Abstract
Thermal performance curves (TPCs) are used to predict changes in species interactions, and hence, range shifts, disease dynamics and community composition, under forecasted climate change. Species interactions might in turn affect TPCs. Here, we investigate how temperature-dependent changes in a microbial host–parasite interaction (the bacterium Pseudomonas fluorescens, and its lytic bacteriophage, SBW) changes the host TPC and the ecological and evolutionary mechanisms underlying these changes. The bacteriophage had a narrower thermal tolerance for infection, with their critical thermal maximum ~6 °C lower than those at which the bacteria still had high growth. Consequently, in the presence of phage, the host TPC changed, resulting in a lower maximum growth rate. These changes were not just driven by differences in thermal tolerance, with temperature-dependent costs of evolved resistance also playing a major role: the largest cost of resistance occurred at the temperature at which bacteria grew best in the absence of phage. Our work highlights how ecological and evolutionary mechanisms can alter the effect of a parasite on host thermal performance, even over very short timescales.
Subject terms: Climate-change ecology, Evolution, Bacteriophages
Introduction
An often overlooked concern surrounding climate change is its impacts on host–parasite interactions [1]. The effect of temperature on species interactions is likely widespread, as temperature influences the physiology, ecology and evolution of both hosts and parasites [2–5]. However, the sign and strength of the effects of warming on host–parasite interactions may be context dependent, changing with the host, parasite and environmental conditions in question [6]. One approach to predict the potential impacts of warming on host–parasite interactions has been based around thermal performance curves (TPCs) of, and differences between, key host and parasite traits [2, 6, 7]. For example, it has been argued that as hosts generally have a narrower thermal range and lower thermotolerance than their parasites [8–10], they are more susceptible to disease at temperatures further away from their optimum temperature.
A probable consequence of temperature-dependent changes in host–pathogen interactions [11] is a change in the host’s TPC in the presence, versus the absence, of the parasite. For example, if the largest impact of the parasite occurs at the host’s optimum growth temperature, key traits such as the maximum growth rate, and optimum temperature of the host could change. In addition to the ecological feedbacks resulting from differences in the thermal performance of host and parasite traits, rapid (co)evolution of resistance and infectivity traits could play a major role in altering TPCs [12, 13]. Crucially, TPCs of hosts and parasites are typically assumed to be fixed across time and in different abiotic and biotic environments [6, 8, 14, 15], but the presence of a predator can alter the TPC of the prey [16] and the prey’s evolutionary response to warming [17]. If parasites affect the thermal performance of their host, this may alter some of the predictions of range shifts and disease dynamics expected under climate change.
To date, most experimental and theoretical work on the thermal performance of organisms is done on single species under highly controlled conditions, where naturally occurring parasites, symbionts and microbiota are greatly or completely removed [18–20]. Consequently, it is unknown if parasites alter the TPC of host fitness and influence key species-level traits such as the optimal, Topt, and cardinal (critical thermal maximum, CTmax, and minimum, CTmin) temperatures of host growth. Understanding these potential impacts is critical to assess the effect of climate change on ecological and evolutionary dynamics of host–parasite pairs, as well as predicting the consequences of novel host–parasite interactions that will occur in a warmer world. Here, we explicitly determine how and why interactions with a parasite affect host thermal performance in arguably the most common host–pathogen interaction on the planet: that between bacteria and their viruses (bacteriophage) [21].
We focus on a well-studied system, the bacterium Pseudomonas fluorescens SBW25 and its lytic phage, SBW. This system has been used extensively for studying host–parasite ecological and evolutionary interactions [22–25]. Over a wide range of temperatures, we measured the replication rate of the phage and the growth rate of the bacteria in the presence and absence of the phage. We utilised the ‘traits’ that underpin TPCs to compare biologically meaningful parameters [15]. We hypothesised that any large difference in thermal performance of bacteria and phage would change the thermal performance of bacteria in the presence vs. the absence of phage. Given the importance of evolution occurring over ecological timescales [26, 27], especially in microbial populations with large population sizes and short generation times, we also investigated evolutionary changes in host populations to determine whether resistance evolution explained any changes in host thermal performance.
Materials and methods
Measuring bacterial growth in the presence and absence of phage
Isogenic P. fluorescens SBW25 was cultured overnight (from a frozen stock) at 28 °C in 6 mL of M9 minimal salts media (M9), supplemented with 5 g of glycerol and 10 g of peptone (50 % concentration of King’s medium B) in glass vials at 180 r.p.m. Overnight stocks were then diluted to ~ 50,000 cells per 10 µL (5 × 106 cells per mL). Growth curves were measured in 96 well plates, with 180 µL of altered M9 (described above). We inoculated wells with 10 µL of bacteria and either 10 µL of M9 or 10 µL of phage (~50 phage) giving a multiplicity of infection (MOI) of 0.001. We used this low MOI and low starting densities to ensure rapid bacterial growth. Six wells were left free for both bacteria and bacteria plus phage treatments at each temperature as blank controls. We set up six replicates of bacteria and bacteria plus phage simultaneously at 8 temperatures (15, 20, 25, 28, 30, 33, 35 and 37 °C). Each plate was placed in a plastic box with a moist sponge at the bottom to prevent evaporation of media from the wells, which may confound measurements of optical density (OD). OD (600 nm wavelength) was measured as a proxy for density of P. fluorescens using a plate reader (Biotek Instruments Ltd). Readings of OD were taken with the lid off at an average of every 3 hr for up to 75 hr.
