: 10.1021/nn507035g. [DOI] [PubMed] [Google Scholar]
- 54.Douglas SM, et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–8. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tikhomirov G, Petersen P, Qian L. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns. Nature. 2017;552:67–71. doi: 10.1038/nature24655. [DOI] [PubMed] [Google Scholar]
- 56.Woo S, Rothemund PW. Programmable molecular recognition based on the geometry of DNA nanostructures. Nat Chem. 2011;3:620–7. doi: 10.1038/nchem.1070. [DOI] [PubMed] [Google Scholar]
- 57.Gerling T, Wagenbauer KF, Neuner AM, Dietz H. Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science. 2015;347:1446–52. doi: 10.1126/science.aaa5372. [DOI] [PubMed] [Google Scholar]
- 58.Han D, et al. Single-stranded DNA and RNA origami. Science. 2017;358 doi: 10.1126/science.aao2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geary C, Rothemund PW, Andersen ES. A single-stranded architecture for cotranscriptional folding of RNA nanostructures. Science. 2014;345:799–804. doi: 10.1126/science.1253920. [DOI] [PubMed] [Google Scholar]
- 60.Praetorius F, Dietz H. Self-assembly of genetically encoded DNA-protein hybrid nanoscale shapes. Science. 2017;355 doi: 10.1126/science.aam5488. [DOI] [PubMed] [Google Scholar]
- 61.Ke Y, Ong LL, Shih WM, Yin P. Three-dimensional structures self-assembled from DNA bricks. Science. 2012;338:1177–83. doi: 10.1126/science.1227268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei B, Dai M, Yin P. Complex shapes self-assembled from single-stranded DNA tiles. Nature. 2012;485:623–6. doi: 10.1038/nature11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yin P, et al. Programming DNA tube circumferences. Science. 2008;321:824–6. doi: 10.1126/science.1157312. [DOI] [PubMed] [Google Scholar]
- 64.Ong LL, et al. Programmable self-assembly of three-dimensional nanostructures from 10,000 unique components. Nature. 2017;552:72–77. doi: 10.1038/nature24648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin C, et al. In vivo cloning of artificial DNA nanostructures. Proc Natl Acad Sci U S A. 2008;105:17626–31. doi: 10.1073/pnas.0805416105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kick B, Praetorius F, Dietz H, Weuster-Botz D. Efficient Production of Single-Stranded Phage DNA as Scaffolds for DNA Origami. Nano Lett. 2015;15:4672–6. doi: 10.1021/acs.nanolett.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Engelhardt FAS, et al. Custom-Size, Functional, and Durable DNA Origami with Design-Specific Scaffolds. ACS Nano. 2019 doi: 10.1021/acsnano.9b01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ducani C, Kaul C, Moche M, Shih WM, Hogberg B. Enzymatic production of 'monoclonal stoichiometric' single-stranded DNA oligonucleotides. Nat Methods. 2013;10:647–52. doi: 10.1038/nmeth.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmidt TL, et al. Scalable amplification of strand subsets from chip-synthesized oligonucleotide libraries. Nat Commun. 2015;6 doi: 10.1038/ncomms9634. 8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kishi JY, Schaus TE, Gopalkrishnan N, Xuan F, Yin P. Programmable autonomous synthesis of single-stranded DNA. Nat Chem. 2018;10:155–164. doi: 10.1038/nchem.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Praetorius F, et al. Biotechnological mass production of DNA origami. Nature. 2017;552:84–87. doi: 10.1038/nature24650. [DOI] [PubMed] [Google Scholar]
- 72.Gu H, Furukawa K, Weinberg Z, Berenson DF, Breaker RR. Small, highly active DNAs that hydrolyze DNA. J Am Chem Soc. 2013;135:9121–9. doi: 10.1021/ja403585e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li M, et al. In vivo production of RNA nanostructures via programmed folding of single-stranded RNAs. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04652-4. 2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Elbaz J, Yin P, Voigt CA. Genetic encoding of DNA nanostructures and their self-assembly in living bacteria. Nat Commun. 2016;7 doi: 10.1038/ncomms11179. 11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chi Q, Wang G, Jiang J. The persistence length and length per base of single-stranded DNA obtained from fluorescence correlation spectroscopy measurements using mean field theory. Physica A: Statistical Mechanics and its Applications. 2013;392:1072–1079. [Google Scholar]
