Abstract
Type 2 diabetes mellitus (T2DM) is a systemic disease, predisposing patients to other inflammatory conditions including periodontitis. The subgingival microbiome, a key player in periodontitis pathogenesis, is not well characterized in T2DM population. To better understand whether the subgingival microbiome is different between T2DM and systemically healthy, nondiabetic (ND) subjects, we performed a longitudinal analysis of the subgingival microbiome in T2DM patients (n = 15) compared with ND subjects (n = 16). Using metagenomic shotgun sequencing, we investigated the microbiome in the healthy periodontal state, periodontitis state, and resolved state after treatment. We found that in the periodontitis state, the shift in the subgingival microbiome from the healthy state was less prominent in T2DM compared with ND subjects, yet the clinical signs of disease were similar for both. Furthermore, we revealed highly correlated presence of pathogenic species in relative abundance not only in the periodontitis state, but also in the healthy state in T2DM, suggesting an elevated risk of progression to periodontitis in this cohort. We further investigated the functional potentials of the subgingival microbiome and identified a set of microbial marker genes associated with the clinical states. These genes were significantly enriched in 21 pathways, some of which are associated with periodontitis and some potentially link T2DM and periodontitis. This study identified the longitudinal changes of the subgingival microbiome associated with periodontitis in T2DM and suggests that T2DM patients are more susceptible to shifts in the subgingival microbiome toward dysbiosis, potentially due to impaired host metabolic and immune regulation.
Subject terms: Microbiome, Diseases
Introduction
Diabetes mellitus is a systemic disease with global epidemic proportions. Clinical evidence suggests that type 2 diabetes mellitus (T2DM) increases the risk of developing inflammatory diseases including periodontitis [1], a significant oral disease affecting half of American adults [2, 3]. Diabetes is associated with host metabolic dysregulation, which upregulates inflammatory response and promotes tissue destruction, while periodontal inflammation adversely affects glycemic control [4–6]. Inflammation is the primary focus of previous studies that attempted to link diabetes and periodontitis [1]. It has been suggested by clinical and animal model studies that cytokines such as interleukin-1β, tumor necrosis factor-α, and receptor activator of nuclear factor κB ligand may mediate periodontitis in diabetes. Interactions between advanced glycation end products (AGEs) and their receptor RAGE (the receptor for AGEs) may contribute to increased inflammation and periodontal tissue destruction in patients with diabetes [5, 6]. A recent study using a mouse model suggested that diabetes increased the pathogenicity of the oral microbiome through pro-inflammatory cytokine IL-17 mediated mechanism. The dysregulated immune response in diabetes leads to dysbiosis in the subgingival microbiome, which predisposes the host to periodontitis [7].
The subgingival microbiome plays a key role in periodontitis pathogenesis. Multiple studies using 16S ribosomal RNA (rRNA) gene analysis have identified the taxonomic composition of the subgingival microbiome in systemically healthy, nondiabetic (ND) subjects [8–14]. These studies revealed distinct differences in the subgingival microbiome between the healthy periodontal state and periodontitis state [8–12], and characterized the longitudinal changes in the microbiome after periodontal treatment [13, 14]. In addition, the functional potentials encoded in the subgingival microbiome have been analyzed in ND subjects based on metagenomic shotgun sequencing [9, 12, 15], which revealed several virulence factors and metabolic pathways associated with the periodontitis state.
While the taxonomic composition of the subgingival microbiome in T2DM patients was characterized in a few case-control studies using 16S rRNA analysis [16–18], longitudinal analysis of the subgingival microbiome associated with periodontitis before and after treatment is lacking. The functional compositions of the microbiome in different states and the comparison with those in ND have not been investigated. In this study, we present the first metagenomic shotgun sequencing analysis of the subgingival microbiome in periodontal health, periodontitis, and resolved states after treatment of periodontitis in T2DM patients. We determined the species compositions and the correlations among species, and investigated the functional potentials encoded in the microbiome associated with periodontitis in T2DM. We also compared the subgingival microbiome in different states in T2DM patients to ND subjects.
Materials and methods
Subject recruitment and sample collection
The study was approved by the Institutional Review Board at University of California, Los Angeles. Written informed consent was obtained from all participants. Subjects with antibiotic treatment in the past 6 months or with a history of smoking were excluded.
