Abstract
Background
Histoplasmosis pulmonary nodules often present in computed tomography (CT) imaging with characteristics suspicious for lung cancer. This presents a work-up decision issue for clinicians in regions where histoplasmosis is an endemic fungal infection, when a nodule suspicious for lung cancer is detected. We hypothesize the application of radiomic features extracted from pulmonary nodules and perinodular parenchyma could accurately distinguish between suspicious histoplasmosis lung nodules and non-small cell lung cancer (NSCLC).
Methods
A retrospective clinical cohort of pulmonary nodules with a confirmed diagnosis of histoplasmosis or NSCLC was collected from the University of Iowa Hospitals and Clincs. Radiomic features were extracted describing characteristics of the nodule and perinodular parenchyma regions and used to build a machine learning tool. These cases were assessed by four expert clinicians who gave a blinded risk prediction for NSCLC. Tool and observer performance were assessed by calculating the area under the curve for the receiver operating characteristic (AUC-ROC) and interclass correlation coefficient (ICC).
Results
A cohort of 71 subjects with confirmed histopathology (40 NSCLC, 31 histoplasmosis) were case-matched based on age, sex, and smoking history. Superior performance (AUC-ROC =0.89) was demonstrated using leave-one-subject out validation in the tool that incorporated radiomics from the nodule and perinodular parenchyma region extended to 100% nodule diameter. Observers had perfect intra-repeatability (ICC =1.0) and demonstrated fair inter-reader variability (ICC =0.52).
Conclusions
Radiomics have potential utility in the challenging task of differentiation between lung cancer and histoplasmosis. Expert clinician readers have high intra-repeatability but demonstrated inter-reader variability which could provide context for a supplemental radiomics-based tool.
Keywords: Granuloma, machine learning, perinodular, risk assessment
Introduction
Histoplasmosis is a fungal infection endemic to parts of the Americas and Caribbean, with cases reported world-wide from Southern Europe, Southeast Asia, Central Africa, and Oceania (1). This disease often presents as a pulmonary nodule (granulomas) via radiographic imaging with X-ray or computed tomography (CT) with attributes resembling lung cancer including laminated calcific rings on CT and increased avidity on fluorodeoxyglucose-positron emissions tomography (FDG-PET) (2,3). As CT is becoming more widely used for the detection and management of pulmonary nodules, this presents a clinical challenge to physicians in endemic regions when faced with decisions for patient immediate and follow-up care. Difficulty distinguishing between benign granulomas and malignant nodules could lead to either a delay in treatment for lung cancer and/or unnecessary invasive diagnostic/intervention procedures for histoplasmosis. While severe cases can result in life-threatening conditions and morbidity, the majority of patients with pulmonary histoplasmosis present with mild to moderate disease, which often resolves without treatment (4). Additionally, the higher incidence of suspicious lung nodules in regions of endemic histoplasmosis may result in increased variability in clinician interpretation.
There is little prior work on identifiable qualitative or quantitative imaging features that are indicative of a histoplasmosis nodule (3). Typically, nodule composition and size are used as the main criteria for quantifying cancer risk (5,6). However, traditional nodule follow-up guidelines such as the Fleischner Society Guidelines for pulmonary nodule management may not apply to populations in endemic regions as malignancy rates may be lower in larger nodules. Therefore, less aggressive interventions are more appropriate (7). Case reports from human observers have detailed some potential radiological characteristics that could assist in the imaging-based determination of histoplasmosis in suspicious lung lesions (8-12). Furthermore, the experience and location of training for clinicians may play a part in follow-up management biases. A small trend towards lower-false positives in lung cancer screening from clinicians at institutions in areas endemic to histoplasmosis infection was seen in a retrospective assessment of the National Lung Cancer Screening Trial (13).
The ability to distinguish histoplasmosis from lung cancer could be enhanced by extending information extraction from CT beyond nodule size to include more advanced radiomic features. Such automatically extracted features have been used previously to develop classification tools for lung nodule applications including differentiation between malignant-benign nodules (14-20). We hypothesize that the application of radiomic features from the nodule and surrounding perinodular parenchyma could accurately distinguish between suspicious histoplasmosis lung nodules and non-small cell lung cancer (NSCLC). This study evaluates the utility of top predictive radiomic features for the distinction of histoplasmosis from NSCLC in suspicious pulmonary nodules and compares the performance to performance from four blinded clinician observer predictions.
