Abstract
Background
Extracellular vesicles (EV) have been proven to contain double-stranded DNA reflecting the mutational status of the parental tumor cells in non-small cell lung cancer (NSCLC), which can be translated into clinically useful EV-based liquid biopsy for Epidermal growth factor receptor (EGFR) genotyping using bronchoalveolar lavage fluid (BALF) obtained from tumor site.
Methods
Patients subjected for an initial lung cancer work-up underwent bronchoscopy and BALF was obtained from tumor site. After isolating EVs from BALF by ultracentrifugation, EV-derived DNA (EV DNA) was extracted for subsequent EGFR genotyping performed through peptide nucleic acid (PNA)-mediated Real-Time PCR. The sensitivity, specificity, and concordance rate of BALF EV-based EGFR genotyping were calculated in comparison to tissue genotyping.
Results
The average sensitivity and specificity of BALF EV-based EGFR genotyping were 76% and 87%, respectively, while the sensitivity significantly increased as the stage progressed. Especially, in stage IV, BALF EV-based EGFR typing identified all tissue-proven EGFR mutant cases (n=31) and detected 6 additional mutant cases. The concordance rate was 79% in stage I, 100% in stage II, 74% in stage III, and 92% in stage IV. As TNM stage advanced, especially in the presence of metastasis, concordance rate significantly increased (P<0.05).
Conclusions
The use of BALF for the collection of EV DNA in lung cancer patients resulted in a highly accurate diagnosis. The establishment of a fast and reliable method to identify target genes using EV DNA illustrated that it can overcome the problems of low sensitivity and instability in using cell-free DNA (cfDNA).
Keywords: Extracellular vesicles (EV), bronchoalveolar lavage fluid (BALF), liquid biopsy, EGFR genotyping, non-small cell lung cancer (NSCLC)
Introduction
Epidermal growth factor receptor (EGFR) genotyping in non-small cell lung cancer (NSCLC), mostly done by tissue-based molecular testing, has been a routine practice when making therapeutic decisions, especially in advanced diseases. However, obtaining adequate tissue for a biopsy is sometimes challenging, as most of advanced NSCLC patients are pathologically diagnosed by small biopsy specimens obtained through either bronchoscopy or percutaneous needle biopsy. Approximately 20–30% of cases rely only on cytology specimen for diagnosis and an insufficient sample is a frequently encountered obstacle (1,2). This inadequate biopsy issue became unavoidable in the era of precision medicine (3). As an alternative, plasma EGFR genotyping using tumor-derived cell-free DNA (cfDNA) has been introduced into the clinical practice (4,5). Although it is an attractive non-invasive option, its clinical usefulness is limited due to varied and low sensitivity caused by low copy number of detectable circulating tumor DNA (ctDNA) in the blood and short half-life of less than 2 hours (6,7).
In order to circumvent these practical issues, we have set up a novel strategy for EGFR genotyping using extracellular vesicles (EV)-derived DNA isolated from the supernatant of bronchoalveolar lavage fluid (BALF). EV are membrane-bound and nanometer-sized particles shed from most types of cells in our body and found in circulation, containing cell-derived biomolecules (e.g., RNA, protein, and metabolite) (8). Recently, we demonstrated that EVs successfully isolated from BALF and pleural effusion (PE) of NSCLC patients contain abundant double-stranded DNA (dsDNA) and that liquid biopsies for EGFR genotyping using BALF and PE EV-derived DNA (EV DNA) are tissue-specific and extremely sensitive when compared with cfDNA (9,10). Other studies also have shown that EV DNA is superior to cfDNA in plasma for the detection of mutations in NSCLC and pancreatic cancer (11,12). Bronchoalveolar lavage (BAL) is a conventional and safe diagnostic technique through the bronchoscope for patients with a variety of pulmonary diseases including lung cancer. BAL has been originally referred to as “liquid lung biopsy,” especially in diffuse interstitial lung diseases, because it is possible to obtain the cellular and non-cellular contents from distal airways and alveoli, directly from the disease-located site (13,14). It is well known that tumor cells shed an abundance of EVs into tumor microenvironment (TME) (8). BAL procedure could be designated as a unique gateway to access TME of lung cancer itself. Therefore, translating EV isolated from BALF into a novel liquid biopsy tool for EGFR genotyping could improve performance by increasing sensitivity and accuracy, compared to liquid biopsy using plasma cfDNA. On these backgrounds, we set up a prospective clinical investigation to verify the usefulness of BALF EV-based EGFR genotyping at the initial diagnostic work-up stage in comparison with standard tissue/cytology-based genotyping and demonstrate a successful translation of EV DNA into clinically useful EGFR genotyping in lung cancer.
Methods
Study population
Among patients diagnosed with NSCLC at Konkuk University Medical Center between October 1, 2016, and December 31, 2017, one hundred thirty-seven patients were included in the analysis. The patients were selected by favorable demographic data for EGFR mutation positivity, such as peripheral location and female while excluding heavy smokers. Bronchoscopic examination was performed at the initial lung cancer work-up stage and the patients with endobronchial obstruction by tumor were excluded. This study was conducted in accordance with the amended Declaration of Helsinki. The study protocol was approved by the institutional review board of Konkuk University Medical Center (KUH1010812), and written informed consents were obtained from all patients. Disease stages were based on the 8th TNM classification criteria (15). Clinical data of the enrolled patients were reviewed and patient demographics and clinical characteristics are detailed in Table 1. Among 137 patients, stage distribution was 26.3% in stage I, 4.4% in stage II, 16.8% in stage III, and 52.6% in stage IV. The distribution of histologic subtype revealed that most cases were adenocarcinoma (81.0%) and 10.2% were NSCLC, and 8.7% not otherwise specified. EGFR mutated cases identified by tissue-based genotyping were 54 patients (39.4%).