- 76.Rechendorff K, Witz G, Adamcik J, Dietler G. Persistence length and scaling properties of single-stranded DNA adsorbed on modified graphite. J Chem Phys. 2009;131 doi: 10.1063/1.3216111. 095103. [DOI] [PubMed] [Google Scholar]
- 77.Zhang DY, Winfree E. Control of DNA strand displacement kinetics using toehold exchange. J Am Chem Soc. 2009;131:17303–14. doi: 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]
- 78.Andersen ES, et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009;459:73–6. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 79.Pan J, Li F, Cha TG, Chen H, Choi JH. Recent progress on DNA based walkers. Curr Opin Biotechnol. 2015;34:56–64. doi: 10.1016/j.copbio.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 80.Thubagere AJ, et al. A cargo-sorting DNA robot. Science. 2017;357 doi: 10.1126/science.aan6558. [DOI] [PubMed] [Google Scholar]
- 81.Gu H, Chao J, Xiao SJ, Seeman NC. A proximity-based programmable DNA nanoscale assembly line. Nature. 2010;465:202–5. doi: 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Valero J, Pal N, Dhakal S, Walter NG, Famulok M. A bio-hybrid DNA rotor-stator nanoengine that moves along predefined tracks. Nat Nanotechnol. 2018;13:496–503. doi: 10.1038/s41565-018-0109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Han D, Pal S, Liu Y, Yan H. Folding and cutting DNA into reconfigurable topological nanostructures. Nat Nanotechnol. 2010;5:712–7. doi: 10.1038/nnano.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang F, Nangreave J, Liu Y, Yan H. Reconfigurable DNA origami to generate quasifractal patterns. Nano Lett. 2012;12:3290–5. doi: 10.1021/nl301399z. [DOI] [PubMed] [Google Scholar]
- 85.Wei B, Ong LL, Chen J, Jaffe AS, Yin P. Complex reconfiguration of DNA nanostructures. Angew Chem Int Ed Engl. 2014;53:7475–9. doi: 10.1002/anie.201402437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Choi Y, Choi H, Lee AC, Lee H, Kwon S. A Reconfigurable DNA Accordion Rack. Angew Chem Int Ed Engl. 2018;57:2811–2815. doi: 10.1002/anie.201709362. [DOI] [PubMed] [Google Scholar]
- 87.Marras AE, Zhou L, Su HJ, Castro CE. Programmable motion of DNA origami mechanisms. Proc Natl Acad Sci U S A. 2015;112:713–8. doi: 10.1073/pnas.1408869112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.List J, Falgenhauer E, Kopperger E, Pardatscher G, Simmel FC. Long-range movement of large mechanically interlocked DNA nanostructures. Nat Commun. 2016;7 doi: 10.1038/ncomms12414. 12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ketterer P, Willner EM, Dietz H. Nanoscale rotary apparatus formed from tight-fitting 3D DNA components. Sci Adv. 2016;2:e1501209. doi: 10.1126/sciadv.1501209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Turek VA, et al. Thermo-Responsive Actuation of a DNA Origami Flexor. 2018;28 1706410. [Google Scholar]
- 91.Song J, et al. Reconfiguration of DNA molecular arrays driven by information relay. Science. 2017;357 doi: 10.1126/science.aan3377. [DOI] [PubMed] [Google Scholar]
- 92.Kopperger E, et al. A self-assembled nanoscale robotic arm controlled by electric fields. Science. 2018;359:296–301. doi: 10.1126/science.aao4284. [DOI] [PubMed] [Google Scholar]
- 93.Maier AM, et al. Magnetic Propulsion of Microswimmers with DNA-Based Flagellar Bundles. Nano Lett. 2016;16:906–10. doi: 10.1021/acs.nanolett.5b03716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kuzyk A, et al. A light-driven three-dimensional plasmonic nanosystem that translates molecular motion into reversible chiroptical function. Nat Commun. 2016;7 doi: 10.1038/ncomms10591. 10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang Y, et al. A Photoregulated DNA-Based Rotary System and Direct Observation of Its Rotational Movement. Chemistry. 2017;23:3979–3985. doi: 10.1002/chem.201605616. [DOI] [PubMed] [Google Scholar]
- 96.Liu N, Liedl T. DNA-Assembled Advanced Plasmonic Architectures. Chem Rev. 2018;118:3032–3053. doi: 10.1021/acs.chemrev.7b00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhou C, Duan X, Liu N. DNA-Nanotechnology-Enabled Chiral Plasmonics: From Static to Dynamic. Acc Chem Res. 2017;50:2906–2914. doi: 10.1021/acs.accounts.7b00389. [DOI] [PubMed] [Google Scholar]
- 98.Samanta A, Banerjee S, Liu Y. DNA nanotechnology for nanophotonic applications. Nanoscale. 2015;7:2210–20. doi: 10.1039/c4nr06283c. [DOI] [PubMed] [Google Scholar]
