Abstract
Background
Non-small cell lung cancer (NSCLC) remains the first leading cause of death in malignancies worldwide. Despite the early screening of NSCLC by low-dose spiral computed tomography (CT) in high-risk individuals caused a 20% reduction in the mortality, there still exists imperative needs for the identification of novel biomarkers for the diagnosis and treatment of lung cancer.
Methods
mRNA microarray datasets GSE19188, GSE33532, and GSE44077 were searched, and the differentially expressed genes (DEGs) were obtained using GEO2R. Functional and pathway enrichment analyses were performed for the DEGs using DAVID database. Protein-protein interaction (PPI) network was plotted with STRING and visualized by Cytoscape. Module analysis of the PPI network was done through MCODE. The overall survival (OS) analysis of genes from MCODE was performed with the Kaplan Meier-plotter.
Results
A total of 221 DEGs were obtained, which were mainly enriched in the terms related to cell division, cell proliferation, and signal transduction. A PPI network was constructed, consisting of 221 nodes and 739 edges. A significant module including 27 genes was identified in the PPI network. Elevated expression of these genes was associated with poor OS of NSCLC patients, including UBE2T, UNF2, CDKN3, ANLN, CCNB2, and CKAP2L. The enriched functions and pathways included protein binding, ATP binding, cell cycle, and p53 signaling pathway.
Conclusions
The DEGs in NSCLC have the potential to become useful targets for the diagnosis and treatment of NSCLC.
Keywords: Non-small cell lung cancer (NSCLC), differentially expressed genes (DEGs), functional enrichment analysis, protein-protein interaction (PPI), Kaplan-Meier plotter
Introduction
Despite the recent progress in the targeted therapies of advanced lung cancer, lung cancer remains the most common cancer with the highest mortality worldwide (1,2). Among all types of lung cancer, non-small cell lung cancer (NSCLC) accounts for approximately 80–85% and is mainly adenocarcinoma and squamous cell carcinoma (3). The treatment of NSCLC has significantly improved over the past decades with the development and progress of new technologies, especially the next-generation sequencing. A large number of genes involved in the receptor tyrosine kinase (RTK) pathway have been identified, and the alterations in a number of genes, including EGFR, ALK, ROS1, BRAF, KRAS, have been demonstrated to be closely related to the carcinogenesis and poor prognosis of lung cancer (4-8).
One of the reasons for the poor prognosis of advanced NSCLC is the low diagnosis rate. In one study, results show that early screening of NSCLC by low-dose spiral computed tomography (CT) in high-risk individuals causes a 20% reduction in the mortality (9). The 5-year survival is closely related to the tumor stage at the initial diagnosis. Tremendous efforts have been put in the investigation and development of biomarkers for the early diagnosis of NSCLC. Several biomarkers have emerged as predictive and prognostic markers for NSCLC, especially in the targeted therapies. For instance, ALK fusion oncogenes and ROS1 rearrangements are identified as predictive markers and can benefit NSCLC patients from crizotinib therapy; while patients with KRAS mutations appear to have a shorter survival than those with wild-type KRAS, which serves as a negative prognostic marker (10-12). Currently, serum biomarkers (such as CEA, SCC and TPA) have been widely used to assist the diagnosis of NSCLC (13-15). The clinical application of these serum biomarkers, however, is limited due to their low sensitivity and specificity. For example, CEA is non-specific for NSCLC and also detectable in other types of cancer or even in case of non-cancer related diseases (16). Thus, further studies are required for the development and identification of more specific and sensitive biomarkers for the early diagnosis of NSCLC.
The bioinformatics analysis has emerged as a promising strategy to comprehensively analyze large quantities of data, including the complicated gene information. In our study, the well-established bioinformatic tools were employed to screen potential biomarkers for the early diagnosis of NSCLC. The Gene Expression Omnibus (GEO) database, an open-access database, was used in which appropriate mRNA profiles were selected. The online analysis tools assisted in analyzing differentially expressed genes (DEGs) between tumor group and normal group. In our study, three mRNA microarray datasets, namely GSE19188, GSE33532, and GSE44077, were downloaded from GEO. DEGs were obtained using GEO2R. Functional and pathway enrichment analysis was performed for DEGs using the DAVID database. Protein-protein interaction (PPI) network was established by using STRING and visualized with Cytoscape. Module analysis of the PPI network was done by using MCODE. Subsequently, overall survival (OS) analysis of genes from MCODE was performed with the online Kaplan Meier-plotter. Eventually, several NSCLC-related modules were screened out for further development of early NSCLC diagnosis system.
Methods
Subjects and gene information
The GEO is a national center for genetic information database, including microarray and next-generation sequencing data, and is freely open to researchers (17). In this study, three gene expression profiles (GSE19188, GSE33532 and GSE44077) were searched and selected from the GEO database. In brief, there were 91 early NSCLC samples and 65 non-cancerous samples from 156 patients in the GSE19188. GSE33532 was composed of 80 early NSCLC samples and 20 non-cancerous samples from 20 patients. GSE44077 consisted of 55 early NSCLC samples and 66 non-cancerous samples from 20 patients. The characteristics of these patients are shown in Table 1. The ethics approval from the ethics committee was waived.