Table 1. Patient demographics and clinical characteristics.
| Characteristics | Value (N=137) |
|---|---|
| Age, median (range), years | 70 (59.5–75.0) |
| Sex, No. (%) | |
| Male | 75 (54.7) |
| Female | 62 (45.3) |
| Smoking status, No. (%) | |
| Never-smoker | 64 (46.7) |
| Ex-smoker | 36 (26.3) |
| Current smoker | 37 (27.0) |
| Stage at diagnosis, No. (%) | |
| Stage I | 36 (26.3) |
| Stage II | 6 (4.4) |
| Stage III | 23 (16.8) |
| Stage IV | 72 (52.6) |
| Tissue/cytology sampling method, No. (%) | |
| Bronchoscopy | 33 (24.1) |
| EBUS-TBNA | 18 (13.1) |
| PCNA or PCNB | 27 (19.7) |
| Pleural effusion cell block | 9 (6.6) |
| Excisional biopsy | 4 (2.9) |
| Surgery | 46 (33.6) |
| Tissue EGFR genotype, No. (%) | |
| Wild typea | 83 (60.6) |
| Mutant | |
| Exon 19 deletionb | 32 (23.4) |
| L858Rc | 20 (14.6) |
| G719Xd | 2 (1.5) |
| Histology, No. (%) | |
| Adenocarcinoma | 111 (81.0) |
| Non-small cell lung cancer, NOS | 14 (10.2) |
| Squamous cell carcinoma | 10 (7.3) |
| Large cell carcinoma | 1 (0.7) |
| Sarcomatoid carcinoma | 1 (0.7) |
a, one with Exon 20 insertion; b, one with Exon 19del + T790M; c, one with L858R+C797S, one with L858R+S768I; d, one with G719C+S768I; PCNA, percutaneous needle aspiration; PCNB, percutaneous needle biopsy.
BALF processing and isolation of EV
A sample of 1ml BALF was used for the isolation of EVs. Cells and debris were removed using centrifugation at 1,000 g for 10 min at 4 °C. Cells and debris free BALF were spun in ultracentrifuge tube at 200,000 g for 1 h at 4 °C using a Beckman rotor (Beckman Coulter, Brea, CA, USA). The supernatant was carefully removed and the pellet was resuspended in 200 μL of Phosphate-buffered saline (PBS). The size of purified EV was analyzed using Nanosight NS300 (Malvern Instruments, Worcestershire, UK). The EV fractions in BALF were visualized by negative stain transmission electron microscopy (TEM). For negative stain TEM, purified EVs were fixed in 2% (vol/vol) paraformaldehyde for 5 minutes at room temperature. After fixation, 10 μL EV suspension was applied to formvar-/carbon-coated grids (200 mesh) for 1 minutes and was stained with 2% uranyl acetate. Excess uranyl format was removed using a filter paper, and the grids were examined using a TEM H7600 (Hitachi, Tokyo, Japan) at 80 kV.
EV DNA extraction and EGFR genotyping
Lysis buffer (10 mM Tris-HCl, 20% Triton X-100) was used to lyse EVs and the DNA from EV was purified using the High-Pure PCR Template Preparation Kit (Roche Diagnostics, Mannheim, Germany). The quality and length of the purified DNA were analyzed using a 4200 Tapestion and Genomic DNA ScreenTape (Agilent Technologies, Santa Clara, CA, USA). The concentration and purity of DNA samples were measured using the NanoDrop (Thermo Scientific, Waltham, MA, USA). For detecting EGFR mutations and genotyping, PANAMutyper™ R EGFR kit (Panagene, Daejeon, Korea) and CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, USA) were used. All reactions had a total volume of 25 μL containing 70 ng of template DNA, the primer and peptide nucleic acid (PNA) probe set along with a PCR master mix. PCR and the melting curve step were performed according to the manufacturer’s protocol. Fluorescence was measured on all four channels (FAM, ROX, Cy5, and HEX). With the use of a mutant-type DNA specific PNA detection probe that had a fluorescent dye and quencher, EGFR mutations could be genotyped by melting peak analysis (16,17). EGFR copy number were calculated by quantification standard curve drawn by EGFR internal control Ct value from serial dilution of EGFR standard materials (Horizon Discovery, Cambridge, UK).
Statistical analysis
Categorical variables were summarized by calculating frequencies and percentages. Means, standard deviations, and ranges, including minimal and maximal values, were used to determine numerical variables. For correlation statistics, the Spearman’s correlation test was used. Wilson score method was used to calculate 95% confidence intervals for concordance, sensitivity, and specificity. The agreement of EGFR genotype between BALF EV and tumor tissue/cytology was measured by Cohen’s kappa coefficient. Data were summarized as medians with interquartile range (IQR) with non-normal distribution. We used Pearson chi-square and Fisher’s exact tests to show the significance of differences in EGFR mutation rate, sensitivity trends of BALF EV-based EGFR genotyping and comparison with clinical parameters between the concordant group and discordant group. Sensitivity trends of BALF EV-based EGFR genotyping according to the TNM stage indicate significant differences after the Bonferroni correction was applied. All statistical analyses were carried out using SPSS Statistics version 25.0 (IBM Corp, Chicago, IL, USA), and a P value <0.05 was regarded as statistically significant.
