Abstract
Background
Circular RNAs (circRNAs) belong to a new type of endogenous non-coding RNA and plays a key role in carcinogenesis. Circ-ZKSCAN1 (hsa_circ_0001727) has been proven to be a tumor-dependent circRNA. However, its role in non-small cell lung cancer (NSCLC) has been underreported.
Methods
The expression patterns of circ-ZKSCAN1 were determined using qRT-PCR in NSCLC samples and cell lines. Cell proliferation was examined utilizing the CCK-8 assay. Cell migration and invasion were evaluated using the Transwell assay. The combination of circ-ZKSCAN1 and miR-330-5p in NSCLC cells was analyzed by RNA pull-down and luciferase reporter assay. We used the bioinformatics software circbank, CircInteractome, TargetScan and Miranda to predict circRNA-miRNA and miRNA-mRNA interactions.
Results
Our results showed that circ-ZKSCAN1 was significantly up-regulated in NSCLC, closely related to malignant characteristics and poor prognosis, and clinically related to tumor size and clinical stage. Subsequent experiments showed that circ-ZKSCAN1 could inhibit the growth of NSCLC cells in vitro and in vivo. Importantly, circ-ZKSCAN1 can act as a sponge of carcinogenic miR-330-5p to increase the expression of FAM83A, resulting in the inhibition of MAPK signal transduction pathway, thus promoting the progress of NSCLC. Interestingly, the increase in FAM83A expression caused by circ-ZKSCAN1 overexpression could in turn promote the expression of circ-ZKSCAN1.
Conclusions
Circ-ZKSCAN1 is a key positive regulator of NSCLC, and clarifies the potential molecular mechanism of the new circ-ZKSCAN1/miR-330-5p/FAM83A feedback loop in promoting the progress of NSCLC.
Keywords: Non-small cell lung cancer (NSCLC), circ-ZKSCAN1, FAM83A, prognosis, miR-330-5p
Introduction
Lung cancer is one of the most common malignant tumors and the main killer of cancer-related mortality in the world (1). Non-small cell lung cancer (NSCLC) is the most common histopathological subtype of lung cancer (about 80% of cases) (2). Although great progress has been made in the treatment of NSCLC including surgical resection, chemotherapy, and radiotherapy, the overall 5-year survival rate of NSCLC patients is low due to the lack of early diagnosis, and the frequency of metastasis and recurrence. Therefore, it is necessary to have a more comprehensive understanding of the molecular mechanism of NSCLC development in order to find reliable and effective therapeutic targets and early biomarkers.
Zinc finger with KRAB and SCAN domains 1 (ZKSCAN1), also known as ZNF139, belongs to the Kruppel C2H2-type zinc-finger family of proteins which contributes to many distinct cellular processes, including proliferation, differentiation, and apoptosis, in addition to the development and homeostasis of several types of tissue; it is also involved in forming stable and abundant circular RNAs (circRNAs) in cancer (3-5). ZKSCAN1 is a carcinogenic driver of hepatocellular carcinoma, its activation and overexpression is associated with the occurrence, development, and invasion of hepatocellular carcinoma (4). ZKSCAN1 can significantly increase drug resistance in gastric cancer (6).
CircRNA, a newly discovered type of non-coding RNA, is widely expressed in cells, and has gene regulation effects (7). It is acknowledged that circRNA has no 5' to 3' polarity or polyadenylated tail, and has a covalently closed loop structure; these features make circRNA very stable and highly resistant to RNA enzyme activity (7,8). There is growing evidence that circRNA acts in a variety of biological functions, including “miRNA sponge”, regulation of transcription, binding protein, and translation (9). Most circRNA molecular mechanisms can be classified under “miRNA sponge”. Most notably, Hansen et al. reported that circRNA CDR1as, also known as ciRS-7, has more than 70 conservative binding sites for miR-7 (10). Extensive subsequent research has confirmed the view that circRNA can be used as a sponge for miRNA to regulate gene expression, especially in cancer (11). For example, circ-AGFG1 promotes triple-negative breast cancer progression by regulating CCNE1 expression through sponge miR-195-5p (12). In hepatocellular carcinoma, has_circ_0078710 acts as a miR-31 sponge to promote cell proliferation, migration, invasion, and tumor growth (13). Circ-HIPK3 plays a carcinogenic role in lung cancer through direct interaction with miR-124 (14).
Circ-ZKSCAN1 (hsa_circ_0001727), a circRNA derived from the second and third exons of the ZKSCAN1 gene (full length is 668bp), was thought to be associated with the progress of some cancers, including brain (15) and hepatocellular carcinoma (4,16). However, its regulatory effect in NSCLC is unknown. In this study, we aimed to explore the clinical significance of circ-ZKSCAN1 and its molecular regulatory mechanism in NSCLC.
Methods
Tissue samples
We collected specimens of tumor tissues and matched adjacent normal tissues from 107 NSCLC patients who completed surgical operation from May 2014 to April 2015. The samples were retrospectively obtained from the sample bank of the Department of Thoracic Surgery, Affiliated Hospital of Qingdao University. Immediately after operation, the tissue samples were frozen in a −80 °C refrigerator until they were used. The patients were not treated with radio- or chemical therapy. The medical ethics for this research were approved by the Ethic Committee of the Affiliated Hospital of Qingdao University (No. QYFYKY 2018-10-11-2).
Predicting the target miRNA and mRNA of circ-ZKSCAN1
In order to use bioinformatics analysis to predict the miRNA-binding sites of circ-ZKSCAN1, we used different data analysis tools, including circbank (http://www.circbank.cn/) and CircInteractome (https://circinteractome.nia.nih.gov/). Next, we chose 4 potential miRNAs. To further reduce the objective range, we identified the miRNA expression level in both NSCLC tissues and matched paired-adjacent normal tissues with quantitative real-time PCR (qRT-PCR) and discovered that only miR-330-5p showed significantly higher expression (P<0.05). Therefore, we focused on miR-330-5p in this study.
To predict the target of miR-330-5p, we found a possible association between miR-330-5p and FAM83A by using TargetScan (http://www.targetscan.org/vert_72/) and Miranda (http://www.microrna.org/).
Cell culture and transfection
According to the instructions of the manufacturer, all NSCLC cell lines, including A549, H1299, H460, H1650, NCI-2228, and human bronchial epithelial cells (HBE) were grown in RPMI1640 or DMEM complete medium supplemented with 10% fetal bovine serum (FBS). Cells usually grow in incubators at 37 °C and under an atmosphere containing 5% CO2. The circ-ZKSCAN1 and FAM83A PUC57 expression vectors (BGI, Beijing, China), miR-330-5p mimics, and FAM83A small interference RNA (Gene Pharmacy, Shanghai, China) were each transfected into A549 and H1299 cells with Polyplus-transfection® (Illkirch, France) alone or jointly.