For each participant, an initial full-mouth examination was conducted at the first visit to assess the clinical parameters of the periodontium including the gingival index, recession of gums, attachment level, pocket depth, and bleeding on probing. For each periodontitis patient, two additional clinic visits were required. Tooth sites identified during the first assessment visit presenting with a probing depth of ≥5 mm, gingival index of ≥1 and with bleeding on probing were considered diseased sites. Affected tooth sites were sampled during the second visit before the initial therapy. The therapy consisted of mechanical periodontal treatment (localized scaling and root planing) without antibiotics. In the third clinic visit, which was 4–7 weeks after the initial therapy, patients with at least one tooth site that was resolved, defined as probing depth ≤4 mm and absence of bleeding on probing or other signs of inflammation, were resampled at the resolved sites for microbiome analysis. For each subject with healthy periodontium, defined as probing depth ≤4 mm and minimal (less than 10–15% of all sites) bleeding on probing, one tooth site with no bleeding on probing was sampled during the second visit following the initial assessment. Majority of the samples (83%) were taken from molar or premolar tooth sites in T2DM and ND subjects. All subjects were required to follow an abstinence protocol prior to the sampling visits—abstain from any mouth rinse 48 h prior to sampling and abstain from any chemical or mechanical oral hygiene measures for 24 h prior to sampling.
The T2DM and ND subjects with periodontitis or healthy periodontium were matched in gender (Table 1). The average age was not significantly different between T2DM and ND subjects with periodontitis (p = 0.29), between T2DM subjects with periodontitis vs. with healthy periodontium (p = 0.43), and between ND subjects with periodontitis vs. with healthy periodontium (p = 0.42), but was different between T2DM and ND subjects with healthy periodontium (p = 0.021).
Table 1.
Demographic and clinical data of the study participants
| Characteristics | Subjects with type 2 diabetes mellitus | Nondiabetic subjects | ||
|---|---|---|---|---|
| Periodontitis (n = 8) | Healthy periodontium (n = 7) | Periodontitis (n = 8) | Healthy periodontium (n = 8) | |
| Age (year) | 51.5 ± 2.9 | 55.6 ± 3.4 | 44.0 ± 4.8 | 37.4 ± 4.6 |
| Gender (% male) | 38% | 57% | 38% | 50% |
| PD in periodontitis state (mm) | 5.5 ± 0.3 | – | 6.0 ± 0.3 | – |
| PD in resolved/healthy state (mm) | 2.9 ± 0.1 | 2.7 ± 0.2 | 3.4 ± 0.2 | 2.8 ± 0.2 |
| BOP of all sites (%) in periodontitis state | 45 ± 11% | – | 34 ± 8% | – |
| BOP of all sites (%) in resolved/healthy state | 2 ± 2% | 0 ± 0.1% | 3 ± 1% | 1 ± 0.4% |
| HbA1c level (%) in periodontitis state | 7.6 ± 0.5% | – | 5.3 ± 0.1% | – |
| HbA1c level (%) in resolved/healthy state | 7.7 ± 0.5% | 7.4 ± 0.5% | 5.4 ± 0.1% | 5.4 ± 0.1% |
Data are presented as mean ± s.e.m unless otherwise indicated
PD probe depth of the sampling site, BOP bleeding on probing
For all T2DM and ND subjects, the glycated hemoglobin (HbA1c) assay was performed. Patients that received the initial periodontal therapy had HbA1c tested at both initial therapy and periodontal reevaluation visits. In our T2DM cohort, there was no significant difference in HbA1c level before and after periodontal treatment (p = 0.36). All ND subjects had an HbA1C level <6.
The subgingival plaques were collected using sterile curettes, suspended directly in ATL buffer (Qiagen, Valencia, CA) containing 0.1 mm glass beads, and immediately transported to the laboratory for further processing.
Genomic DNA extraction
Genomic DNA was extracted from the plaque samples using the QIAamp DNA Micro kit (Qiagen, Valencia, CA) with a modified “Isolation of Genomic DNA from Tissues” protocol. Bead beating for maximal bacterial cell lysis was added. The extracted genomic DNA was eluted with EB buffer and stored at −20 °C for short term (within a week) and −80 °C for long term.