Results
Characterization of isolated EVs and EV DNA from BALF
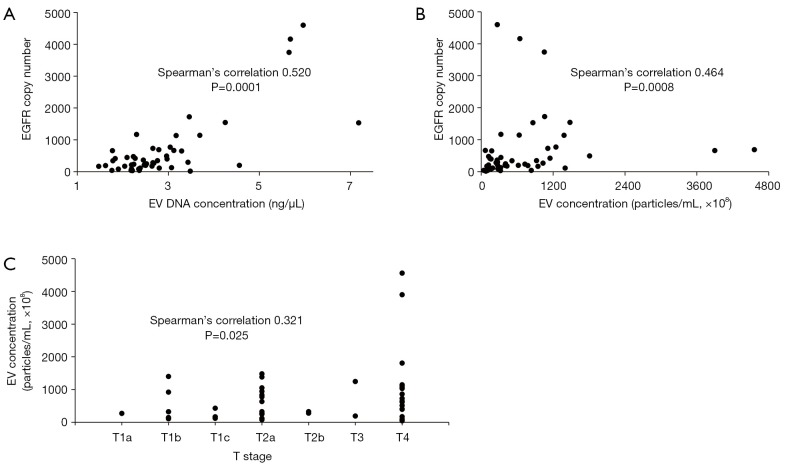
The isolated EVs from BALF were visualized by negative stain TEM (Figure S1). The size and concentration of EVs isolated from BALF were analyzed by Nanosight using nanoparticle tracking analysis (NTA) from randomly selected 49 patients out of 137 (Figure S2). Isolated EVs showed a mean size of 201.0±50.1 nm and concentration of 7.1×1010±8.6×1010 particles/mL. A mean concentration of EV DNA was 2.9±1.2 ng/μL by TapeStation analysis (Figure S3) and EGFR copy number was 661±999 copys/μL in EV DNA. EGFR copy number correlates with the EV DNA concentration (Figure 1A, Spearman’s correlation 0.520, P=0.0001) and EV concentration (Figure 1B, Spearman’s correlation 0.464, P=0.0008), but the correlation was not found between EGFR copy number and EV size (Spearman’s correlation 0.265, P>0.05). EGFR copy number increased as T stage progressed (Figure 1C, Spearman’s correlation 0.321, P=0.025). But EV concentration, size, and EV DNA concentration did not show any trend.
Figure S1.

Electron microscopy (EM) image of BALF EVs. Samples for EM analysis were negatively stained. BALF, bronchoalveolar lavage fluid; EV, extracellular vesicles.
Figure S2.
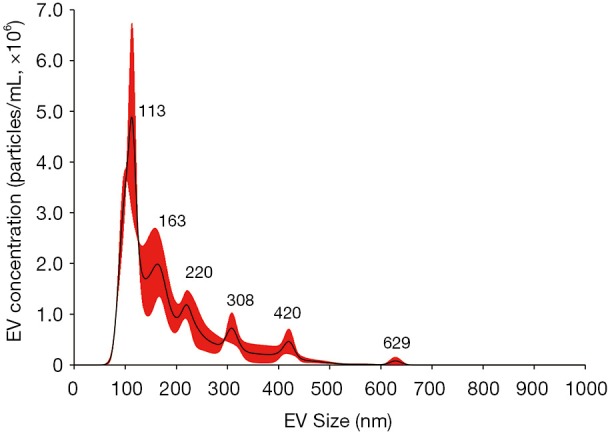
Size distribution of BALF EV. Sizes of purified EVs were determined using Nanosight NS300. Average size distribution from three separate measurements is plotted in concentration (particles/mL) according to their size. BALF, bronchoalveolar lavage fluid; EV, extracellular vesicles.
Figure S3.
Gel-like images of EV DNA from BALF. Gel-like images show the size and amount of EV DNA determined using the TapeStation. First lane shows the standard size ladder distribution, and numbers on the left indicate corresponding sizes (bp).BALF, bronchoalveolar lavage fluid; EV, extracellular vesicles.
Figure 1.
Characterization of isolated EVs and EV DNA. (A) The correlation between EV DNA concentration and EGFR copy number; (B) the correlation between EV concentration and EGFR copy number; (C) the correlation between EV concentration and T stage.
Sensitivity trend according to disease stages
The overall average sensitivity and specificity of BALF EV-based EGFR genotyping in all patients (n=137) revealed to be 75.9% (95% CI, 62.1–86.1%) and 86.7% (95% CI, 77.1–92.9%), respectively (Table 2). The sensitivity of BALF EV-based EGFR genotyping showed a significant increase according to the advancement of TMN staging factors (Figure 2). The sensitivity in the T1 stage was 40% which increased to 75% in T2 and then to 100% in T3 and T4 stages. N staging also significantly affected the sensitivity with 63.3% at N0, 75% at N1/N2 and reaching 100% at the N3 stage. The presence of metastasis regardless of intrathoracic (M1a) or extrathoracic (M1b and M1c) event was the most powerful determinant for reaching 100% sensitivity (Figure 2). These results suggest that the increased release of tumor-specific EVs containing oncogenic EGFR mutant DNA, depends on the increment of tumor extent and metastatic events.
Table 2. Performance of BALF EV-based EGFR genotyping: sensitivity, specificity, positive and negative predictive value.