Table 1. Summary of patient characteristics of three GEO datasets.
| GEO accession | GSE19188 | GSE33532 | GSE44077 |
|---|---|---|---|
| No. of patients | |||
| Health | 65 | 0 | |
| Tumor | 91 | 20 | |
| Mean age (years) | 62.9 | 63 | NA |
| Gender | |||
| Male | 109 | 16 | 11 |
| Female | 47 | 4 | 9 |
| Histological type | |||
| ADC | 32 | 11 | 14 |
| SCC | 27 | 4 | 5 |
| LCC | 13 | 0 | 0 |
| Other | 19 | 5 | 1 |
| Stage | |||
| I | 57 | 14 | 14 |
| II | 29 | 6 | 4 |
| III | 5 | 0 | 2 |
GEO, Gene Expression Omnibus.
Data analysis
Large quantities of high-throughput functional genomic researches have been collected in the GEO database. Various methods can be applied to process and normalize all these data. GEO2R is one of the powerful tools to screen the differentially expressed mRNAs between NSCLC samples and non-cancerous samples. Almost any GEO profile can be analyzed with GEO2R to compare two groups under the same experimental condition (18). The P value <0.05 and |logFC| >1.5 were used as the selection criteria.
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
GO and KEGG pathway enrichment analysis were performed for the identified DEGs using the Database for Annotation, Visualization and Integrated Discovery (DAVID) database, aiming to provide a comprehensive set of functional annotation tools for the classification of different genes into several groups with distinct annotations, which thereby leads to a better understanding of the biological functions of genes (19). P value <0.05 was used to distinguish significant enriched genes.
PPI network visualization
PPI network of DEGs was simulated to evaluate the functional interactions among proteins. The Search Tool for the Retrieval of Interacting Genes (STRING) database and Cytoscape were utilized for the visualization of PPI network (20,21). In the following, the Molecular Complex Detection (MCODE) was conducted to select appropriate modules of PPI network. In detail, cutoff value was 2 for the degree and 0.2 for the node score, the k-score was 2, and the maximum depth was 100 (22).
Kaplan-Meier survival analysis of DEGs
The influence of selected DEGs on the survival can be analyzed with an online tool, namely Kaplan Meier-plotter (KM plotter) that contains the information of a large number of patients with different cancers, including breast cancer, gastric cancer and ovarian cancer (23). The NSCLC patients were divided into two groups based on the individual expression of a particular gene (high versus low expression). The OS of NSCLC patients was obtained and plotted on this website. The hazard ratio (HR) with 95% confidence interval (CI) and log rank P value were calculated and displayed on the plot.
Results
Screening of DEGs
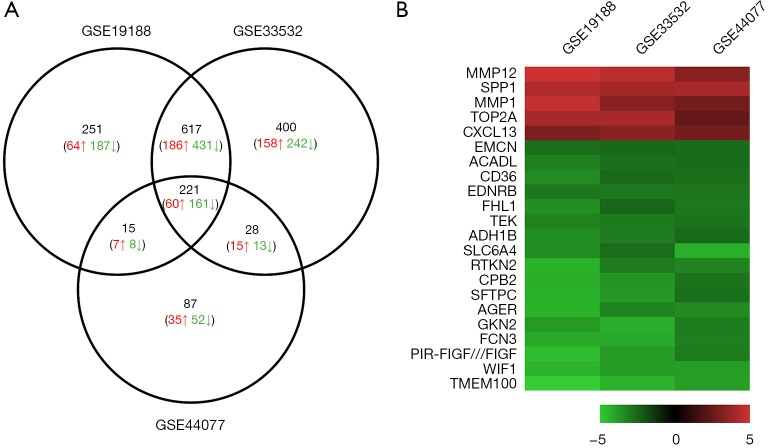
A total of 1,104, 1,266 and 351 DEGs were identified from the GSE19188, GSE33532, and GSE44077 datasets, respectively. A total of 221 genes were found in all three datasets (Figure 1A). Interestingly, the trends of these gene expressions were similar in three datasets. Among them, 60 genes showed up-regulated expression and 161 had down-regulated expression in the NSCLC as compared to non-cancerous samples. 22 genes were shown on the heatmap as the top regulated genes which were defined by |logFC| >2.5 and P<0.05 in all three datasets (Figure 1B).
Figure 1.
Identification of differentially expressed genes in mRNA expression profiling datasets (GSE19188, GSE33532, and GSE44077). (A) Venn diagram of DEGs (P<0.05 and |logFC| >1.5); (B) heatmap of top DEGs (P<0.05 and |logFC| >2.5). DEG, differentially expressed gene.