Sequencing and data cleaning
Shotgun metagenomic sequencing was performed using the procedure described in our previous study [15]. Briefly, genomic sequencing libraries were prepared using Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA) according to the manufacturer’s manual. The libraries were sequenced using the shotgun method with paired-end reads of 100 bp on Illumina HiSeq sequencing platforms 2000 and 2500 (Illumina, San Diego, CA). A data cleaning process was applied to all sequence data prior to analysis. Human sequences were removed using the procedure (BMTagger) as described in the Human Microbiome Project [19] and by mapping to the human genome using Bowtie2 [20]. Low-quality bases with a Phred quality value below 20 were trimmed off at the ends of the reads. The read pairs were removed if any of the two reads was trimmed to shorter than 60 bp or had ≥3% uncertain bases. The Nextera adapter and primer sequences were trimmed off using Cutadapt [21].
Analyses of taxonomic and functional compositions of the microbiome
To determine the taxonomic composition of the subgingival microbiome, using a method similar to Schloissnig et al. [22], we mapped the metagenomic shotgun sequences against microbial reference genomes using Bowtie2 [20]. To improve the mapping resolution and efficiency of the analyses, we constructed a nonredundant genomic reference genome set consisting of the pan-genomes of 375 bacterial species belonging to 124 genera. A collection of 1,309 human oral bacterial genomes from the Human Oral Microbiome Database [23] and Genbank [24] was included in the constructions of the pan-genomes using the methods described previously [25, 26]. To avoid mapping artifacts, we defined the presence of a bacterial species in the samples with stringent criteria by using an 80% sequence similarity threshold and requiring at least 80% of the genome covered by sequencing reads. The abundance of the organisms in each sample was calculated by counting the number of base pairs (bp) covering the pan-genome followed by normalization to the average genome size of the species.
We annotated the microbial functional genes using the KEGG Orthology (KO) [27] and the Virulence Factors DataBase (VFDB) [28] based on sequence similarity using BLAST [29]. The abundance of the KO genes was calculated in copy numbers by summing the number of bp covering the microbial genes that were annotated as KO genes in the pan-genomes of all the identified species. The relative abundance of the KO genes was then normalized by the gene length and the sequencing depth (per 108 microbial bp) of each sample. We determined the statistical significance of the enrichment of KO genes with differential prevalence between clinical states in microbial pathways using the hypergeometric distribution test. A pathway was considered present if it is consisted of more than five KO genes that were identified. Pathways that were significantly enriched (p < 0.05) in at least half of the samples in any clinical state were presented.
Microbiome index
We used a microbiome index to quantitatively compare the microbiome across different clinical states or subject groups. We compared the relative abundances of all the identified bacterial species in each sample to the mean values of the species from an independent dataset obtained from ND subjects [15]. The microbiome index is then calculated based on the statistic score of each species using the formula below as described in the previous sample classification studies [30–32],
where MI is the microbiome index, Xg is the relative abundance of the species (g) in the sample, μ1g and μ2g are the means of the relative abundance of the species (g) in the periodontitis state and the resolved state from an independent dataset [15], and tg is the statistic t score used as the weighted factor of the species (g) when its relative abundance is compared between the two states.
Statistical analysis
The microbial community similarity analysis based on Weighted UniFrac distance and principal coordinate analysis (PCoA) was performed using QIIME [33]. Nonparametric multivariate analysis of changes in the microbial community (ANOSIM) was conducted using Mothur [34, 35] with default parameters to test whether the microbiome similarities within states are significantly different from the similarities between states. Wilcoxon rank-sum test with two-tailed distribution was used in the statistical comparisons between states unless otherwise indicated, and paired Wilcoxon rank-sum test was applied to the longitudinally paired samples. The p-values were adjusted for multiple testing with p.adjust in R using false discovery rate [36]. The correlations between species in each state were calculated using Pearson correlation coefficient based on their relative abundances in the samples.
Data access
The sequence data generated in this study have been deposited to the NCBI BioProject under accession PRJNA255922.