| Stage (N=137) | Sensitivity, % (95% CI) | Tissue/cytology (+) | Specificity, % (95% CI) | Tissue/cytology (−) | Positive predictive value, % (95% CI) | Negative predictive value, % (95% CI) | ||
|---|---|---|---|---|---|---|---|---|
| BALF EV (+)a | BALF EV (−)b | BALF EV (+)c | BALF EV (−)d | |||||
| Stage I (N=36) | 35.3 (15.2−61.4) | 6 | 11 | 94.7 (71.9−99.7) | 1 | 18 | 85.7 (42.0−99.2) | 62.1 (42.4−78.7) |
| Stage II (N=6) | 100 (5.5−100) | 1 | 0 | 100 (46.3−100) | 0 | 5 | 100 (5.5−100) | 100 (46.3−100) |
| Stage III (N=23) | 60 (17.0−92.7) | 3 | 2* | 77.8 (51.9−92.6) | 4 | 14 | 42.9 (11.8−79.8) | 87.5 (60.4−97.8) |
| Stage IV (N=72) | 100 (86.3−100) | 31 | 0 | 85.4 (70.1−93.9) | 6 | 35 | 83.8 (67.3−93.2) | 100 (87.7−100) |
| All stages (N=137) | 75.9 (62.1−86.1) | 41 | 13 | 86.7 (77.1−92.9) | 11 | 72 | 78.8 (64.9−88.5) | 84.7 (74.9−91.3) |
a, BALF EV (+) indicates positive results for EGFR-TKI sensitive mutation when tested with both tissue/cytology and BALF; b, BALF EV (−) indicates positive result for EGFR-TKI sensitive mutation when tested with tissue/cytology but negative with BALF; c, BALF EV (+) indicates negative results for EGFR-TKI sensitive mutation when tested with tissue/cytology but positive BALF; d, BALF EV (−) indicates negative results for EGFR-TKI sensitive mutation when tested with both tissue/cytology and BALF. *, these two patients were up-staged cases after surgery from clinical stage I disease because mediastinal lymph node dissection revealed minimal N2 disease at single nodal station. TKI, tyrosine kinase inhibitor; BALF, bronchoalveolar lavage fluid; EV, extracellular vesicle.
Figure 2.
Sensitivity trends of BALF EV-based EGFR genotyping according to TNM stage. (A) T stage; (B) N stage; (C) M stage. *, indicates significant differences after the Bonferroni correction.
Comparison of EGFR genotyping using BALF EV and tumor tissue/cytology
The concordance rates or agreements between BALF EV-based and tissue/cytology-based EGFR genotyping of each stage are 79% (95% CI, 51.3–82.1%) in stage I (n=36), 100% in stage II (n=6), 74% (95% CI, 56.0–91.9%) in stage III (n=23), and 92% (95% CI, 85.3–98.1%) in stage IV (n=72) (Figure 3). In stage IV disease, all tissue-proven EGFR-mutated patients (n=31) were detected by BALF EV-based genotyping (20 patients of exon 19 del, 9 patients of exon 21 L858R, and 2 patients of exon 18 G719X). In one case, a patient identified as having in exon 19 deletion in tissue genotyping, later shown to have exon 21 L858R mutation with BALF EV. It also newly identified six new mutant cases with exon 19 deletion. Even though the sensitivity in stage IV was 100%, the concordance rate was slightly decreased to 92% due to the additional detection of these six cases (kappa =0.834, P=0.000). In early stages I and II, the concordance rate was significantly high, each with 79% (kappa =0.310, P=0.023) and 100% (kappa =1.0, P=0.014), respectively. Among 36 stage I patients, 17 were with solid nodule, 19 were patients with GGN showed 30% (95% CI, 6.7–65.3%) sensitivity and 88.9% (95% CI, 51.8–99.7%) specificity (Table S1). This finding is suggestive of the potential usefulness of BALF EV-based EGFR genotyping in early-stage NSCLC patients.
Figure 3.
Comparison of EGFR genotyping using BALF EV and tumor tissue/cytology. a, exon 20 insertion or T790M was regarded as wild type.
Table S1. Performance of BALF EV-based EGFR genotyping at stage I: sensitivity, specificity, positive and negative predictive value.
| Stage I (N= 36) | Sensitivity, % (95% CI) | Tissue/cytology (+) | Specificity, % (95% CI) | Tissue/cytology (−) | Positive predictive value, % (95% CI) | Negative predictive value, % (95% CI) | ||
|---|---|---|---|---|---|---|---|---|
| BALF EV (+) | BALF EV (−) | BALF EV (+) | BALF EV (−) | |||||
| Solid nodule (N=17) | 42.9 (9.9–81.6) | 3 | 4 | 100 (69.2–100) | 0 | 10 | 100 | 71.4 (56.8–82.6) |
| GGN (N=19) | 30 (6.7–65.3) | 3 | 7 | 88.9 (51.8–99.7) | 1 | 8 | 75 (27.3–96.0) | 53.3 (41.7–64.6) |
Detecting EGFR mutations in stage IV could provide late-stage patients with a more effective and tolerable EGFR-tyrosine kinase inhibitor (TKI) treatment opportunities. In our study, BALF EV-based EGFR genotyping significantly increased the detection of mutant cases compared to conventional tissue/cytology-based EGFR genotyping (Figure 4, 51.4% vs. 43.1%, P<0.05), especially during the late stages. In stage III, there was no statistically significant difference between two methods, but BALF EV-based genotyping is the superior method excluding two irregular cases with stage discrepancies before and after surgery. Although BALF EV-based genotyping showed slightly decreased detection rate in stage I, it is noteworthy that about 20% of cases can be pre-operatively identified as EGFR-mutant lung cancer without a tissue biopsy (Table 2 and Figure 3).
Figure 4.

EGFR mutation detecting rate comparison according to each stage. *P<0.05.
Discordant cases between BALF-EV based genotyping and tissue-based genotyping
In all 137 patients, we found a total of 24 discordant cases from all four stages (Table 2 and Figure 3), which include BALF EV-based genotyping detecting 11 more EGFR-mutant cases and failed to detect 13 tissue-proven cases (Tables S2,S3). Comparison of clinical characteristics between the concordant group (n=113) and discordant group (n=24) did not reveal any statistical difference except for the presence of metastasis and TNM stage (Table S4). The demographic and clinical factors including sex, smoking status, histologic subtype, tissue vs. cytology, a location of BAL, and open bronchus sign did not affect the concordance (Table S4). All 13 patients who failed to be detected by BALF EV-based genotyping had clinical stage I disease with two unusual cases of up-staging to IIIA after surgery due to minimal N2 disease at a single nodal station (Table 2 and Table S2).