Measuring phage replication rate
Replication of the lytic phage SBW was measured using methods similar to Knies et al. [28, 29], at the same temperatures as the bacterial growth curves, with the addition of 3 additional temperatures (22.5 °C, 26 °C and 27 °C) to better characterise temperatures around the optimum of phage replication. First, isogenic P. fluorescens was grown overnight in conditions described above. The bacteria were transferred into fresh media at 28 °C and allowed to grow for 6 hr while shaking to increase density (~107 cells). We then added 20 µL of phage (~106; MOI ~0.02, N0) to each tube (six replicates per temperature). Vials were left static for 4 hr at each temperature, after which phage was extracted using chloroform extraction. 100 µL of chloroform was added to 900 µL of culture, then vortexed and centrifuged at 10,000 × g for 5 min. The supernatant was removed and placed in fresh Ependorf tubes. Final phage titres, N4, were measured using plaque assays against the ancestral bacteria at 28 °C. Phage replication rate, r, was then calculated as .
Measuring resistance of bacteria
To investigate the mechanism behind any effect of phage on bacterial growth, we measured the resistance of bacteria within a single growth curve. We set up 18 wells of 96 well plates at 8 temperatures that contained ~50,000 cells and ~50 phage (as described above). We then destructively sampled 6 wells at three time points through the growth curve (after 12, 24 and 48 hr). To do this, 20 µL of each well was placed in 180 µL of M9. These were then serially diluted and plated onto KB agar. Twelve colonies from each replicate were taken per time point and grown overnight in 150 µL of altered M9, shaking at 28 °C. Each clone was then checked for resistance against the ancestral phage using a phage streak assay. Phage streak assays were incubated overnight at 28 °C.
Measuring the cost of resistance
To determine whether any effect of phage was due to a cost of resistance, we grew 12 replicates of P. fluorescens in the presence and absence of phage for 12 hr at 28 °C. After 12 hr, each population was plated onto KB agar and grown for 2 days at 28 °C. Three clones were isolated from each replicate and grown for two days in modified M9 media. Each isolate was checked for resistance against the ancestral phage. Growth curves of each clone were done using the methods described above, but inoculate density was ~500,000 cells to reduce the lag time and no phage was added.
Statistical analyses
Calculating exponential growth rate for bacteria
For bacterial growth, we wanted to estimate exponential population growth rate in the presence and absence of phage, and for resistant and susceptible clones. In the presence and absence of phage, prior to model fitting, we removed 3.42% of points (Figs. S1–S8) in order to obtain the best estimate of exponential growth at each temperature. The results were qualitatively unchanged by the data cleaning procedure (Fig. S9). For a full explanation of the data cleaning procedure please see the supplementary methods section. After this initial data cleaning, we fitted the Gompertz model [30] to measurements of log10OD600 through time, t, in hours, using code extracted from the R package ‘nlsMicrobio’ [31]:
| 1 |
where log10n0 is the starting density, log10nmax is carrying capacity, r is the exponential growth rate (hr−1) and lag is the lag time in hours. Model fitting was done using non-linear least squares regression using the R package ‘nls.multstart’ [32]. This method of model fitting involved running up to 500 iterations of the fitting process with start parameters drawn from a uniform distribution and retaining the fit with the lowest Akaike Information Criterion score (AIC). The parameters of the model (r, log10n0, log10nmax and lag) can be seen as population-level growth ‘traits’, which may vary with both temperature and the presence and absence of phage. In this study, r is defined as exponential growth rate of the population and lag is likely determined by the time it takes until growth is detected by the OD reader. Consequently, lag time confounds any actual lag phase with decreases in abundance and slower growth rates that increase the time it takes for abundance to be detected. Other growth models were fitted (e.g. Baranyi, Buchanan; Table S1), but the Gompertz model returned lower AIC scores for the majority of model fits (Fig. S10).
For susceptible and resistant clones, we cleaned the data by removing the first measurement (where bubbles due to pipetting could alter the OD reading) and setting time zero to the time at which the first optical density measurement was detected for each clone. We initially used the same modelling approach, but this time the Baranyi model without lag was the model most selected using AIC scores (Fig. S11). However, after examining the predictions and residuals of the model fits (Figs. S12 and S13), we found that exponential growth rate was underestimated at temperatures where bacteria grew best, and at these temperatures there was a significantly greater underestimation of growth rate in susceptible, rather than resistant, bacteria (Fig. S14). Consequently, exponential growth rate per clone was calculated here using rolling regression, taking the steepest slope of the linear regression between ln OD600 and time in hours in a shifting window of every 4 time points (~7 hr) as the estimate of exponential growth. Average growth rate per replicate was calculated by taking the mean clonal growth rate. After data cleaning and model fitting, every growth curve had estimates of exponential growth rate that were then used to model the thermal performance of bacteria.
Fitting thermal performance curves to phage and bacteria
Thermal performance curves were fitted for phage replication rate and r of bacteria in the presence and absence of phage, and for resistant and susceptible bacterial clones. We used the Sharpe–Schoolfield equation for high-temperature inactivation [33], which extends the original Boltzmann equation to incorporate a decline in growth rate beyond the optimum.
| 2 |
b(T) is the rate of phage replication or bacterial growth at temperature, T, in Kelvin (K). Instead of the intercept being at 0 K (−273.15 °C), b(Tc) is the rate at a common temperature, Tc = 20 °C (293.15 K) [34]. E (eV) describes the thermal sensitivity of the biological rate, k is Boltzmann’s constant (8.62 × 10−5 eV K−1), Eh (eV) characterises the decline in the rate past the optimum temperature and Th (K) is the temperature at which half the rate is reduced due to high temperatures. Equation 2 yields an optimum temperature, Topt, (K).
