Abstract
Background
Lung cancer is the leading cause of cancer-related death worldwide, with 5-year overall survival less than 15%. Therefore, it is essential to find biomarkers for early detection and prognosis. Aberrant DNA methylation is a common feature of human cancers and its utility is already recognized in cancer management. The aim of this study was to explore the diagnostic and prognostic value of the promoter methylation status of the ASC/TMS1/PYCARD and MyD88 genes, key adaptor molecules in the activation of the innate immune response and apoptosis pathways.
Methods
A total of 50 non-small cell lung cancer (NSCLC) patients were enrolled in the study. Methylation of bisulphite converted DNA was quantified by pyrosequencing in fresh frozen malignant tissues and adjacent non-malignant tissues. Associations between methylation and lung function, tumor grade and overall survival were evaluated using receiver-operating characteristics (ROC) analysis and statistical tests of hypothesis.
Results
Methylation level of tested genes is generally low but significantly decreased in tumor tissues (ASC/TMS1/PYCARD, P<0.0001; MyD88, P<0.0002), which correlates with increased protein expression. Three CpG sites were identified as promising diagnostic marker candidates; CpG11 (-63 position) in ASC/TMS1/PYCARD and CpG1 (-253 position) and 2 (-265 position) in MyD88. The association study showed that the methylation status of the ASC/TMS1 CpG4 site (-34 position) in malignant and non-malignant tissues is associated with the overall survival (P=0.019) and the methylation status of CpG8 site (-92 position) is associated with TNM-stage (P=0.011).
Conclusions
The methylation status of the ASC/TMS1/PYCARD and MyD88 promoters are promising prognostic biomarker candidates. However, presented results should be considered as a preliminary and should be confirmed on the larger number of the samples.
Keywords: ASC/TMS1/PYCARD, MyD88, methylation status, association, overall survival, TNM, non-small cell lung cancer (NSCLC)
Introduction
Lung cancer is the leading cause of cancer-related death worldwide which makes it a serious health and economical problem. In addition, the 5-year survival rate is still less than 15%. Since lung cancer is mostly diagnosed at the advanced stage of disease and known therapeutic approaches are still not efficient enough, it is of particular importance to identify new biomarkers that could be used for early detection and prognosis assessment. Since epigenetic alterations are more frequent than genetic mutations in cancer genome, a promising approach for early cancer diagnosis emerged in form of epigenetic biomarkers (1). Epigenetic changes can be described as stable and heritable chromatin changes which could have an effect on gene expression, without changing the DNA sequence itself (2). Epigenetic alterations that include variability in DNA methylation contribute to the overall phenotypic characteristics of an individual and can participate in the risk for malignancy development (3). If CpG islands of the gene promoter region are affected by aberrant methylation, this can significantly contribute to tumor promotion (3,4). Thus, tumor suppressor gene hypermethylation is recognized as a hallmark of lung cancer and tends to occur as an early event in the carcinogenic process (5,6). In contrast to hypermethylation, genomic hypomethylation is thought to occur late in carcinogenesis in NSCLC, and gene-specific hypomethylation is also recorded for several genes involved in the promotion of lung cancer (1,7). Additionally, it is well known that lung cancer development is strongly associated with chronic inflammation that could be influenced by DNA methylation (8). If acute inflammation is not regulated, it can lead to chronic inflammation and may contribute to cancer initiation, progression and dissemination (9). During the last decade multiple studies have shown that respiratory immune cells, epithelial and mesothelial cells, upon endogenous or exogenous injury, act together to trigger inflammation via activation of pattern recognition receptors (PRRs) (10-12). Toll-like receptors (TLRs), one of the best characterized PRRs receptors are known to be associated with immune and inflammatory diseases through dysregulated production and release of IFNs and proinflammatory cytokines (13,14). TLRs are activated either by pathogen-associated molecular patterns (PAMPs), or danger-associated molecular patterns (DAMPs), released at sites of infection or by tissue damage (15). Critical adaptor protein in TLR signaling is MyD88 (Myeloid differentiation primary response protein 88). Initiation of the TLR signaling lead to a complex signaling cascade culminating in activation of different transcription factors, such as nuclear factor κB (NF-κB) and interferon regulatory factors (IRFs) (16,17). Besides TLRs other PPRs, NLRs (NOD-like receptors), can also be activated by specific ligands and form a multiprotein platform called inflammasome upon ligand binding (18). Inflammasome formation is required for activation of caspase-1 and subsequent secretion of proinflammatory cytokines, IL-18 and IL-1β, a process that promote carcinogenesis (19) and support tumor survival by triggering secretion of several factors, such as VEGF, FGF2 or STAT3 (20). A critical adaptor molecule in inflammasome assembly and activation through homologous interactions is ASC (apoptosis-associated speck-like protein containing a caspase recruitment domain), a bipartite intracellular signalling molecule composed of both N-terminal pyrin domain (PYD) and the C-terminal caspase-recruitment domain (CARD) (21). A synonym for ASC used in the literature is TMS1 (Target of Methylation induced Silencing protein 1). The inflammasome complex can activate caspase-8 dependent apoptotic cell death, but also caspase-1 dependent pyroptotic cell death (22). Given the important role of ASC/TMS1/PYCARD gene in activation and subsequent regulation of inflammation in the tumor and non-tumor tissues, methylation of the CpG islands in the promoter region was the subject of many studies. Aberrant hypermethylation of the promoter region of ASC/TMS1/PYCARD has been reported in many different human neoplasms such as renal carcinoma (23), breast cancer (24), colorectal cancer (25), glioblastoma (26), hepatocellular carcinoma (27), melanoma (28), neuroblastoma (29), non-small cell lung and small cell lung cancer (30), ovarian tumors (31), prostate cancer (32) and thyroid cancer (33). This could lead to the conclusion that ASC/TMS1/PYCARD might be a tumor suppressor gene, and its silencing could promote carcinogenesis of some tumor types. Thereby, it is assumed that the ASC/TMS1/PYCARD tumor-associated methylation can serve as a potential target for the development of improved therapeutic treatments, or as a diagnostic and prognostic predictor. Since alteration of MyD88 expression is associated with the constitutive activation of NF-κB signaling, MyD88 is supposed to have a role in carcinogenesis as well. Several groups have shown that increased protein expression of MyD88 is associated with generally worse outcome in different tumor types. It has been shown that increased MyD88 expression is linked to poor prognosis of patients with colorectal cancer (34) and TLR4-mediated paclitaxel chemoresistance in ovarian cancer (35). In breast cancer there is an association with increased MyD88 protein expression and metastasis, TNM stage and poor overall survival (35), and similar findings were noticed in NSCLC (36). However, no published data could be found for the methylation status of MyD88 promoter region. On the other hand, several studies have been published dealing with the evaluation of the methylation status of ASC/TMS1/PYCARD, measured by methyl-specific PCR approach (MSP), and relationship between methylation status and protein expression and different clinical outcomes in different tumor types, including lung cancer. Many of them, if not all of them, detected that aberrant hypermethylation of CpG islands, in the ASC/TMS1/PYCARD promoter region is linked with silencing of gene expression in various cancers including prostate cancer (32), breast cancer (24), gastric cancer (37) and NSCLC (38). For example, Virmani et al. (30) found that promoter hypermethylation (147 bp upstream of ATG site) is the cause of loss of gene expression in SCLC and breast cancer. They also reported that the ASC/TMS1/PYCARD promoter was methylated in 41% of SCLC and in 32% of breast tumor tissues. Furthermore, Zhang et al. (38) reported hypermethylation of ASC/TMS1/PYCARD gene in NSCLC and Machida et al. (39) found that hypermethylation of ASC/TMS1/PYCARD occurs at late stages of lung cancer, not present at earlier stages. The DNA methylation status of promoter sites of the specific genes may represent a promising biomarker for early detection, precise diagnosis and treatment of several human cancers. Using DNA methylation status as a biomarker would have potential advantages, comparing to other markers, since it can be detected with a broad spectrum of affordable techniques (1,7). It is worth to mention that widely used non-quantitative technology, such as MSP, usually failed to quantify methylation status correctly because significant proportion of lowly methylated samples are recognized as methylated indicating a very high sensitivity even for low levels of DNA methylation (40). This might lead to overestimation of DNA methylation. Therefore, in the current study, we aim to re-evaluate the methylation status of ASC/TMS1/PYCARD and MyD88 genes in the NSCLC tumor samples and paired non-tumor tissue using a pyrosequencing approach, highly sensitive quantitative method. The aim of the study was to evaluate if methylation status of tested genes possess the potential to serve as diagnostic or prognostic biomarkers. We investigated the correlation of methylation of the aforementioned gene promoters with overall survival and tumor grade (TNM stage).
Methods
Tissue samples
Resected, early-stage NSCLC tissues (adenocarcinoma and squamous cell carcinomas) with the adjacent non-malignant lung parenchyma from treatment-naïve patients (N=50) were obtained during surgery at Clinical Hospital Center Zagreb, Department for Respiratory Diseases Jordanovac. Tissue samples were snap frozen in liquid nitrogen and kept stored at −80 °C for further analysis. The pathologic diagnosis of each case was confirmed by the review of hematoxylin and eosin stained slides, according to the WHO 2015 (REF). Only sections with a minimum of 70% tumor cells advanced to phase of DNA/RNA/protein extraction. Tumors were staged according to the International Union Against Cancer (UICC) TNM staging system, 8th edition (41). Clinical and pathological features of the patients are shown in the Table 1. All patients signed informed consent and the study was approved by the local ethic committee (University Hospital Centre Zagreb, Department for Respiratory Diseases).
Table 1. Clinicopathologic characteristics of lung cancer patients.
| Characteristic | N (%) |
|---|---|
| Age at diagnosis, years, median (range) | 63 [38–79] |
| Histologic type | |
| Adenocarcinoma | 33 (66.0) |
| Squamous-cell carcinoma | 17 (34.0) |
| T-stage | |
| 1 | 7 (14.0) |
| 2 | 34 (68.0) |
| 3 | 7 (14.0) |
| 4 | 2 (4.0) |
| N-stage | |
| 0 | 31 (62.0) |
| 1 | 14 (28.0) |
| 2 | 5 (10.0) |
| TNM-stage | |
| 1 | 20 (40.8) |
| 2 | 20 (40.8) |
| 3 | 8 (16.3) |
| 4 | 1 (2.0) |
| †FEV1, %, mean (SD) | 81.21±19.84 |
Data are presented as number (%), unless otherwise indicated †FEV1, forced expiratory volume in one second.
DNA isolation, bisulphite conversion, PCR and pyrosequencing
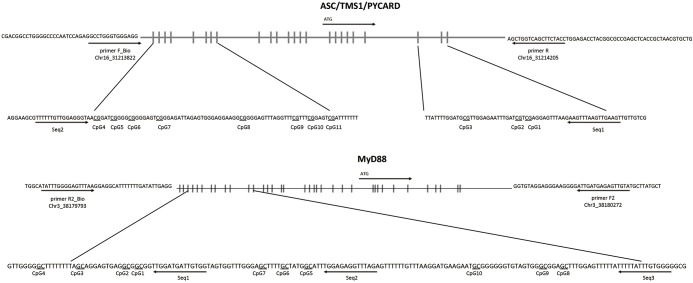
Genomic DNA was isolated from tumor and non-tumor tissues with the DNeasy blood and tissue kit (Qiagen) according to the manufacturer’s instructions and quantified using a NanoDrop 2000 spectrophotometer. Bisulphite conversion was performed on 450 ng DNA of each sample using the Epitect 96 bisulphite kit (Qiagen) according to the manufacturer’s instructions. Pyrosequencing primers were designed using PyroMark Assay Design Software 2.0 (Qiagen). The primers were designed to cover a total of 11 CpG sites located in the 5'-flanking promoter region and the surrounding area of exon 1 of the ASC/TMS1/PYCARD gene and a total of 10 CpG sites in the 5' flanking promoter region of the MyD88 gene (Figure 1). A prediction analysis (AliBaba2.1) has shown that both analysed ASC/TMS1/PYCARD and MYD88 regions contain a high density of CpG sites and transcription factors binding sites characteristic for regulatory domains. PCR primers for ASC/TMS1/PYCARD: F_Bio-5'-GAGGTTTGGGTGGGAGG, R-5'-AATCTCCAAATAAAAACTAACCAAC; SeqPrimer 1_5'-GTTTTTTGTTGGAGGGTAA; SeqPrimer 2_5'-CAACTTCAACTTAAACTTCTT. PCR primers for MyD88: F_5'-TATGTTGAGAGTAGTTAGGG; R_Bio_5'-TATAAACCCCTCAAATTCCTC; SeqPrimer 1_5'-TGGTGATGGTGTTAGTA; SeqPrimer 2_5'- GAGATTTGGAGAGGTT; SeqPrimer 3_5'-GGGGTGTTTATTTTTATT. PCR reactions were carried out in 25 µL final volume containing 2.5 µL of reaction buffer, 0.5 µL (10 mmol) dNTP mix, 0.2 µL FastStartTaq DNA polymerase (1U, Roche), 1 µL of forward and reverse primers (10 pmol final concentration), 18.8 µL of UltraPure Nuclease-Free Water and 1 µL of bisulphite-treated DNA. Pyrosequencing was performed on a PyroMark Q96 ID platform (Qiagen) with PyroMark Gold Q96 reagent kit (Qiagen) according to manufacturer’s instructions. Data were analysed using the PyroMark CpG Software 1.0.11 (Qiagen).