Methods
Study population
Subjects included were part of a larger cohort collected retrospectively with from the University of Iowa Hospitals and Clinics, located in a region endemic for Histoplasmosis (21,22). With Institutional Board Approval (IRB #201202740), radiology reports from thoracic CT scans were text searched for the terms “pulmonary nodule” or “lung nodule”. The electronic medical records (Epic, WI, USA) from identified patients were manually searched for inclusion criteria of having diagnosis of pulmonary nodule through histopathology and CT imaging of solitary pulmonary nodule (4–30 mm) prior to diagnosis. A subject was defined as having pulmonary histoplasmosis or lung cancer if the pathology examination of the lung biopsy (obtained by surgery, bronchoscopy, or CT guided needle biopsy) showed the presence of granulomas or malignant cells compatible with NSCLC respectively. These subjects were matched based on age, sex, and smoking history.
Machine learning tool application
A pipeline for machine learning tool development, recently published by Uthoff et al., was applied with slight modifications to the features to accommodate the high variability in CT acquisition protocol from the retrospective, clinically acquired data (17). To summarize, the nodule and surrounding parenchyma were segmented semi-automatically using a seed-click method as described in Mukhopadhyay et al. (23). The perinodular region identified was segmented into rings that were nodule size-standardized through a nodule mask dilation procedure at 0%, 25%, 50%, 75%, and 100% the nodule diameter for five candidate tools, Nodule, Margin, Immediate, Extended, and Extended+ respectively. One hundred and one quantitative imaging characteristics describing the intensity and 2D texture were extracted from the nodule and perinodular regions; 17 features describing border, size, and shape features were also extracted from the nodule mask. Highly correlated features were clustered using k-medoids clustering, and the resulting medoids were sent through information theory-based feature set selection. The selected feature set was used to build an ensemble of artificial neural networks (ENNs) to differentiate between Histoplasmosis and NSCLC using leave-one-subject-out cross-validation for performance measure assessment.
Observer assessment
We performed a controlled observer study on the full cohort of 71 plus 29 repeated cases (total of 100 cases provided to the observer) to examine the inter- and intra- observer variability. Four observers (2 radiologists, 2 pulmonologists) were each provided de-identified CT data and accompanying basic clinical information in a manner blinded to diagnosis. The clinical information provided included, subject age, sex, FDG-PET avidity, and if the radiology report noted the presence of cavitation or calcification. The observers were asked to provide a categorical risk (low, medium, high) for NSCLC and a continuous analog risk between 0 (likely histoplasmosis) and 1 (likely NSCLC).
Statistical assessment and performance measures
Machine learning and observer continuous analog risk assessment performance were measured using the area under the curve for the receiver operating characteristic (AUC-ROC) (Delong) and Youden’s J statistic. McNemar’s test was used to compare binary classification differences. Interclass correlation coefficient (ICC) was used as the assessment of consistency or reproducibility of continuous (0–1) risk made by different observers on the same nodule, the guidelines put forth by Cicchetti were used for interpretation (24). Weighted Cohen Kappa and percent agreement were used to assess the categorical agreement among readers (25).
Results
Matching reduces demographic and size bias in cohort
A total of 151 suitable subjects with histopathology confirmed diagnosis (49 histoplasmosis, 102 NSCLC) were retrospectively identified from the University of Iowa. Cases were matched between histoplasmosis and NSCLC based on subject: (I) sex; (II) age within ± 3-years; (III) self-reported smoking history. As pack-years were significantly different between groups and the accurate collection of pack-year information is difficult in long-term smokers, smoking history was split into three categories (I) never smokers; (II) <30 pack-year history—not smoking eligible for low-dose CT screening; (III) ≥30 pack-year history—smoking eligible for low-dose CT screening. This resulted in 71 unique subjects (31 histoplasmosis, 40 NSCLC) and 94 total matches (some subjects matched to more than one other subject). Table 1 indicates demographical variables for the matched cohort. No statistical demographic difference was found between diagnosis groups and nodule size was not significant (P=0.40).
Table 1. Demographics of matched cohort along with p-value comparison between histoplasmosis and NSCLC.
| Demographic Variable | Histoplasmosis | NSCLC | p |
|---|---|---|---|
| N | 31 | 40 | – |
| Sex | 23 F:8 M | 23 F:17 M | 0.14 |
| Age (years), mean, range | 57±8, 46–76 | 59±7.6, 47–79 | 0.12 |
| BMI, mean, range | 30.7±8.2, 19.1–51.8 | 27.9±8.2, 16.0–44.5 | 0.11 |
| Pack-years, mean, range | 14.5±17.8, 0–66 | 26.5±26.8, 0–92 | 0.03 |
| Smoking tertiary | |||
| Non | 10 | 7 | 0.1 |
| NSE | 16 | 18 | |
| SE | 5 | 15 | |
| Location | 9 | 20 | 0.05 |
| RUL | 9 | 20 | 0.05 |
| RML | 4 | 2 | |
| RLL | 11 | 4 | |
| LUL | 5 | 10 | |
| LLL | 2 | 4 | |
| Nodule diameter, mean, range | 11.4±3.5, 4.7–20.6 | 12.1±3.3, 4.4–17.1 | 0.40 |
NSCLC, non-small cell lung cancer; N, number of subjects; F, female; M, male; BMI, body mass index; Non, never smoker; NSE, less than 30 pack-year history; SE, 30 or greater pack-year history; RUL, right upper lobe; RML, right middle lobe; RLL, right lower lobe; LUL, left upper lobe; LLL, left lower lobe.