Results
Sample and data collection
We recruited 32 participants, which were assigned to one of the four groups: T2DM patients with chronic periodontitis (n = 8) or periodontal health (n = 8), and ND subjects with chronic periodontitis (n = 8) or periodontal health (n = 8) (Table 1). The healthy/disease state of the periodontium was determined by certified periodontists based on clinical parameters (see “Materials and methods”). For each periodontitis patient, we sequenced the subgingival plaque samples collected from one affected tooth site before initial therapy (periodontitis state) and after the site was resolved (resolved state). For each subject with a healthy periodontium, one subgingival sample (healthy state) was sequenced. All samples were sequenced using metagenomic shotgun approach and analyzed. One sample collected from a T2DM subject with healthy periodontium was excluded in the final data analysis due to a high percentage of human reads and thus low sequencing coverage of the microbiome. A total of 93.7 billion bp of microbial sequences were obtained from 47 samples with an average of 2.0 billion bp per sample.
Changes in the subgingival microbiome among different periodontal states in T2DM and ND subjects
We compared the taxonomic composition of the subgingival microbiome in different states in T2DM and ND subjects. A total of 129 bacterial species from 45 genera were identified in our dataset. We found that, in ND subjects, the subgingival microbiome compositions obtained in this study were consistent with those from our previous study [15] and from the Human Microbiome Project [19] (Supplementary Fig. 1).
We investigated the associations of the microbiome with clinical states. We found that there was no significant difference in the subgingival microbiome between resolved state and healthy state in either T2DM or ND (Fig. 1), suggesting that the subgingival microbiome in the resolved state largely resembles the healthy state. The subgingival microbiome in the periodontitis state was significantly different from that in the healthy state in ND (ANOSIM, p < 0.001), consistent with previous findings. However, the difference between the disease state and healthy state in T2DM patients was not significant (p = 0.26).
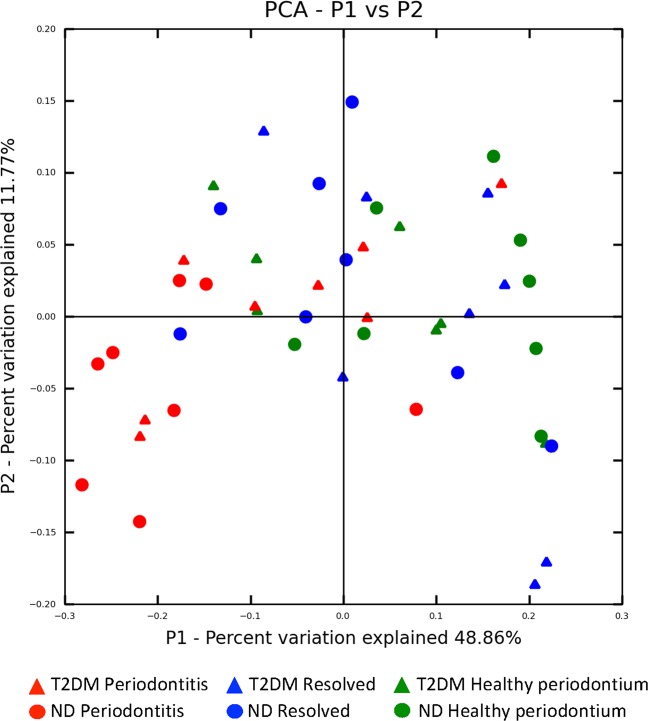
Fig. 1.