Table S2. Demographic characteristics associated with patients who yielded BALF EV-based EGFR genotyping (−)a.
| No | Sex | Age | Histological subtype | Tumor location | Size (mm) | Sample collection method (tissue/cytology) | Tumor EGFR genotyping | BFS cytology | Location of BAL | Open bronchus sign | TNM stage |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 78 | ADC | LLL | 20 | Surgery | L858R | No malignant cell | LB6 | No | IA2 |
| 2 | M | 73 | ADC | LUL | 20 | Surgery | L858R | No malignant cell | LB2 | Yes | IA2 |
| 3 | M | 73 | ADC | LLL | 12 | Surgery | L858R, C797S | No malignant cell | LB6 | No | IA2 |
| 4 | F | 65 | ADC | RUL | 13 | Surgery | Exon 19 del | No malignant cell | RB3 | No | IA2 |
| 5 | F | 77 | ADC | LUL | 15 | Surgery | Exon 19 del | No malignant cell | LB4 | Yes | IA2 |
| 6 | M | 64 | ADC | RUL | 20 | Surgery | Exon 19 del | No malignant cell | RB1 | No | IA2 |
| 7 | F | 63 | ADC | RUL | 25 | Surgery | Exon19 del | No malignant cell | RB1 | Yes | IA3 |
| 8 | F | 73 | ADC | RUL | 25 | Surgery | L858R | No malignant cell | RB1 | Yes | IA3 |
| 9 | M | 75 | ADC | RLL | 40 | Surgery | Exon 19 del | No malignant cell | RB10 | Yes | IB |
| 10 | F | 70 | ADC | LUL | 29 | Surgery | L858R | No malignant cell | LB1 | No | IB |
| 11 | M | 62 | ADC | LUL | 19 | Surgery | Exon 19 del | No malignant cell | LB3 | No | IB |
| 12 | F | 66 | ADC | RLL | 29 | Surgery | L858R | No malignant cell | RB8 | Yes | IIIA |
| 13 | M | 68 | ADC | LLL | 38 | PCNB | Exon 19 del | Atypical cell | LB9 | Yes | IIIA |
a, BALF EV-based genotyping (−) indicates positive results for EGFR-TKI sensitive mutation when tested with tissue/cytology but negative BALF. BALF, bronchoalveolar lavage fluid; EV, extracellular vesicles; ADC, adenocarcinoma; PCNB, percutaneous needle biopsy; BFS, bronchofiberscopy; RB, right bronchus; LB, left bronchus; del, deletion.
Table S3. Demographic characteristics associated with patients who yielded BALF EV-based EGFR genotyping (+)a.
| No | Sex | Age | Histological subtype | Tumor location | Size (mm) | Sample collection method (tissue/cytology) | BFS cytology | Location of BAL | Open bronchus sign | BALF EV-based genotype | TNM stage | Treatment | EGFR-TKI | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 75 | ADC | RUL | 20 | Surgery | No malignant cell | RB1 | Yes | L861Q | IA2 | Surgery | Not done | |
| 2 | M | 70 | ADC | LLL | 12 | Surgery | No malignant cell | LB8 | Yes | Exon 19 del | IIIA | Surgery, adjuvant chemoRx | Not done | |
| 3 | F | 67 | NSCLC | RUL | 51 | EBUS-TBNA | No malignant cell | RB2 | No | Exon 19 del | IIIB | Afatinib | Afatinib 14 days | Transfer |
| 4 | M | 67 | SQCC | RUL | 34 | EBUS-TBNA | SQCC | RB1 | Yes | G719S | IIIB | CCRT | Not done | |
| 5 | M | 85 | SQCC | Lingular | 20 | Bronchoscopy | SQCC | LB4 | No | Exon 19 del | IIIC | Tx refuse | Not done | |
| 6 | F | 77 | NSCLC | LUL | 55 | PCNB | No malignant cell | LB3 | Yes | Exon 19 del | IVA | Tx refuse | Not done | |
| 7 | M | 80 | ADC | RUL | 11 | Bronchoscopy | ADC | RB1 | Yes | Exon 19 del | IVA | ChemoRx | Not done | |
| 8 | F | 61 | NSCLC | RUL, RLL | 39, 21 | PCNB | No malignant cell | RB2 | Yes | Exon 19 del | IVB | After ChemoRx, Erlotinib | Erlotinib 42 days | Transfer |
| 9 | M | 83 | ADC | Lingular | 60 | PCNB | No malignant cell | LB4 | Yes | Exon 19 del | IVB | After ChemoRx, Gefitinib | Gefitinib 5 days | Death |
| 10 | M | 77 | NSCLC | LUL | 70, 42 | PCNB | No malignant cell | LB2 | No | Exon 19 del | IVB | ChemoRx | Not done | Transfer |
| 11 | M | 56 | NSCLC | RUL | 25 | EBUS-TBNA | No malignant cell | RB2 | Yes | Exon 19 del | IVB | ChemoRx | Not done | Transfer |
a, BALF EV-based genotyping (+) indicates negative results for EGFR-TKI sensitive mutation when tested with tissue/cytology but positive BALF. BALF, bronchoalveolar lavage fluid; EV, extracellular vesicles; ADC, adenocarcinoma; EBUS-TBNA, endobronchial ultrasound guided transbronchial needle aspiration; PCNB, percutaneous needle biopsy; BFS, bronchofiberscopy; RB, right bronchus; LB, left bronchus; del, deletion.