Information theory-based feature set selection illustrates features from the parenchyma are informative of disease
Following a rule of thumb of one feature per five training subjects, a maximum of 14 features were allowed for the development of the candidate tools. The four parenchymal inclusion tools used between 9 and 12 features while the Nodule tool used 10 (Table 2). Neither RECIST diameter nor nodule volume was selected in any candidate tools but the water-equivalent diameter was selected in the Nodule and Margin tools. No single feature was selected in all five candidate tools. Parenchymal ring Long Run High Gray-Level Emphasis was selected in all candidates with perinodular signal inclusion. There was little overlap in the selected nodule vs. parenchyma features (i.e., nodular Low Gray-Level Zone Emphasis and parenchymal ring Low Gray-Level Zone Emphasis were not both selected in the same tool) indicating that the signals detected from these regions are unique. In all, 42 of the total 51 features selected in at least one of the candidate tools were textural with the most candidate translational features (included in multiple tools) being run-length or size-zone features.
Table 2. Selected features from among the five candidate tools.
| Feature type | Feature name | Nodule | Margin | Immediate | Extended | Extended+ |
|---|---|---|---|---|---|---|
| Nodule | ||||||
| BS | Mean Sphere Difference | 4 | ||||
| SH | Sphericity | 1 | 10 | |||
| SZ | H2O Equivalent Diameter | 3 | 8 | |||
| IH | 25th HU Percentile | 9 | 7 | 7 | ||
| LTEM | Mean-5 | 6 | ||||
| NGTD | Kurtosis-16 | 8 | 10 | 3 | ||
| Strength | 10 | 3 | 9 | 8 | ||
| GLRL | Gray-Level Variance | 1 | 1 | |||
| High Gray-Level Run Emphasis | 5 | 4 | 11 | 5 | ||
| GLSZ | Gray-Level Non-Uniformity | 2 | 7 | |||
| Low Gray-Level Zone Emphasis | 2 | |||||
| Small Zone Emphasis | 7 | |||||
| Zone-Size Non-Uniformity | 3 | 2 | ||||
| Small Zone High Gray-Level Emphasis | 7 | 8 | ||||
| Parenchyma | ||||||
| IH | Full-Width-At-Half-Maximum | 5 | ||||
| LTEM | Mean-1 | 11 | 9 | |||
| NGTD | Busyness | 1 | 9 | |||
| Contrast | 9 | 4 | 6 | 2 | ||
| GLRL | Gray-Level Non-Uniformity | 12 | 6 | 8 | 6 | |
| Low Gray-Level Run Emphasis | 4 | |||||
| Long Run High Gray-Level Emphasis | 5 | 5 | 3 | 1 | ||
| GLSZ | Low Gray-Level Zone Emphasis | 2 | ||||
| Small Zone High-Gray Level Emphasis | 6 | 4 | ||||
| Total | 10 | 12 | 11 | 9 | 10 | |
BS, border sphere comparison; SH, shape; Sz, size; IH, intensity histogram; LTEM, law’s texture energy measure; NGTD, neighborhood gray-tone difference; GLRL, gray-level run length; GLSZ, gray-level size zone.