Subgingival microbiome comparison among periodontal states in T2DM and ND subjects. Principal coordinate analysis (PCoA) based on weighted UniFrac distance shows no significant difference between the healthy state and resolved state in both T2DM and ND subjects, however, the extent of the microbiome shift from the healthy to the periodontitis state was less in T2DM (p = 0.26) compared with ND subjects (p < 0.001). The same PCoA plot with subject IDs labeled is shown in Supplementary Fig. 2 for visualization of related samples
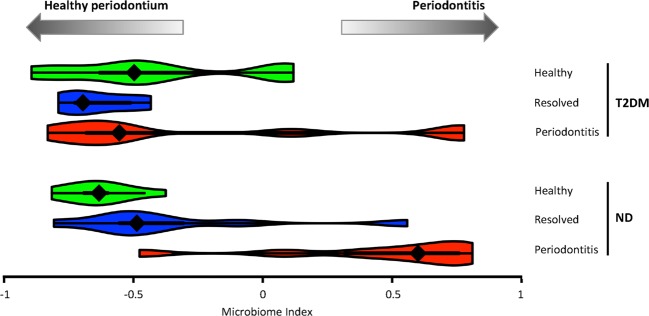
To better quantitatively compare the subgingival microbiome between T2DM and ND and its association with periodontitis, we formulated a “microbiome index” based on the metagenomic sequence data (see “Materials and methods”) to define the state of the subgingival microbiome. We found that while the microbiome index in ND subjects was distinct between the periodontitis state (median = 0.60) and the resolved state (median = −0.49, p = 0.0069) or the healthy state (median = −0.63, p = 0.0003), in T2DM patients the microbiome index in the periodontitis state (median = −0.55) was not significantly different from the resolved state (median = −0.70, p = 0.382) or the healthy state (median = −0.50, p = 1). It is also apart from the periodontitis state in ND (p = 0.08) (Fig. 2). This indicates that in T2DM patients the subgingival microbiome had not yet entirely shifted to a “pathogenic” periodontitis state as seen in ND, however, the periodontal tissue was already inflamed and clinically manifested signs of disease.
Fig. 2.
The microbiome index indicates subgingival microbiome differences among periodontal states and between T2DM and ND subjects. The microbiome index was represented in a violin plot in each clinical state in T2DM and ND subjects. In T2DM, the microbiome in the periodontitis state had not shifted to the extent as observed in the disease state of ND subjects, while the periodontal tissue clinically manifested signs of periodontitis, suggesting that T2DM subjects are more susceptible to shifts in the subgingival microbiome toward the disease state
Differences in the taxonomic composition and correlations among bacterial species between T2DM and ND subjects
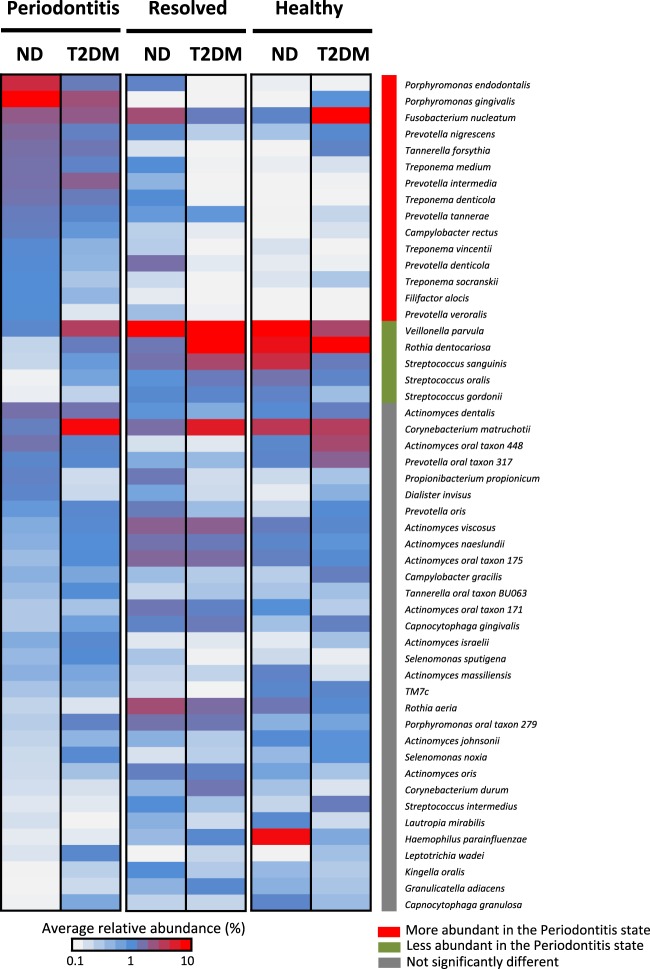
We next investigated the microbiome differences between T2DM and ND subjects at the level of individual subgingival microorganisms. We identified a total of 51 prevalent species with greater than 1% in average relative abundance and with at least 1% relative abundance in more than two samples in any clinical state (Fig. 3). Our results for ND subjects are consistent with our previous study based on 16S rRNA gene sequences [15], with a higher resolution to the species level. Fifteen species were significantly more abundant in the periodontitis state in ND, including periodontal pathogens of the red complex (Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola) and potential opportunistic pathogens in the orange complex (Fusobacterium nucleatum, Campylobacter rectus, Prevotella intermedia, and Prevotella nigrescens) defined by Socransky et al. [37], as well as Filifactor alocis [38]. On the other hand, five species were significantly more abundant in the healthy state in ND, including the yellow complex Streptococcus species (S. sanguinis, S. gordonii, and S. oralis), the purple complex species (Veillonella parvula), and Rothia dentocariosa. In contrast, in T2DM patients, although the prevalent species had similar trends as seen in ND, none of them showed statistically significant differences in relative abundance, consistent with our finding at the microbial community level described above.