RNA extraction and qRT-PCR
According to the manufacturer’s protocol, TRIzol reagent (Invitrogen, CA, USA) was used to extract total RNA from human frozen tissues and cultured cells. The concentration and purity of RNA samples were evaluated with Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.) and cDNA was produced by commercial cDNA synthesis kit (Takara, Dalian, China). QRT-PCR for circRNA, miRNA, and mRNA were performed by SYBR Green PCR Kit (Takara, Dalian, China) and using the primers (Tsingke, Qingdao, China) that are listed in Table S1. GAPDH and U6 were used as internal controls, and all procedures were repeated 3 times. The relative expression of circRNA, miRNA, and mRNA were compared with internal controls and analyzed by the 2-ΔΔCt method.
Table S1. PCR primer and siRNA sequence.
| Gene name | Primer sequence |
|---|---|
| Circ-ZKSCAN1 | F: GAAACCCCGCCTCTTACA |
| R: TTCCTCTTCCACCTTCACTATTA | |
| FAM83A | F: AAGCACAACAACATCAGAGACCT |
| R: ATGGAAATGTTCTTGAGGTGACTG | |
| GAPDH | F: CTGACTTCAACAGCGACACC |
| R: TGCTGTAGCCAAATTCGTTGT | |
| hsa-miR-330-5p | F: TCTCTGGGCCTGTGTCTTAG |
| R: CAGTGCGTGTCGTGGAGT | |
| U6 | F: GTGCTCGCTTCGGCAGCACATATAC |
| R: AAAAATATGGAACGCTTCACGAATTTG | |
| Si-circ-ZKSCAN1-1 | GUUUCUUUACUAUUCCUCGU |
| Si-circ-ZKSCAN1-2 | UAUUCCUCGUGACUGUAAGA |
Cell proliferation and colony formation assays
For the determination of cell proliferation, the transfected cells were inoculated into 96-well plate at a density of 1×104 cells per well. After incubation for 0, 24, 48, and 72 hours, 10 µL Cell Counting Kit-8 (CCK-8) (Beyotime, Shanghai, China) solution was seeded to each well, and 96-well plates were cultured in the medium in the dark for 4 h at 37 °C. The optical density of each hole was measured by microporous plate spectrophotometer obtained from BioTek Instruments, Inc. (Winooski, VT, USA) under 450 nm. For the determination of colony formation, 0.5×103 cells were inoculated into the 6-well plates. After culturing for 14 days, the cells were washed with PBS 3 times, fixed with absolute methanol, and stained with 0.1% crystalline purple. The cell colonies were subsequently photographed and counted.
Wound healing assay
After the treatment, cells were inoculated into 6-well plates until they were reconnected and achieved 80% subconfluence. The single-cell layer was scraped with the tip of a 10 µL pipette, washed by PBS to remove the cell fragments, and fresh medium was then added. The wound healed for 48 h. Images were obtained at 0 and 48 h at the same wound location, and ImageJ software calculated the wound width.
Migration assays
The migration ability of cells was measured by Transwell chambers (SproutStrong Biotech, Beijing, China) with polycarbonate filter and 8 µm aperture. Briefly, the medium (containing 10% FBS) was added to the lower chamber as a chemical attractant. About 5×104 of transfected cells were incubated with 200 µL serum-free medium in the upper chamber, and then incubated with 5% CO2 at 37 °C. After 24 h, the cells were fixed with 70% ethanol and stained with 0.1% crystal violet, and the migrated cells were photographed by a microscope (Nikon Ti-s, Shanghai, China).
Cell apoptosis assays
According to the manufacturer’s instructions, apoptosis rates of A549 and H1299 were measured using the Annexin V-FITC Apoptosis Kit (JiaMay Biotech, Beijing, China). The cold 1× PBS was used to briefly wash transfected cells which were then suspended in Annexin V binding buffer. The cells were collected and resuspended in the mixture of annexin V-FITC and PI in a dark at room temperature for 15 minutes. Finally, the apoptosis rate was detected by flow cytometer.
Western blot (WB) analysis
The total proteins in transfected A549 and H1299 cells were isolated by AmyJet cell protein extraction kit (Wuhan, Hubei, China), separated on 10% SDS-PAGE gel, and transferred to PVDF membrane. The membrane with 5% skim milk was blocked at room temperature for 1 h, and then immunoblotted overnight at 4 °C with the following antibodies: anti-FAM83A (Abcam, Shanghai, China), anti-JNK, p38, and ERK (Abcam, Shanghai, China), and β-actin (Abcam, Shanghai, China). The membranes were then incubated with second antibodies and visualized.
Luciferase reporter assay
The full-length sequences of circ-ZKSCAN1 and FAM83A 3'-UTR with or without mutant miR-330-5p binding sites were cloned downstream of the Renilla luciferase gene in the dual plasmid psiCHECK2 vector to construct the psiCHECK2-circZKSCAN1 vector or psiCHECK2-FAM83A-3'-UTR vector, respectively (Promega). Subsequently, 10 pmol miR-330-5p or control mimics were co-transfected with the luciferase vector into A549 and H1299 cells by 40 ng Lipofectamine® 3000 (Invitrogen). After transfection for 48 h, luciferase activity was examined by luciferase report analysis kit (Promega, Beijing, China). All experiments were conducted 3 times to ensure the reliability of the data.
Xenografts in mice
About 1×106 of A549 cells were injected subcutaneously into the lower part of the left armpit of BALB/C male nude mice purchased from Qingdao University Laboratory Animal Center (age, about 4 weeks; weight, 18 to 22 g; 5 mice per group). The tumor volume was measured every 4 days, and the weight was calculated until the mice were sacrificed.
Immunohistochemistry (IHC)
Fresh tumor tissues from nude mice were fixed with 4% paraformaldehyde for 24 h, and the ethanol solution was dehydrated and embedded in paraffin. The paraffin-embedded tissue was cut at a thickness of 4 µm slides and immunohistochemical staining was performed according to the procedure described earlier (4). The sections were incubated with the anti-FAM83A antibody to measure the expression of FAM83A. The image was captured at an appropriate magnification in the microscope (Nikon Microsystems, Shanghai, China).