Tool assessment performance improved with the inclusion of surrounding parenchyma
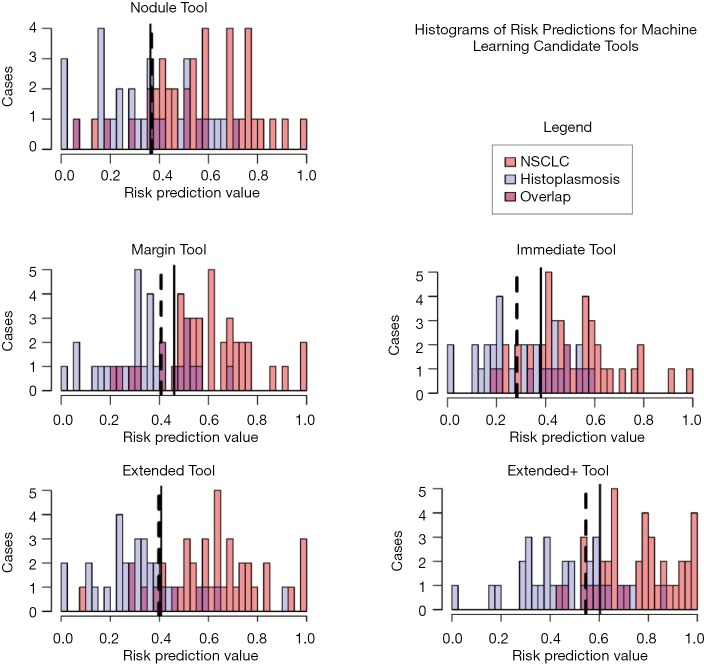
The five candidate tools were run through feature-set selection and development on ENN using leave-one-out; Figure 1 demonstrates the range of predictions from the five candidate tools. The performance measures are summarized in Table 3. Pairwise Delong assessment showed no statistical difference between ROC curves (P value between 0.12–0.99). Of the candidate tools, the Extended+ (incorporating parenchymal ring at 100% diameter) achieved the highest AUC-ROC using leave-one-out (AUC-ROC =0.89) utilizing ten features, 4 from the nodule and 6 from the perinodular parenchyma. The top-ranked feature was the parenchymal Long Run High Gray-Level Emphasis. Applying the Youden threshold (0.60), the Extended+ tool achieved 84% specificity and 83% sensitivity. Applying the 90%-sensitivity threshold (0.55) to the Extended+ tool achieved a specificity of 61%.
Figure 1.
Overlay histogram visualization of five candidate tools applied to the Histoplasmosis-NSCLC cohort. Solid lines indicate Youden threshold, dashed lines indicate threshold for 90% sensitivity. NSCLC, non-small cell lung cancer.
Table 3. Performance measures of machine learning candidate tools applied using leave-one-subject out cross validation and performance measures of quantitative (analog) risk assessment from the four human readers.
| Assessment Method | Expert | AUC-ROC | Youden | Specificity | Sensitivity |
|---|---|---|---|---|---|
| ML-tool candidate | Nodule | 0.78 | 0.36 | 0.61 | 0.90 |
| Margin | 0.86 | 0.46 | 0.77 | 0.85 | |
| Immediate | 0.79 | 0.38 | 0.65 | 0.80 | |
| Extended | 0.88 | 0.41 | 0.81 | 0.88 | |
| Extended+ | 0.89 | 0.60 | 0.84 | 0.83 | |
| Observer (Analog) | Reader 1 | 0.76 | 0.63 | 0.62 | 0.88 |
| Reader 2 | 0.80 | 0.67 | 0.73 | 0.79 | |
| Reader 3 | 0.74 | 0.74 | 0.88 | 0.65 | |
| Reader 4 | 0.65 | 0.75 | 0.31 | 0.94 | |
| Average reader | 0.74 | NA | 0.63 | 0.82 |
ML, machine learning; AUC-ROC, area-under-curve of receiver-operator characteristic; NA, not applicable.
Observer categorical and continuous quantitative assessments demonstrate variation between readers
All four readers agreed on the categorical risk (low, medium or high) in 23 of the 71 cases; of those, 5 were scored low, 3 medium, and 15 high (Figure 2). All agreed-upon low-risk scored nodules were benign and all agreed-upon high-risk nodules were NSCLC. This indicates that for 38% of the NSCLC cases there was high confidence in lung cancer classification among all observers and for 16% of the histoplasmosis cases there was high confidence in benign classification among all observers. Categorical assessment agreement was an average of 0.49 in weighted Cohen Kappa for all readers. The pulmonologists had higher level of agreement between each other (0.62) and the radiologists had lower level of agreement (0.36), however, some of the ‘disagreement’ could be due to the risk-aversity of readers (i.e., one radiologist decided on an ‘extreme’ category—low/high while the other chose medium). In fact, categorical percentage of agreement hovered at random chance 32.4%—given there are three categories there is the unbiased draw likelihood that any rater will agree with another 33% of the time. On the quantitative assessment of risk, the ICC was 0.52 between all four raters indicating a fair level of agreement between raters. While differences existed between readers (inter-reader), assessment of the intra-reader differences in categorical assessments showed readers were 100% repeatable in category assignment. Continuous quantitative assessment was slightly less repeatable with a range in the difference between <0.01 and 0.13.
Figure 2.