Figure 1.
Sequence map of ASC/TMS1/PYCARD and MyD88 gene promoter. (A) The analysed CpG sites are located at 134–65 bp upstream or 33–52 bp downstream of ASC/TMS1/PYCARD transcription start site (ATG); (B) the analysed CpG sites are located between −278 and −146 bp upstream of the MyD88 transcription start site. The vertical lines in the upper panels represent individual promotor CpG sites. Arrows indicate transcription start sites. Underlined CpG sites were analysed.
RNA extraction and RT-Qpcr
RNA was isolated from tumor and adjacent non-malignant tissues using a commercial RNeasy Mini Kit (Qiagen), according to the manufacturer’s protocol. Quality and integrity of the RNA was determined by RIN number using a 2100 Bioanalyzer and the RNA 6000 Nano Kit (Agilent Technologies). Samples with RIN numbers of 5 and higher were used for downstream analysis. In order to reduce the cost of the analysis, the same amount of the RNAs, with highest RIN scores, isolated from three tumor sample tissues were used for cDNA synthesis of the tumor mix (200 ng total RNA input; RT2 First Strand Kit) according to manufacturer’s instruction (Qiagen). cDNAs were tested by RT2PCR assay (PAHS-097ZA-2, Qiagen) according to manufacturer’s instructions. Adjacent non-malignant tissues were processed in the same way. CT values were exported and analysed by the data analysis web portal (http://www.qiagen.com/geneglobe).
Protein extraction and immunoblot analysis
Proteins were isolated from tissue slices (10 µm). Briefly, tissue slices were incubated for 30 minutes on ice in Passive Lysis buffer (Promega), followed by centrifugation on +4 °C at 21000 G for 30min (Eppendorf Centrifuge 5810R). After centrifugation, supernatant with proteins was carefully collected. Protein concentration was determined using BCA assay (PierceTM BCA Protein Assay Kit, Thermo scientific), according to manufacturer’s instructions. Proteins (100 µg) were denatured for 10 min at 95 °C in sample buffer (2ME, glycerol, bromophenol blue, Tris-HCl) and loaded on 10% SDS gels. SDS-PAGE electrophoresis was performed at 180 V. Proteins were transferred to a 0.2 µm nitrocellulose membrane in case of MyD88, or to a PVDF membrane in case of ASC/TMS1 (BIO-RAD), each using the Trans Blot TurboTM Transfer System (BIO-RAD). Membranes were incubated overnight at +4 °C with monoclonal mouse anti-MyD88 (NBP2-27369-Novus Biologicals, 1:100) and polyclonal goat anti-ASC/TMS1 (AF3805- R&D Systems, 1:200) primary antibodies, respectively. Monoclonal mouse anti-Vinculin [(7F9): sc-73614, Santa Cruz-Biotechnology, 1:1,000] was used as housekeeping control. Secondary antibodies used were HRP-labelled anti-mouse (170-6516-BIO-RAD, 1:10,000) or anti-goat (P0449, DAKO-Agilent Technologies, 1:5,000). Luminol based substrate was used for protein imaging (Clarity™ Western ECL, BIO-RAD) with Uvitec Imager (UVItec, Cambridge, UK). Intensity of the bands was quantified using the ImageJ program.
Statistical analysis
Differences in methylation status and protein expression between tumor and healthy tissue samples were tested using a paired t-test. Associations of methylation status and clinical variables were tested using an independent samples t-test and one-way analysis of variance (ANOVA) with Tukey-Kramer’s post-hoc test for pairwise comparison. Data are presented as mean ± standard error of the mean (SEM). To assess the diagnostic properties of indicated biomarker, receiver operating characteristic (ROC) curves were plotted and the areas under curves (AUC) together with 95% confidence intervals (95% CIs). Impact of clinical variables and methylation status on overall survival (OS) was assessed by the Kaplan-Meier method and survival curves were compared by the log-rank test. Median was used as a cut-off to dichotomize continuous variables. Association between methylation status and pulmonary function was assessed by calculating Spearman’s rank correlation coefficient (ρ). Only P values ≤0.05 were considered statistically significant. Statistical analyses were performed using GraphPad Prism version 5.0 for Windows (GraphPad Software, La Jolla, California, USA) and MedCalc for Windows, version 19.0.5 (MedCalc Software, Ostend, Belgium). AliBaba2.1 (http://gene-regulation.com/pub/programs/alibaba2/) program was used for predicting potential binding sites of transcription factor binding sites.
Results
Methylation status of the ASC/TMS1/PYCARD and MyD88 in the tumor and adjacent non-malignant tissues of NSCLC patients
We quantified the overall promoter methylation status and the methylation status of individual CpG sites located in the ASC/TMS1 and MyD88 promoter regions. The aim of this analysis was to define the methylation levels of the NSCLC patient’s tumor and adjacent non-malignant tissue and to see if any of the tested sites could be considered as a potential tumor-tissue marker. The methylation status of all tested CpG sites in all tested samples is represented as an average value of the methylation (%) of at least three individual PCR/pyrosequencing reactions (Tables S1,S2). Pyrosequencing analysis showed an overall methylation of the ASC/TMS1/PYCARD gene promoter in tumor tissues is significantly decreased (average 6.58%; 4.43–13.54%) compared to non-malignant tissues (average 7.41%; 6.50–9.09%, P<0.0001) (Figure 2A, left panel). Overall methylation of the MyD88 promoter region was also significantly lower in tumor tissues (average 6.26%; 0.71–9.20%) compared adjacent non-tumor tissues (average 7.88%; 2.35–10.02%, P<0.0002) (Figure 2A, right panel).
Table S1. Methylation status (%) of ASC/TMS1/PYCARD promoter in tumour and adjacent non-tumour tissues of the NSCLC patients.
| Sample ID | CpG1 (+52)† | CpG2 (+49) | CpG3 (+33) | CpG4 (−134) | CpG5 (−129) | CpG6 (−124) | CpG7 (−116) | CpG8 (−92) | CpG9 (−76) | CpG10 (−71) | CpG11 (−65) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H | T | H | T | H | T | H | T | H | T | H | T | H | T | H | T | H | T | H | T | H | T | |||||||||||
| 1-080 | 3.73 | 6.51 | 8.29 | 11.32 | 2.57 | 5.17 | 4.63 | 6.77 | 9.67 | 9.89 | 10.59 | 17.51 | 7.275 | 10.39 | 6.435 | 10.39 | 4.435 | 3.675 | 7.24 | 5.25 | 10.51 | 5.7 | ||||||||||
| 1-083 | 5.59 | 3.97 | 9.27 | 8.67 | 2.38 | 3.25 | 5.28 | 4.87 | 9.43 | 5.89 | 11.33 | 12.70 | 9.345 | 7.47 | 7.445 | 6.745 | 3.685 | 5.085 | 4.5 | 5.795 | 5.57 | 5.13 | ||||||||||
| 1-085 | 4.70 | 4.89 | 9.33 | 11.58 | 2.39 | 4.23 | 5.02 | 6.85 | 8.30 | 9.47 | 12.80 | 11.10 | 7.85 | 8.92 | 7.63 | 5.885 | 5.5 | 8.895 | 6.36 | 7.075 | 5.92 | 7.33 | ||||||||||
| 1-091 | 3.63 | 0.00 | 7.21 | 8.66 | 2.57 | 8.25 | 6.26 | 0.00 | 8.78 | 7.56 | 9.61 | 13.03 | 8.735 | 0 | 7.675 | 0 | 4.505 | 3.88 | 10.9 | 5.48 | 12.66 | 5.35 | ||||||||||
| 1-092 | 4.89 | 19.08 | 9.18 | 21.01 | 2.37 | 15.96 | 4.18 | 11.12 | 7.62 | 21.32 | 11.82 | 25.19 | 8.71 | 11.73 | 8.085 | 11.16 | 15.26 | 4.07 | 8.035 | 5.325 | 16.83 | 2.975 | ||||||||||
| 1-097 | 4.61 | 3.88 | 10.17 | 8.41 | 3.47 | 2.89 | 6.52 | 5.40 | 10.53 | 7.09 | 15.41 | 11.80 | 10.75 | 6.605 | 8.655 | 7.195 | 6.515 | 4.435 | 5.995 | 5.71 | 11.79 | 5.69 | ||||||||||
| 1-101 | 6.09 | 2.51 | 10.70 | 6.17 | 3.39 | 2.10 | 4.39 | 3.57 | 8.31 | 4.74 | 11.45 | 9.53 | 7.9 | 6.145 | 6.74 | 5.81 | 8.88 | 4.17 | 5.16 | 5.145 | 10.75 | 5.13 | ||||||||||
| 1-103 | 5.30 | 3.68 | 11.17 | 9.74 | 2.25 | 6.84 | 6.17 | 6.71 | 8.64 | 6.68 | 14.90 | 13.75 | 10.03 | 6.54 | 8.33 | 5.705 | 5.13 | 4.415 | 3.775 | 5.58 | 9.16 | 4.98 | ||||||||||
| 1-104 | 5.50 | 2.83 | 12.49 | 5.98 | 3.35 | 1.77 | 6.79 | 4.13 | 10.90 | 4.53 | 17.10 | 11.21 | 9.855 | 6.945 | 10.37 | 5.98 | 4.85 | 5.765 | 5.7 | 6.465 | 9.125 | 5.875 | ||||||||||
| 1-108 | 4.10 | 3.64 | 8.36 | 7.97 | 3.93 | 3.27 | 5.74 | 4.62 | 7.48 | 6.69 | 13.54 | 11.23 | 8.595 | 9.62 | 7.83 | 5.63 | 5.65 | 6.46 | 6.135 | 7.56 | 7.705 | 7.61 | ||||||||||