Data analysis
Data were analyzed by SPSS software (Version 22.0) and GraphPad Prism software (Version 7.0), with P<0.05 considered to be statistically significant. All continuous data are shown as mean ± standard deviation (SD). Chi-square test was performed to evaluate the difference of classification variables.
Results
Increased circ-ZKSCAN1 expression was observed in NSCLC cells and tissues
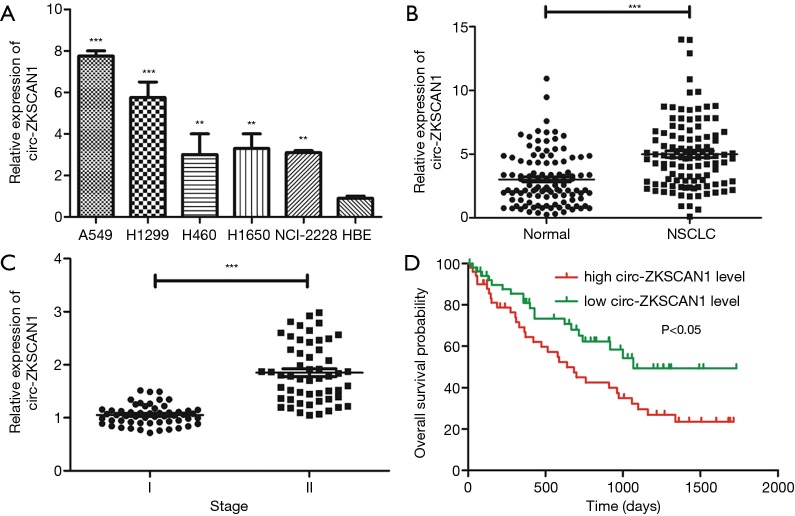
Previous studies have demonstrated the specificity and the site of cyclization of circ-ZKSCAN1 (4). Circ-ZKSCAN1 was significantly increased in NSCLC cell lines (A549, H1299, H460, H1650, NCI-2228) compared with normal HBE cells (Figure 1A). Similarly, as exhibited in Figure 1B, the expression of circ-ZKSCAN1 was higher in NSCLC tissues than that in adjacent normal tissues (P<0.001). Interestingly, increased expression of circ-ZKSCAN1 was found to be associated with advanced clinical stages of NSCLC in patients (P<0.001) (Figure 1C). In addition, we further evaluated the relationship between circ-ZKSCAN1 expression and prognosis in patients with NSCLC. Kaplan-Meier analysis showed that the overall time of NSCLC patients with high circ-ZKSCAN1 expression was significantly shorter than that of patients with high circ-ZKSCAN1 expression (Figure 1D). In conclusion, these results suggest that the imbalance of circ-ZKSCAN1 may be closely related to the progress of NSCLC.
Figure 1.
Circ-ZKSCAN1 is highly up-expressed in NSCLC tissues and cells. (A) Relative circ-ZKSCAN1 expression in NSCLC tumor tissues compared with adjacent normal tissues (n=107) by qRT-PCR analysis. (B) Relative circ-ZKSCAN1 expression in NSCLC cell lines. (C) Relative circ-ZKSCAN1 expression in different clinical stages (I vs. II). (D) The overall survival (OS) probability of NSCLC patients with low- and high-circ-ZKSCAN1 expression. (**P<0.01, ***P<0.001). NSCLC, non-small cell lung cancer.
Abnormally expressed circ-ZKSCAN1 promoted the growth of NSCLC cells both in vitro and in vivo
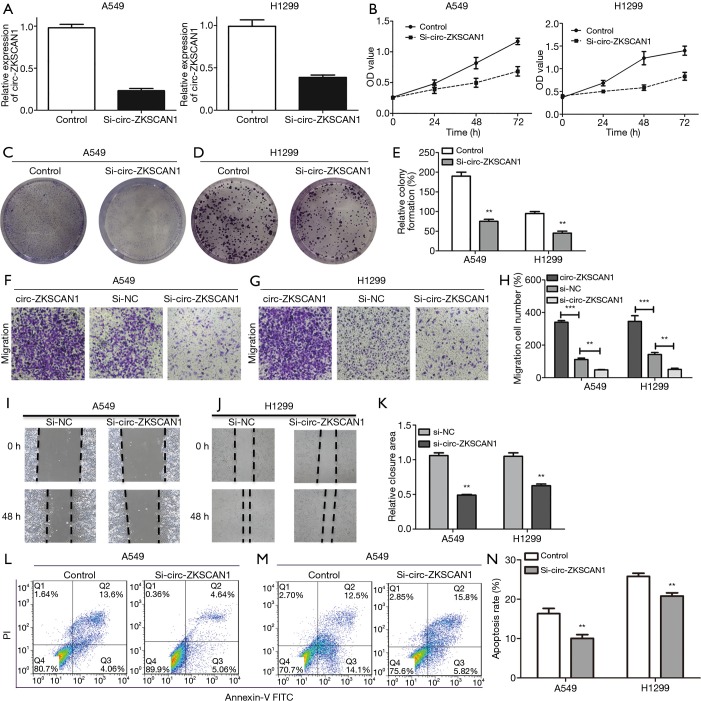
Subsequently, we evaluated the cellular function of circ-ZKSCAN1 in NSCLC, and selected the two NSCLC cell lines (A549 and H1299) with the highest expression of circ-ZKSCAN1 to inhibit circ-ZKSCAN1 (Figure 2A). The siRNA suppression efficiency is shown in Figure S1A. The CCK-8 assay showed that the proliferation rate of A549 and H1299 cells knocking down circ-ZKSCAN1 was slower than that of the normal cells (Figure 2B). Similarly, there were fewer clones formed in the circ-ZKSCAN1 knockdown of A549 and H1299 cells (Figure 2C,D,E). Functionally, transwell migration assays demonstrated that circ-ZKSCAN1 depletion led to decreased cell proliferation and migration, while overexpression of circ-ZKSCAN1 promoted cell proliferation and migration in A549 and H1299 cells (Figure 2F,G,H). Consistently, the cells with circ-ZKSCAN1 inhibition also displayed defects in wound healing compared to the controls (Figure 2I,J,K). Additionally, we also stably transferred circ-ZKSCAN1 into normal human lung epithelial cells to verify the effect of this circRNA on normal cells (Figure S1B,C,D,E,F), but this revealed no obvious difference.
Figure 2.