- 99.Lan X, Wang Q. DNA-programmed self-assembly of photonic nanoarchitectures. Npg Asia Materials. 2014;6:e97. [Google Scholar]
- 100.Zheng J, et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature. 2009;461:74–7. doi: 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Simmons CR, et al. Construction and Structure Determination of a Three-Dimensional DNA Crystal. J Am Chem Soc. 2016;138:10047–54. doi: 10.1021/jacs.6b06508. [DOI] [PubMed] [Google Scholar]
- 102.Simmons CR, et al. Tuning the Cavity Size and Chirality of Self-Assembling 3D DNA Crystals. J Am Chem Soc. 2017;139:11254–11260. doi: 10.1021/jacs.7b06485. [DOI] [PubMed] [Google Scholar]
- 103.Stahl E, Praetorius F, de Oliveira Mann CC, Hopfner KP, Dietz H. Impact of Heterogeneity and Lattice Bond Strength on DNA Triangle Crystal Growth. ACS Nano. 2016 doi: 10.1021/acsnano.6b04787. [DOI] [PubMed] [Google Scholar]
- 104.McNeil R, Jr, Paukstelis PJ. Core-Shell and Layer-by-Layer Assembly of 3D DNA Crystals. Adv Mater. 2017;29 doi: 10.1002/adma.201701019. [DOI] [PubMed] [Google Scholar]
- 105.Zhang T, et al. 3D DNA Origami Crystals. Adv Mater. 2018;30:e1800273. doi: 10.1002/adma.201800273. [DOI] [PubMed] [Google Scholar]
- 106.Rinker S, Ke Y, Liu Y, Chhabra R, Yan H. Self-assembled DNA nanostructures for distance-dependent multivalent ligand-protein binding. Nat Nanotechnol. 2008;3:418–22. doi: 10.1038/nnano.2008.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tokura Y, et al. Fabrication of Defined Polydopamine Nanostructures by DNA Origami-Templated Polymerization. Angew Chem Int Ed Engl. 2018;57:1587–1591. doi: 10.1002/anie.201711560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kershner RJ, et al. Placement and orientation of individual DNA shapes on lithographically patterned surfaces. Nat Nanotechnol. 2009;4:557–61. doi: 10.1038/nnano.2009.220. [DOI] [PubMed] [Google Scholar]
- 109.Hung AM, et al. Large-area spatially ordered arrays of gold nanoparticles directed by lithographically confined DNA origami. Nat Nanotechnol. 2010;5:121–6. doi: 10.1038/nnano.2009.450. [DOI] [PubMed] [Google Scholar]
- 110.Funke JJ, Dietz H. Placing molecules with Bohr radius resolution using DNA origami. Nat Nanotechnol. 2016;11:47–52. doi: 10.1038/nnano.2015.240. [DOI] [PubMed] [Google Scholar]
- 111.Fu J, et al. Assembly of multienzyme complexes on DNA nanostructures. Nat Protoc. 2016;11:2243–2273. doi: 10.1038/nprot.2016.139. [DOI] [PubMed] [Google Scholar]
- 112.Ke G, et al. Directional Regulation of Enzyme Pathways through the Control of Substrate Channeling on a DNA Origami Scaffold. Angew Chem Int Ed Engl. 2016;55:7483–6. doi: 10.1002/anie.201603183. [DOI] [PubMed] [Google Scholar]
- 113.Zhang Y, Tsitkov S, Hess H. Proximity does not contribute to activity enhancement in the glucose oxidase-horseradish peroxidase cascade. Nat Commun. 2016;7 doi: 10.1038/ncomms13982. 13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhao Z, et al. Nanocaged enzymes with enhanced catalytic activity and increased stability against protease digestion. Nat Commun. 2016;7 doi: 10.1038/ncomms10619. 10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sharonov A, Hochstrasser RM. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc Natl Acad Sci U S A. 2006;103:18911–6. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jungmann R, et al. Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami. Nano Lett. 2010;10:4756–61. doi: 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- 117.Dai M, Jungmann R, Yin P. Optical imaging of individual biomolecules in densely packed clusters. Nat Nanotechnol. 2016;11:798–807. doi: 10.1038/nnano.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jungmann R, et al. Quantitative super-resolution imaging with qPAINT. Nat Methods. 2016;13:439–42. doi: 10.1038/nmeth.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zanacchi FC, et al. A DNA origami platform for quantifying protein copy number in super-resolution. Nat Methods. 2017;14:789–792. doi: 10.1038/nmeth.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Douglas SM, Bachelet I, Church GM. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012;335:831–4. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 121.Bastings MMC, et al. Modulation of the Cellular Uptake of DNA Origami through Control over Mass and Shape. Nano Lett. 2018;18:3557–3564. doi: 10.1021/acs.nanolett.8b00660. [DOI] [PubMed] [Google Scholar]