| 3 |
Maximal growth rate, rmax, was calculated by using the estimated model parameters to predict the rate at Topt. As in previous studies [18, 19], these ‘traits’ were then used to look for differences between (1) bacteria in the presence and absence of phage, and (2) resistant and susceptible bacteria. Similar species-level ‘traits’ are used in climate change research to explain range shift dynamics [15, 35], but how they are influenced by species interactions remains relatively unknown [16]. As phage replication was negative at high temperatures, an offset was added to the equation to raise all rates above 0 to allow model fitting. This invalidated any interpretation of the thermal sensitivities of phage replication. However, this was already difficult as phage replication is partially determined by bacteria growth rate, which is also temperature dependent and could cause differences in the number of susceptible hosts across temperatures. Consequently, for phage replication we concentrated on the optimum temperature (Topt) and critical thermal maximum (CTmax), which is the temperature at which phage replication became negative at high temperatures.
For phage and bacteria, Eq. 2 was fitted to the data using non-linear regression in a Bayesian framework using the R package ‘brms’ [36]. This method allows for prior information on suitable parameter values and the estimation of uncertainty around predictions and parameters, including derived parameters not present in the original model formulation such as Topt, CTmax and rmax. Different models were fitted for phage replication rate, exponential growth rate of bacteria in the presence and absence of phage, and exponential growth rate of resistant and susceptible bacterial clones. For the analysis including resistant and susceptible clones, a random effect was added to account for the non-independence of measurements of the same clone across temperatures. For bacteria exponential growth rate, phage presence/absence or susceptible/resistance was added as a factor that could alter each parameter of the model. Models were run for 5000 iterations and 3 chains were used with uninformative priors. Model convergence was assessed using posterior predictive checks, Rhat values (all values were 1) and manually checking of chain mixing. Differences between parameter estimates are described using 95% credible intervals. Credible intervals of predictions and parameters were calculated from the posterior distribution using the R package ‘tidybayes’ [37]. Non-overlapping 95% credible intervals indicate statistical significance at (at least) the p = 0.05 level.
Using predictions from the model for bacterial growth, the relative fitness of bacteria in the presence of phage was estimated across the continuous temperature range (15–37 °C). The difference was calculated as a selection coefficient, where relative fitness at each temperature, , was calculated as:
| 4 |
where is the growth rate at a given temperature in the presence of phage and is the growth rate in the absence of phage. When the 95% credible intervals of the predictions do not cross 1, it indicates that phage significantly altered bacterial growth rate. When there is overlap with the predictions and 1, it means there is no significant change in relative fitness. An identical statistical approach was taken for analysing the growth rates of susceptible and resistant clones. In this instance, the relative fitness across temperatures, , represented the cost of resistance.
Analysing phage resistance assays
A logistic regression was used to analyse the proportion of resistance through time and across temperatures. A binomial model was fitted to the number of resistant and susceptible individuals per replicate at each temperature and time point using the logit transformation. As there were many populations where all clones were completely susceptible or resistant (resulting in zero and one inflated data), we added one to both the number of resistant and susceptible individuals in each population and used a quasibinomial error structure to control for overdispersion. By adding one to both susceptible and resistant totals, it meant that the model tended to produce slight underestimates for resistance in fully resistant populations, and slight overestimates of resistance in fully susceptible populations, while having little effect on populations with intermediate resistance. This led to the model giving conservative estimates of differences in resistance between temperatures and through time. We fitted a model that combined the number of resistant and susceptible clones in a population as the response variable and included temperature and time (in hours) as discrete predictor variables. Model selection was done through likelihood ratio tests using F tests. Pairwise post-hoc comparisons were done on the response scale using the R package ‘emmeans’ [38]. All analyses were done using the statistical programming language R (v3.5.1) [39] and all plots were made using the R package ‘ggplot2’ [40].
Results
Bacteria and phage had mismatches in their thermal performance
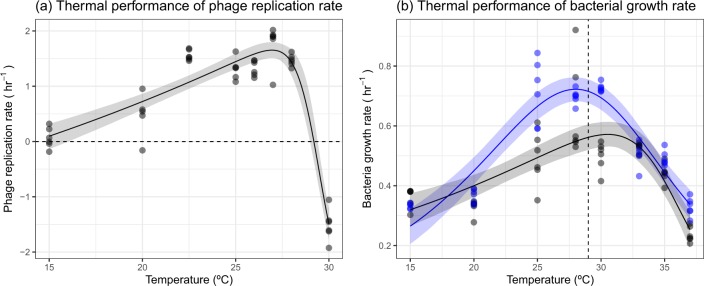
We measured phage replication rate and bacterial growth rate across eight temperature (15–37 °C) to determine whether there were mismatches in the thermal performance of the host and its parasite. To do this, we modelled the thermal performance curve of each rate and used estimated and derived parameters of the model (see Eq. 2 in Methods) as traits that we used to compare the thermal responses of bacteria and phage. Phage replication rate increased to a thermal optimum, , of 27 °C (95% credible intervals [CI]: 26.5–27.5 °C) before rapidly declining to a negative replication rate by 30 °C (Fig. 1a). The critical thermal maximum, , of phage replication was 29.2 °C (95% CI: 29.0–29.4 °C), beyond which phage decreased in abundance over 4 h (Fig. 1a). This indicated that phage struggled to infect the host at temperatures beyond their , which was similar to previous work that measured the coevolution of this bacteria–phage system across temperatures [25]. The bacteria, P. fluorescens, had a similar optimum temperature (Fig. 1b [blue]; = 28 °C; 95% CI: 27.1–29.0 °C), but growth was maintained well beyond , with high growth rates still occurring at 35 °C (Fig. 1b), >6 °C above the of the phage. This could act as a high-temperature refuge for the bacteria as phage infection at these temperatures is extremely low. Due to these mismatches in the thermal performance of phage infection and bacterial growth, it was expected that the parasite would alter the thermal performance of its host.