Circ-ZKSCAN1 inhibition retards NSCLC cells’ biological process. (A) Relative circ-ZKSCAN1 expression in A549 and H1299 cells after transfection with si-circ-ZKSCAN1 or control. CCK-8 assays (B) and colony formation assays (C,D,E) in A549 and H1299 cells treated with or without circ-ZKSCAN1 inhibition. (F,G,H) A549 and H1299 cells in transwell chambers were applied to cell migration assays (100×). (I,J,K) Cells treated with si-NC or si-circ-ZKSCAN1 to conduct wound healing assays. (L,M,N) Flow cytometry to determine cell apoptosis of A549 and H1299 after si-circ-ZKSCAN1 or si-NC. (**P<0.01, ***P<0.001). NSCLC, non-small cell lung cancer.
Due to the inhibition of proliferation of NSCLC cells after knockdown of circ-ZKSCAN1, we speculated that this process may be involved in apoptosis, which was analyzed and verified by flow cytometry. The results showed that the circ-ZKSCAN1 depletion significantly promoted the apoptosis of NSCLC cells (Figure 2L,M,N).
In conclusion, as shown in Table 1, we analyzed the relationship between the expression of circ-ZKSCAN1 and clinicopathological factors in 107 patients with NSCLC. We discovered that the expression of circ-ZKSCAN1 was positively correlated with clinical stage (P=0.043) and tumor size (P=0.017), but not with age, sex, and smoking.
Table 1. The relationship between the expression of circ-ZKSCAN1 and various clinicopathological variables.
| Characteristics | Total | circ-ZKSCAN1 expression | P value | |
|---|---|---|---|---|
| Low | High | |||
| Total | 107 | 37 | 70 | |
| Age (years) | 0.360 | |||
| <60 | 63 | 24 | 39 | |
| ≥60 | 44 | 13 | 31 | |
| Sex | 0.514 | |||
| Male | 59 | 22 | 37 | |
| Female | 48 | 15 | 33 | |
| Clinical stage | 0.043* | |||
| I | 55 | 24 | 31 | |
| II | 52 | 13 | 39 | |
| Tumor size (cm) | 0.017* | |||
| <1 | 44 | 21 | 23 | |
| ≥1 | 63 | 16 | 47 | |
| Smoking | 0.326 | |||
| Yes | 48 | 19 | 29 | |
| No | 59 | 18 | 41 | |
| Differentiation | 0.893 | |||
| High | 36 | 13 | 23 | |
| Moderately | 48 | 17 | 31 | |
| Poorly | 23 | 7 | 16 | |
| Lymph node metastasis | 0.193 | |||
| Yes | 32 | 14 | 18 | |
| No | 75 | 23 | 52 | |
*P<0.05.
Circ-ZKSCAN1 promoted tumor growth in vitro
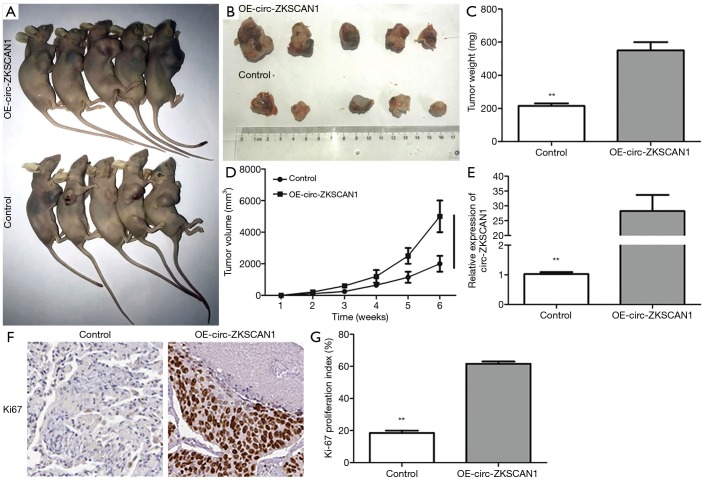
In order to verify whether circ-ZKSCAN1 also plays a role in vivo, we used subcutaneous injection of stable overexpressed-circ-ZKSCAN1 A549 cells into nude mice to establish a xenograft tumor model. Four weeks later, the nude mice were killed, and the tumor was carefully taken out (Figure 3A,B). The results confirmed the increasing circ-ZKSCAN1 results in bigger tumor weight and volume (Figure 3C,D), and improving efficiency of circ-ZKSCAN1 in vivo through qRT-PCR (Figure 3E). The circ-ZKSCAN1 expression was up-regulated in the OE-CIRC-ZKSCAN1 group as indicated by IHC staining. Ki-67 showed an increased amount of Ki-67-positive cells in the circ-ZKSCAN1-overexpressing group compared with the control group (Figure 3F,G).
Figure 3.
Circ-ZKSCAN1 can promote tumor formation in vivo. (A,B) After inoculating A549 cells for 40 days in nude mice. (C,D) The weight and volume of tumors in the control group and the OE-circ-ZKSCAN1 group. (E) Relative circ-ZKSCAN1 expression in tumors of nude mice. (F,G) Ki-67 proliferative marker compared controls with circ-ZKSCAN1-overexpressing xenografts, as determined by IHC staining. (**P<0.01) (200×). IHC, immunohistochemistry.