- 122.Wang P, et al. Visualization of the Cellular Uptake and Trafficking of DNA Origami Nanostructures in Cancer Cells. J Am Chem Soc. 2018;140:2478–2484. doi: 10.1021/jacs.7b09024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang D, Paukstelis PJ. Enhancing DNA Crystal Durability through Chemical Crosslinking. Chembiochem. 2016;17:1163–70. doi: 10.1002/cbic.201500610. [DOI] [PubMed] [Google Scholar]
- 124.Gerling T, Kube M, Kick B, Dietz H. Sequence-programmable covalent bonding of designed DNA assemblies. Sci Adv. 2018;4 doi: 10.1126/sciadv.aau1157. eaau1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhao J, et al. Post-Assembly Stabilization of Rationally Designed DNA Crystals. Angew Chem Int Ed Engl. 2015;54:9936–9. doi: 10.1002/anie.201503610. [DOI] [PubMed] [Google Scholar]
- 126.Perrault SD, Shih WM. Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano. 2014;8:5132–40. doi: 10.1021/nn5011914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Auvinen H, et al. Protein Coating of DNA Nanostructures for Enhanced Stability and Immunocompatibility. Adv Healthc Mater. 2017;6 doi: 10.1002/adhm.201700692. [DOI] [PubMed] [Google Scholar]
- 128.Agarwal NP, Matthies M, Gur FN, Osada K, Schmidt TL. Block Copolymer Micellization as a Protection Strategy for DNA Origami. Angew Chem Int Ed Engl. 2017;56:5460–5464. doi: 10.1002/anie.201608873. [DOI] [PubMed] [Google Scholar]
- 129.Ponnuswamy N, et al. Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat Commun. 2017;8 doi: 10.1038/ncomms15654. 15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee AJ, Endo M, Hobbs JK, Walti C. Direct Single-Molecule Observation of Mode and Geometry of RecA-Mediated Homology Search. ACS Nano. 2018;12:272–278. doi: 10.1021/acsnano.7b06208. [DOI] [PubMed] [Google Scholar]
- 131.Endo M, Katsuda Y, Hidaka K, Sugiyama H. Regulation of DNA methylation using different tensions of double strands constructed in a defined DNA nanostructure. J Am Chem Soc. 2010;132:1592–7. doi: 10.1021/ja907649w. [DOI] [PubMed] [Google Scholar]
- 132.Nickels PC, et al. Molecular force spectroscopy with a DNA origami-based nanoscopic force clamp. Science. 2016;354:305–307. doi: 10.1126/science.aah5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Funke JJ, et al. Uncovering the forces between nucleosomes using DNA origami. Sci Adv. 2016;2:e1600974. doi: 10.1126/sciadv.1600974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kilchherr F, et al. Single-molecule dissection of stacking forces in DNA. Science. 2016;353 doi: 10.1126/science.aaf5508. [DOI] [PubMed] [Google Scholar]
- 135.Krishnan S, et al. Molecular transport through large-diameter DNA nanopores. Nat Commun. 2016;7 doi: 10.1038/ncomms12787. 12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Langecker M, et al. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science. 2012;338:932–6. doi: 10.1126/science.1225624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ketterer P, et al. DNA origami scaffold for studying intrinsically disordered proteins of the nuclear pore complex. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03313-w. 902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fisher PDE, et al. A Programmable DNA Origami Platform for Organizing Intrinsically Disordered Nucleoporins within Nanopore Confinement. ACS Nano. 2018;12:1508–1518. doi: 10.1021/acsnano.7b08044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang Z, Yang Y, Pincet F, Llaguno MC, Lin C. Placing and shaping liposomes with reconfigurable DNA nanocages. Nat Chem. 2017;9:653–659. doi: 10.1038/nchem.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Grome MW, Zhang Z, Pincet F, Lin C. Vesicle Tubulation with Self-Assembling DNA Nanosprings. Angew Chem Int Ed Engl. 2018;57:5330–5334. doi: 10.1002/anie.201800141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yang Y, et al. Self-assembly of size-controlled liposomes on DNA nanotemplates. Nat Chem. 2016;8:476–83. doi: 10.1038/nchem.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Franquelim HG, Khmelinskaia A, Sobczak JP, Dietz H, Schwille P. Membrane sculpting by curved DNA origami scaffolds. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03198-9. 811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ohmann A, et al. A synthetic enzyme built from DNA flips 10(7) lipids per second in biological membranes. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04821-5. 2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Akbari E, et al. Engineering Cell Surface Function with DNA Origami. Adv Mater. 2017;29 doi: 10.1002/adma.201703632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Balzani V, Credi A, Venturi M. Light powered molecular machines. Chem Soc Rev. 2009;38:1542–50. doi: 10.1039/b806328c. [DOI] [PubMed] [Google Scholar]