Fig. 1.
Thermal performance of phage and bacteria. a Phage replication increased with temperature up to an optimum before declining rapidly to a negative replication rate at 30 °C. b Bacteria growth showed unimodal responses to temperature in the presence (black) and absence of phage (blue). However, phage changed the shape of the thermal response. Points represent an independent replicate at each temperature. Solid lines represent the mean prediction and shaded bands represent the 95% credible interval of predictions. In a the dashed line represents 0 growth, below which phage abundance decreased. In b, the dashed line represents the CTmax of the phage, beyond which phage abundance decreased
Phage altered the thermal performance of its bacterial host
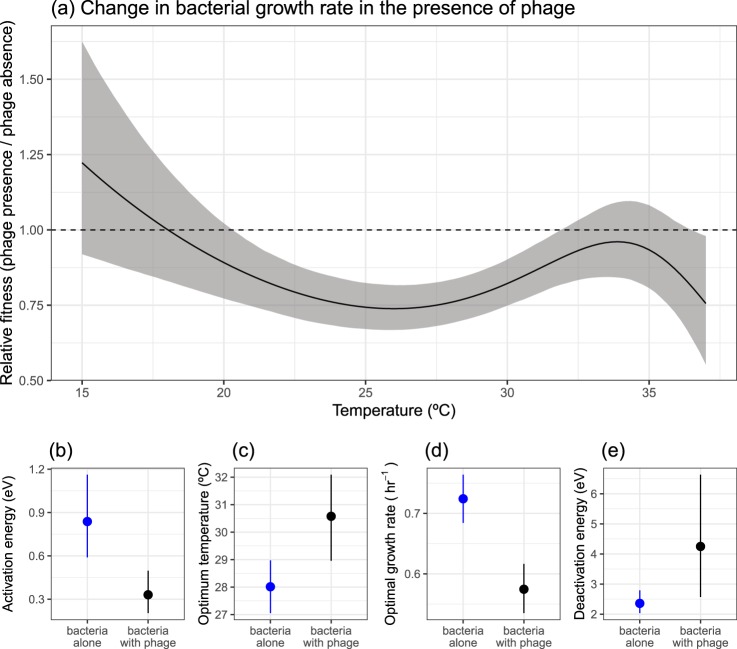
Due to the thermal mismatches between bacteria and phage, we explored whether phage altered the thermal performance of its host. To do this, we measured the growth rate of bacteria in the presence and absence of the phage and compared key traits that underpinned the thermal performance curve (see Methods). We observed marked differences in the response of bacteria to temperature when in the presence of its phage (Fig. 1b and Table S2). Phage presence changed the optimum temperature of bacterial growth (Fig. 2c), shifting from 28 °C (95% CI: 27.1–29.0 °C) to 30.6 °C (95% CI: 29.0–32.1 °C). Moreover, phage presence resulted in a 20.1% (95% CI: 13–27.3%) decline in the maximal growth rate, , in the presence of phage (Fig. 2d). To better understand the non-linear, temperature-dependent effect of phage on bacterial growth, we calculated the relative fitness of bacteria in the presence of phage across temperatures (see Methods; Fig. 2a). The largest impacts of phage on bacterial growth occurred at intermediate temperatures where growth in the absence of phage was highest (Fig. 2a, where relative fitness was <1), whereas no significant change in growth rate was observed at the low and high temperatures measured (credible intervals of predictions overlap 1). The non-linear changes to bacterial growth also resulted in differences in other key traits (Table S2) such as the thermal sensitivity of the rate before (E; Fig. 2b) and after (Eh; Fig. 2e) the optimum temperature.
Fig. 2.
Effect of phage on the thermal performance of bacteria. a Phage altered the growth rate of bacteria (calculated as relative fitness) in a non-linear fashion with increasing temperatures. b–e The effect of phage on key thermal performance traits. Phage altered the b activation energy, c optimum temperature, d optimal growth rate and e deactivation energy. In a the solid line represents the mean prediction and shaded band represents the 95% credible interval of predictions. The dashed line at y = 1 would indicate that phage do not alter growth rate. Below 1, phage reduces the growth rate of the bacteria. In b–e points and lines represent the mean and 95% credible intervals of the estimated parameters
The evolution and cost of resistance was temperature dependent
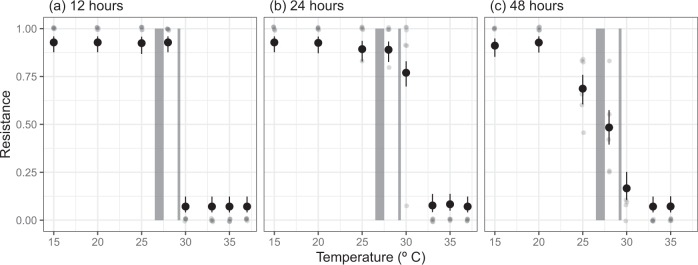
It is possible that the change in thermal performance of P. fluorescens could have resulted simply from the mismatches in thermal performances of the host and parasite. Up to of the phage (~27 °C), phage presence reduced the abundance and thus population growth rate of the bacteria. Consequently, the rapid decline of phage replication at temperatures above 30 °C, while bacteria still had high growth rates, could explain observed shift in the thermal performance of the bacteria. However, bacteria can rapidly evolve resistance to phage within the timescales of our assays, and this has been demonstrated in our host–parasite pair [41, 42]. If, as expected, resistance is costly, and resistance does not evolve at temperatures beyond the phage , the effect of phage on the thermal performance of the host may in part be driven by evolutionary change. To investigate this, we measured the resistance of bacteria through a single logistic growth curve at each temperature (Fig. 3). The evolution of phage resistance changed across temperatures and through time, and there was a significant time × temperature interaction (likelihood ratio test comparing models with and without time × temperature interaction: Δd.f. = 13, F = 11.56, p < 0.001). There was no measurable resistance in the ancestral bacteria, but after just 12 hr, all populations at 28 °C (close to of phage replication [~27 °C]) or lower were close to 100% resistant (Fig. 3a), consistent with a selective sweep in which susceptible cells are lysed and resistant mutants reach fixation. Moreover, after 12 hr, bacterial abundance was much lower than expected at temperatures where phage infection occurred, indicative of a phage epidemic that wiped out susceptible hosts. In contrast, resistance rarely, or never, evolved at temperatures well above those of the critical thermal maximum of phage replication rate (33 °C and higher, Fig. 3). Where resistance did evolve at these temperatures, it was at very low frequency (1 clone out of 12). We found no bacteria still living at 37 °C after 48 hr, indicating that although growth occurs at those temperatures, this is quickly proceeded by death.