Circ-ZKSCAN1 acted as a sponge of miR-330-5p in NSCLC
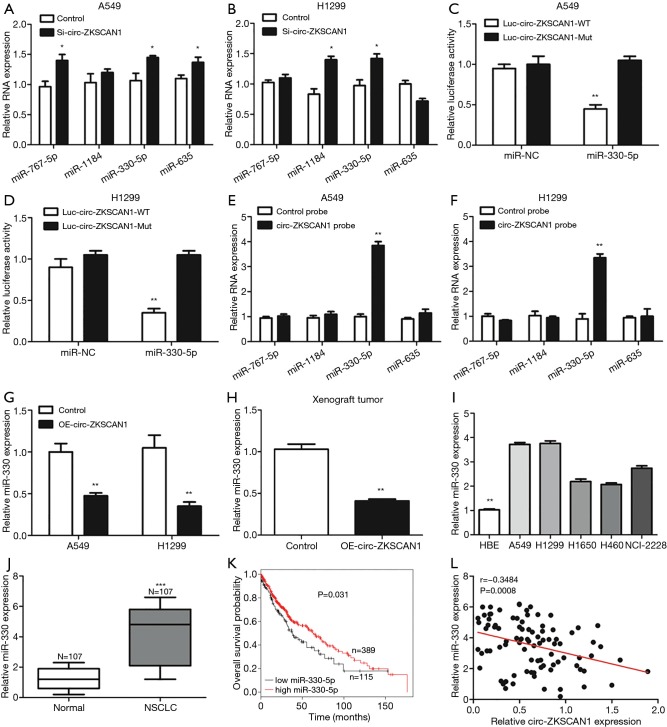
By searching online databases CircInteractome and circBank, we examined 4 miRNAs—miR-767-5p, miR-1184, miR-330-5p and miR-635—that are hypothesized to bind to circ-ZKSCAN1 and whose function and expression have been related to NSCLC (Figure S2A). As shown in Figure 4A,B, the qRT-PCR results demonstrated that knock down of circ-ZKSCAN1 increased miR-767-5p, miR-330-5p, and miR-635 expression in A549 cells, and up-regulated miR-1184 and miR-330-5p expression in H1299 cells. Because miR-330-5p showed common expression changes, we selected it for the next verification. Inhibiting miR-330-5p in A549 and H1299 cells significantly increased luciferase activity of the wild type circ-ZKSCAN1 luciferase carrier but did not increase luciferase activity of the mutant one (Figures 4C,D,S2B). RNA pull-down assay demonstrated that circ-ZKSCAN1 probe enriched miR-330-5p and not miR-767-5p, miR-1184, and miR-635 in A549 and H1299 cells (Figure 4E,F). In addition, knock down the expression of circ-ZKSCAN1 significantly increased the expression of miR-330-5p with this result being confirmed in the xenograft model (Figure 4G,H). Additionally, we confirmed that miR-330-5p was significantly increased in NSCLC cells and tissues (Figure 4I,J), and the total survival time of NSCLC patients with low miR-330-5p expression was shorter than that of patients with high miR-330-5p expression (Figure 4K) (data from Kaplan-Meier plotter). More interestingly, evidence indicated that circ-ZKSCAN1 expression was negatively correlated with miR-330-5p expression in 107 NSCLC tissues (r=−0.3484, P=0.0008) (Figure 4L). In general, the above data mean that miR-330-5p is a critical downstream target for circ-ZKSCAN1 in NSCLC.
Figure 4.
Circ-ZKSCAN1 can sponge and suppress miR-330-5p in NSCLC cells. (A,B) The relative indications of 4 miRNA expressions in NSCLC cells after transfection with the knock-down circ-ZKSCAN1 or control group. (C,D) Circ-ZKSCAN1-wild- or -mutant-type luciferase vector after transfection with miR-330-5p or control mimics in A549 and H1299 cells by luciferase activity. (E,F) Four indicated miRNA expressions after RNA pull-down assay with circ-ZKSCAN1 probe and control probe in A549 and H1299 cells. (G) MiR-330-5p expression in control or OE-circ-ZKSCAN1 A549 and H1299 cells. (H, I, J) Relative expression levels of miR-330-5p in xenografts of nude mice (H), NSCLC cells (I), and tissues (J). (K) The OS probability of NSCLC patients with low- and high-miR-330-5p expression. (L) The correlation between circ-ZKSCAN1 and miR-330-5p expression in NSCLC tissues. (*P<0.05, **P<0.01, ***P<0.001). NSCLC, non-small cell lung cancer.
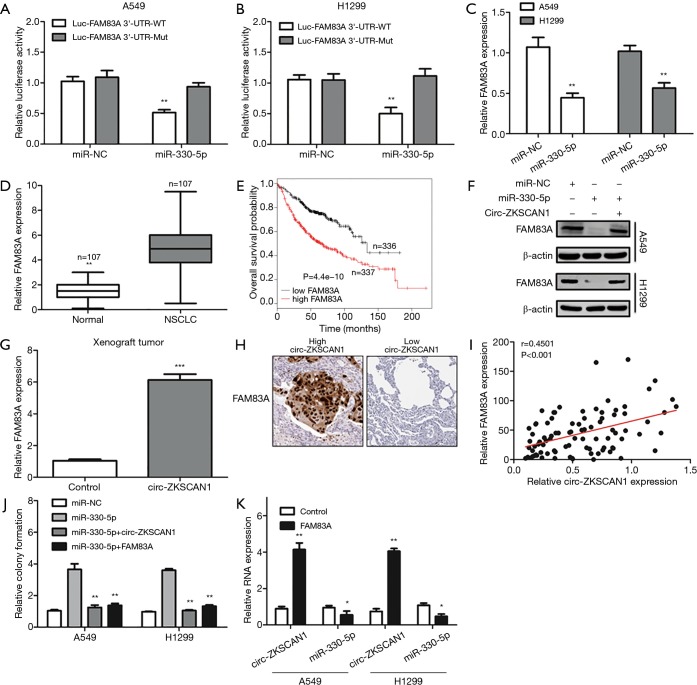
Circ-ZKSCAN1 up-regulated FAM83A expression by sponging miR-330-5p in NSCLC
We identified FAM83A to be a potential target gene of miR-330-5p through using the TargetScan database. As shown in Figure 5A, luciferase reporter assay revealed that miR-330-5p mimics could decrease the luciferase activity of the wild-type FAM83A 3'-UTR luciferase vector but had no impact at the mutant one (Figure 5B). FAM83A was down-expressed after A549 and H1299 cells were transfected with miR-330-5p mimics. Also, FAM83A was significantly increased in NSCLC tissues (Figure 5C,D), and its elevation was closely related to worse prognosis as confirmed by data from the Kaplan-Meier plotter (Figure 5E). It is worth noting that miR-330-5p-induced down-regulation of FAM83A was rescued by the overexpression of circ-ZKSCAN1 (Figure 5F), and the anomalous expression of circ-ZKSCAN1 increased the expression of FAM83A in the xenograft model (Figure 5G). Commonly, the high expression of FAM83A was detected in NSCLC patients with a high expression of circ-ZKSCAN1 (r=0.4501, P<0.001) (Figure 5H,I). In addition, the overexpression of circZKSCAN1 or FAM83A can counteract the increased proliferation induced by miR-330-5p mimics (Figure 5J). Interestingly, we discovered that circ-ZKSCAN1 expression could also be controlled by FAM83A. The overexpression of FAM83A significantly increased the expression of circ-ZKSCAN1 and decreased the expression of miR-330-5p in A549 and H1299 cells (Figure 5K). In summary, these findings suggest that circ-ZKSCAN1, miR-330-5p, and FAM83A form a feedback loop in NSCLC.
Figure 5.