GO and KEGG pathway enrichment analysis
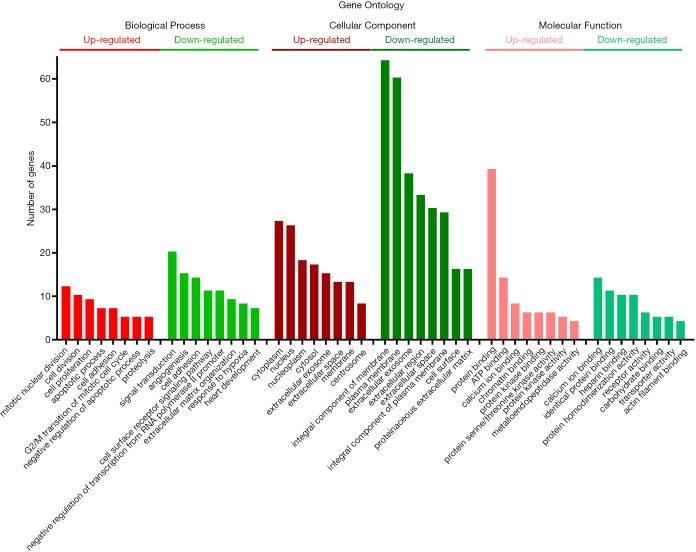
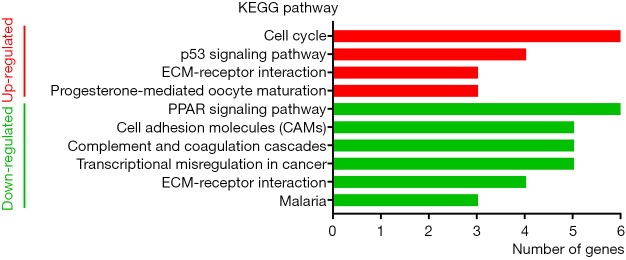
GO analysis of individual DEGs and KEGG pathway enrichment analysis were performed on the DAVID platform to obtain more insightful details into the diverse functions of particular DEGs. The up-regulated genes were mainly related to the mitotic nuclear division, cell division, and cell proliferation; the down-regulated genes were mainly involved in signal transduction, angiogenesis, and cell adhesion (Figure 2). In addition, the up-regulated DEGs were related to four KEGG pathways, including cell cycle, p53 signaling pathway, ECM-receptor interaction, and progesterone-mediated oocyte maturation, while six KEGG pathways including PPAR signaling pathway and cell adhesion molecule were enriched in the down-regulated genes (Figure 3). Surprisingly, the enriched pathways were observed in both up-regulated and down-regulated genes, such as ECM-receptor interaction, which results in a direct or indirect control of cellular activities, including adhesion, proliferation and migration.
Figure 2.
Histogram of gene ontology analysis including biological process, cellular component, and molecular function with up-regulated and down-regulated DEGs. DEG, differentially expressed gene.
Figure 3.
Histogram of KEGG pathway analysis with up-regulated and down-regulated DEGs. KEGG, Kyoto Encyclopedia of Genes and Genomes; DEG, differentially expressed gene.
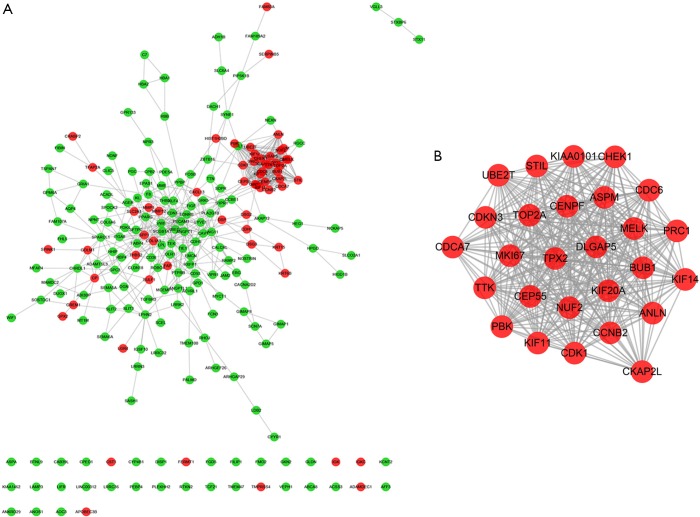
PPI network construction and module selection
A total of 221 nodes and 739 edges were selected to plot the PPI network, which consisted of 60 up-regulated genes and 161 down-regulated genes (Figure 4A). Subsequently, a pivotal module of 27 genes (TOP2A, ANLN, DLGAP5, TPX2 and ASPM) was identified with the degree ≥10 as the cut-off value by using MCODE (Figure 4B). Moreover, it is worth noting that, in the PPI network, cyclin B2 (CCNB2) could interact with kinesin family member 11 (KIF11) and centromere protein F (CENPF). Functional and KEGG pathway enrichment analysis revealed that genes in this module were mainly associated with protein binding, ATP binding, cell cycle, and p53 signaling pathway (Table 2).