| 1-113 | 3.17 | 4.09 | 6.73 | 8.22 | 3.19 | 0.00 | 5.66 | 3.57 | 8.59 | 7.78 | 13.80 | 15.03 | 8.445 | 8.605 | 6.35 | 8.62 | 4.675 | 4.995 | 6.32 | 5.33 | 8.5 | 2.745 | ||||||||||
| 1-116 | 3.67 | 4.41 | 7.51 | 12.54 | 3.24 | 4.34 | 5.79 | 5.51 | 8.65 | 8.04 | 13.79 | 9.70 | 8.72 | 9.895 | 8.65 | 11.05 | 4.59 | 3.92 | 5.695 | 4.73 | 8.32 | 6.915 | ||||||||||
| 1-120 | 3.61 | 7.83 | 8.56 | 10.79 | 3.27 | 10.88 | 6.28 | 6.37 | 10.96 | 11.46 | 12.58 | 16.91 | 8.335 | 8.355 | 5.335 | 7.94 | 3.795 | 4.815 | 6.09 | 5.215 | 6.89 | 4.475 | ||||||||||
| 1-124 | 3.33 | 3.20 | 7.85 | 5.75 | 3.56 | 3.01 | 3.25 | 3.99 | 10.42 | 5.09 | 12.45 | 10.23 | 8.48 | 6.815 | 6.025 | 4.725 | 3.645 | 3.72 | 5.175 | 4.255 | 7.36 | 4.69 | ||||||||||
| 1-126 | 4.74 | 2.69 | 8.85 | 4.69 | 3.28 | 2.60 | 5.50 | 4.20 | 9.61 | 3.66 | 13.90 | 7.13 | 8.795 | 4.14 | 5.705 | 3.17 | 3.705 | 4.925 | 6.45 | 5.89 | 7.485 | 5.65 | ||||||||||
| 1-127 | 4.58 | 3.30 | 7.44 | 6.71 | 4.43 | 2.25 | 7.91 | 5.30 | 7.84 | 5.48 | 12.35 | 9.32 | 6.585 | 6.135 | 6.86 | 4.67 | 5.835 | 6.83 | 5.47 | 6.005 | 8.59 | 6.93 | ||||||||||
| 1-135 | 5.99 | 4.16 | 8.99 | 9.41 | 4.18 | 4.74 | 5.70 | 5.25 | 7.81 | 7.22 | 12.72 | 11.30 | 9.47 | 8.865 | 9.315 | 6.425 | 5.865 | 3.185 | 3.66 | 4.265 | 5.335 | 6.97 | ||||||||||
| 1-143 | 3.90 | 2.90 | 8.65 | 8.46 | 3.01 | 2.37 | 4.76 | 4.05 | 8.32 | 7.13 | 13.11 | 10.98 | 8.045 | 6.395 | 7.525 | 6.4 | 4.155 | 3.065 | 4.805 | 1.455 | 7.51 | 5.58 | ||||||||||
| 1-144 | 4.99 | 3.78 | 11.18 | 7.03 | 2.94 | 2.40 | 6.79 | 4.04 | 12.26 | 6.08 | 16.70 | 10.66 | 9.585 | 5.095 | 8.815 | 5.63 | 3.305 | 2.58 | 7.265 | 2.63 | 8.855 | 5.275 | ||||||||||
| 1-152 | 5.20 | 3.42 | 9.23 | 8.50 | 4.31 | 2.50 | 4.72 | 5.93 | 8.17 | 7.69 | 11.57 | 10.16 | 9.48 | 8.015 | 6.785 | 5.045 | 3.88 | 2.935 | 5.07 | 4.055 | 4.13 | 2.795 | ||||||||||
| 1-155 | 7.67 | 4.40 | 9.92 | 8.63 | 5.47 | 3.46 | 9.40 | 4.54 | 13.35 | 10.06 | 11.96 | 12.37 | 11.45 | 8.4 | 13.12 | 6.41 | 5.125 | 2.655 | 5.995 | 3.065 | 6.575 | 4.865 | ||||||||||
| 1-163 | 3.46 | 6.49 | 7.70 | 9.04 | 5.21 | 6.70 | 5.96 | 5.75 | 8.43 | 7.32 | 14.74 | 14.64 | 6.015 | 8.39 | 8.215 | 10.18 | 4.565 | 3.73 | 3.93 | 4.645 | 5.265 | 5.355 | ||||||||||
| 1-166 | 7.26 | 4.25 | 9.79 | 10.38 | 5.57 | 4.18 | 6.23 | 5.14 | 8.41 | 7.47 | 14.70 | 11.64 | 11.26 | 8.905 | 7.455 | 7.995 | 4.05 | 1.605 | 3.83 | 1.825 | 5.165 | 0 | ||||||||||
| 1-170 | 4.85 | 4.09 | 9.66 | 8.68 | 3.97 | 3.26 | 6.35 | 6.91 | 8.47 | 7.04 | 14.44 | 13.79 | 9.07 | 7.455 | 7.945 | 5.09 | 4.435 | 3.25 | 4.32 | 3.275 | 4.045 | 4.03 | ||||||||||
| 1-171 | 5.06 | 3.58 | 9.73 | 7.92 | 4.08 | 3.72 | 5.46 | 3.76 | 9.08 | 7.59 | 13.77 | 13.00 | 8.3 | 6.075 | 7.18 | 7.9 | 3.78 | 2.435 | 3.735 | 2.35 | 5.805 | 1.655 | ||||||||||
| 1-178 | 3.70 | 4.53 | 10.44 | 9.54 | 3.20 | 2.26 | 6.07 | 6.43 | 9.57 | 8.30 | 14.75 | 10.50 | 9.55 | 8.25 | 9.285 | 5.805 | 3.88 | 1.745 | 5.155 | 4.16 | 6.705 | 4.91 | ||||||||||
| 1-184 | 4.06 | 5.75 | 9.86 | 11.28 | 4.47 | 3.63 | 7.23 | 6.53 | 9.57 | 11.17 | 13.49 | 17.20 | 9.35 | 10.85 | 8.835 | 8.215 | 4.825 | 3.51 | 5.76 | 5.515 | 8.395 | 7.05 | ||||||||||
| 1-195 | 4.92 | 10.05 | 9.06 | 14.19 | 3.74 | 10.82 | 6.44 | 12.50 | 10.06 | 13.07 | 14.06 | 18.19 | 8.985 | 12.82 | 6.35 | 11.32 | 4.78 | 7.61 | 5.5 | 7.535 | 7.805 | 9.415 | ||||||||||
| 1-197 | 4.89 | 4.02 | 9.26 | 8.63 | 4.88 | 2.77 | 5.06 | 6.39 | 8.03 | 6.09 | 12.82 | 12.02 | 8.43 | 6.965 | 5.575 | 6.71 | 4.08 | 1.885 | 5.415 | 2.23 | 7.365 | 1.15 | ||||||||||
| 1-209 | 4.07 | 3.33 | 12.11 | 7.85 | 5.25 | 2.77 | 5.20 | 6.01 | 8.79 | 7.42 | 12.93 | 11.70 | 7.47 | 8.295 | 6.84 | 6.64 | 3.73 | 2.46 | 4.225 | 2.89 | 5.77 | 3.38 | ||||||||||
| 1-213 | 5.41 | 4.52 | 12.58 | 10.07 | 3.58 | 5.08 | 6.28 | 6.87 | 9.33 | 9.40 | 18.60 | 13.55 | 10.85 | 8.17 | 10.3 | 7.21 | 3.325 | 1.01 | 3.53 | 3.965 | 5.87 | 1.63 | ||||||||||
| 1-215 | 4.59 | 4.53 | 9.38 | 9.02 | 4.86 | 2.83 | 7.52 | 5.80 | 10.91 | 6.35 | 15.46 | 13.43 | 8.745 | 7.32 | 8.645 | 7.575 | 3.2 | 2.99 | 4.995 | 1.84 | 5.605 | 0 | ||||||||||
| 1-225 | 4.26 | 3.46 | 9.80 | 6.38 | 3.59 | 3.55 | 6.16 | 4.40 | 9.29 | 5.39 | 12.10 | 9.68 | 9.14 | 5.535 | 7.3 | 5.525 | 4.695 | 2.915 | 6.52 | 3.6 | 6.455 | 3.69 | ||||||||||
| 1-228 | 5.79 | 9.14 | 10.35 | 11.71 | 2.79 | 10.22 | 6.43 | 10.37 | 8.32 | 10.86 | 14.38 | 16.29 | 9.11 | 12.72 | 7.68 | 10.61 | 5.305 | 6.86 | 6.415 | 4.835 | 9.195 | 0 | ||||||||||
| 1-233 | 4.59 | 2.69 | 10.64 | 6.55 | 4.28 | 3.53 | 7.58 | 4.59 | 7.85 | 6.99 | 14.70 | 10.27 | 12.34 | 5.51 | 6 | 4.32 | 3.89 | 6.91 | 4.895 | 6.485 | 6.08 | 0 | ||||||||||
| 1-238 | 4.16 | 4.26 | 9.98 | 5.56 | 4.47 | 1.87 | 6.55 | 3.45 | 9.29 | 5.66 | 14.15 | 10.80 | 8.84 | 5.78 | 7.585 | 5.35 | 4.175 | 6.91 | 7.335 | 9.025 | 6.44 | 9.415 | ||||||||||
†, represents distance from transcriptional start site. Each number represents an average value of the methylation (%) of at least 3 individual PCR/Pyrosequencing reactions, from the same bisulphite-converted DNA. SDs for triplicates should be less than 1%.
Table S2. Methylation status (%) of MyD88 promoter in tumour and adjacent non-tumour tissues of the NSCLC patients.
| Sample ID | CpG1 (−253)† | CpG2 (−256) | CpG3 (−267) | CpG4 (−278) | CpG5 (−210) | CpG6 (−216) | CpG7 (−222) | CpG8 (−146) | CpG9 (−151) | CpG10 (−167) | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H | T | H | T | H | T | H | T | H | T | H | T | H | T | H | T | H | T | H | T | ||||||||||
| 1-001 | 16.09 | 11.47 | 23.97 | 14.95 | 13.57 | 10.61 | 21.55 | 17.4 | 6.2 | 3.68 | 5.805 | 7.435 | 5.77 | 9.83 | 5.425 | 3.725 | 1.975 | 2.33 | 4.88 | 2.415 | |||||||||
| 1-003 | 13.08 | 11.41 | 19.9 | 18.85 | 11.48 | 6.33 | 16.8 | 12.78 | 11.13 | 8.69 | 5.655 | 11.88 | 3.07 | 3.735 | 6.72 | 6.535 | 2.325 | 0 | 0 | 0 | |||||||||
| 1-004 | 12.6 | 6.055 | 19.17 | 13.29 | 6.59 | 9.095 | 11.22 | 12.19 | 10.29 | 6.235 | 11.8 | 0 | 3.44 | 11.42 | 9.865 | 0 | 2.84 | 0 | 0 | 0 | |||||||||
| 1-005 | 12.57 | 8.51 | 19.2 | 14.58 | 9.715 | 6.225 | 16.79 | 14.43 | 6.36 | 4.55 | 7.205 | 10.23 | 9.21 | 13.4 | 5.47 | 0 | 8.585 | 0 | 4.83 | 0 | |||||||||
| 1-006 | 12.96 | 7.09 | 19.07 | 10.23 | 5.17 | 4.955 | 13.62 | 7.98 | 3.45 | 0 | 5.535 | 10.37 | 6.41 | 11.25 | 11.34 | 2.845 | 9.725 | 0 | 3.61 | 6.54 | |||||||||
| 1-008 | 3.495 | 12.07 | 13.36 | 18.94 | 6.525 | 0 | 16.15 | 12.2 | 4.295 | 7.27 | 11.16 | 0 | 0 | 0 | 10.11 | 3.865 | 6.055 | 0 | 3.84 | 3.055 | |||||||||
| 1-009 | 13.04 | 12.76 | 18.27 | 17.37 | 13.6 | 8.945 | 19.06 | 14.51 | 7.125 | 9.39 | 14.33 | 3.165 | 0 | 0 | 7.7 | 0 | 6.885 | 0 | 6.975 | 3.625 | |||||||||
| 1-010 | 12.55 | 8.68 | 20.42 | 16.27 | 4.965 | 3.47 | 16.15 | 12.49 | 7.615 | 0 | 15.64 | 16.04 | 0 | 19.59 | 7.895 | 9.82 | 0 | 2.96 | 3.65 | 6.09 | |||||||||
| 1-012 | 9.315 | 7.09 | 12.79 | 15.9 | 3.54 | 6.45 | 9.775 | 9.36 | 10.01 | 9.415 | 6.465 | 0 | 9.31 | 0 | 3.39 | 9.865 | 4.4 | 3.22 | 3.8 | 6.455 | |||||||||
| 1-014 | 9.08 | 7.625 | 14.16 | 13.93 | 7.435 | 0 | 12.46 | 0.005 | 8.14 | 3.575 | 3.945 | 11.17 | 2.51 | 9.65 | 7.765 | 7.67 | 0 | 0 | 0 | 8.055 | |||||||||
| 1-015 | 12.37 | 11.4 | 14.05 | 14.24 | 7.78 | 3.3 | 8.85 | 8.575 | 1.74 | 12.19 | 6.655 | 14.56 | 0 | 0 | 4.7 | 6.955 | 6.52 | 4.295 | 9.18 | 6.635 | |||||||||
| 1-016 | 13.15 | 7.63 | 20.79 | 9.52 | 4.79 | 8.68 | 16.17 | 12.18 | 0.745 | 0 | 6.155 | 5.545 | 0 | 2.2 | 5.185 | 9.03 | 0 | 2.745 | 3.345 | 0 | |||||||||
| 1-080 | 10.42 | 14.15 | 18.2 | 21.08 | 7.4 | 7.765 | 19.05 | 11.49 | 8.91 | 0 | 5.235 | 5.34 | 8.695 | 4.63 | 5.76 | 4.92 | 0 | 0 | 0 | 0 | |||||||||
| 1-082 | 11.89 | 3.9 | 18.19 | 8.495 | 4.275 | 6.09 | 13.29 | 15.47 | 5.13 | 0 | 6.94 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 5.25 | 0 | |||||||||