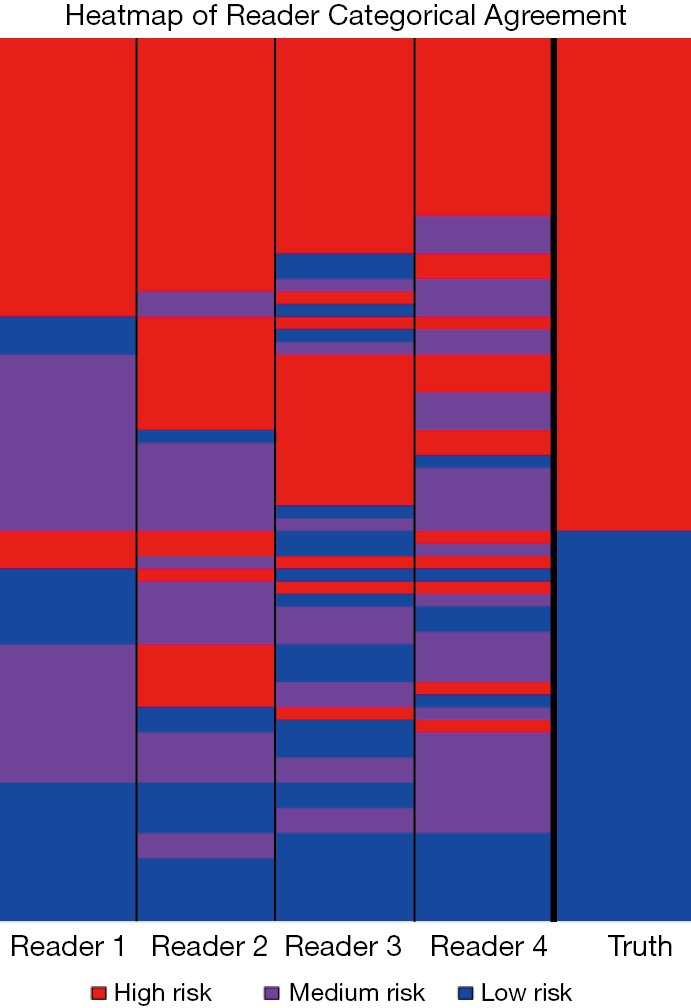
Heatmap of categorical agreement among readers.
Observer continuous quantitative assessment demonstrates benefit of human observer, potential for simple self-trained tool
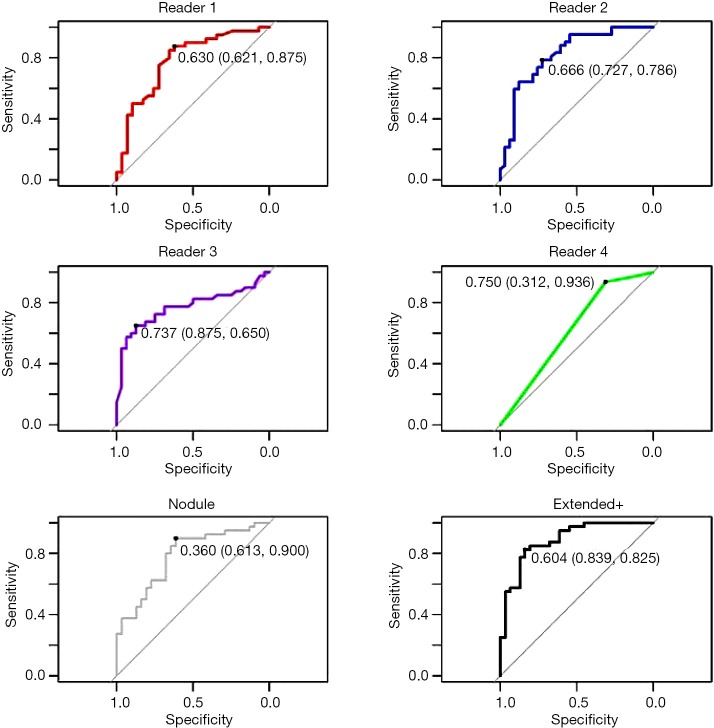
The observers’ continuous quantitative risk scores (between 0–1) were assessed for direct comparison to the machine learning tool. Observers ranged in AUC-ROC =0.65–0.80 (power 0.54–0.99) and Youden-threshold based sensitivity and specificity between 0.65–0.94 and 0.31–0.88 respectively (Table 3, Figure 3). Youden-threshold based sensitivity indicates the potential aid a simple ‘self-training’ tool could be assistive with compared to the categorical threshold. When compared to the average performance across all four observer readings, the Extended+ tool had comparable sensitivity with improved specificity.
Figure 3.
Charts display the Youden Threshold (specificity, sensitivity).
Discussion
In this study we observed that clinician’s intra-observer risk predictions are stable, however, we recorded high inter-observer variability in these challenging cases. This disparity could be due to personal subjective biases (level of conservativeness, experience in endemic histoplasmosis region) and could result in a lack of standardization in the way lung nodule cases are managed in the clinic. As this variability exists, a test or tool that would lend additional information about subject risk could be beneficial to increasing standardization of care between providers. Recently, a minimally invasive biomarker using serum enzyme immunoassay analysis has been developed to address this problem (sensitivity 42%, specificity 85%) (26). A radiomic approach is potentially advantageous as it utilizes existing data – requiring no additional sample collection or work-up process. We found that the Extended+ prediction tool (sensitivity 83%, specificity 84%) had higher performance than the averaged clinician practicing in an endemic region (sensitivity 82%, specificity 63%). Using a deterministic prediction method could present a valuable support tool for clinicians to reduce inter-observer variability in suspicious nodule management in locations with endemic histoplasmosis.