Table S4. Comparison of clinical characteristics between concordant group and discordant group.
| Characteristics | Concordant groupa (N=113, %) | Discordant groupb (N=24, %) | P value |
|---|---|---|---|
| Sex | 0.950 | ||
| Male | 62 (54.9) | 13 (54.2) | |
| Female | 51 (45.1) | 11 (45.8) | |
| Smoking status | 0.279 | ||
| Never-smoker | 53 (46.9) | 11 (45.8) | |
| Ex-smoker | 27 (23.9) | 9 (37.5) | |
| Current smoker | 33 (29.2) | 4 (16.7) | |
| Histological subtype | 0.164 | ||
| ADC | 94 (83.2) | 17 (70.8) | |
| Non-ADC | 19 (16.8) | 7 (29.2) | |
| Cytology versus tissue | 1.000 | ||
| Cytology | 12 (10.6) | 2 (8.3) | |
| Tissue | 101 (89.4) | 22 (91.7) | |
| Location of BAL | 0.644 | ||
| Dependent position | 46 (40.7) | 11 (45.8) | |
| Non-dependent position | 67 (59.3) | 13 (54.2) | |
| Open bronchus sign | 0.845 | ||
| Yes | 73 (64.6) | 15 (62.5) | |
| No | 40 (35.4) | 9 (37.5) | |
| TNM classification | |||
| T stage | 0.059 | ||
| T1 | 19 (16.8) | 11 (45.9) | |
| T2 | 35 (31.0) | 5 (20.8) | |
| T3 | 11 (9.7) | 2 (8.3) | |
| T4 | 48 (42.5) | 6 (25.0) | |
| N stage | 0.145 | ||
| N0 | 46 (40.7) | 14 (58.3) | |
| N1 | 7 (6.2) | 1 (4.2) | |
| N2 | 16 (14.2) | 5 (20.8) | |
| N3 | 44 (38.9) | 4 (16.7) | |
| M stage | 0.027 | ||
| M0 | 47 (41.6) | 18 (75.0) | |
| M1 | 65 (58.4) | 6 (25.0) | |
| TNM stage | 0.014 | ||
| I | 24 (21.2) | 12 (50.0) | |
| II | 6 (5.3) | 0 (0) | |
| III | 17 (15.1) | 6 (25.0) | |
| IV | 66 (58.4) | 6 (25.0) | |
a, concordant group indicates all agreement for EGFR genotyping test between tissue/cytology and BALF; b, disconcordant group indicates inconsistency for EGFR genotyping test between both tissue/cytology and BALF. ADC, adenocarcinoma; BALF, bronchoalveolar lavage fluid.
BALF EV-based genotyping noticeably outperformed in clinically advanced stages, specifically in clinical stage III and IV. BALF EV-based genotyping detected 10 more patients that include four cases from stage III, and six cases from stage IV as having EGFR-mutants (Table 2 and Figure 3). Also, one case from stage IA2 was identified by BALF EV-based genotyping making total of 11 additional findings. Subtypes of these cases were mostly an exon 19 deletion mutation (n=9), one case of single mutation L861Q and one case of G719S mutation. Histologic typing of these patients showed a relatively high incidence of NSCLC (n=5; 45.5%) and squamous cell carcinoma (n=2; 18.2%) with four cases of conventional adenocarcinoma (36.4%) (Table S3). This implies that BALF EV-based genotyping could increase the opportunity of EGFR-TKI treatment in the first-line setting to more than 10% of advanced NSCLC patients.
Unfortunately, the drug response to EGFR-TKIs of these patients could not be verified due to various reasons (Table S3). Therefore, further investigation is required to identify the reason for the relative high detection rate while using BALF EV in histologically unfavorable EGFR mutations. It seems that the heterogeneity of tumors makes detection of cancer with tissue biopsy difficult. Therefore, introducing BALF EV-based genotyping to cancer diagnosis is clinically significant by helping patients begin treatment sooner in the way identifying cancers otherwise undetectable.
Turn-around time (TAT) for EGFR mutations testing
Conventionally, the decision is made based on tissue/cytology-based EGFR mutations testing, which usually takes about 9.9 working days in Konkuk University Medical Center (95% CI, 9.4–10.5 days) and up to 12 working days according to others (18). In comparison, the BALF EV-based EGFR mutation testing method only takes about 1.9 working days (95% CI, 1.8–2.1 days) at our facility. The time-line of tissue/cytology takes 13–19 hours for making Formalin-Fixed Paraffin-Embedded (FFPE) tissue specimens and 3 hours for the DNA extraction from the FFPE tissue specimens, which adds up to 16–22 h when worked on a sample non-stop. On the other hand, EV isolation takes about 2 h and DNA extraction from the EV takes about an hour which only requires total of 3 hours. There is no significant sample delivery time difference between tissue/cytology or EV DNA routine. For example, in the case of a 57-year-old female EGFR-mutant metastatic adenocarcinoma patient, tissue-based genotyping takes 14 days just to get the results. On the contrary, BALF EV-based EGFR genotyping is shown to demonstrate much higher efficiency by producing results in a single working day leading to treatment the next day. Therefore, effects of the Gefitinib therapy can be observed after only 21 days (Figure S4). Altogether, we have shown that EV-based EGFR genotyping is faster and more accurate method in the genotyping of advanced NSCLC patients.
Figure S4.
Turn-around time (TAT): a case demonstration of 57-year-old female metastatic EGFR mutant adenocarcinoma. BALF, bronchoalveolar lavage fluid; EV, extracellular vesicles.