A feedback loop in NSCLC cells comprising Circ-ZKSCAN1, miR-330-5p, and FAM83A. (A,B) The relative luciferase activity of FAM83A 3’-UTR-wild- or -mutant-type luciferase vector after transfection with miR-330-5p mimics or control in A549 and H1299 cells. (C) Relative expression of FAM83A in miR-330-5p or control mimics in A549 and H1299 cells. (D) Relative FAM83A expression in NSCLC and paired normal tissues. (E) The OS probability of low and high expression of FAM83A in NSCLC patients. (F) WB analysis for the protein expression of FAM83A in control or miR-330-5p mimics A549 and H1299 cells after transfection with OE-circ-ZKSCAN1 vector and control. β-actin as an internal reference. (G) Relative FAM83A expression in tumors of nude mice. (H) FAM83A in NSCLC tissues by IHC staining. (I) The correlation between FAM83A and circ-ZKSCAN1 expression in NSCLC tissues. (J) Colony formation assays for overexpressed-miR-330-5p or control in NSCLC cells after transfection with OE-circ-ZKSCAN1 or OE-FAM83A vector. (K) Relative expression of circ-ZKSCAN1 and miR-330-5p in control or OE-FAM83A of A549 and H1299 cells. (*P<0.05, **P<0.01, ***P<0.001). NSCLC, non-small cell lung cancer.
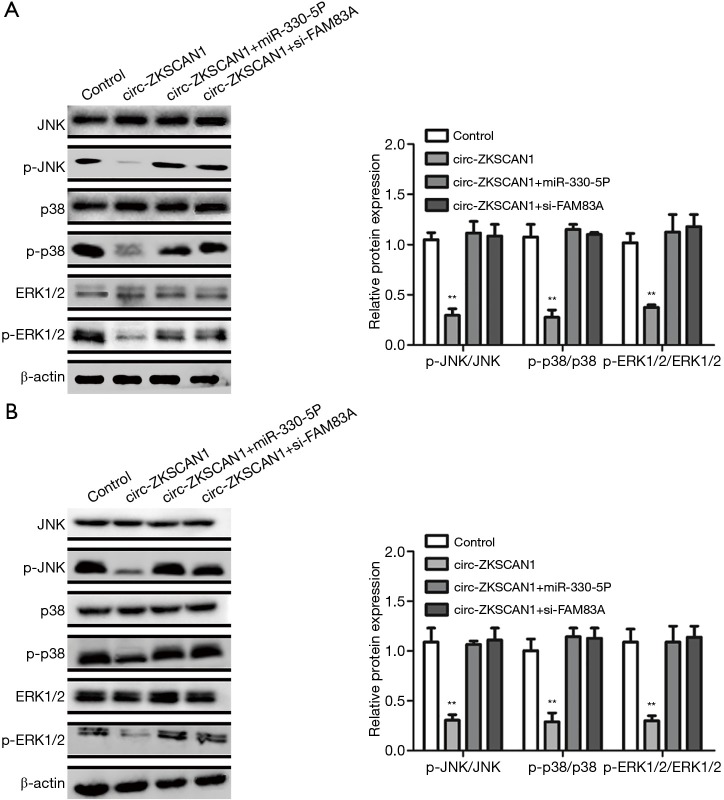
The regulatory circuit of circ-ZKSCAN1/miR-330-5p/FAM83A functioned via the inactivation of MAPK signaling pathway
It has been reported that FAM83A plays its role by inhibiting MAPK signaling pathway (17). Therefore, we speculated that circ-ZKSCAN1 can change the MAPK signaling pathway by regulating the expression of FAM83A. As expected, overexpression of circZKSCAN1 significantly decreased the expression of JNK, p38, and ERK (3 crucial downstream genes of MAPK signaling pathway) in A549 and H1299 cells (Figure 6A,B). Importantly, the suppression of MAPK pathway caused by circ-ZKSCAN1 overexpression was blocked by miR-330-5p mimics or FAM83A silencing (Figure 6A,B). Overall, the above data show that the circ-ZKSCAN1-miR-330-5p-FAM83A feedback loop is the key negative regulator of MAPK/ERK signal transduction.
Figure 6.
The overexpression of miR-330-5p or silencing of FAM83A could block the suppression of MAPK pathway caused by circ-ZKSCAN1 overexpression in NSCLC cells. (A,B) WB analysis for the expression of JNK, p38, and ERK1/2 in control or OE-circ-ZKSCAN1 A549 (A) and H1299 (B) cells with miR-330-5p mimics or si-FAM83A. (**P<0.01). NSCLC, non-small cell lung cancer.
Discussion
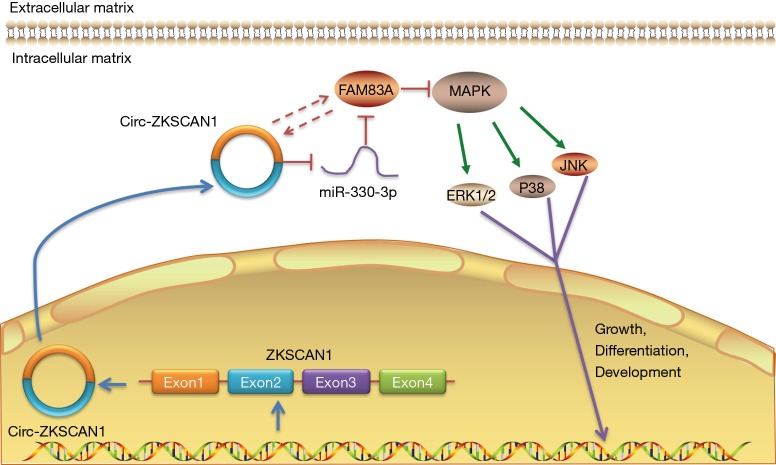
In this study, we demonstrated that circ-ZKSCAN1 promotes the development of miR-330-5p by maintaining the expression of FAM83A and inactivating the MAPK signaling pathway, as indicated by functional and molecular assays. This study is the first to show that the expression of FAM83A in NSCLC is regulated by circular RNA, and the mechanism and clinical significance of circ-ZKSCAN1 in NSCLC are also reported for the first time here (Figure 7).
Figure 7.
Schematic diagram of the mechanism by which the circ-ZKSCAN1-miR-330-5p-FAM83A regulatory loop regulates lung cancer proliferation by inactivating MAPK signaling pathway.