Figure 4.
Protein-protein interaction network and a significant module. (A) Protein-protein interaction network of DEGs. (B) A significant module selected from protein-protein interaction network. Red nodes are up-regulated genes, green nodes are down-regulated genes, while blue nodes are genes from STRING database. The lines represent the interaction relationship between nodes. DEG, differentially expressed gene.
Table 2. Function and pathway enrichment analysis of genes in module.
| Term | Term description | Count | P value |
|---|---|---|---|
| GO:0005515 | Protein binding | 21 | 0.001470438 |
| GO:0005524 | ATP binding | 14 | 1.93E−08 |
| GO:0004674 | Protein serine/threonine kinase activity | 6 | 1.60E−04 |
| GO:0003682 | Chromatin binding | 5 | 0.002085892 |
| GO:0004672 | Protein kinase activity | 5 | 0.001526096 |
| GO:0019901 | Protein kinase binding | 5 | 0.001808275 |
| GO:0008017 | Microtubule binding | 4 | 0.003083292 |
| GO:0016301 | Kinase activity | 3 | 0.045588858 |
| GO:0003777 | Microtubule motor activity | 3 | 0.00572113 |
| GO:0008022 | Protein C-terminus binding | 3 | 0.027310961 |
| GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed | 2 | 0.023907438 |
| GO:0004693 | Cyclin-dependent protein serine/threonine kinase activity | 2 | 0.047266579 |
| hsa04110 | Cell cycle | 6 | 1.02E−08 |
| hsa04115 | p53 signaling pathway | 3 | 0.001354866 |
| hsa04914 | Progesterone-mediated oocyte maturation | 3 | 0.002274714 |
Survival analysis
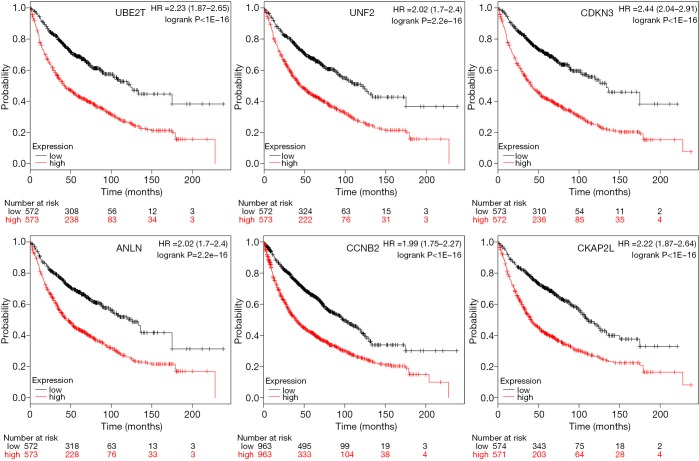
Twenty-seven genes in PPI network were evaluated for their prognostic value on the Kaplan-Meier plotter. Nevertheless, the expression of most genes failed to display a dramatic difference based on the prognosis. Only six genes exhibited their potential in the prediction of survival based on their expression. The OS for NSCLC patients was determined based on the expression level of each gene (low vs. high). As shown in Figure 5, high mRNA expression of UBE2T (HR 2.23, 95% CI: 1.87–2.65, P<1E−16) was associated with a poorer OS for NSCLC patients, and this association was also noted in UNF2 (HR 2.02, 95% CI: 1.7–2.4, P=2.2e−16), CDKN3 (HR 2.44, 95% CI: 2.04–2.91, P<1E−16), ANLN (HR 2.02, 95% CI: 1.7–2.4, P=2.2e−16), CCNB2 (HR 1.99, 95% CI: 1.75–2.27, P<1E−16), CKAP2L (HR 2.22, 95% CI: 1.87–2.64, P<1E−16). It should be noted that the data on KM plotter summarized the survival of all lung cancer patients, not just for NSCLC. Besides, the histological features and combined diseases were not clearly clarified. However, these results still shed light on the importance of these genes to a certain extent, since NSCLC accounts for over 80% lung cancer (3).
Figure 5.
Prognostic estimation of genes including UBE2T, UNF2, CDKN3, ANLN, CCNB2, and CKAP2L in NSCLC patients. NSCLC, non-small cell lung cancer.