Fig. 3.
Comparison of the relative abundances of the prevalent subgingival bacterial species among clinical states and between T2DM and ND subjects. The average relative abundances of the 51 prevalent bacterial species are shown for each clinical state in T2DM and ND. In ND, 15 species indicated by the red bar on the right were significantly more abundant in periodontitis state than in the healthy state, while five species indicated by the green bar were significantly less abundant. In T2DM, the differences between the periodontitis and healthy states had a similar trend as seen in ND, but none of the species differed with statistical significance
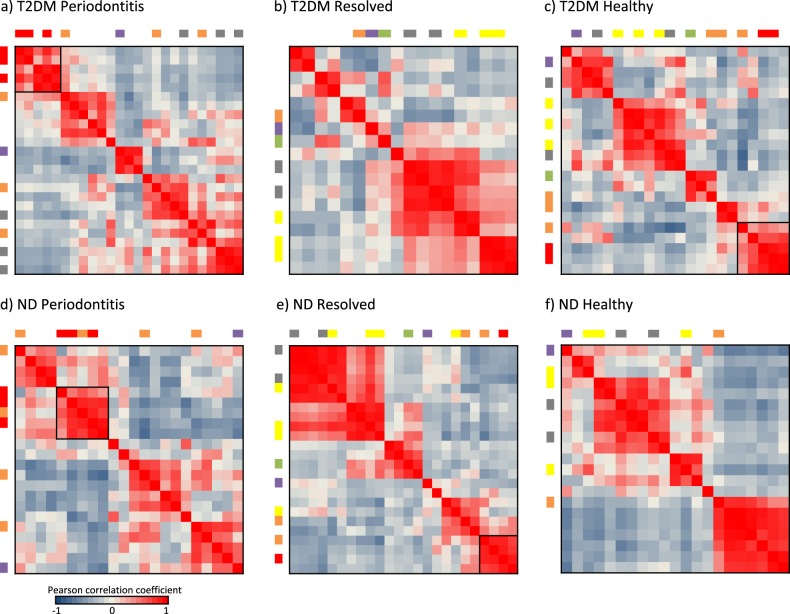
Periodontitis is a polymicrobial disease. We investigated potential inter-species interactions based on correlations in relative abundance of the species in different states and between T2DM and ND (Fig. 4). The red complex species were highly correlated in relative abundance and were all present in the periodontitis state in both T2DM and ND subjects, forming a tight module (Fig. 4a–d). In addition, two of the three red complex species were found in the healthy state of T2DM patients (Fig. 4c) with correlated relative abundances, suggesting an elevated risk of progression to periodontitis in T2DM patients with healthy periodontium compared with systemically healthy subjects.
Fig. 4.