Fig. 3.
Levels of resistance of Pseudomonas fluorescens to phage through time and across temperatures. After 12 hr, populations were completely resistant at temperatures of 28 °C or lower. After 24 hr, most bacteria populations at 30 °C, close to the estimated critical thermal maxima (CTmax) of the phage, evolved resistance, but populations beyond the CTmax of phage infection remained susceptible. After 48 hr, at temperatures where phage impacted bacterial growth, intermediate levels of resistance were observed. Small points represent the observed level of resistance for a population. Large points represent the predicted levels of resistance (of transformed data [see Methods]) from a binomial regression with 95% confidence intervals. Shaded regions represent the upper and lower confidence intervals of the optimum temperature and critical thermal maxima of the phage
At temperatures where phage altered the growth rate of bacteria (25, 28 and 30 °C), we observed significant changes in the proportion of resistance through time (see Table S3 for pairwise differences of resistance through time for each temperature). Resistance evolved and was at high proportions after 12 or 24 hr where populations were still in exponential growth phase. However, after 48 hr, when populations had reached stationary phase at all temperatures apart from 15 and 20 °C (Fig. S15), the proportion of resistance decreased significantly (Fig. 3c). From 24 to 48 hr, 25 °C resistance fell from 0.89 (95% CI: 0.83–0.94) to 0.69 (95% CI: 0.60–0.78), at 28 °C from 0.89 (95% CI: 0.83–0.93) to 0.48 (95% CI: 0.40–0.57) and at 30 °C from 0.77 (95% CI 0.68–0.83) to 0.17 (95% CI: 0.11–0.25). This temporal effect did not occur at low and high temperatures where there was little effect of phage on bacterial growth rate (Fig. 2a and Table S3), suggesting that there was a non-linear cost of resistance across the temperature range.
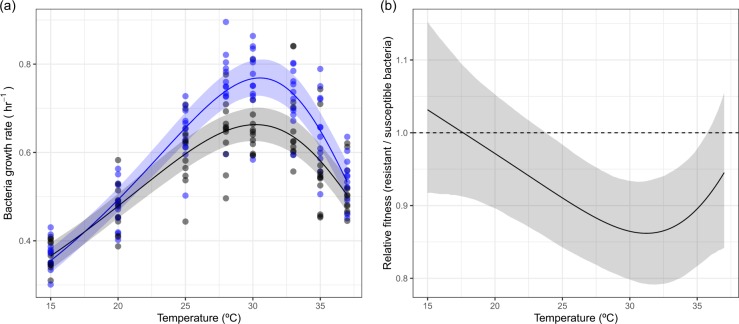
To confirm whether there was a cost of resistance and if any cost varied with temperature, we isolated clones that were either resistant or susceptible to the phage and measured their thermal performance in the absence of phage. The thermal performance of resistant clones differed from that of susceptible clones (Fig. 4), closely matching the patterns observed when bacteria were grown with phage (Figs. 1b and 2a). At low and high temperatures, there were no differences in the growth rate of resistant and susceptible clones (Fig. 4). However, at temperatures where growth of susceptible clones was highest (25–30 °C), there was a cost of resistance (Fig. 4b), resulting in a 13.4% (95% CI: 6.8–20.2%) reduction in maximal growth rate. This temperature-dependent cost of resistance was qualitatively similar to the effect of phage on bacteria growth, being greatest at intermediate temperatures (Figs. 2a and 4b).
Fig. 4.