More specifically, we have found a new type of circRNA, called circ_ZKSCAN1, which is significantly upregulated in human NSCLC and related to clinical stage and tumor size. Functionally, the decrease of circ_ZKSCAN1 can inhibit the migration and proliferation of NSCLC cells and the growth of tumor in vivo. In terms of mechanism, we proved, through RNA pull-down assay and luciferase reporter assays, that circ_ZKSCAN1 binds to miR-330-5p. WB and functional experiments showed the correlation between circ-ZKSCAN1, miR-330-5p, and FAM83A. We thus proposed that circ-ZKSCAN1 could act as a sponge for miR-330-5p to reduce the suppression of FAM83A. Accordingly, our experimental data show that circ-ZKSCAN1 may play a regulatory participator in the development and pathogenesis of NSCLC.
CircRNA as a type of competing endogenous RNA (ceRNA) is common in cells and has the function of gene regulation (18). Most of the circRNAs are positioned within the cytoplasm while some are in the nucleus; the circRNA is quite stable and not easily degraded by RNase R (7). Recent research has reported that circRNAs play pivotal roles in regulating gene expression, from binding microRNA to adsorbing RNA binding proteins and regulating transcription factors (9,19). Through the development of bioinformatics technology and high throughput sequencing, circRNA is more frequently being found in tumors. CircRNAs are characterized by a long half-life, stable expression, and specific expression in different tumors, thus making them a new type of tumor biomarker that can be used for early diagnosis and even treatment of tumors (20,21).
CircRNA usually acts as a miRNA sponge to eliminate the activity of miRNA, which leads to the decrease of the expression and function of miRNA and the expression of target genes regulated by miRNA (22,23). So far, some studies have verified the relationship between NSCLC and circRNA. Liu et al. found that circ-FOXM1 may be a new biomarker which can regulate NSCLC through the circFOXM1-miR-1304-5p-PPDPF/MACC1 axis (24). CircABCB10 promotes NSCLC cell migration and proliferation by regulating the miR-1252/FOXR2 axis (25). Circ-PTK2 inhibits TGF-beta-induced EMT (epithelial-mesenchymal transition) and metastasis by controlling TIF1γ in NSCLC (26). Zhou et al. demonstrated that hsa_circ_0004015 regulates the proliferation, invasion, and TKI drug resistance of NSCLC by miR-1183/PDPK1 signaling pathway (27). In addition, we observed that circ-ZKSCAN1 regulated the expression of FAM83A through sponging miR-330-5p to promote the progress of NSCLC.
The MAPK/ERK signaling pathway is highly involved in the invasion and metastasis of NSCLC, with some studies showing that MAPK/ERK is continuously activated in NSCLC (28,29). In families with sequence similarity, the family with sequence similarity 129, member B and FAK signaling pathway can activate or inhibit the pathway through the corresponding binding, and promote the invasion and proliferation of NSCLC cells by promoting the phosphorylation of FAK (30). The binding of MAPK/ERK to FAM83A has been validated previously (17), and FAM83 has been closely related to many diseases. Recently, one study has shown that mutations in FAM83 proteins that prevent them from binding to CK1 interfere with the right subcellular localization and cell characteristic of FAM83 proteins and their CK1-binding ligands, which regulate cell division and apoptosis, thereby helping cancer development (31). Therefore, drugs targeting this consistent cancer-promoting factor can control tumor growth, while the discovery and production of FAM83A inhibitors may be beneficial to cancer patients. Our results show that the circ-ZKSCAN1/miR-330-5p/FAM83A axis may play a vital role in the development of NSCLC and could represent a new strategy to suppress NSCLC.
Conclusions
We have shown that circ-ZKSCAN1 is highly expressed and plays a carcinogenic role in NSCLC tissues and cell lines. In addition, circ-ZKSCAN1 is used as a sponge of miR-330-5p to eliminate the inhibition of its target gene FAM83A and inhibit MAPK signal transduction. The signal axis promotes the progress of NSCLC and can be used as a potential new biomarker and therapeutic target of NSCLC.
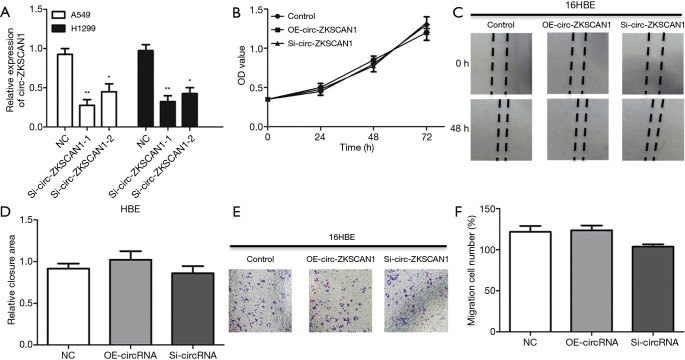
Figure S1.
Circ-ZKSCAN1 in HBE cells. (A) Expression levels of circ-ZKSCAN1 in NSCLC cells treated with siRNAs. (B) CCK8 analysis of HBE cells with silencing or stably overexpressing circ-ZKSCAN1. (C,D) Cells treated with control, OE-circ-ZKSCAN1, and si-circ-ZKSCAN1 to perform wound healing assays. (E,F) HBE cells in transwell chambers were applied to cell migration assays (100×). (*P<0.05, **P<0.01). HBE, human bronchial epithelial.
Figure S2.
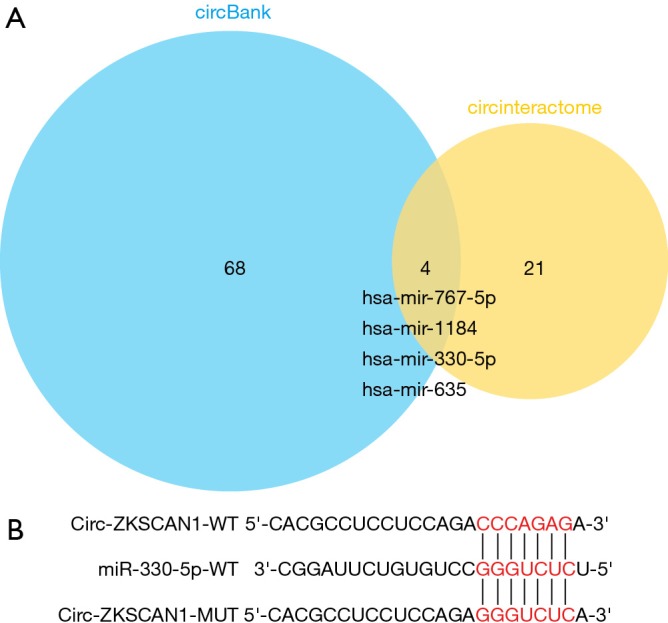
Circ-ZKSCAN1 targeting miRNA. (A) Intersection of miRNAs predicted by circBank and CircInteractome databases. (B) The binding sites of circ-ZKSCAN1 and miR-330-5p.