In this study, we have demonstrated the transferability of the previously described pipeline by Uthoff et al. to a cohort of retrospectively collected clinical CT scans (17). This study applied machine learning techniques previously developed and tuned on a large research cohort with malignant/benign distinction, to the problem of histoplasmosis/NSCLC distinction. It incorporated consideration of the impact from surrounding perinodular region’s signal on this distinction. It further compared predictive results from the model to observers with significant experience in distinction of histoplasmosis from other nodule forming pulmonary diseases. The results indicate high observer performance (sensitivity 82%, specificity 63%), however, broad diversity across readers (range: sensitivity 64–94%, specificity 31–88%). In this study, we demonstrate the model developed on a small cohort and utilizing only CT extracted features, could achieve high predictive performance (AUC-ROC 0.89, sensitivity 83%, specificity 84%) in line with the average observer, with the added benefit of no subjective variability.
To our knowledge, this is the first study to interrogate quantitative imaging features from histoplasmosis nodules. In an early publication, Ayers and Huang compared CT-based features for the differentiation between benign and malignant lung tumors and included a case study for a histoplasmosis subject who displayed a pattern similar to other benign cases (27). Gazzoni et al. recently presented a review of the radiological presentations of pulmonary nodular fungal infections describing of centralized or laminated calcification in histoplasmosis and the presence of bilateral and mediastinal hilar lymph node enlargement (3). However, these imaging characteristics may not be present in all histoplasmosis pulmonary nodules. Rolston et al. showed in a 3-year retrospective analysis of subjects referred to their endemic-region institution for biopsy based on chest radiography findings found no clinical or radiological features indicative of infection versus lung cancer (12).
Previously it has been shown that incorporation of the perinodular signal significantly improves discriminatory ability on a large cohort of CT scans with less variation in acquisition protocol (17). In the current study, the inclusion of the perinodular signal also improved the performance at a level that approached significance between the Nodule and Extended/Extended+ tools. In the tool with the highest AUC-ROC, Extended+, six of the features were selected from the perinodular region including five texture and one intensity histogram measure. This investigation which incorporated various perinodular zones revelated some insight into feature stability. Several features—Contrast, Gray-Level Non-Uniformity in Runs, Long Run High Gray-Level Emphasis—were selected in all four candidate tools incorporating perinodular features, indicating this textural signal is beneficial to the classification problem at multiple zones of the size-standardization amount. Several features were selected in only the Extended+ tool, including the perinodular Small Zone High Gray-Level Emphasis and Full-Width-at-Half-Maximum, indicating the usefulness of these features at a greater distance away from the lung nodule.
It is likely that increased performance could be achieved with this method with CT protocol standardization. For example, in the retrospective clinical cases used in this investigation, subjects were not coached to a particular lung volume and differences in lung inflation likely reduce signal integrity of the perinodular features extracted (28). Also, a large proportion (66/71) subjects in this cohort had iodine contrast-enhanced scans which likely affect the values of some measures—particularly intensity histogram features (29). As slice thickness was much larger in this cohort (mean =3.30 mm), we adapted the feature extraction pipeline for two-dimensional textural features extracted from the slices containing nodule and perinodular region. It has been previously shown in the classification of lung cancer brain metastases that three-dimensional textural features are more descriptive than two-dimensional features (30). The CT data used here were acquired between October 2007 and December 2014; with increased technological advances making their way into the clinical setting, such as faster-acquisition, low-dose CT (LDCT) and improved reconstruction algorithms, there will be improvements over time with the z-plane thickness which would make three-dimensional features a more powerful option.
While the observers did differ in their categorical and continuous risk scores on a per nodule basis, they did perform well in distinguishing histoplasmosis to NSCLC and their categorical risk scores were 100% repeatable on the intra-reader analysis. The experience level of these individuals is high in the given task while the machine learning tool was built only using the looped leave-one-out training cases (subjects =70, per run) the number of true histoplasmosis and NSCLC cases the observers have been trained on is orders of magnitude larger. We did see potential improvement in observers when using a simple linear discriminant (Youden threshold of risk score) implying that it may improve observer (intra) repeatability and consistency/accuracy to use a continuous risk that has been tuned to their own level of ‘risk percentage application’ as opposed to categorical assessment.