Temperature-dependent cost of resistance in Pseudomonas fluorescens in the absence of phage. a The thermal performance of susceptible (blue) and resistant (black) clones. Resistant clones had a lower maximum growth rate. b The derived selection coefficient of resistance across temperatures. The cost of resistance changed across temperatures, being greatest at 30 °C and other temperatures where growth in the absence of phage is high. In a points represent individual clones, solid lines represent the mean prediction and shaded bands represent the 95% credible interval of predictions. In b the dashed line at y = 1 would indicate that phage do not alter growth rate. Below 1, phage reduces the growth rate of the bacteria
Discussion
Here, we show that the presence of a parasite can profoundly impact the thermal performance of its host. Notably, phage reduced bacterial growth most at temperatures where the bacteria grew fastest, close to the bacterial , while having little or no impact at cold or high temperatures well beyond (Figs. 1 and 2). This resulted in changes to the thermal performance curve of bacterial growth in the presence of phage (Fig. 2b). These results can be explained by a combination of ecological and evolutionary processes. Ecologically, at temperatures below the critical thermal maxima of the phage, phage presence vastly reduced bacterial abundance (i.e. increased lag time in the logistic growth curve, Figs. S1–S4). In contrast, phage could not infect above 30 °C, but bacteria still had high growth rates. However, rapid evolution also played an important role in altering the thermal performance of P. fluorescens. While phage resistance evolved rapidly and was at high levels at all temperatures below the phage , at higher temperatures there was no selection for resistance (Fig. 3). Crucially, there were costs associated with resistance, but these costs changed non-linearly with temperature (Fig. 4). At low temperatures and temperatures far beyond the bacteria , there was no measurable cost of resistance, but significant costs of resistance at intermediate temperatures where bacteria growth was highest (Fig. 4). At some temperatures, susceptible bacteria re-emerged after resistance had evolved (Fig. 3) during stationary phase, which could be a result of nutrient limitation or reduced phage infection of susceptible bacteria in stationary phase [43], both of which would alter the fitness cost of resistance. Overall, these results demonstrate that phage alter the TPC of their host (Fig. 1b) through both ecological (due to differences in thermal tolerance between phage infection and bacterial growth) and evolutionary processes (temperature-dependent costs of resistance), resulting in a shift in the TPC for the host in the presence of the phage (Fig. S16). It is worth noting that costs of phage resistance were also greatest at the optimum temperature in another well-studied bacteria–phage system; Escherichia coli and bacteriophage T4 [44, 45].
How general are these results likely to be? We suggest that parasites (and symbionts more generally) impacts on host TPCs are likely widespread, because no change in host TPC would occur only when host and parasite traits respond equivalently with temperature. In reality, there are almost certainly mismatches between host and parasite TPCs and differences in local adaptation to prevailing temperatures appears to be the norm [7, 46]. Here, we observed rapid evolutionary interactions between our bacteria–phage pair because of the strong parasite-imposed selection and the large population size and short generation time of P. fluorescens (~14 generations after 12 hr at 30 °C). As this is true of most micro-organisms, we expect that evolutionary mechanisms could frequently drive changes in population-level TPCs, although the selection for resistance is likely to be lower in more heterogeneous environments and with different parasitic lifecycles.
Across other host–parasite systems, similar genotype × genotype × environment interactions (G × G × E) occur in different traits, but may be driven more by ecological, rather than evolutionary, processes. For example, in larger, longer-lived hosts, individuals may experience substantial variation in temperature and parasitism over the course of a single generation. In such instances, the individual-level cost of parasitism can still be highest at intermediate temperatures [47] and variation in critical thermal maxima between different host species [8] and thermal mismatches between host and parasite [6] can drive temperature-dependent changes in host susceptibility. Consequently, the effect of parasites on the thermal performance of the host may be widespread across many host–parasite systems, driven by ecological or rapid evolutionary processes depending on host lifespan and magnitude of parasite-imposed selection.
However, as with the effect of changing temperature on disease severity, precisely how TPCs will change will be context dependent, changing with, amongst other factors, the host–parasite pair and the biological traits measured. For example, phage replication across temperature depends on the thermal sensitivity of multiple processes such as latency period, burst size and thermal stability [48], such that the limiting factor for phage replication may also differ across temperature. Moreover, the effect of any of these phage traits in isolation may result in a different impact on the host TPC. Marine phage are generally more thermally stable than their hosts [9], but, as shown here, it that does not mean that the phage can infect at all temperatures [49]. Across ectotherms, thermal breadth across multiple traits is generally wider in smaller organisms [10], but whether this impacts host or parasite TPCs (parasites are generally smaller than their host) in the presence of each other remains to be seen.
In conclusion, our study demonstrated that host–parasite interactions change in non-linear ways with temperature (G × G × E interaction), and this had a significant impact on the thermal performance of the host. By measuring the thermal performance of the host and the parasite simultaneously, and also examining the evolution and cost of resistance, we identified the mechanisms through which phage altered the thermal performance of the host. Our results highlight that TPCs measured under axenic conditions should be interpreted with caution; measuring TPCs in the absence of their parasites (and other associated microbiota) may not be reflective of the host’s TPC in nature where such interactions are ubiquitous. Future work should investigate the longer term evolutionary and coevolutionary consequences of climate warming [13] and in a broader, more realistic ecological context, to determine how this impacts host–parasite interactions. In an era of human-induced climate change, it is more important than ever to gain a deeper understanding of how evolutionary and ecological processes can indirectly impact thermal performance and how host–parasite interactions will change with temperature.
Data accessibility
All data and R code used in the analysis will be made available on GitHub and archived on Zenodo. All data and analyses are available on GitHub (https://git.io/Je4nz) and archived on Zenodo (10.5281/zenodo.3492192).
Supplementary information
Acknowledgements
We thank three anonymous referees for their insightful comments. This work was funded by NERC.
Author contributions
DP and AB conceived the study and designed the experimental work. DP and MC conducted the experiments. DP analysed the data. All authors contributed significantly to the first draft of the manuscript and to revisions.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-019-0526-5) contains supplementary material, which is available to authorized users.