| 1-083 | 8.845 | 13.22 | 17.95 | 23.99 | 4.035 | 33.01 | 10.9 | 24.5 | 5.14 | 2.655 | 3.425 | 2.455 | 4.225 | 2.42 | 3.665 | 17.04 | 0 | 0 | 5.9 | 0 | |||||||||
| 1-091 | 5.86 | 10.3 | 29.58 | 16.64 | 18.76 | 5.595 | 14.3 | 12.67 | 3.13 | 2.935 | 13.02 | 4.47 | 5.46 | 4.7 | 4.57 | 2.26 | 4.2 | 0 | 5.81 | 4.93 | |||||||||
| 1-092 | 14.42 | 8.79 | 11.91 | 17.44 | 10.16 | 3.2 | 45.77 | 26.82 | 13.01 | 0 | 16.97 | 8.195 | 15.21 | 7 | 14.57 | 0 | 0 | 0 | 12.58 | 2.695 | |||||||||
| 1-097 | 14.88 | 4.13 | 26.24 | 6.435 | 10.82 | 5.655 | 20.79 | 10.46 | 6.81 | 2.465 | 6.07 | 1.895 | 4.895 | 7.365 | 6.725 | 1.84 | 3.275 | 0 | 10.17 | 0 | |||||||||
| 1-101 | 14.33 | 6.46 | 23.23 | 11.94 | 7.155 | 4.428 | 27.15 | 18.64 | 8.36 | 1.233 | 7.595 | 5.045 | 0 | 7.183 | 4.7 | 0.92 | 7.16 | 0 | 5.33 | 1.348 | |||||||||
| 1-104 | 17.13 | 9.205 | 24.87 | 9.965 | 5.185 | 5.43 | 12.33 | 16.36 | 5.82 | 4.24 | 4.98 | 8.005 | 4.645 | 6.45 | 2.635 | 8.505 | 0 | 0 | 7.385 | 2.485 | |||||||||
| 1-108 | 13.92 | 8.05 | 17.28 | 15.98 | 3.335 | 6.525 | 11.3 | 5.41 | 1.92 | 3.455 | 2.885 | 5.99 | 4.04 | 4.99 | 4.72 | 6.485 | 0 | 1.57 | 6.085 | 2.03 | |||||||||
| 1-113 | 10.59 | 4.67 | 18.45 | 17.9 | 7.935 | 6.275 | 17.52 | 12.56 | 3.185 | 2.73 | 0 | 6.955 | 1.54 | 0 | 1.97 | 0 | 0 | 0 | 1.975 | 5.1 | |||||||||
| 1-116 | 8.685 | 7.19 | 17.62 | 11.51 | 5.25 | 0 | 13.3 | 8.895 | 0 | 3.46 | 0 | 6.31 | 0 | 5.07 | 2.195 | 0 | 0 | 0 | 5.08 | 0 | |||||||||
| 1-120 | 11.21 | 11.02 | 16.41 | 4.86 | 5.375 | 0 | 15.48 | 2.685 | 2.26 | 5.615 | 1.62 | 3.605 | 1.55 | 6.98 | 4.915 | 0 | 1.685 | 0 | 4.48 | 3.695 | |||||||||
| 1-126 | 16.76 | 10.47 | 21.04 | 9.965 | 6.485 | 6.98 | 14.98 | 9.505 | 1.805 | 2.87 | 0 | 5.535 | 0 | 0 | 4.275 | 3.58 | 0 | 0 | 4.535 | 2.715 | |||||||||
| 1-127 | 11.68 | 8.42 | 13.66 | 11.49 | 7.745 | 4.375 | 9.715 | 20.08 | 0 | 4.01 | 0 | 0 | 0 | 2.585 | 4.245 | 11.2 | 0 | 0 | 5.395 | 0 | |||||||||
| 1-135 | 22.35 | 3.215 | 30.23 | 10.18 | 13.13 | 6.96 | 27.05 | 14.48 | 6.745 | 3.255 | 3.83 | 6.86 | 6.8 | 2.82 | 5.155 | 9.395 | 0 | 3 | 5.28 | 0 | |||||||||
| 1-143 | 15.03 | 5.76 | 17.89 | 7.36 | 8.36 | 7.54 | 12.53 | 9.765 | 1.62 | 2.625 | 2.46 | 2.675 | 2.375 | 2.945 | 5.4 | 6.485 | 0 | 0 | 6.205 | 0 | |||||||||
| 1-144 | 9.42 | 7.135 | 14.14 | 7.13 | 8.35 | 2.05 | 20.83 | 8.78 | 5.515 | 2.005 | 5.73 | 5.67 | 0 | 4.32 | 1.81 | 3.715 | 0 | 0 | 7.745 | 0 | |||||||||
| 1-152 | 13.24 | 2.895 | 17.07 | 7.845 | 8.46 | 12.21 | 8.98 | 6.94 | 6.515 | 6.81 | 6.435 | 6.275 | 0 | 4.225 | 2.11 | 18.02 | 0 | 0 | 5.955 | 0 | |||||||||
| 1-155 | 11.97 | 7.605 | 16.02 | 14.73 | 5.91 | 2.52 | 9.14 | 12.93 | 2.5 | 4.32 | 0 | 1.07 | 3.615 | 3.035 | 6.845 | 7.205 | 0 | 2.41 | 2.85 | 4.5 | |||||||||
| 1-163 | 15.45 | 13.6 | 18.39 | 9.695 | 7.23 | 2.805 | 10.86 | 9.985 | 1.86 | 1.31 | 2.68 | 5.375 | 0 | 3.44 | 4.73 | 5.59 | 0 | 1.59 | 2.945 | 4.955 | |||||||||
| 1-166 | 13.61 | 2.745 | 21.74 | 5.025 | 6.71 | 3.735 | 15.53 | 8.71 | 2.565 | 1.55 | 2.14 | 1.405 | 3.985 | 1.885 | 4.495 | 5.4 | 0 | 0 | 2.805 | 6.16 | |||||||||
| 1-170 | 15.02 | 8.335 | 16.02 | 14.9 | 9.635 | 6.02 | 12.76 | 10.46 | 2.765 | 4.445 | 0 | 1.045 | 2.43 | 3.775 | 0 | 7.46 | 0 | 0 | 8.33 | 1.875 | |||||||||
| 1-171 | 8.62 | 10.32 | 17.72 | 6.765 | 0 | 2.02 | 8.81 | 9.9 | 4.955 | 0 | 0 | 8.235 | 2.92 | 6.12 | 9.02 | 0 | 4.9 | 0 | 0 | 6.4 | |||||||||
| 1-184 | 12.23 | 7.855 | 17.78 | 13.75 | 5.36 | 7.245 | 14.26 | 12.72 | 8.82 | 2.645 | 6.3 | 6.67 | 1.61 | 2.565 | 5.45 | 8.625 | 0 | 2.445 | 6.71 | 0 | |||||||||
| 1-193 | 14.61 | 11.73 | 16.75 | 13.35 | 4.23 | 8.37 | 9.635 | 14.76 | 0 | 2.465 | 5.525 | 2.51 | 0 | 2.255 | 0 | 3.53 | 0 | 0 | 6.995 | 5.175 | |||||||||
| 1-195 | 15.69 | 8.055 | 19.28 | 13.52 | 11.78 | 6.555 | 26.52 | 10.17 | 0 | 4.475 | 0 | 2.905 | 0 | 0 | 7.06 | 7.3 | 5.01 | 0 | 4.11 | 6.75 | |||||||||
| 1-197 | 14.05 | 24.24 | 23.09 | 33.77 | 7.995 | 14.11 | 13.55 | 28.45 | 2.975 | 1.755 | 0 | 0 | 7.465 | 6.57 | 0 | 10.96 | 0 | 0 | 2.955 | 18.62 | |||||||||
| 1-207 | 16.1 | 2.92 | 21.99 | 8.3 | 7.615 | 4.205 | 14.45 | 4.97 | 0 | 5.74 | 0 | 6.82 | 2.535 | 2.455 | 2.855 | 0 | 0 | 0 | 3.57 | 0 | |||||||||
| 1-209 | 12.63 | 6.14 | 20.14 | 11.66 | 5.32 | 2.285 | 12.45 | 11.02 | 3.19 | 1.2 | 1.13 | 5.27 | 3.755 | 5.675 | 4.775 | 7.4 | 4.245 | 0 | 4.63 | 7.535 | |||||||||
| 1-213 | 10 | 6.905 | 15.78 | 9.6 | 5.13 | 6.615 | 15.35 | 12.13 | 1.81 | 1.95 | 2.175 | 1.495 | 1.725 | 1.885 | 4.96 | 2.81 | 0 | 4.975 | 8.115 | 0 | |||||||||
| 1-225 | 11.41 | 7.44 | 16 | 12.75 | 4.655 | 9.69 | 9.405 | 11.57 | 1.94 | 1.705 | 1.805 | 0 | 2.18 | 5.545 | 3.48 | 3.395 | 3.355 | 0 | 4.765 | 7.4 | |||||||||
†, represents distance from transcriptional start site. Each number represents an average value of the methylation (%) of at least 3 individual PCR/Pyrosequencing reactions, from the same bisulphite-converted DNA. SDs for triplicates should be less than 1%.
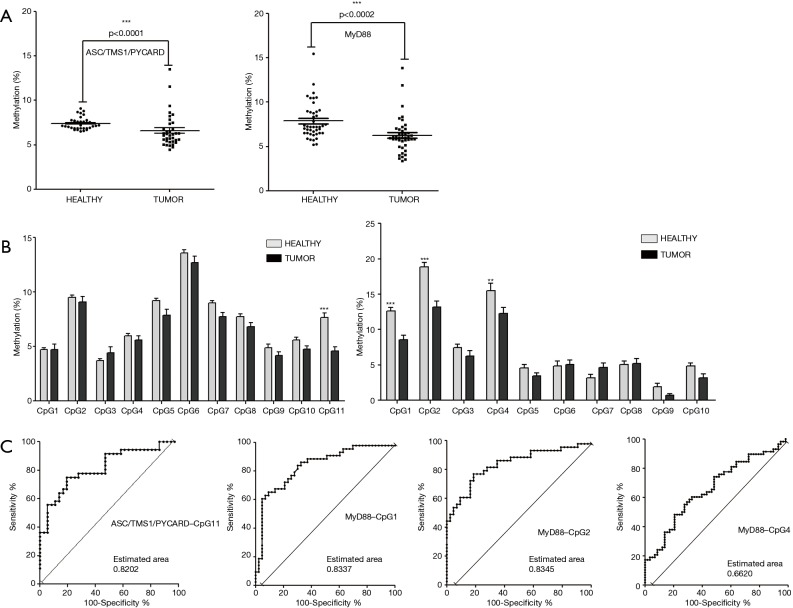
Figure 2.
Methylation status of the ASC/TMS1/PYCARD and MyD88 in the tumor and adjacent non-malignant tissues. (A) Correlation analysis of overall promoter methylation in non-tumor vs. tumor tissue from NSCLC patients. Each data point represents the mean percentage methylation (%) across the tested region of 11 tested CpG sites in the ASC/TMS1/PYCARD gene promotor (left panel) and 10 CpG sites in the MyD88 promoter (right panel). (B) Methylation status of individual CpG sites in the ASC/TMS1/PYCARD and MyD88 gene promoters. Each bar represents mean percentage methylation (%) of individual CpG sites. A grey bar represents healthy tissues and dark bars tumor tissues. The left panel corresponds to ASC/TMS1/PYCARD and right panel to MyD88. For (A) and (B) data were analysed using the paired t-test; P≤0.05 is considered significant. Data are presented as mean ± SEM. **, P≤0.01; ***, P≤0.001. (C) ROC analysis. The panels show the ROC analysis for ASC/TMS1/PYCARD CpG11 (panel 1) and MyD88 CpG1, 2 and 4 (panels 2, 3 and 4) to differentiate tumor from non-tumor tissue.
Aiming to define the diagnostic value of the tested CpG sites, we compared the methylation status (%) of individual CpG sites in tumor and adjacent non-malignant tissues. For ASC/TMS1/PYCARD we found that decreased methylation of CpG site 11, located at position -63, upstream of the transcription start site (TSS), is common trait of all tested NSCLC tumor samples (average 4.56%; 0–9.42%), compared to adjacent non-tumor tissue (average 7.65%; 4.05–16.83%; P<0.001) (Figure 2B, left panel). For MyD88, we found that decreased methylation of CpG site 1 (located at position -253 upstream of the TSS; average 8.57%; 2.75–24.24%), CpG site 2 (located −256 upstream of TSS; average 13.16%; 4.86–33.76%) and CpG site 4 (located at −278 upstream of TSS; average 12.26%; 0–28.45%) are common trait of all tested NSCLC tumor samples compared to non-tumor samples (average CpG1 12.61%, 3.49–22.35%, P<0.001; average CpG2 18.83%, 11.91–30.25%, P<0.001; average CpG4 15.50%; 8.81–45.77%, P<0.01) (Figure 2B, right panel).
In order to check if differentially methylated CpG sites could have some diagnostic applications, we performed ROC curve analysis. The ROC analysis indicated that CpG sites in ASC/TMS1 (CpG11) and MyD88 (CpG1 and CpG2) are promising biomarker candidates for differentiation of NSCLC tissue vs surrounding non-malignant tissue because corresponding areas under curve (AUC) were 80–90% (Figure 2C). Since an AUC ≥0.75 generally indicates a marker that could potentially have clinical utility (42), we classified those CpG sites as a marker with good accuracy. This suggests that methylation status of specific CpG sites in the ASC/TMS1 and MyD88 genes are promising biomarker candidates for differentiation of NSCLC from surrounding non-malignant tissue.