Discussion
Regardless of the progression in the treatment of NSCLC, NSCLC remains to be the most common cause of cancer-related death in the past few years. The high mortality of NSCLC is partially due to the lack of sufficient screening methods for early stage NSCLC with high sensitivity and specificity. Therefore, it is imperative to identify the potential biomarkers for the early screening of NSCLC. Fortunately, microarray technology and next-generation sequencing have emerged as pivotal tools to provide comprehensive genetic information of NSCLC samples and to reveal the detailed alterations in the progression of diseases. In this study, the well-established online bioinformatic approaches were employed to investigate the biomarkers for the early NSCLC diagnosis. Three GEO datasets were downloaded from the website and a total of 221 DEGs were identified, including 60 up-regulated genes and 161 down-regulated genes. These up-regulated genes were mainly involved in 4 pathways, among which cell cycle, P53 signaling pathway and ECM-receptor interaction had close relationships with cancer, while the down-regulated genes were mainly enriched in another 6 pathways, among which PPAR signaling pathway, cell adhesion molecules, complement and coagulation cascades, transcriptional misregulation in cancer, and ECM-receptor interaction. In addition, among these DEGs, 27 genes showed the highest degrees in the PPI network. Further survival analysis revealed that 6 DEGs of these 27 modules were closely related to a shorter survival time of NSCLC patients, including UBE2T, UNF2, CDKN3, ANLN, CCNB2, and CKAP2L.
Studies have shown that ubiquitin-conjugating enzyme E2T (UBE2T) overexpression was observed in various types of cancer, including gastric cancer, prostate cancer, hepatocellular cancer, bladder cancer, breast cancer as well as lung cancer (24-29). In a study, UBE2T was suggested to activate the PI3K/Akt signaling pathway, playing a crucial role in the tumor progression, invasion and metastasis. The down-regulation of UBE2T led to the inactivation of PI3K/Akt signaling pathway, which implied UBE2T might be a potential therapeutic target for cancers (30). Another study demonstrates that UBE2T suppresses the progression of hepatocellular carcinoma via ubiquitinating p53. Apart from the investigation of molecular mechanism of UBE2T, high UBE2T expression has also been found to be related to higher pathological grade, advanced TNM stage, tumor vascular invasion, poor OS and poor disease-free survival in two independent cohorts containing 827 patients with HCC (24). Collectively, UBE2T can not only serve as a potential therapeutic target, but also may be used as a promising prognostic marker. In the breast cancer and lung cancer, the UBE2T expression is elevated in around 7% of patients and related to the disease recurrence after surgical resection (26).
Cyclin-dependent kinase inhibitor 3 (CDKN3) has been shown to dephosphorylate CDK2 kinase, thereby preventing the activation of CDK2 kinase. The deletion, mutation, or over-expression of CDKN3 is relevant to the oncogenesis and metastasis (31,32). It has been reported that silencing of CDKN3 gene inhibits the proliferation of epithelial ovarian cancer cells by arresting cell cycle in G1 phase to lower expression of DNA replication and cell cycle related proteins, decreases cell invasion and promotes cells apoptosis. These findings suggest that CDKN3 may serve as a useful potential target for the treatment of ovarian cancer (33,34). Similar results are also reported in the prostate cancer, gastric cancer, hepatocellular cancer, colorectal cancer, and renal cancer (35-37). In addition, there is evidence showing that the CDKN3 mRNA expression also emerges as a biomarker for the survival and prognosis of cervical cancer patients. In a study, CDKN3 mRNA expression was measured in 134 cervical cancer patients and 25 controls by quantitative PCR. Results showed the CDKN3 mRNA expression was on average 6.4-fold higher in cervical cancer than in normal tissues, and patients with higher CDKN3 expression had a shorter OS as compared to those with lower CDKN3 expression (38). A meta-analysis reveals CDKN3 is a prognostic biomarker of lung adenocarcinoma. Higher CDKN3 expression is associated with poorer survival of patients with adenocarcinoma, but not squamous cell carcinoma. In addition, CDKN3 may also be a promising candidate as early diagnostic marker (39).
ANLN, a homologue of anillin (an actin-binding protein in Drosophila), seems to play a significant role in the carcinogenesis. It has been reported that the expression of ANLN as a transcript was significantly up-regulated in the BLCA as compared to normal tissues (40). Bladder cancer patients with elevated ANLN expression have poorer cancer-specific, progression-free and recurrence-free survivals as compared to those with low ANLN expression (41). In addition, ANLN expression is also associated with the survival of breast cancer patients as well as the clinical outcome of patients receiving anthracycline-based chemotherapy. Breast cancer patients with high ANLN expression will experience a poor clinical outcome to anthracycline-based chemotherapy and have shorter survival time (42). In another study, transfection of siRNAs against ANLN in NSCLC cells suppresses ANLN expression, thereby inhibiting cell proliferation. Interestingly, the inhibition of PI3K/AKT pathway in NSCLC cells reduces the ANLN expression in the nuclei. Immunohistochemistry shows that high ANLN expression is associated with a poor survival of NSCLC patients, indicating that ANLN may serve as a prognostic marker (43). ANLN is over-expressed in the colorectal cancer and ANLN expression is positively associated with tumor invasion and enlarged tumor size. Furthermore, Kaplan-Meier survival analysis reveals patients with higher expression of ANLN have a shorter OS (44).