Discussion
Recently, detecting T790M mutation in re-biopsy and obtaining tissue samples for PCR or NGS analysis has become important in the diagnosis of lung cancer (19,20). Consequently, so-called ‘tissue is the issue’ emerged as a problem in the time of precision medicine. To overcome the obstacle of obtaining tissue samples, liquid biopsy using ctDNA in blood has been introduced into clinical settings but with major shortcomings of low sensitivity and instability, which limits clinical usability. In this aspect, lipid membrane-bound EV can be a great alternative source for liquid biopsy increasing stability and sensitivity. cfDNA are usually small fragments of 200–400 bp, while EV DNA are longer than 1 kb dsDNA (21-23). Along with abundant new discoveries in various tumor-derived EVs and EV DNA, they have great potential in cancer biomarker discoveries as well as providing a platform for personalized medicine (9-12,23-25).
According to a previous study, we used BALF from the tumor site to obtain enriched tumor-derived EVs, thereby increasing the sensitivity of EGFR genotyping (9). As a result, BALF EV-based genotyping produced results with high accuracy and sensitivity. BALF EV-based EGFR genotyping not only verified all patients previously found by the conventional tissue/cytology-based EGFR but identified 11 additional EGFR mutation positive patients (Table 2 and Table S3). In addition, another study proved clinical usefulness of genotyping using BALF EV DNA, as it exceeded tissue genotyping in the detection T790M mutation for the prescription of the 3rd generation EGFR-TKIs such as osimertinib (9).
The detection sensitivity of BALF EV-based genotyping increases according to the spread of cancer, reaching 100% in any stages of cancer when the patient is T3, N3, or M1 (Figure 2) demonstrated that in cases where biopsy is inaccessible for various reasons such as poor performance status or the position of the tumor being in difficult position for percutaneous needle biopsy, BALF EV-based EGFR genotyping could replace conventional diagnostic methods.
On the other note, in stage I when the tumor is less than 4 cm in size and has not spread to the regional lymph nodes, sensitivity and specificity were 35.3% and 94.7%, respectively (Table 2), sensitivity being particularly low. It seems that the difference between the early-stage and more advanced lung cancer patient arises from the fact that EVs are secreted more abundantly in the tumor microenvironment as the tumor progresses and metastasis occurs. Despite low sensitivity, the ability to screen for EGFR mutations in the early stage disease without an invasive biopsy is promising, as EGFR genotyping can be informative for setting up therapeutic strategies before the surgery. Though most people do not go through EGFR testing in the early surgical stage at present, this result is implicative of pursuing further research of BALF EV-based EGFR genotyping in the early-stage NSCLC.
In addition, we have investigated if the position of the tumor such as in the middle lobe or anterior segment and the presence of open bronchus sign have any effect on the results of BALF-EV based EGFR genotyping, but did not identify any significant association (Table S4). In comparison, a recent study the sensitivity and specificity of plasma genotyping in early lung cancer were 25.7% and 96.6%, respectively, showing that BAL EV DNA based genotyping in the early stages of lung cancer is a more superior method (11).
Liquid biopsy using BAL to screen for mutant EGFR DNA is an easier and safer way of diagnosing patients compared to lung biopsy. Transforming diagnostic methods from tissue- or cell-based diagnosis to genotype- or molecule-based diagnosis is an innovative paradigm shift that would advance clinical diagnostics. Especially in Asian countries where approximately 40% of patients have EGFR mutation associated lung cancer (26), along with the development of targeted therapy drugs, having a reliable and repeatable diagnostic method is becoming ever more important. Also, in the case of ground glass nodules (GGNs), the occurrence of EGFR mutation is high and biopsy is often difficult with low yields (27). Therefore, during clinical follow up surgical resection often becomes the only option if the tumor grows, which makes the opportunity of non-invasive EFGR genotyping even more appealing and valuable.
Another advantage of BALF EV-based EGFR genotyping is the short TAT. Identification of EGFR-mutations is the first step in the diagnosis and treatment selection between EGFR-TKIs and cytotoxic chemotherapy in treatment-naïve and newly diagnosed advanced NSCLC patients. Conventional tissue EGFR mutation testing usually takes about 10 working days (18), which could be painstakingly long for patients, but performing BAL and obtaining EV DNA genotyping only takes about two working days to get the result. In most cases, patients have to wait for the result of EGFR genotyping before they can begin the treatment, which could seem especially long for patients who have high probability of having EGFR mutations such as female, never or minimal smokers, East Asians and patients with peripheral tumors. Liquid biopsy using plasma circulating tumor DNA is an option, but the sensitivity is relatively low (28,29).
Future studies are required for BALF EV-based EGFR genotyping of patients with pulmonary nodules and consider combining next-generation sequencing (NGS) based tests with liquid biopsy to increase the sensitivity in early lung cancer detection.
Acknowledgments
We thank Dr. Min Kyo Jung for the electron microscopy image of BALF EVs.