RNA and protein expression of ASC/TMS1/PYCARD and MyD88 in tumor and non-tumor tissue samples from NSCLC patients
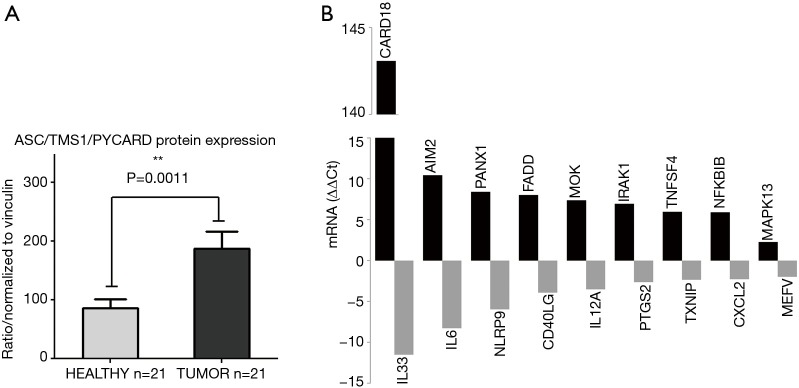
In order to evaluate if the significantly decreased methylation of the ASC/TMS1/PYCARD and MYD88 promoters in tumor tissues, detected in this study, could impact protein expression, we isolated whole proteins from tumor and adjacent non-malignant pairs of tissue lysates. Samples with low whole protein concentration were excluded. We quantified the protein expression level and found that ASC/TMS1/PYCARD and MyD88 are significantly higher expressed in NSCLC tumor tissues compared to non-tumor tissues (P=0.0011 for ASC and P=0.0207 for MyD88) (Figure 3A). These data indicate that the slightly decreased methylation of the ASC/TMS1/PYCARD and MyD88 genes in tumor tissues, observed in this study, could be potentially associated with increased protein expression.
Figure 3.
Protein expression and mRNA expression profile of ASC/TMS1/PYCARD in tumor and non-tumor tissues. (A) Protein expression levels in tumor tissue are significantly higher than in non-tumor tissue for both ASC/TMS1/PYCARD (P=0.0011) and MyD88 (P=0.0207). Tissue lysates were analysed by Western blot with anti-ASC/TMS1/PYCARD, anti-MyD88 and anti-Vinculin antibodies. Intensity of the bands was measured using ImageJ software. Graphs represent the intensity of the respective bands, expressed as relative intensity normalized to Vinculin. Data are presented as mean ± SEM. (B) Profiles of 84 inflammasome-related genes on mRNA level. Total RNA was extracted, transcribed to cDNA and analysed for the expression of 84 genes involved in inflammasome signaling pathway measured with the RT2 Profiler PCR Array-PAHS-097ZA-2. Shown are the fold change values in arbitrary units. Data were normalized to B2M housekeeping gene.
Aiming to investigate the impact of decreased methylation on mRNA expression level in tumor and adjacent non-tumor tissue pairs, we performed RT-qPCR, using a predesigned array comprising 84 inflammasome-related genes, including ASC/TMS1and MyD88. Out of 84 tested panel genes, 77 showed normal fluorescence curves in both runs. Out of these 77 successfully amplified genes, 36 genes were up-regulated in tumor tissue while 9 were downregulated, in comparison to non-tumor tissue. All tested genes and the detailed report of gene expression are shown in Table S3. Tumor specific genes exhibited the highest expression level, with fold-change higher than 5 were: CARD18 (Caspase Recruitment Domain family member 18), AIM2 (Absent In Melanoma 2), PANX1 (Pannexin 1), FADD (Fas Associated via Death Domain), MOK (MOK protein kinase), IRAK1 (Interleukin 1 Receptor Associated Kinase 1), TNFSF4 (Tumor Necrosis Factor SuperFamily member 4), NFKBIB (NFκB Inhibitor Beta), MAPK13 (Mitogen-Activated Protein Kinase 13) and NOD2 (Nucleotide Binding Oligomerization Domain Containing 2). ASC/TMS1/PYCARD was also up-regulated (Fold Up-regulation 2.27), while expression of MyD88 was not changed. Among the most significant tumor-specifically downregulated genes were IL33 (Interleukin 33), IL6 (Interleukin 6) and NLRP9 (NLR Family Pyrin Domain Containing 9) (Figure 3B).
Table S3. Expression of 84 genes involved in inflammasome signaling (RT2 Profiler PCR Array-PAHS-097ZA-2) on mRNA level.
| Gene symbol | Gene name | Level of regulation (fold-change) |
|---|---|---|
| Up-regulated genes | ||
| CARD18 | Caspase recruitment domain family member 18 | 143.07 |
| AIM2 | Absent in melanoma 2 | 10.44 |
| PANX1 | Pannexin 1 | 8.4 |
| FADD | Fas associated via death domain | 8.02 |
| MOK | MOK protein kinase | 7.37 |
| IRAK1 | Interleukin 1 receptor associated kinase 1 | 6.93 |
| TNFSF4 | Tumor necrosis factor superfamily member 4 | 5.96 |
| NFKBIB | NFKB inhibitor beta | 5.9 |
| MAPK13 | Mitogen-activated protein kinase 13 | 5.44 |
| NOD2 | Nucleotide binding oligomerization domain containing 2 | 5.14 |
| HSP90AB1 | Heat shock protein 90 alpha family class B member 1 | 4.88 |
| NLRX1 | NLR family member X1 | 4.83 |
| HSP90B1 | Heat shock protein 90 beta family member 1 | 4.37 |
| MAPK12 | Mitogen-activated protein kinase 12 | 4.37 |
| RELA | RELA proto-oncogene. NF-kB subunit | 3.76 |
| BCL2L1 | BCL2 like 1 | 3.74 |
| XIAP | X-linked inhibitor of apoptosis | 3.51 |
| MAP3K7 | Mitogen-activated protein kinase 7 | 3.36 |
| PEA15 | Phosphoprotein enriched in astrocytes 15 | 3.13 |
| HSP90AA1 | Heat shock protein 90 alpha family class A member 1 | 3.05 |
| MAPK9 | Mitogen-activated protein kinase 9 | 3.03 |
| MAPK1 | Mitogen-activated protein kinase 1 | 2.95 |
| TRAF6 | TNF receptor associated factor 6 | 2.9 |
| TAB2 | TGF-beta activated kinase 1/MAP3K7 binding protein 2 | 2.86 |
| CXCL1 | C-X-C motif chemokine ligand 1 | 2.83 |
| SUGT1 | SGT1 homolog. MIS12 kinetochore complex assembly cochaperone | 2.72 |
| TAB1 | TGF-beta activated kinase 1/MAP3K7 binding protein 1 | 2.65 |
| IL18 | Interleukin 18 | 2.59 |
| IKBKG | Inhibitor of kappa light polypeptide gene enhancer in B-cells. kinase gamma | 2.46 |
| RIPK2 | Receptor interacting serine/threonine kinase 2 | 2.33 |
| CCL2 | C-C motif chemokine ligand 2 | 2.3 |
| PYCARD | PYD and CARD domain containing | 2.27 |
| CHUK | Conserved helix-loop-helix ubiquitous kinase | 2.16 |
| IL1B | Interleukin 1 beta | 2.05 |
| MAPK8 | Mitogen-activated protein kinase 8 | 2.02 |
| CTSB | Cathepsin B | 2.02 |
| Down-regulated genes | ||
| IL33 | Interleukin 33 | −11.52 |
| IL6 | Interleukin 6 | −8.29 |
| NLRP9 | NLR family pyrin domain containing 9 | −5.98 |
| CD40LG | CD40 ligand | −3.93 |
| IL12A | Interleukin 12A | −3.53 |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | −2.64 |
| TXNIP | Thioredoxin interacting protein | −2.36 |
| CXCL2 | C-X-C motif chemokine ligand 2 | −2.29 |
| MEFV | Mediterranean fever | −2 |
| Genes with no change in expression | ||
| BIRC2 | Baculoviral IAP repeat containing 2 | 1.95 |
| MAPK3 | Mitogen-activated protein kinase 3 | 1.89 |
| IRF2 | Interferon regulatory factor 2 | 1.8 |
| NFKB1 | Nuclear factor kappa B subunit 1 | 1.75 |
| CASP5 | Caspase 5 | 1.65 |
| CASP1 | Caspase 1 | 1.62 |
| IL12B | Interleukin 12B | 1.51 |
| NFKBIA | NFKB inhibitor alpha | 1.5 |
| IFNG | Interferon gamma | 1.49 |
| IFNB1 | Interferon beta 1 | 1.46 |
| TIRAP | TIR domain containing adaptor protein | 1.39 |
| IKBKB | Inhibitor of kappa light polypeptide gene enhancer in B-cells. kinase beta | 1.39 |
| CARD6 | Caspase recruitment domain family member 6 | 1.27 |
| NLRP6 | NLR family pyrin domain containing 6 | 1.21 |
| IRF1 | Interferon regulatory factor 1 | 1.2 |
| MYD88 | Myeloid differentiation primary response 88 | 1.17 |
| NLRP1 | NLR family pyrin domain containing 1 | 1.08 |
| NOD1 | Nucleotide binding oligomerization domain containing 1 | −1.04 |
| NLRC5 | NLR family CARD domain containing 5 | −1.06 |
| P2RX7 | Purinergic receptor P2X 7 | −1.25 |
| CASP8 | Caspase 8 | −1.31 |
| NLRP12 | NLR family pyrin domain containing 12 | −1.39 |
| NLRP3 | NLR family pyrin domain containing 3 | −1.41 |
| CCL5 | C-C motif chemokine ligand 5 | −1.41 |
| MAPK11 | Mitogen-activated protein kinase 11 | −1.45 |
| TNF | Tumor necrosis factor | −1.49 |
| CFLAR | CASP8 and FADD like apoptosis regulator | −1.53 |
| NLRC4 | NLR family CARD domain containing 4 | −1.64 |
| CIITA | Class II major histocompatibility complex transactivator | −1.71 |
| PSTPIP1 | Proline-serine-threonine phosphatase interacting protein 1 | −1.73 |
| NAIP | NLR family apoptosis inhibitory protein | −1.76 |
| BIRC3 | Baculoviral IAP repeat containing 3 | −1.79 |
Shown are the fold change values in arbitrary units. Data were normalized to B2M housekeeping gene.
Association of ASC/TMS1/PYCARD and MyD88 promoter methylation with overall survival and tumor stage
In order to evaluate if differentially methylated CpG sites could be associated with any prognostic or diagnostic characteristic, we performed an association study on overall survival (OS), TNM status and lung function.
Association of methylation levels and clinical variables
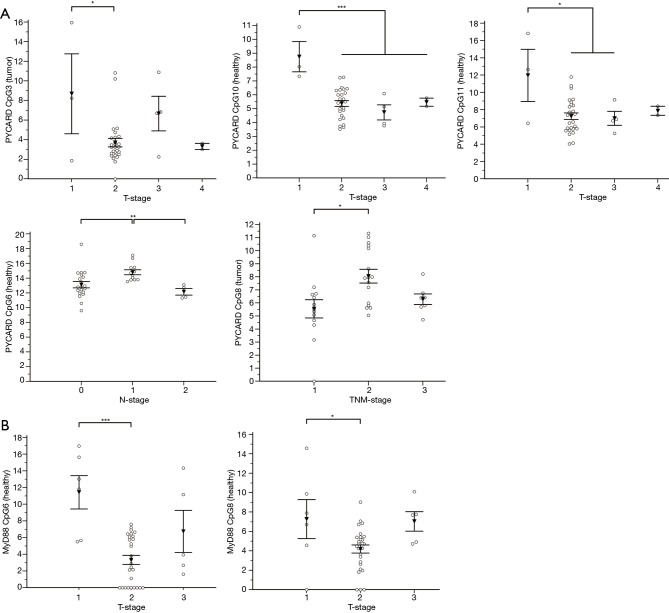
Association analysis of the methylation status of ASC/TMS1/PYCARD promoter CpG sites pointed out that methylation level of certain sites is associated with TNM stage of the NSCLC patients. Post hoc analysis (Tukey-Kramer) showed that CpG3 is significantly higher methylated in tumor samples with T-stage 1 when compared to T-stage 2 (P=0.023). CpG10 is significantly higher methylated in non-malignant samples with T-stage 1 when compared to all other T-stage samples (T2, 3 and 4) (P<0.001). CpG11 is significantly higher methylated in T-stage 1 non-malignant tissues when compared to T-stage 2 and 3 (P=0.016). CpG6 is significantly higher methylated in adjacent non-malignant tissue samples with N-stage 1 when compared to N-stage 2 (P=0.007). M-stage was not analyzed because only 2 patient samples were positive for metastasis. CpG8 is significantly higher methylated in malignant tissues with TNM-stage 2 when compared to TNM-stage 1 (P=0.011) (Figure 4A). Association analysis of the methylation status of MyD88 promoter CpG sites indicated that CpG6 is significantly higher methylated in T-stage 1 malignant samples when compared to T-stage 2 (P<0.001). CpG8 in non-malignant tissues is significantly higher methylated in T-stage 1 samples when compared with T-stage 2 (P<0.016) (Figure 4B). To conclude, it seems that loss of methylation of the specific CpG sites in the ASC/TMS1/PYCARD and MyD88 genes might be indicator of the advanced tumor stage and the regional lymph node status of the NSCLC patients. All results of the association analyses of ASC/TMS1/PYCARD and MYD88 methylation status and investigated clinical variables are presented in Table 2.