References
- 1.Harvell CD, Mitchell CE, Ward JR, Altizer S, Dobson AP, Ostfeld RS, et al. Climate warming and disease risks for terrestrial and marine biota. Science. 2002;296:2158–62. doi: 10.1126/science.1063699. [DOI] [PubMed] [Google Scholar]
- 2.Demory D, Arsenieff L, Simon N, Six C, Rigaut-Jalabert F, Marie D, et al. Temperature is a key factor in Micromonas–virus interactions. ISME J. 2017;11:601. doi: 10.1038/ismej.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paull SH, LaFonte BE, Johnson PT. Temperature-driven shifts in a host-parasite interaction drive nonlinear changes in disease risk. Glob Change Biol. 2012;18:3558–67. [Google Scholar]
- 4.Paull SH, Raffel TR, LaFonte BE, Johnson PT. How temperature shifts affect parasite production: testing the roles of thermal stress and acclimation. Funct Ecol. 2015;29:941–50. [Google Scholar]
- 5.Molnár PK, Kutz SJ, Hoar BM, Dobson AP. Metabolic approaches to understanding climate change impacts on seasonal host-macroparasite dynamics. Ecol Lett. 2013;16:9–21. doi: 10.1111/ele.12022. [DOI] [PubMed] [Google Scholar]
- 6.Cohen JM, Venesky MD, Sauer EL, Civitello DJ, McMahon TA, Roznik EA, et al. The thermal mismatch hypothesis explains host susceptibility to an emerging infectious disease. Ecol Lett. 2017;20:184–93. doi: 10.1111/ele.12720. [DOI] [PubMed] [Google Scholar]
- 7.Gehman Alyssa-Lois M., Hall Richard J., Byers James E. Host and parasite thermal ecology jointly determine the effect of climate warming on epidemic dynamics. Proceedings of the National Academy of Sciences. 2018;115(4):744–749. doi: 10.1073/pnas.1705067115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nowakowski AJ, Whitfield SM, Eskew EA, Thompson ME, Rose JP, Caraballo BL, et al. Infection risk decreases with increasing mismatch in host and pathogen environmental tolerances. Ecol Lett. 2016;19:1051–61. doi: 10.1111/ele.12641. [DOI] [PubMed] [Google Scholar]
- 9.Mojica KD, Brussaard CP. Factors affecting virus dynamics and microbial host–virus interactions in marine environments. FEMS Microbiol Ecol. 2014;89:495–515. doi: 10.1111/1574-6941.12343. [DOI] [PubMed] [Google Scholar]
- 10.Rohr JR, Civitello DJ, Cohen JM, Roznik EA, Sinervo B, Dell AI. The complex drivers of thermal acclimation and breadth in ectotherms. Ecol Lett. 2018;21:1425–39. doi: 10.1111/ele.13107. [DOI] [PubMed] [Google Scholar]
- 11.Thomas MB, Blanford S. Thermal biology in insect-parasite interactions. Trends Ecol Evolution. 2003;18:344–50. [Google Scholar]
- 12.Tseng M., Bernhardt Joey R., Chila Alexander E. Species interactions mediate thermal evolution. Evolutionary Applications. 2019;12(7):1463–1474. doi: 10.1111/eva.12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorter FA, Scanlan PD, Buckling A. Adaptation to abiotic conditions drives local adaptation in bacteria and viruses coevolving in heterogeneous environments. Biol Lett. 2016;12:20150879. doi: 10.1098/rsbl.2015.0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinclair BJ, Marshall KE, Sewell MA, Levesque DL, Willett CS, Slotsbo S, et al. Can we predict ectotherm responses to climate change using thermal performance curves and body temperatures? Ecol Lett. 2016;19:1372–85. doi: 10.1111/ele.12686. [DOI] [PubMed] [Google Scholar]
- 15.Sunday JM, Bates AE, Dulvy NK. Thermal tolerance and the global redistribution of animals. Nat Clim Change. 2012;2:686–90. [Google Scholar]
- 16.Luhring TM, DeLong JP. Predation changes the shape of thermal performance curves for population growth rate. Curr Zool. 2016;62:501–5. doi: 10.1093/cz/zow045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tseng M, O’Connor MI. Predators modify the evolutionary response of prey to temperature change. Biol Lett. 2015;11:20150798. doi: 10.1098/rsbl.2015.0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Padfield D, Yvon-Durocher G, Buckling A, Jennings S, Yvon-Durocher G. Rapid evolution of metabolic traits explains thermal adaptation in phytoplankton. Ecol Lett. 2016;19:133–42. doi: 10.1111/ele.12545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaum C-E, Barton S, Bestion E, Buckling A, Garcia-Carreras B, Lopez P, et al. Adaptation of phytoplankton to a decade of experimental warming linked to increased photosynthesis. Nat Ecol Evol. 2017;1:0094. doi: 10.1038/s41559-017-0094. [DOI] [PubMed] [Google Scholar]
- 20.Angilletta Jr MJ. Thermal adaptation: a theoretical and empirical synthesis. Oxford: Oxford University Press; 2009.