CCNB2 acts as an important mediator during the G2/M transition of cell cycle and is a regulatory subunit in the mitosis-promoting factor (MPF). The cyclin B conjugates with cyclin-dependent kinase (CDK) to form MPF (45). It has been shown that CCNB2 expression is higher in the bladder urothelial carcinoma than in the normal bladder mucosa, especially in the invasive bladder cancer. The down-regulation of CCNB2 expression in the bladder cancer greatly inhibits the cell invasion and metastatic abilities. In vivo study also reveals that blocking CCNB2 expression is able to prolong survival time (46). In addition to the tissue CCNB2 expression, Mo et al. detected circulating CCNB2 mRNA expression in 103 cancer patients, 19 healthy controls, and 40 benign disease patients by real-time quantitative PCR. Their results showed circulating CCNB2 mRNA expression in cancer patients was significantly higher than that in healthy controls and benign disease subjects and related to the cancer stage and metastasis status. The receiver operating characteristic (ROC) analysis showed the area under the curve (AUC) was 0.87 and 0.83 in diagnosis of lung metastasis and gastric metastasis, respectively. These findings suggest that circulating CCNB2 mRNA expression has potential clinical application in screening and monitoring the progression and metastasis of cancers (47). Qian et al. detected the CCNB2 mRNA and protein expressions by RT-PCR and immunohistochemistry in NSCLC and normal lung tissues. Both mRNA and protein expressions of CCNB2 increased in the NSCLC tissues, which was positively related to the clinical stage and metastasis status. In addition, CCNB2 protein over-expression was an independent unfavorable prognostic factor for the OS of NSCLC patients. In summary, CCNB2 overexpression is associated with cancer progression and poor prognosis in NSCLC patients (48). In our study, CCNB2 was also found as a diagnostic marker and may be used as in the early screening of NSCLC.
Currently, little is known about the role of UNF2 and CKAP2L in the cancers. Our study showed UNF2 and CKAP2L could serve as specific targets for the early NSCLC screening. However, there is still discrepancy between the predicted data and the clinical outcomes. Further investigations are needed to confirm the clinical application of both UNF2 and CKAP2L in cancers. Moreover, the simulated survival data from KM plotter is only for reference because it includes both NSCLC and SCLC data. More attention should be paid to the NSCLC alone in future studies. Sub-category analysis is required for clarifying the features of biomarkers in each group. In our future studies, a cohort of NSCLC patients will be recruited to investigate the sensitivity and specificity of these biomarkers in the early screening of NSCLC patients, which may promotes the clinical application of markers.
Conclusions
In summary, a total of 221 DEGs are identified by comparing three GEO profiles, and UBE2T, UNF2, CDKN3, ANLN, CCNB2, and CKAP2L may be the key genes relevant to NSCLC. Comprehensive bioinformatics analysis is a powerful tool for the biomarker screening. The interaction among selected proteins provides important information on the progression of NSCLC, and mechanism underlying the occurrence, development, metastasis and drug resistance of cancers. However, the predictive values of these markers should be confirmed in more studies before their clinical application. The mechanism underlying the activities of these genes is also needed to be elucidated, which is helpful for the delineation of the predicted network of these genes. We optimize the parameters of integrated bioinformatics analysis to screen the potential biomarkers for the early NSCLC diagnosis and we hope our study provides new approaches for the early diagnosis and prediction of NSCLC.
Acknowledgments
We sincerely express our gratitude to Polaris Biology (Shanghai, China) and Dr. Zhi Yang for their heartful and prompt assistance in this study.
Funding: This study was supported by the National Natural Science Fund (81573937) and Shanghai Science and Technology Fund (15401930900, 18401901500).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Sun K, Zheng R, et al. Cancer incidence and mortality in China, 2014. Chin J Cancer Res 2018;30:1-12. 10.21147/j.issn.1000-9604.2018.01.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oser MG, Niederst MJ, Sequist LV, et al. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol 2015;16:e165-72. 10.1016/S1470-2045(14)71180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang R, Wang L, Li Y, et al. FGFR1/3 tyrosine kinase fusions define a unique molecular subtype of non-small cell lung cancer. Clin Cancer Res 2014;20:4107-14. 10.1158/1078-0432.CCR-14-0284 [DOI] [PubMed] [Google Scholar]