Figure 4.
Statistically significant associations of ASC/TMS1/PYCARD and MYD88 methylation levels and clinical variables. (A) Methylation levels of specific ASC/TMS1/PYCARD CpG sites significantly associated with T-, N- and TNM-stage; (B) methylation levels of two MyD88 CpG sites are significantly associated with T-stage. Black rectangle represents mean value and bars represent ± SEM. *, P≤0.05; **, P≤0.01; ***, P≤0.001.
Table 2. Association of ASC/TMS1/PYCARD and MYD88 methylation levels and clinical variables.
| CpG site | Tissue type | ASC/TMS1/PYCARD | MYD88 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| T-stage | N-stage | TNM-stage | Age | T-stage | N-stage | TNM-stage | Age | |||
| CpG1 | H | 0.101 | 0.195 | 0.763 | 0.531 | 0.198 | 0.409 | 0.737 | 0.968 | |
| T | 0.275 | 0.665 | 0.596 | 0.175 | 0.202 | 0.257 | 0.625 | 0.557 | ||
| CpG2 | H | 0.735 | 0.874 | 0.781 | 0.482 | 0.281 | 0.535 | 0.694 | 0.588 | |
| T | 0.381 | 0.771 | 0.702 | 0.095 | 0.458 | 0.369 | 0.404 | 0.672 | ||
| CpG3 | H | 0.642 | 0.455 | 0.436 | 0.091 | 0.387 | 0.864 | 0.909 | 0.795 | |
| T | 0.023 | 0.682 | 0.810 | 0.457 | 0.193 | 0.070 | 0.232 | 0.414 | ||
| CpG4 | H | 0.768 | 0.168 | 0.162 | 0.495 | 0.483 | 0.805 | 0.763 | 0.264 | |
| T | 0.865 | 0.511 | 0.590 | 0.062 | 0.094 | 0.147 | 0.153 | 0.357 | ||
| CpG5 | H | 0.688 | 0.411 | 0.474 | 0.703 | 0.067 | 0.963 | 0.502 | 0.472 | |
| T | 0.173 | 0.469 | 0.630 | 0.025 | 0.339 | 0.620 | 0.662 | 0.796 | ||
| CpG6 | H | 0.322 | 0.007 | 0.170 | 0.913 | <0.001 | 0.345 | 0.810 | 0.824 | |
| T | 0.146 | 0.612 | 0.703 | 0.116 | 0.366 | 0.332 | 0.509 | 0.816 | ||
| CpG7 | H | 0.828 | 0.481 | 0.943 | 0.048 | 0.183 | 0.935 | 0.799 | 0.718 | |
| T | 0.531 | 0.479 | 0.100 | 0.064 | 0.065 | 0.581 | 0.999 | 0.641 | ||
| CpG8 | H | 0.994 | 0.739 | 1.000 | 0.784 | 0.016 | 0.964 | 0.894 | 0.306 | |
| T | 0.764 | 0.132 | 0.011 | 0.474 | 0.309 | 0.267 | 0.182 | 0.832 | ||
| CpG9 | H | 0.052 | 0.721 | 0.557 | 0.428 | 0.598 | 0.488 | 0.330 | 0.358 | |
| T | 0.815 | 0.754 | 0.868 | 0.922 | 0.643 | 0.188 | 0.326 | 0.522 | ||
| CpG10 | H | <0.001 | 0.218 | 0.114 | 0.808 | 0.625 | 0.447 | 0.964 | 0.521 | |
| T | 0.275 | 0.775 | 0.868 | 0.976 | 0.623 | 0.590 | 0.458 | 0.128 | ||
| CpG11 | H | 0.016 | 0.776 | 0.990 | 0.536 | – | – | – | – | |
| T | 0.615 | 0.588 | 0.456 | 0.248 | – | – | – | – | ||
| Average | H | 0.450 | 0.203 | 0.553 | 0.453 | 0.076 | 0.727 | 0.847 | 0.850 | |
| T | 0.386 | 0.656 | 0.584 | 0.146 | 0.424 | 0.241 | 0.389 | 0.919 | ||
Data are presented as P values. Statistically significant P values <0.05 are in italic. H stands for non-tumor tissues and T for tumor tissues.
Association of overall survival with clinical variables and methylation levels
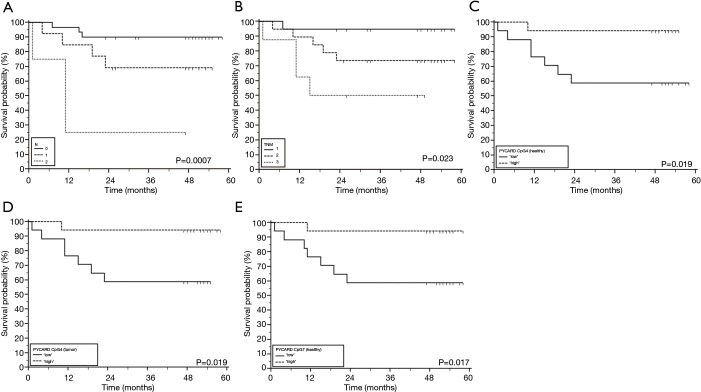
Follow-up data used for OS were obtained from medical records. Survival data was obtained for 49 patients, of whom 12 (24.5%) died during the follow-up period. Two of them died immediately after diagnosis and were excluded from survival analyses. The median follow-up time was 48 months (range, 1–58 months). OS time was measured from the date of diagnosis to the time of death by any cause. Detailed results of survival analysis are presented in Tables 3,4. Among clinicopathological characteristics, both highest N-stage and TNM-stage were significantly associated with shorter OS of lung cancer patients (P=0.0007 and P=0.023, respectively) (Figure 5A,B). TNM-stage 3 patients have five times higher chances to die compared to stage 2 patients (HR 5.31, 95% CI 1.39–20.25) and twelve times higher chances compared to stage 1 patients (HR 12.39, 95% CI 1.88–81.83). When we compared methylation level of the tested CpG sites with OS we found that lower methylation levels of ASC/TMS1/PYCARD CpG4 (below the median value), in both non-tumor and tumor tissue, are associated with shorter OS (P=0.019 for both) (Figure 5C,D). These patients have an eight times higher probability to die compared to patients with higher ASC/TMS1/PYCARD CpG4 methylation levels (HR 8.11, 95% CI 2.02–32.56). Similarly, lower methylation of ASC/TMS1/PYCARD CpG8 in tumor tissue is also associated with shorter overall survival (P=0.017, HR 8.38, 95% CI 2.08–33.70) (Figure 5E). In conclusion, it seems that decreased methylation of the specific CpG sites in the ASC/TMS1/PYCARD gene could be a predictor of poor OS in the NSCLC patients.
Table 3. Impact of clinical variables on overall survival of patients with lung cancer.
| Variable | P value |
|---|---|
| Age at diagnosis | 0.172 |
| Histologic type | 0.558 |
| T-stage | 0.354 |
| N-stage | 0.0007 |
| TNM-stage | 0.023 |
| FEV1 | 0.587 |
Statistically significant P values for log-rank test <0.05 are in italic.
Table 4. Impact of ASC/TMS1/PYCARD and MYD88 methylation levels on overall survival of patients with lung cancer.
| Methylation/CpG site | Tissue type | ASC/TMS1/PYCARD | MYD88 |
|---|---|---|---|
| CpG1 | H | 0.278 | 0.927 |
| T | 0.485 | 0.114 | |
| CpG2 | H | 0.399 | 0.510 |
| T | 0.120 | 0.927 | |
| CpG3 | H | 0.915 | 0.560 |
| T | 0.136 | 0.333 | |
| CpG4 | H | 0.019 | 0.925 |
| T | 0.019 | 0.925 | |
| CpG5 | H | 0.385 | 0.497 |
| T | 0.107 | 0.974 | |
| CpG6 | H | 0.438 | 0.484 |
| T | 0.334 | 0.994 | |
| CpG7 | H | 0.017 | 0.991 |
| T | 0.427 | 0.409 | |
| CpG8 | H | 0.414 | 0.962 |
| T | 0.991 | 0.896 | |
| CpG9 | H | 0.858 | 0.398 |
| T | 0.427 | 0.405 | |
| CpG10 | H | 0.163 | 0.927 |
| T | 0.324 | 0.945 | |
| CpG11 | H | 0.935 | – |
| T | 0.989 | – | |
| Average | H | 0.120 | 0.940 |
| T | 0.427 | 0.135 |
Statistically significant P values for log-rank test <0.05 are in italic. H stands for non-tumor tissues and T for tumor tissues.
Figure 5.
Clinicopathologic characteristics and methylation of ASC/TMS1/PYCARD CpG sites with significant impact on overall survival in patients with lung cancer: (A) N-stage; (B) TNM-stage; (C) ASC/TMS1/PYCARD CpG4 in non-malignant tissue; (D) ASC/TMS1 CpG4 in tumor tissue; (E) ASC/TMS1/PYCARD CpG7 in non-malignant tissue. Tick marks indicate censored cases.
Correlation between pulmonary function and methylation levels
Correlation analysis between FEV1 (lung function measured in forced expiratory volume in 1 second) and ASC/TMS1/PYCARD or MyD88 methylation levels have shown weak positive correlations with level of MyD88 CpG1 site methylation in the tumor tissue (ρ=0.36, P=0.030), CpG2 (ρ=0.49, P=0.002) and average methylation (ρ=0.34, P=0.044). On the contrary, weak negative correlation was observed between ASC/TMS1/PYCARD methylation level of CpG9 in non-tumor tissue and FEV1 (ρ=−0.40, P=0.032). All the results are presented in the Table 5.
Table 5. Correlation between pulmonary function expressed by forced expiratory volume in one second (FEV1) and ASC/TMS1/PYCARD and MYD88 methylation levels.
| CpG site | Tissue type | ASC/TMS1/PYCARD | MYD88 | |||
|---|---|---|---|---|---|---|
| ρ* | P value | ρ* | P value | |||
| CpG1 | H | 0.03 | 0.872 | −0.28 | 0.094 | |
| T | 0.13 | 0.507 | 0.36 | 0.030 | ||
| CpG2 | H | −0.20 | 0.302 | −0.21 | 0.219 | |
| T | 0.22 | 0.246 | 0.49 | 0.002 | ||
| CpG3 | H | −0.29 | 0.131 | 0.09 | 0.590 | |
| T | 0.12 | 0.551 | 0.10 | 0.571 | ||
| CpG4 | H | −0.21 | 0.265 | −0.17 | 0.316 | |
| T | 0.00 | 0.988 | 0.10 | 0.557 | ||
| CpG5 | H | 0.17 | 0.379 | −0.02 | 0.933 | |
| T | 0.24 | 0.212 | 0.27 | 0.113 | ||
| CpG6 | H | −0.17 | 0.367 | −0.14 | 0.417 | |
| T | −0.01 | 0.962 | −0.05 | 0.756 | ||
| CpG7 | H | 0.00 | 0.988 | −0.09 | 0.608 | |
| T | 0.16 | 0.395 | −0.08 | 0.648 | ||
| CpG8 | H | −0.32 | 0.090 | −0.11 | 0.514 | |
| T | 0.12 | 0.538 | −0.08 | 0.650 | ||
| CpG9 | H | −0.40 | 0.032 | 0.05 | 0.776 | |
| T | 0.07 | 0.720 | −0.14 | 0.402 | ||
| CpG10 | H | 0.35 | 0.064 | 0.07 | 0.673 | |
| T | 0.08 | 0.679 | −0.08 | 0.646 | ||
| CpG11 | H | 0.06 | 0.767 | – | – | |
| T | −0.15 | 0.432 | – | – | ||
| Average | H | −0.11 | 0.571 | −0.23 | 0.176 | |
| T | 0.00 | 0.988 | 0.34 | 0.044 | ||
*, statistically significant Spearman’s rank correlation coefficients (ρ) with P values <0.05 are in italic. H stands for non-tumor tissues and T for tumor tissues.