This study contained limitations. First, it was a retrospective study collected from a clinical cohort, leading to diversity in scanning protocols—including contrast enhancement—and selection biases. Second, the sample size collected was small (n=71) and due to case-control matching study design, the clinical proportions of the two disease states were not maintained. In true clinical practice, it is unknown the actual rate of pulmonary histoplasmosis as often patients with pulmonary histoplasmosis nodules are not symptomatic and likely many are not definitively diagnosed as histoplasmosis. The histoplasmosis cases in this study were histopathologically diagnosed, such that only cases clinically warranting invasive procedure were included. In practice, clinicians consider multiple factors in determining if a subject’s nodule is worrisome and if a subject is a good candidate for more invasive procedures. In addition, the presentation of data with only a two-class outcome (histoplasmosis versus NSCLC) does not reflect clinical practice, in which multiple benign and malignant categories exist. Interpretation of the Extended+ tool and human readers performance should acknowledge the targeted approach of this study and not infer clinical practice performance from these results. Additionally, the small cohort size limits both the number of features we allowed each machine learning tool to implement in ENN and increases the potential effects of scanning variability and inter-subject biological variance irrelated to pulmonary nodule pathology.
Conclusions
In this study, we have demonstrated the potential utility of a machine learning tool to the challenging task of lung cancer differentiation from histoplasmosis in clinical-quality, case-matched scans. We have further compared the developed tool to four blinded expert readers, demonstrating the potential value of machine learning, particularly in the context of inter-reader variability. Further studies incorporating this machine learning tool in the management algorithm of prospective cohorts in patients with lung nodules from endemic areas of histoplasmosis would be needed to assess its clinical utility.
Acknowledgments
Funding: This work was funded by the American Lung Association (LH-574107). The authors would like to thank Samantha K.N. Dilger, Kimberly Schroeder and Patrick Ten Eyck.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics Committee of University of Iowa Hospitals and Clinics (IRB #201202740), with a waiver of documentation of consent.
Footnotes
Conflicts of Interest: J Uthoff reports grants from American Lung Association, during the conduct of the study. JC Sieren reports personal fees from VIDA Diagnostics, grants from NIH/NHLBI, outside the submitted work. P Nagpal reports a grant from RSNA outside the submitted work.The other authors have no conflicts of interest to declare.
References
- 1.Azar MM, Hage CA. Clinical Perspectives in the Diagnosis and Management of Histoplasmosis. Clin Chest Med 2017;38:403-15. 10.1016/j.ccm.2017.04.004 [DOI] [PubMed] [Google Scholar]
- 2.Dall Bello AG, Severo CB, Guazzelli LS, et al. Histoplasmosis mimicking primary lung cancer or pulmonary metastases. J Bras Pneumol 2013;39:63-8. 10.1590/S1806-37132013000100009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gazzoni FF, Severo LC, Marchiori E, et al. Fungal diseases mimicking primary lung cancer: radiologic-pathologic correlation. Mycoses 2014;57:197-208. 10.1111/myc.12150 [DOI] [PubMed] [Google Scholar]
- 4.Wheat LJ, Freifeld AG, Kleiman MB, et al. Clinical practice guidelines for the management of patients with histoplasmosis: 2007 update by the Infectious Diseases Society of America. Clin Infect Dis 2007;45:807-25. 10.1086/521259 [DOI] [PubMed] [Google Scholar]
- 5.Radiology ACo. Lung CT Screening Reporting and Data System (Lung-RADS). Available online: https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/Lung-Rads
- 6.MacMahon H, Naidich DP, Goo JM, et al. Guidelines for Management of Incidental Pulmonary Nodules Detected on CT Images: From the Fleischner Society 2017. Radiology 2017;284:228-43. 10.1148/radiol.2017161659 [DOI] [PubMed] [Google Scholar]
- 7.Warren WA, Markert RJ, Stewart ED. Pulmonary nodule tracking using chest computed tomography in a histoplasmosis endemic area. Clin Imaging 2015;39:417-20. 10.1016/j.clinimag.2014.11.002 [DOI] [PubMed] [Google Scholar]
- 8.Baum GL, Green RA, Schwarz J. Enlarging pulmonary histoplasmoma. Am Rev Respir Dis 1960;82:721-6. [DOI] [PubMed] [Google Scholar]
- 9.Palayew MJ, Frank H. Benign progressive multinodular pulmonary histoplasmosis. A radiological and clinical entity. Radiology 1974;111:311-4. 10.1148/111.2.311 [DOI] [PubMed] [Google Scholar]