- 5.Rothschild SI. Targeted Therapies in Non-Small Cell Lung Cancer-Beyond EGFR and ALK. Cancers (Basel) 2015;7:930-49. 10.3390/cancers7020816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmermann S, Peters S. Going beyond EGFR. Ann Oncol 2012;23 Suppl 10:x197-203. 10.1093/annonc/mds319 [DOI] [PubMed] [Google Scholar]
- 7.Blumenschein GR, Jr, Smit EF, Planchard D, et al. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC) dagger. Ann Oncol 2015;26:894-901. 10.1093/annonc/mdv072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivalland G, Mitchell P. Combined BRAF and MEK inhibition in BRAF-mutant NSCLC. Lancet Oncol 2016;17:860-2. 10.1016/S1470-2045(16)30203-0 [DOI] [PubMed] [Google Scholar]
- 9.Humphrey LL, Deffebach M, Pappas M, et al. Screening for lung cancer with low-dose computed tomography: a systematic review to update the US Preventive services task force recommendation. Ann Intern Med 2013;159:411-20. 10.7326/0003-4819-159-6-201309170-00690 [DOI] [PubMed] [Google Scholar]
- 10.Gainor JF, Tan DS, De Pas T, et al. Progression-Free and Overall Survival in ALK-Positive NSCLC Patients Treated with Sequential Crizotinib and Ceritinib. Clin Cancer Res 2015;21:2745-52. 10.1158/1078-0432.CCR-14-3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solomon BJ MT, Kim DW, Wu YL, Nakagawa K, Mekhail T, Felip E, Cappuzzo F, Paolini J, Usari T, et al. First-Line Crizotinib versus Chemotherapy in ALK-Positive Lung Cancer. N Engl J Med 2015;373:1582. [DOI] [PubMed] [Google Scholar]
- 12.Ju L, Han M, Zhao C, et al. EGFR, KRAS and ROS1 variants coexist in a lung adenocarcinoma patient. Lung Cancer 2016;95:94-7. 10.1016/j.lungcan.2016.03.005 [DOI] [PubMed] [Google Scholar]
- 13.Arrieta O, Villarreal-Garza C, Martinez-Barrera L, et al. Usefulness of serum carcinoembryonic antigen (CEA) in evaluating response to chemotherapy in patients with advanced non small-cell lung cancer: a prospective cohort study. BMC Cancer 2013;13:254. 10.1186/1471-2407-13-254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma S, Shen L, Qian N, et al. The prognostic values of CA125, CA19.9, NSE, AND SCC for stage I NSCLC are limited. Cancer Biomark 2011-2012;10:155-62. 10.3233/CBM-2012-0246 [DOI] [PubMed] [Google Scholar]
- 15.Qin HF, Qu LL, Liu H, et al. Serum CEA level change and its significance before and after Gefitinib therapy on patients with advanced non-small cell lung cancer. Asian Pac J Cancer Prev 2013;14:4205-8. 10.7314/APJCP.2013.14.7.4205 [DOI] [PubMed] [Google Scholar]
- 16.Alsabti EA, Kamel A. Carcinoembryonic antigen (CEA) in patients with malignant and non-malignant diseases. Neoplasma 1979;26:603-9. [PubMed] [Google Scholar]
- 17.Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 2013;41:D991-5. 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu S, Wu F, Jiang Z. Identification of hub genes, key miRNAs and potential molecular mechanisms of colorectal cancer. Oncol Rep 2017;38:2043-50. 10.3892/or.2017.5930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 20.Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. 10.1186/1471-2105-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lánczky A, Nagy A, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat 2016;160:439-46. 10.1007/s10549-016-4013-7 [DOI] [PubMed] [Google Scholar]
- 24.Liu LP, Yang M, Peng QZ, et al. UBE2T promotes hepatocellular carcinoma cell growth via ubiquitination of p53. Biochem Biophys Res Commun 2017;493:20-7. 10.1016/j.bbrc.2017.09.091 [DOI] [PubMed] [Google Scholar]
- 25.Yu H, Xiang P, Pan Q, et al. Ubiquitin-Conjugating Enzyme E2T is an Independent Prognostic Factor and Promotes Gastric Cancer Progression. Tumour Biol 2016;37:11723-32. 10.1007/s13277-016-5020-3 [DOI] [PubMed] [Google Scholar]
- 26.Perez-Peña J, Corrales-Sanchez V, Amir E, et al. Ubiquitin-conjugating enzyme E2T (UBE2T) and denticleless protein homolog (DTL) are linked to poor outcome in breast and lung cancers. Sci Rep 2017;7:17530. 10.1038/s41598-017-17836-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen M, Kwon Y, Wang Y, et al. Elevated expression of UBE2T exhibits oncogenic properties in human prostate cancer. Oncotarget 2015;6:25226-39. 10.18632/oncotarget.4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ueki T, Park JH, Nishidate T, et al. Ubiquitination and downregulation of BRCA1 by ubiquitin-conjugating enzyme E2T overexpression in human breast cancer cells. Cancer Res 2009;69:8752-60. 10.1158/0008-5472.CAN-09-1809 [DOI] [PubMed] [Google Scholar]