Discussion
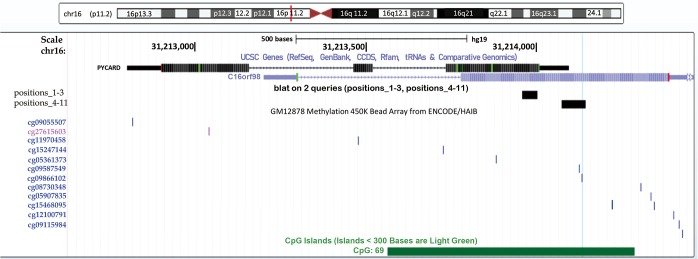
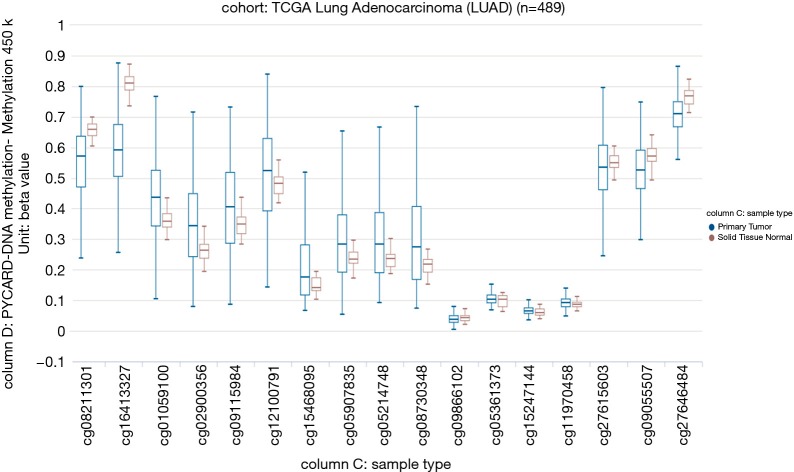
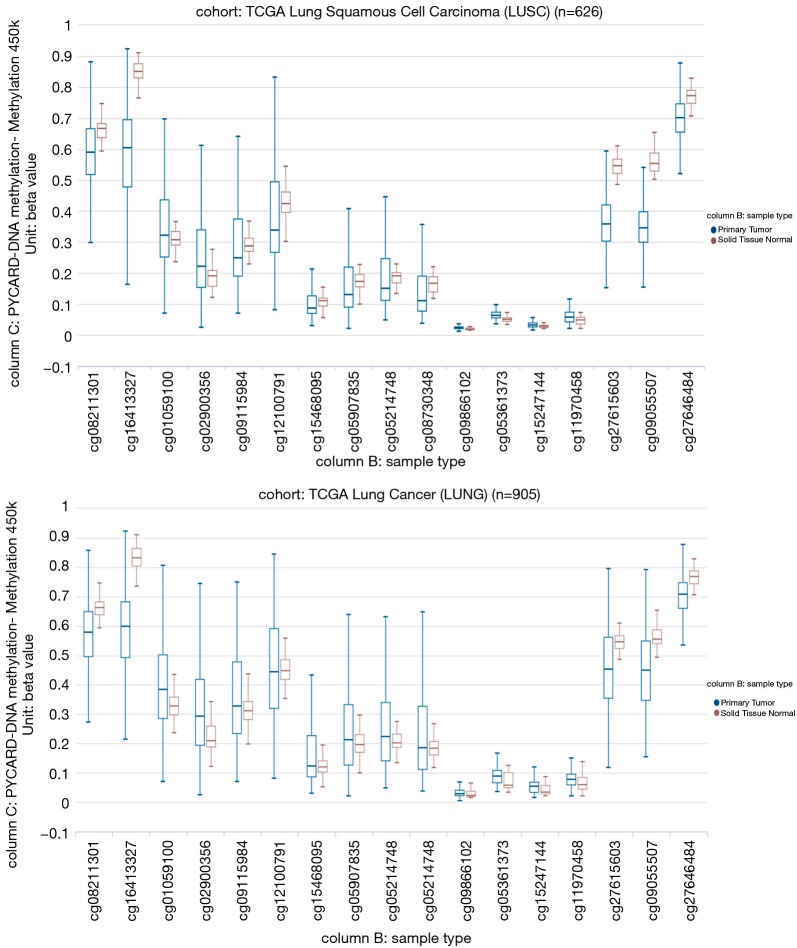
Our study aimed to quantify the methylation status of ASC/TMS1/PYCARD and MyD88 genes by pyrosequencing approach and to evaluate if this could be utilized as potential biomarkers in diagnosis and prognosis of lung cancer. To the best of our knowledge, there is no published data on quantification of the overall methylation status of the selected CpG sites, as well as the methylation at specific loci by pyrosequencing approach. The analysis was performed for ASC/TMS1/PYCARD and MyD88 promoter regions in the tumor and adjacent non-malignant tissues of the NSCLC patient. Here we report decreased methylation of the ASC/TMS1/PYCARD and MyD88 in NSCLC. We found that the overall methylation level of tested genes in NSCLC samples is low, in both tumor and adjacent non-tumor tissue. However, we observed a significantly decreased methylation of the tumor tissues compared to non-malignant tissues that result in increased mRNA and protein expression. All of the above mentioned studies, in the Introduction part of this study, reported aberrant hypermethylation of the ASC/TMS1/PYCARD promoter. However methylation status in those studies was analysed by semi-quantitative approach, predominantly by MSP. In our study we went beyond previously published data and, for the first time, quantified the methylation status of ASC/TMS1/PYCARD and MyD88 promoter regions in NSCLC by a bisulphite pyrosequencing approach. To the best of our knowledge, there is only one study published so far by Wong et al., where the methylation status of ASC/TMS1/PYCARD was quantified by pyrosequencing (43). They quantified methylation status of six tumor suppressor genes, including ASC/TMS1/PYCARD, in exfoliated epithelial cells isolated from breast milk of healthy women. They found that the overall methylation status of 12 tested CpG sites in the ASC/TMS1/PYCARD promoter is 3.67%, which is low and is consistent with our results. In the case of MyD88 we are the first to report both overall methylation and methylation at specific CpG sites, as previously mentioned. As already discussed, published data on ASC/TMS1/PYCARD promoter methylation status does is not consistent with our results. Aiming to resolve the apparent discrepancy, first we have examined publicly available whole genome methylation data (450k Infinium chip) obtained through The Cancer Genome Atlas project (TCGA) (44). The data was visualized through the recently developed USCS XENA (https://www.biorxiv.org/content/early/2018/08/28/326470) viewer (45). Out of 13 CpG sites included in the whole genome 450k Infinium methylation array, only 2 fell within the immediate vicinity of ASC/TMS1/PYCARD start site and overlapped with the pyrosequenced regions (Figure S1). However, the TCGA data was only available for CpG6 (corresponding to cg09866102; Illumina 450K ID) while for CpG7 (corresponding to cg09587549; Illumina 450K ID) no data was available as this probe was probably filtered out during the initial analysis of the TCGA data by the consortium. The data from lung adenocarcinoma subset (LUAD; Figure S2) indicated that cancer tissue was slightly hypomethylated at the cg09866102 probe position (mean beta value 0.0386; n=457) compared to patient matched normal/non-tumor tissue (mean beta value 0.0432; n=32). Although small, the difference was significant (P=2.441e-8). On the other hand the situation in both the TCGA lung squamous cell carcinoma (LUSC) primary carcinoma subset and the combined TCGA lung cancer (LUNG) primary carcinoma dataset (Figure S3) was opposite. In the case of squamous cell cancer subset, normal tissue had a mean of 0.0191 (n=43) and cancer a mean of 0.0239 (n=372) beta methylation value while in the combined set mean normal tissue methylation was 0.0236 (n=75) and cancer 0.0284 (n=830). Taken together, TCGA data indicate that different lung cancer subgroups do show some differences in methylation patterns with adenocarcinoma exhibiting decreased methylation near the ASC/TMS1/PYCARD gene transcription start site, while squamous carcinoma show slightly increased methylation upstream of the start site. Interestingly, this finding is supported by our study; when we stratified the samples in different histopathological subgroups (adenocarcinoma and squamous cell cancer) we observed above mentioned pattern—adenocarcinoma tumors were slightly, but statistically significantly hypomethylated (data not shown). The TCGA data also indicated that methylation varied considerably depending on the position relative to the start site (from almost zero in the immediate vicinity of the start to 60–70% on either side of the gene). On the other hand, it has also been shown that DNA hypomethylation is the important and most constant companion to hypermethylation of the genome in many cancers (46), including lung cancer (47). It seems that DNA hypomethylation is the initial epigenetic abnormality in human tumors (48). DNA hypomethylation in repetitive sequences has been reported to occur in early stages of squamous cell lung cancer, and individuals with hypomethylation in repetitive elements are at a high risk of developing and dying from cancer (49). Indeed, a result of our association study shown that hypomethylation of CpG4 and CpG8 sites in the ASC/TMS1/PYCARD promoter is associated with shorter overall survival. Also, it is very important to emphasize that common use of semi-quantitative approach for methylation analysis, like MSP, is prone to produce false positive results (50). As it was shown by Claus et al., when direct comparison of MSP and pyrosequencing data was conducted on two genes epigenetically silenced in acute myeloid leukemia (AML), ID4 and SFRP, they determine significant overestimation of DNA methylation data by MSP. Therefore, they suggested quantitative approaches, like pyrosequencing, for precise characterization and reliable biomarker detection of aberrant DNA methylation in patient samples (51). With the aim of further confirmation of our results, we analysed mRNA and protein expression and found that decreased methylation status, in our cohort study, correlate with increased protein expression, while increased mRNA expression was confirmed only for ASC/TMS1/PYCARD. These results are mainly consistent with the results of the methylation status, however, one should keep in mind that difference in methylation status, between tumor and non-malignant tissues, although statistically significant, are very small. Therefore, further functional studies that are out of scope of presented study are required to explore if such a small differences could impact mRNA/protein expression. Beside the quantification of the methylation status, we also analysed the correlation of methylation status of each tested CpG site with overall survival and tumor growth of NSCLC patients. We found that hypomethylated CpG4 and CpG8 sites in ASC/TMS1/PYCARD promoter are associated with reduced OS of NSCLC patients. We further speculate that hypomethylation of ASC/TMS1/PYCARD promoter region may correlate with tumor growth. Statistical analysis showed that higher methylation of CpG8 in the ASC/TMS1/PYCARD promoter is associated with TNM grade 2. As for overall survival, this was independent of tumor type. Potential translational usage of this finding could be the detection of transformation of tumor tissue from early stage to more severe forms of tumors. For Myd88 we did not find any correlation of methylation status with overall survival and tumor growth for any of the tested CpG sites. In the end, although we demonstrate here that differentially methylated CpG sites are promising prognostic and diagnostic biomarker candidates, the present work was carried out on a relatively small group of participants and was focused only on two genes. The results we present here require further replication in a larger cohort. Our study is mainly a demonstration of the concept that highlights the utility of the methylation status of certain genes that can be translated into the clinical practice as reliable diagnostic markers, when differentiation of tumor and non-tumor tissues is difficult, and as potential predictors for different clinical outcome in the NSCLC patients.
Figure S1.
Genomic position of the ASC/TMS1/PYCARD gene with locations of pyrosequencing regions and Infinium 450K whole methylation array CPG probes visualized in USCS Genome browser. CPG island is indicated in green while thin blue vertical line indicates CpG position assayed by both methods. Note that ASC/TMS1/PYCARD is transcribed in the opposite direction indicated by small arrows within the gene region.
Figure S2.
Beta values of the TCGA Lung Adenocarcinoma samples in the vicinity of ASC/TMS1/PYCARD gene visualized in XENA USCS browser. Note that for the CpG probe cg09866102 assayed by both methods data indicates small reduction of mean methylation value in cancer compared to matched control tissue of the same patient. Non primary tumors and samples with no measurements were excluded. TCGA stands for The Cancer Genome Atlas project.
Figure S3.
Beta values of the TCGA Lung Squamous Cell Carcinoma samples (top) and combined TCGA Lung Cancer dataset (bottom) visualized in XENA USCS browser. In both cases non primary tumors and samples with no measurements were excluded. TCGA stands for The Cancer Genome Atlas project.