- 10.Khoo T, Clarke G, Psevdos G. Lung Cancer Screening Reveals a Nonspiculated Nodule: Histoplasmosis. J Glob Infect Dis 2018;10:226-7. 10.4103/jgid.jgid_92_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye C, Zhang G, Wang J, et al. Histoplasmosis presenting with solitary pulmonary nodule: two cases mimicking pulmonary metastases. Niger J Clin Pract 2015;18:304-6. 10.4103/1119-3077.151075 [DOI] [PubMed] [Google Scholar]
- 12.Rolston KV, Rodriguez S, Dholakia N, et al. Pulmonary infections mimicking cancer: a retrospective, three-year review. Support Care Cancer 1997;5:90-3. 10.1007/BF01262563 [DOI] [PubMed] [Google Scholar]
- 13.Pinsky PF, Gierada DS, Nath PH, et al. National lung screening trial: variability in nodule detection rates in chest CT studies. Radiology 2013;268:865-73. 10.1148/radiol.13121530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dilger SK, Uthoff J, Judisch A, et al. Improved pulmonary nodule classification utilizing quantitative lung parenchyma features. J Med Imaging (Bellingham) 2015;2:041004. 10.1117/1.JMI.2.4.041004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Causey JL, Zhang J, Ma S, et al. Highly accurate model for prediction of lung nodule malignancy with CT scans. Scientific Reports 2018;8:9286. 10.1038/s41598-018-27569-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang P, Park S, Yan R, et al. Added Value of Computer-aided CT Image Features for Early Lung Cancer Diagnosis with Small Pulmonary Nodules: A Matched Case-Control Study. Radiology 2018;286:286-95. 10.1148/radiol.2017162725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uthoff J, Stephens MJ, Newell JD, Jr, et al. Machine learning approach for distinguishing malignant and benign lung nodules utilizing standardized perinodular parenchymal features from CT. Med Phys 2019;46:3207-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhara AK, Mukhopadhyay S, Dutta A, et al. A Combination of Shape and Texture Features for Classification of Pulmonary Nodules in Lung CT Images. J Digit Imaging 2016;29:466-75. 10.1007/s10278-015-9857-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferreira JR, Jr, Oliveira MC, de Azevedo-Marques PM. Characterization of Pulmonary Nodules Based on Features of Margin Sharpness and Texture. J Digit Imaging 2018;31:451-63. 10.1007/s10278-017-0029-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaffar MA, Siddiqui AB, Mushtaq M. Ensemble classification of pulmonary nodules using gradient intensity feature descriptor and differential evolution. Cluster Comput 2018;21:393-407. 10.1007/s10586-017-0876-6 [DOI] [Google Scholar]
- 21.Dilger SKN. Pushing the boundaries: feature extraction from the lung improves pulmonary nodule classification. Univeristy of Iowa, 2016.
- 22.Manos NE, Ferebee SH, Kerschbaum WF. Geographic variation in the prevalence of histoplasmin sensitivity. Dis Chest 1956;29:649-68. 10.1378/chest.29.6.649 [DOI] [PubMed] [Google Scholar]
- 23.Mukhopadhyay S. A Segmentation Framework of Pulmonary Nodules in Lung CT Images. J Digit Imaging 2016;29:86-103. 10.1007/s10278-015-9801-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cicchetti DV. Guidelines, criteria, and rules of thumb for evaluating normed and standardized assessment instruments in psychology. Psychol Assess 1994;6:284-90. 10.1037/1040-3590.6.4.284 [DOI] [Google Scholar]
- 25.Cohen J. Weighted kappa: nominal scale agreement with provision for scaled disagreement or partial credit. Psychol Bull 1968;70:213-20. 10.1037/h0026256 [DOI] [PubMed] [Google Scholar]
- 26.Deppen SA, Massion PP, Blume J, et al. Accuracy of a Novel Histoplasmosis Enzyme Immunoassay to Evaluate Suspicious Lung Nodules. Cancer Epidemiol Biomarkers Prev 2019;28:321-6. 10.1158/1055-9965.EPI-18-0169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayers WR, Huang HK. The use of computerized tomography in the diagnosis of pulmonary nodules. Comput Tomogr 1978;2:55-62. 10.1016/0363-8235(78)90003-0 [DOI] [PubMed] [Google Scholar]
- 28.Oliver JA, Budzevich M, Zhang GG, et al. Variability of Image Features Computed from Conventional and Respiratory-Gated PET/CT Images of Lung Cancer. Transl Oncol 2015;8:524-34. 10.1016/j.tranon.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Kadi OS. Assessment of texture measures susceptibility to noise in conventional and contrast enhanced computed tomography lung tumour images. Comput Med Imaging Graph 2010;34:494-503. 10.1016/j.compmedimag.2009.12.011 [DOI] [PubMed] [Google Scholar]
- 30.Ortiz-Ramón R, Larroza A, Ruiz-Espana S, et al. Classifying brain metastases by their primary site of origin using a radiomics approach based on texture analysis: a feasibility study. Eur Radiol 2018;28:4514-23. 10.1007/s00330-018-5463-6 [DOI] [PubMed] [Google Scholar]