- 29.Gong YQ, Peng D, Ning XH, et al. UBE2T silencing suppresses proliferation and induces cell cycle arrest and apoptosis in bladder cancer cells. Oncol Lett 2016;12:4485-92. 10.3892/ol.2016.5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Leng H, Chen H, et al. Knockdown of UBE2T Inhibits Osteosarcoma Cell Proliferation, Migration, and Invasion by Suppressing the PI3K/Akt Signaling Pathway. Oncol Res 2016;24:361-9. 10.3727/096504016X14685034103310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeh CT, Lu SC, Chen TC, et al. Aberrant transcripts of the cyclin-dependent kinase-associated protein phosphatase in hepatocellular carcinoma. Cancer Res 2000;60:4697-700. [PubMed] [Google Scholar]
- 32.Li Y, Yang Q, Guan H, et al. ZNF677 Suppresses Akt Phosphorylation and Tumorigenesis in Thyroid Cancer. Cancer Res 2018;78:5216-28. 10.1158/0008-5472.CAN-18-0003 [DOI] [PubMed] [Google Scholar]
- 33.Li T, Xue H, Guo Y, et al. CDKN3 is an independent prognostic factor and promotes ovarian carcinoma cell proliferation in ovarian cancer. Oncol Rep 2014;31:1825-31. 10.3892/or.2014.3045 [DOI] [PubMed] [Google Scholar]
- 34.Zhang LP, Li WJ, Zhu YF, et al. CDKN3 knockdown reduces cell proliferation, invasion and promotes apoptosis in human ovarian cancer. Int J Clin Exp Pathol 2015;8:4535-44. [PMC free article] [PubMed] [Google Scholar]
- 35.Dai W, Miao H, Fang S, et al. CDKN3 expression is negatively associated with pathological tumor stage and CDKN3 inhibition promotes cell survival in hepatocellular carcinoma. Mol Med Rep 2016;14:1509-14. 10.3892/mmr.2016.5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng M, Wang J, Chen Y, et al. Silencing cyclin-dependent kinase inhibitor 3 inhibits the migration of breast cancer cell lines. Mol Med Rep 2016;14:1523-30. 10.3892/mmr.2016.5401 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Li Y, Ji S, Fu LY, et al. Knockdown of Cyclin-Dependent Kinase Inhibitor 3 Inhibits Proliferation and Invasion in Human Gastric Cancer Cells. Oncol Res 2017;25:721-31. 10.3727/096504016X14772375848616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrón EV, Roman-Bassaure E, Sanchez-Sandoval AL, et al. CDKN3 mRNA as a Biomarker for Survival and Therapeutic Target in Cervical Cancer. PLoS One 2015;10:e0137397. 10.1371/journal.pone.0137397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zang X, Chen M, Zhou Y, et al. Identifying CDKN3 Gene Expression as a Prognostic Biomarker in Lung Adenocarcinoma via Meta-analysis. Cancer Inform 2015;14:183-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang S, Nguyen LH, Zhou K, et al. Knockdown of Anillin Actin Binding Protein Blocks Cytokinesis in Hepatocytes and Reduces Liver Tumor Development in Mice Without Affecting Regeneration. Gastroenterology 2018;154:1421-34. 10.1053/j.gastro.2017.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng S, Yu X, Ma C, et al. Transcriptome sequencing identifies ANLN as a promising prognostic biomarker in bladder urothelial carcinoma. Sci Rep 2017;7:3151. 10.1038/s41598-017-02990-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z, Chen J, Zhong MZ, et al. Overexpression of ANLN contributed to poor prognosis of anthracycline-based chemotherapy in breast cancer patients. Cancer Chemother Pharmacol 2017;79:535-43. 10.1007/s00280-017-3248-2 [DOI] [PubMed] [Google Scholar]
- 43.Ogata T, Teshima T, Inaoka M, et al. Carbon ion irradiation suppresses metastatic potential of human non-small cell lung cancer A549 cells through the phosphatidylinositol-3-kinase/Akt signaling pathway. J Radiat Res 2011;52:374-9. 10.1269/jrr.10102 [DOI] [PubMed] [Google Scholar]
- 44.Wang G, Shen W, Cui L, et al. Overexpression of Anillin (ANLN) is correlated with colorectal cancer progression and poor prognosis. Cancer Biomark 2016;16:459-65. 10.3233/CBM-160585 [DOI] [PubMed] [Google Scholar]
- 45.Nam HJ, van Deursen JM. Cyclin B2 and p53 control proper timing of centrosome separation. Nat Cell Biol 2014;16:538-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lei CY, Wang W, Zhu YT, et al. The decrease of cyclin B2 expression inhibits invasion and metastasis of bladder cancer. Urol Oncol 2016;34:237.e1-10. 10.1016/j.urolonc.2015.11.011 [DOI] [PubMed] [Google Scholar]
- 47.Mo ML, Chen Z, Li J, et al. Use of serum circulating CCNB2 in cancer surveillance. Int J Biol Markers 2010;25:236-42. 10.5301/JBM.2010.6088 [DOI] [PubMed] [Google Scholar]
- 48.Qian X, Song X, He Y, et al. CCNB2 overexpression is a poor prognostic biomarker in Chinese NSCLC patients. Biomed Pharmacother 2015;74:222-7. 10.1016/j.biopha.2015.08.004 [DOI] [PubMed] [Google Scholar]