Funding: This research was conducted with the support from AstraZeneca Korea Ltd (ISSIRES011, KUH1010613), Hanmi Pharm. (Korea, KUH1010786), Chong Kun Dang Pharm. (Korea, KUH1010868), and Technology Innovation Program-Post Genome Project (development of kit for screening of cancer related target genes with high sensitivity in the liquid biopsy, 10067810) funded by the Ministry of Trade, Industry & Energy (MOTIE, Korea).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the amended Declaration of Helsinki. The study protocol was approved by the institutional review board of Konkuk University Medical Center (KUH1010812), and written informed consents were obtained from all patients.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Shiau CJ, Babwah JP, da Cunha Santos G, et al. Sample features associated with success rates in population-based EGFR mutation testing. J Thorac Oncol 2014;9:947-56. 10.1097/JTO.0000000000000196 [DOI] [PubMed] [Google Scholar]
- 2.Cardarella S, Ortiz TM, Joshi VA, et al. The introduction of systematic genomic testing for patients with non-small-cell lung cancer. J Thorac Oncol 2012;7:1767-74. 10.1097/JTO.0b013e3182745bcb [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Travis WD, Brambilla E, Noguchi M, et al. Diagnosis of lung cancer in small biopsies and cytology: implications of the 2011 International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification. Arch Pathol Lab Med 2013;137:668-84. 10.5858/arpa.2012-0263-RA [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo J, Shen L, Zheng D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: a systematic review and meta-analysis. Sci Rep 2014;4:6269. 10.1038/srep06269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayo-de-Las-Casas C, Jordana-Ariza N, Garzon-Ibanez M, et al. Large scale, prospective screening of EGFR mutations in the blood of advanced NSCLC patients to guide treatment decisions. Ann Oncol 2017;28:2248-55. 10.1093/annonc/mdx288 [DOI] [PubMed] [Google Scholar]
- 6.Diaz LA, Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. 10.1200/JCO.2012.45.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. 10.1038/nrc3066 [DOI] [PubMed] [Google Scholar]
- 8.Shah R, Patel T, Freedman JE. Circulating Extracellular Vesicles in Human Disease. N Engl J Med 2018;379:958-66. 10.1056/NEJMra1704286 [DOI] [PubMed] [Google Scholar]
- 9.Hur JY, Kim HJ, Lee JS, et al. Extracellular vesicle-derived DNA for performing EGFR genotyping of NSCLC patients. Mol Cancer 2018;17:15. 10.1186/s12943-018-0772-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JS, Hur JY, Kim IA, et al. Liquid biopsy using the supernatant of a pleural effusion for EGFR genotyping in pulmonary adenocarcinoma patients: a comparison between cell-free DNA and extracellular vesicle-derived DNA. BMC Cancer 2018;18:1236. 10.1186/s12885-018-5138-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan Y, Liu B, Lei H, et al. Nanoscale extracellular vesicle-derived DNA is superior to circulating cell-free DNA for mutation detection in early-stage non-small-cell lung cancer. Ann Oncol 2018;29:2379-83. 10.1093/annonc/mdy458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allenson K, Castillo J, San Lucas FA, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol 2017;28:741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller RJ, Casal RF, Lazarus DR, et al. Flexible Bronchoscopy. Clin Chest Med 2018;39:1-16. 10.1016/j.ccm.2017.09.002 [DOI] [PubMed] [Google Scholar]
- 14.Calvayrac O, Pradines A, Pons E, et al. Molecular biomarkers for lung adenocarcinoma. Eur Respir J 2017;49. doi: . 10.1183/13993003.01734-2016 [DOI] [PubMed] [Google Scholar]
- 15.Detterbeck FC, Boffa DJ, Kim AW, et al. The Eighth Edition Lung Cancer Stage Classification. Chest 2017;151:193-203. [DOI] [PubMed] [Google Scholar]
- 16.Kim YT, Kim JW, Kim SK, et al. Simultaneous genotyping of multiple somatic mutations by using a clamping PNA and PNA detection probes. Chembiochem 2015;16:209-13. 10.1002/cbic.201402640 [DOI] [PubMed] [Google Scholar]
- 17.Han JY, Choi JJ, Kim JY, et al. PNA clamping-assisted fluorescence melting curve analysis for detecting EGFR and KRAS mutations in the circulating tumor DNA of patients with advanced non-small cell lung cancer. BMC Cancer 2016;16:627. 10.1186/s12885-016-2678-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldman JW, Noor ZS, Remon J, et al. Are liquid biopsies a surrogate for tissue EGFR testing? Ann Oncol 2018;29:i38-46. 10.1093/annonc/mdx706 [DOI] [PubMed] [Google Scholar]
- 19.Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin Cancer Res 2016;22:1103-10. 10.1158/1078-0432.CCR-15-1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malapelle U, Pisapia P, Rocco D, et al. Next generation sequencing techniques in liquid biopsy: focus on non-small cell lung cancer patients. Transl Lung Cancer Res 2016;5:505-10. 10.21037/tlcr.2016.10.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vagner T, Spinelli C, Minciacchi VR, et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J Extracell Vesicles 2018;7:1505403. 10.1080/20013078.2018.1505403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lázaro-Ibáñez E, Lasser C, Shelke GV, et al. DNA analysis of low- and high-density fractions defines heterogeneous subpopulations of small extracellular vesicles based on their DNA cargo and topology. J Extracell Vesicles 2019;8:1656993. 10.1080/20013078.2019.1656993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahlert C, Melo SA, Protopopov A, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem 2014;289:3869-75. 10.1074/jbc.C113.532267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 2014;24:766-9. 10.1038/cr.2014.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balaj L, Lessard R, Dai L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun 2011;2:180. 10.1038/ncomms1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohno T, Nakaoku T, Tsuta K, et al. Beyond ALK-RET, ROS1 and other oncogene fusions in lung cancer. Transl Lung Cancer Res 2015;4:156-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi Y, Mitsudomi T, Sakao Y, et al. Genetic features of pulmonary adenocarcinoma presenting with ground-glass nodules: the differences between nodules with and without growth. Ann Oncol 2015;26:156-61. 10.1093/annonc/mdu505 [DOI] [PubMed] [Google Scholar]
- 28.Singh AP, Li S, Cheng H. Circulating DNA in EGFR-mutated lung cancer. Ann Transl Med 2017;5:379. 10.21037/atm.2017.07.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fenizia F, De Luca A, Pasquale R, et al. EGFR mutations in lung cancer: from tissue testing to liquid biopsy. Future Oncol 2015;11:1611-23. 10.2217/fon.15.23 [DOI] [PubMed] [Google Scholar]