- 21.Clokie MR, Millard AD, Letarov AV, Heaphy S. Phages in nature. Bacteriophage. 2011;1:31–45. doi: 10.4161/bact.1.1.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Pascua LDC, Buckling A. Increasing productivity accelerates host–parasite coevolution. J Evol Biol. 2008;21:853–60. doi: 10.1111/j.1420-9101.2008.01501.x. [DOI] [PubMed] [Google Scholar]
- 23.Buckling A, Rainey PB. Antagonistic coevolution between a bacterium and a bacteriophage. Proc R Soc Lond B Biol Sci. 2002;269:931–6. doi: 10.1098/rspb.2001.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gómez P, Buckling A. Bacteria-phage antagonistic coevolution in soil. Science. 2011;332:106–9. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Q-G, Buckling A. Antagonistic coevolution limits population persistence of a virus in a thermally deteriorating environment. Ecol Lett. 2011;14:282–8. doi: 10.1111/j.1461-0248.2010.01586.x. [DOI] [PubMed] [Google Scholar]
- 26.Ellner SP, Geber MA, Hairston NG., Jr Does rapid evolution matter? Measuring the rate of contemporary evolution and its impacts on ecological dynamics. Ecol Lett. 2011;14:603–14. doi: 10.1111/j.1461-0248.2011.01616.x. [DOI] [PubMed] [Google Scholar]
- 27.Hairston NG, Jr, Ellner SP, Geber MA, Yoshida T, Fox JA. Rapid evolution and the convergence of ecological and evolutionary time. Ecoulogy Lett. 2005;8:1114–27. [Google Scholar]
- 28.Knies JL, Izem R, Supler KL, Kingsolver JG, Burch CL. The genetic basis of thermal reaction norm evolution in lab and natural phage populations. PLoS Biol. 2006;4:e201. doi: 10.1371/journal.pbio.0040201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knies JL, Kingsolver JG, Burch CL. Hotter is better and broader: thermal sensitivity of fitness in a population of bacteriophages. Am Naturalist. 2009;173:419–30. doi: 10.1086/597224. [DOI] [PubMed] [Google Scholar]
- 30.Gompertz B. On the nature of the function expressive of the law of human mortality, and on a new mode of determining the value of life contingencies. Philos Trans R Soc Lond B: Biol Sci. 1825;115:513–83. doi: 10.1098/rstb.2014.0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baty F, Delignette-Muller ML. nlsMicrobio: nonlinear regression in predictive microbiology. R package version 3.2. 2. 2016.
- 32.Padfield D, Matheson G. nls.multstart: robust non-linear regression using AIC scores. R package version 1.0.0. 2018.
- 33.Schoolfield RM, Sharpe PJH, Magnuson CE. Non-linear regression of biological temperature-dependent rate models based on absolute reaction-rate theory. J Theor Biol. 1981;88:719–31. doi: 10.1016/0022-5193(81)90246-0. [DOI] [PubMed] [Google Scholar]
- 34.Padfield Daniel, Buckling Angus, Warfield Ruth, Lowe Chris, Yvon-Durocher Gabriel. Linking phytoplankton community metabolism to the individual size distribution. Ecology Letters. 2018;21(8):1152–1161. doi: 10.1111/ele.13082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sunday JM, Pecl GT, Frusher S, Hobday AJ, Hill N, Holbrook NJ, et al. Species traits and climate velocity explain geographic range shifts in an ocean-warming hotspot. Ecol Lett. 2015;18:944–53. doi: 10.1111/ele.12474. [DOI] [PubMed] [Google Scholar]
- 36.Bürkner P-C. brms: an R package for Bayesian multilevel models using Stan. J Stat Softw. 2017;80:1–28. [Google Scholar]
- 37.Kay M. tidybayes: tidy data and geoms for Bayesian models. 2018.
- 38.Lenth R. Emmeans: estimated marginal means, aka least-squares means. R package version 2018; 1.
- 39.Team RC. R: a language and environment for statistical computing. 2013.
- 40.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer; 2016.
- 41.Buckling A, Rainey PB. The role of parasites in sympatric and allopatric host diversification. Nature. 2002;420:496. doi: 10.1038/nature01164. [DOI] [PubMed] [Google Scholar]
- 42.Lenski RE, Levin BR. Constraints on the coevolution of bacteria and virulent phage: a model, some experiments, and predictions for natural communities. Am Naturalist. 1985;125:585–602. [Google Scholar]
- 43.Bryan D, El-Shibiny A, Hobbs Z, Porter J, Kutter EM. Bacteriophage T4 infection of stationary phase E. coli: life after log from a phage perspective. Front Microbiol. 2016;7:1391. doi: 10.3389/fmicb.2016.01391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quance MA, Travisano M. Effects of temperature on the fitness cost of resistance to bacteriophage T4 in Escherichia coli. Evol Int J Org Evol. 2009;63:1406–16. doi: 10.1111/j.1558-5646.2009.00654.x. [DOI] [PubMed] [Google Scholar]
- 45.Cooper VS, Bennett AF, Lenski RE. Evolution of thermal dependence of growth rate of Escherichia coli populations during 20,000 generations in a constant environment. Evolution. 2001;55:889–96. doi: 10.1554/0014-3820(2001)055[0889:eotdog]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 46.Dell A. I., Pawar S., Savage V. M. Systematic variation in the temperature dependence of physiological and ecological traits. Proceedings of the National Academy of Sciences. 2011;108(26):10591–10596. doi: 10.1073/pnas.1015178108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirk D, Jones N, Peacock S, Phillips J, Molnár PK, Krkošek M, et al. Empirical evidence that metabolic theory describes the temperature dependency of within-host parasite dynamics. PLoS Biol. 2018;16:e2004608. doi: 10.1371/journal.pbio.2004608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sillankorva S, Oliveira R, Vieira MJ, Sutherland I, Azeredo J. Pseudomonas fluorescens infection by bacteriophage ΦS1: the influence of temperature, host growth phase and media. FEMS Microbiol Lett. 2004;241:13–20. doi: 10.1016/j.femsle.2004.06.058. [DOI] [PubMed] [Google Scholar]
- 49.Delisle AL, Levin RE. Characteristics of three phages infectious for psychrophilic fishery isolates ofPseudomonas putrefaciens. Antonie van Leeuwenhoek. 1972;38:1–8. doi: 10.1007/BF02328071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and R code used in the analysis will be made available on GitHub and archived on Zenodo. All data and analyses are available on GitHub (https://git.io/Je4nz) and archived on Zenodo (10.5281/zenodo.3492192).