- 146.Silvi S, Venturi M, Credi A. Light operated molecular machines. Chem Commun (Camb) 2011;47:2483–9. doi: 10.1039/c0cc03829f. [DOI] [PubMed] [Google Scholar]
- 147.Baker D. What has de novo protein design taught us about protein folding and biophysics? Protein Science. 2019;28:678–683. doi: 10.1002/pro.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lin Y-R, et al. Control over overall shape and size in de novo designed proteins. Proceedings of the National Academy of Sciences. 2015;112:E5478–E5485. doi: 10.1073/pnas.1509508112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chevalier A, et al. Massively parallel de novo protein design for targeted therapeutics. Nature. 2017;550:74. doi: 10.1038/nature23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Whitford PC, Onuchic JN. What protein folding teaches us about biological function and molecular machines. Current Opinion in Structural Biology. 2015;30:57–62. doi: 10.1016/j.sbi.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 151.Giri Rao VVH, Gosavi S. Using the folding landscapes of proteins to understand protein function. Current Opinion in Structural Biology. 2016;36:67–74. doi: 10.1016/j.sbi.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 152.Elber R, Kirmizialtin S. Molecular machines. Current Opinion in Structural Biology. 2013;23:206–211. doi: 10.1016/j.sbi.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 153.Astumian RD. Microscopic reversibility as the organizing principle of molecular machines. Nature Nanotechnology. 2012;7:684. doi: 10.1038/nnano.2012.188. [DOI] [PubMed] [Google Scholar]
- 154.Cross RA. Review: Mechanochemistry of the kinesin-1 ATPase. Biopolymers. 2016;105:476–482. doi: 10.1002/bip.22862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang W, Cao L, Wang C, Gigant B, Knossow M. Kinesin, 30 years later: Recent insights from structural studies. Protein Science. 2015;24:1047–1056. doi: 10.1002/pro.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sielaff H, Yanagisawa S, Frasch WD, Junge W, Börsch M. Structural Asymmetry and Kinetic Limping of Single Rotary F-ATP Synthases. Molecules. 2019;24:504. doi: 10.3390/molecules24030504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Junge w, Nelson N. ATP Synthase. Annual Review of Biochemistry. 2015;84:631–657. doi: 10.1146/annurev-biochem-060614-034124. [DOI] [PubMed] [Google Scholar]
- 158.Stewart AG, Laming EM, Sobti M, Stock D. Rotary ATPases—dynamic molecular machines. Current Opinion in Structural Biology. 2014;25:40–48. doi: 10.1016/j.sbi.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 159.Watson MA, Cockroft SL. Man-made molecular machines: membrane bound. Chemical Society Reviews. 2016;45:6118–6129. doi: 10.1039/c5cs00874c. [DOI] [PubMed] [Google Scholar]
- 160.Kassem S, et al. Artificial molecular motors. Chemical Society Reviews. 2017;46:2592–2621. doi: 10.1039/c7cs00245a. [DOI] [PubMed] [Google Scholar]
- 161.Erbas-Cakmak S, Leigh DA, McTernan CT, Nussbaumer AL. Artificial Molecular Machines. Chemical Reviews. 2015;115:10081–10206. doi: 10.1021/acs.chemrev.5b00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Baroncini M, et al. Making and Operating Molecular Machines: A Multidisciplinary Challenge. ChemistryOpen. 2018;7:169–179. doi: 10.1002/open.201700181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Astumian RD, Hänggi P. Brownian Motors. Physics Today. 2002:33–39. [Google Scholar]
- 164.Dogan Merve Y, Can S, Cleary Frank B, Purde V, Yildiz A. Kinesin’s Front Head Is Gated by the Backward Orientation of Its Neck Linker. Cell Reports. 2015;10:1967–1973. doi: 10.1016/j.celrep.2015.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Noji H, Ueno H, McMillan DGG. Catalytic robustness and torque generation of the F1-ATPase. Biophys Rev. 2017;9:103–118. doi: 10.1007/s12551-017-0262-x. [DOI] [PMC free article] [PubMed] [Google Scholar]