Acknowledgments
Funding: This study was funded by Key R & D programs in Shandong Province (grant number: 2018GSF118119).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The medical ethics for this research were approved by the Ethic Committee of the Affiliated Hospital of Qingdao University (No. QYFYKY 2018-10-11-2).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2.Osarogiagbon RU, Veronesi G, Fang W, et al. Early-Stage NSCLC: Advances in Thoracic Oncology 2018. J Thorac Oncol 2019;14:968-78. 10.1016/j.jtho.2019.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X, Yu S, Fan J. Epigenetic diagnostic biomarkers for non-small cell lung cancer: present and future perspectives. Transl Cancer Res 2018;7:796-802. 10.21037/tcr.2018.05.03 [DOI] [Google Scholar]
- 4.Yao Z, Luo J, Hu K, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol 2017;11:422-37. 10.1002/1878-0261.12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ravasi T, Huber T, Zavolan M, et al. Systematic characterization of the zinc-finger-containing proteins in the mouse transcriptome. Genome Res 2003;13:1430-42. 10.1101/gr.949803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan B, Li Y, Zhao Q, et al. ZNF139 increases multidrug resistance in gastric cancer cells by inhibiting miR-185. Biosci Rep 2018. doi: . 10.1042/BSR20181023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Lu T, Wang Q, et al. Circular RNAs: Crucial regulators in the human body (Review). Oncol Rep 2018;40:3119-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu L, Liu Y, Yang Y, et al. CircRNA ZNF609 promotes growth and metastasis of nasopharyngeal carcinoma by competing with microRNA-150-5p. Eur Rev Med Pharmacol Sci 2019;23:2817-26. [DOI] [PubMed] [Google Scholar]
- 9.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol 2015;12:381-8. 10.1080/15476286.2015.1020271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 11.Dong Y, He D, Peng Z, et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol 2017;10:2. 10.1186/s13045-016-0370-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang R, Xing L, Zheng X, et al. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer 2019;18:4. 10.1186/s12943-018-0933-7 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Xie B, Zhao Z, Liu Q, et al. CircRNA has_circ_0078710 acts as the sponge of microRNA-31 involved in hepatocellular carcinoma progression. Gene 2019;683:253-61. 10.1016/j.gene.2018.10.043 [DOI] [PubMed] [Google Scholar]
- 14.Cao Y, Yuan G, Zhang Y, et al. High glucose-induced circHIPK3 downregulation mediates endothelial cell injury. Biochem Biophys Res Commun 2018;507:362-8. 10.1016/j.bbrc.2018.11.041 [DOI] [PubMed] [Google Scholar]
- 15.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 2014;28:2233-47. 10.1101/gad.251926.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu YJ, Zheng B, Luo GJ, et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics 2019;9:3526-40. 10.7150/thno.32796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cipriano R, Miskimen KL, Bryson BL, et al. Conserved oncogenic behavior of the FAM83 family regulates MAPK signaling in human cancer. Mol Cancer Res 2014;12:1156-65. 10.1158/1541-7786.MCR-13-0289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Z, Li P, Fan L, et al. The Potential Role of circRNA in Tumor Immunity Regulation and Immunotherapy. Front Immunol 2018;9:9. 10.3389/fimmu.2018.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Zhang J, Li J, et al. CircRNA_014511 affects the radiosensitivity of bone marrow mesenchymal stem cells by binding to miR-29b-2-5p. Bosn J Basic Med Sci 2019;19:155-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer 2017;16:94. 10.1186/s12943-017-0663-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Liu K, Liu Y, et al. Evaluating the bias of circRNA predictions from total RNA-Seq data. Oncotarget 2017;8:110914-21. 10.18632/oncotarget.22972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol 2016;238:42-51. 10.1016/j.jbiotec.2016.09.011 [DOI] [PubMed] [Google Scholar]
- 23.Panda AC. Circular RNAs Act as miRNA Sponges. Adv Exp Med Biol 2018;1087:67-79. 10.1007/978-981-13-1426-1_6 [DOI] [PubMed] [Google Scholar]
- 24.Liu G, Shi H, Deng L, et al. Circular RNA circ-FOXM1 facilitates cell progression as ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in non-small cell lung cancer. Biochem Biophys Res Commun 2019;513:207-12. 10.1016/j.bbrc.2019.03.213 [DOI] [PubMed] [Google Scholar]
- 25.Tian X, Zhang L, Jiao Y, et al. CircABCB10 promotes nonsmall cell lung cancer cell proliferation and migration by regulating the miR-1252/FOXR2 axis. J Cell Biochem 2019;120:3765-72. 10.1002/jcb.27657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Tong X, Zhou Z, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-beta-induced epithelial-mesenchymal transition and metastasis by controlling TIF1gamma in non-small cell lung cancer. Mol Cancer 2018;17:140. 10.1186/s12943-018-0889-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, Zheng X, Xu B, et al. Circular RNA hsa_circ_0004015 regulates the proliferation, invasion, and TKI drug resistance of non-small cell lung cancer by miR-1183/PDPK1 signaling pathway. Biochem Biophys Res Commun 2019;508:527-35. 10.1016/j.bbrc.2018.11.157 [DOI] [PubMed] [Google Scholar]
- 28.Zhuo H, Zhao Y, Cheng X, et al. Tumor endothelial cell-derived cadherin-2 promotes angiogenesis and has prognostic significance for lung adenocarcinoma. Mol Cancer 2019;18:34. 10.1186/s12943-019-0987-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pradhan R, Singhvi G, Dubey SK, et al. MAPK pathway: a potential target for the treatment of non-small-cell lung carcinoma. Future Med Chem 2019;11:793-5. 10.4155/fmc-2018-0468 [DOI] [PubMed] [Google Scholar]
- 30.Zhou X, Yang F, Zhang Q, et al. FAM129B promoted tumor invasion and proliferation via facilitating the phosphorylation of FAK signaling and associated with adverse clinical outcome of non-small cell lung cancer patients. Onco Targets Ther 2018;11:7493-501. 10.2147/OTT.S161852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fulcher LJ, Bozatzi P, Tachie-Menson T, et al. The DUF1669 domain of FAM83 family proteins anchor casein kinase 1 isoforms. Sci Signal 2018. doi: . 10.1126/scisignal.aao2341 [DOI] [PMC free article] [PubMed] [Google Scholar]