Conclusions
We demonstrated that differentially methylated CpG sites in the promotor region of ASC/TMS1/PYCARD and MyD88 genes could serve for accurate differentiation between malignant and nonmalignant lung tissue specimens. Methylation status of the CpG11 site in the ASC/TMS1/PYCARD promoter as well as CpG1 and 2 in the MyD88 promoter could be considered as good and reliable biomarkers for distinguishing non-tumor from tumor tissue. We also demonstrated that ASC/TMS1/PYCARD CpG4 and CpG8 hypomethylation is associated with decreased overall survival, indicating the prognostic value of tested CpG sites. The published data on this subject is very limited, and for the confirmation of our results a larger number of samples, consisted of different lung tumor types and stages/grades should be analysed.
Acknowledgments
Funding: This work was supported by Croatian Science Foundation; project number IP-2016-06-1441, LungInflaCare (J Knežević) and FP7-REGPOT-2012-2013-1, Grant Agreement Number 316289 InnoMol (O Vugrek).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the local ethic committee at University Hospital Centre Zagreb, Department for Respiratory Diseases (No. 02/21AG). All patients signed informed consent.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Diaz-Lagares A, Mendez-Gonzalez J, Hervas D, et al. A Novel Epigenetic Signature for Early Diagnosis in Lung Cancer. Clin Cancer Res 2016;22:3361-71. 10.1158/1078-0432.CCR-15-2346 [DOI] [PubMed] [Google Scholar]
- 2.Ansari J, Shackelford RE, El-Osta H. Epigenetics in non-small cell lung cancer: from basics to therapeutics. Transl lung cancer Res. Transl Lung Cancer Res 2016;5:155-71. 10.21037/tlcr.2016.02.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan X, Sagara J, Yokoyama T, et al. ASC/TMS1, a caspase-1 activating adaptor, is downregulated by aberrant methylation in human melanoma. Int J Cancer 2003;107:202-8. 10.1002/ijc.11376 [DOI] [PubMed] [Google Scholar]
- 4.Virmani A, Rathi A, Heda S, et al. Aberrant methylation of thecyclin D2 promoter in primary small cell, nonsmall cell lung and breast cancers. Int J Cancer 2003;107:341-5. 10.1002/ijc.11393 [DOI] [PubMed] [Google Scholar]
- 5.Pfeifer GP. Defining Driver DNA Methylation Changes in Human Cancer. Int J Mol Sci 2018;19:1166. 10.3390/ijms19041166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brzeziańska E, Dutkowska A, Antczak A. The significance of epigenetic alterations in lung carcinogenesis. Mol Biol Rep 2013;40:309-25. 10.1007/s11033-012-2063-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsou JA, Hagen JA, Carpenter CL, et al. DNA methylation analysis: a powerful new tool for lung cancer diagnosis. Oncogene 2002;21:5450-61. 10.1038/sj.onc.1205605 [DOI] [PubMed] [Google Scholar]
- 8.Balkwill F, Coussens LM. Cancer: An inflammatory link. Nature 2004;431:405-6. 10.1038/431405a [DOI] [PubMed] [Google Scholar]
- 9.Multhoff G, Molls M, Radons J. Chronic inflammation in cancer development. Front Immunol 2012;2:98. 10.3389/fimmu.2011.00098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hosseinian N, Cho Y, Lockey RF, et al. The role of the NLRP3 inflammasome in pulmonary diseases. Ther Adv Respir Dis 2015;9:188-97. 10.1177/1753465815586335 [DOI] [PubMed] [Google Scholar]
- 11.Maris NA, Dessing MC, de Vos AF, et al. Toll-like receptor mRNA levels in alveolar macrophages after inhalation of endotoxin. Eur Respir J 2006;28:622-6. 10.1183/09031936.06.00010806 [DOI] [PubMed] [Google Scholar]
- 12.Barchet W, Krug A, Cella M, et al. Dendritic cells respond to influenza virus through TLR7- and PKR-independent pathways. Eur J Immunol 2005;35:236-42. 10.1002/eji.200425583 [DOI] [PubMed] [Google Scholar]
- 13.Ospelt C, Gay S. TLRs and chronic inflammation. Int J Biochem Cell Biol 2010;42:495-505. 10.1016/j.biocel.2009.10.010 [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto M, Sato S, Mori K, et al. Cutting Edge: A Novel Toll/IL-1 Receptor Domain-Containing Adapter That Preferentially Activates the IFN-Promoter in the Toll-Like Receptor Signaling. J Immunol 2002;169:6668-72. 10.4049/jimmunol.169.12.6668 [DOI] [PubMed] [Google Scholar]
- 15.Franz KM, Kagan JC. Innate Immune Receptors as Competitive Determinants of Cell Fate. Mol Cell 2017;66:750-60. 10.1016/j.molcel.2017.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Nardo D, Balka KR, Cardona Gloria Y, et al. Interleukin-1 receptor-associated kinase 4 (IRAK4) plays a dual role in myddosome formation and Toll-like receptor signaling. J Biol Chem 2018;293:15195-207. 10.1074/jbc.RA118.003314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 2010;11:373-84. 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- 18.Howrylak JA, Nakahira K. Inflammasomes: Key Mediators of Lung Immunity. Annu Rev Physiol 2017;79:471-94. 10.1146/annurev-physiol-021115-105229 [DOI] [PubMed] [Google Scholar]
- 19.Guo B, Fu S, Zhang J, et al. Targeting inflammasome/IL-1 pathways for cancer immunotherapy. Sci Rep 2016;6:36107. 10.1038/srep36107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottschlich A, Endres S, Kobold S. Can we use interleukin-1β blockade for lung cancer treatment? Transl lung cancer Res 2018;7:S160-4. 10.21037/tlcr.2018.03.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin C, Zhang J. Inflammasomes in Inflammation-Induced Cancer. Front Immunol 2017;8:271. 10.3389/fimmu.2017.00271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He Q, Fu Y, Tian D, et al. The contrasting roles of inflammasomes in cancer. Am J Cancer Res 2018;8:566-83. [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Q, Jin J, Ying J, et al. Epigenetic inactivation of the candidate tumor suppressor gene ASC/TMS1 in human renal cell carcinoma and its role as a potential therapeutic target. Oncotarget 2015;6:22706-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine JJ, Stimson-Crider KM, Vertino PM. Effects of methylation on expression of TMS1/ASC in human breast cancer cells. Oncogene 2003;22:3475-88. 10.1038/sj.onc.1206430 [DOI] [PubMed] [Google Scholar]
- 25.Riojas MA, Guo M, Glöckner SC, et al. Methylation-induced silencing of ASC/TMS1, a pro-apoptotic gene, is a late-stage event in colorectal cancer. Cancer Biol Ther 2007;6:1710-6. 10.4161/cbt.6.11.4829 [DOI] [PubMed] [Google Scholar]
- 26.Martinez R, Schackert G, Esteller M. Hypermethylation of the proapoptotic gene TMS1/ASC: prognostic importance in glioblastoma multiforme. J Neurooncol 2007;82:133-9. 10.1007/s11060-006-9264-4 [DOI] [PubMed] [Google Scholar]
- 27.Zhang C, Li H, Zhou G, et al. Transcriptional silencing of the TMS1/ASC tumour suppressor gene by an epigenetic mechanism in hepatocellular carcinoma cells. J Pathol 2007;212:134-42. 10.1002/path.2173 [DOI] [PubMed] [Google Scholar]
- 28.Schinke C, Mo Y, Yu Y, et al. Aberrant DNA methylation in malignant melanoma. Melanoma Res 2010;20:253-65. 10.1097/CMR.0b013e328338a35a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grau E, Martinez F, Orellana C, et al. Epigenetic alterations in disseminated neuroblastoma tumour cells: influence of TMS1 gene hypermethylation in relapse risk in NB patients. J Cancer Res Clin Oncol 2010;136:1415-21. 10.1007/s00432-010-0796-9 [DOI] [PubMed] [Google Scholar]
- 30.Virmani A, Rathi A, Sugio K, et al. Aberrant methylation ofTMS1 in small cell, non small cell lung cancer and breast cancer. Int J Cancer 2003;106:198-204. 10.1002/ijc.11206 [DOI] [PubMed] [Google Scholar]
- 31.Terasawa K, Sagae S, Toyota M, et al. Epigenetic Inactivation of TMS1/ASC in Ovarian Cancer. Clin Cancer Res 2004;10:2000-6. 10.1158/1078-0432.CCR-0932-03 [DOI] [PubMed] [Google Scholar]
- 32.Collard RL, Harya NS, Monzon FA, et al. Methylation of the ASC gene promoter is associated with aggressive prostate cancer. Prostate 2006;66:687-95. 10.1002/pros.20371 [DOI] [PubMed] [Google Scholar]
- 33.Siraj AK, Hussain AR, Al-Rasheed M, et al. Demethylation of TMS1 Gene Sensitizes Thyroid Cancer Cells to TRAIL-Induced Apoptosis. J Clin Endocrinol Metab 2011;96:E215-24. 10.1210/jc.2010-0790 [DOI] [PubMed] [Google Scholar]
- 34.Wang EL, Qian ZR, Nakasono M, et al. High expression of Toll-like receptor 4/myeloid differentiation factor 88 signals correlates with poor prognosis in colorectal cancer. Br J Cancer 2010;102:908-15. 10.1038/sj.bjc.6605558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang F, Ni Z, Zhan Y, et al. Increased expression of MyD88 and association with paclitaxel resistance in breast cancer. Tumour Biol 2016;37:6017-25. 10.1007/s13277-015-4436-5 [DOI] [PubMed] [Google Scholar]
- 36.Zhu J, Li Q, He J, et al. Expression and significance of myeloid differentiation factor 88 in non-small cell lung carcinoma and normal paracancerous tissues. Genet Mol Res 2015;14:14239-45. 10.4238/2015.November.13.7 [DOI] [PubMed] [Google Scholar]
- 37.Wu L, Zhang C, Wang X, et al. Methylation of ASC/TMS1 promoter is associated with poor prognosis of patients with gastric cancer. Clin Transl Oncol 2016;18:296-303. 10.1007/s12094-015-1367-y [DOI] [PubMed] [Google Scholar]
- 38.Zhang Z, Tan S, Zhang L. Prognostic Value of Apoptosis-Associated Speck-like Protein Containing a CARD Gene Promoter Methylation in Resectable Non-Small-Cell Lung Cancer. Clin Lung Cancer 2006;8:62-5. 10.3816/CLC.2006.n.035 [DOI] [PubMed] [Google Scholar]
- 39.Machida EO, Brock MV, Hooker CM, et al. Hypermethylation of ASC/TMS1 Is a sputum marker for late-stage lung cancer. Cancer Res 2006;66:6210-8. 10.1158/0008-5472.CAN-05-4447 [DOI] [PubMed] [Google Scholar]
- 40.Lee ES, Issa JP, Roberts DB, et al. Quantitative Promoter Hypermethylation Analysis of Cancer-Related Genes in Salivary Gland Carcinomas: Comparison with Methylation-Specific PCR Technique and Clinical Significance. Clin Cancer Res 2008;14:2664-72. 10.1158/1078-0432.CCR-07-1232 [DOI] [PubMed] [Google Scholar]
- 41.Lim W, Ridge CA, Nicholson AG, et al. The 8(th) lung cancer TNM classification and clinical staging system: review of the changes and clinical implications. Quant Imaging Med Surg 2018;8:709-18. 10.21037/qims.2018.08.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radhakrishna U, Vishweswaraiah S, Veerappa AM, et al. Newborn blood DNA epigenetic variations and signaling pathway genes associated with Tetralogy of Fallot (TOF). PLoS One 2018;13:e0203893. 10.1371/journal.pone.0203893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong CM, Anderton DL, Smith-Schneider S, et al. Quantitative analysis of promoter methylation in exfoliated epithelial cells isolated from breast milk of healthy women. Epigenetics 2010;5:645-55. 10.4161/epi.5.7.12961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68-77. 10.5114/wo.2014.47136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res 2002;12:996-1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfeifer GP, Rauch TA. DNA methylation patterns in lung carcinomas. Semin Cancer Biol 2009;19:181-7. 10.1016/j.semcancer.2009.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rauch TA, Zhong X, Wu X, et al. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci USA 2008;105:252-7. 10.1073/pnas.0710735105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gama-Sosa MA, Slagell VA, Trewyn RW, et al. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res 1983;11:6883-94. 10.1093/nar/11.19.6883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 1983;301:89-92. 10.1038/301089a0 [DOI] [PubMed] [Google Scholar]
- 50.Lim AM, Candiloro ILM, Wong N, et al. Quantitative methodology is critical for assessing DNA methylation and impacts on correlation with patient outcome. Clin Epigenetics 2014;6:22. 10.1186/1868-7083-6-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Claus R, Wilop S, Hielscher T, et al. A systematic comparison of quantitative high-resolution DNA methylation analysis and methylation-specific PCR. Epigenetics 2012;7:772-80. 10.4161/epi.20299 [DOI] [PMC free article] [PubMed] [Google Scholar]