Correlations in relative abundance between subgingival species. The correlations in relative abundance among prevalent species (see list in Supplementary Table 1) were calculated based on Pearson correlation coefficients. In each heatmap, the species are listed in the same order in both horizontal and vertical axes, and the colored bars indicate the bacterial complexes based on Socransky’s designation [37] (more details in Supplementary Table 1). Black boxes in the heatmaps indicate the bacterial clusters that comprise red complex species with an average Pearson correlation coefficients >0.7 between the species. Hierarchical clustering was applied to the correlation matrices
Functional potentials of the subgingival microbiome associated with periodontitis and diabetes
We compared the functional potentials encoded in the subgingival microbiome and their associations with periodontitis between T2DM and ND. We identified a total of 3,201 KO genes with at least one copy in any of the samples. 549 KO genes varied across clinical states in prevalence with a standard deviation >0.2. Among them, 373 KO genes (67.9%) were from the genomes of the representative species designated by Socransky et al. [37] (Fig. 5). We found 229 KO genes corresponded to unique microbial complexes [37], while 144 were from two or more microbial complexes. These findings based on the KO gene profiles were consistent with the trends that we observed in the microbiome index of periodontitis (Fig. 2). KO genes from the red complex species were less prevalent in the periodontitis state in T2DM compared with ND. On the other hand, in the periodontal healthy state, T2DM patients had more KO genes from orange complex species, which are often considered opportunistic pathogens. Taken together, we identified a set of marker genes that indicate the differences in the microbiome between different states and between T2DM and ND.
Fig. 5.
KO genes encoded in the subgingival microbiome with different prevalence among clinical states and between T2DM and ND subjects. A total of 373 KO genes with different prevalence among the clinical states in T2DM and ND are shown. The bacterial species, which the KO genes belong to, are indicated by the color bars on the right according to Socransky’s designation of the bacterial complexes [37]. The KO genes labeled in brown are identified in the species from more than one microbial complex (more details in Supplementary Table 2)
We further investigated whether certain microbial functional pathways were associated with periodontitis and T2DM. We found that the KO genes with differential prevalence between clinical states were significantly enriched in 21 functional pathways (hypergeometric distribution test) (Fig. 6). Among them, we identified the microbial pathways associated with periodontitis, including four pathways that were enriched with virulence factors and were more prevalent in the periodontitis state in T2DM and ND. They are pathways associated with cell motility (bacterial motility, flagellar assembly, and bacterial chemotaxis) and a signal transduction pathway (two-component system). Furthermore, we identified differences in prevalence of microbial pathways between T2DM and ND. Two pathways in lipid metabolism (ether lipid metabolism, and arachidonic acid metabolism) and one pathway in carbohydrate metabolism (inositol phosphate metabolism) were more prevalent in periodontitis state only in ND but not in T2DM. In contrast, three pathways in carbohydrate metabolism (butanoate metabolism, pentose and glucuronate interconversions, and ascorbate and aldarate metabolism) were more prevalent in both periodontitis state and healthy state in T2DM than in healthy state in ND.
Fig. 6.
Microbial pathways associated with periodontitis and T2DM. Twenty-one microbial pathways were significantly enriched with KO genes that were different in prevalence between clinical states in T2DM and ND. The proportion of known virulence factors identified in each pathway is shown to the left of the heatmap
Discussion
Both diabetes and periodontitis are among the most prevalent chronic diseases affecting a large adult population in the US [1–3] and many other countries worldwide. The two diseases have intertwined pathogenesis with diabetes increasing the risk for periodontitis and periodontal inflammation adversely affecting glycemic control [4, 5]. While the relationship between the degree of hyperglycemia and severity of periodontitis clearly exists, and systemic inflammation status, immune functioning, neutrophil activity, and cytokine biology have been implicated in the pathogenesis [6], the mechanisms underlying the connection between these two conditions are not well understood.
In this study, we aimed to shed light on the link between T2DM and periodontitis from the microbiome perspective. We investigated the subgingival microbiome differences between T2DM patients and systemically healthy subjects at the metagenomic level in periodontal health, periodontitis, and resolved states. We found that the subgingival microbiome in the periodontitis state differed from that of the healthy state in both groups, albeit to a different extent. Using the microbiome index that we defined based on metagenomic signatures, we revealed that the extent of the microbiome shift from the healthy to the periodontitis state was less prominent in T2DM compared with ND subjects. The subgingival microbiome in T2DM patients had not shifted to a characteristic disease state as seen in ND, yet the clinical signs of periodontitis were evident (Fig. 2). This suggests that T2DM patients are less tolerant to the presence of periodontal pathogens and periodontitis manifests earlier in response to a less severe shift of the subgingival microbiome toward dysbiosis, likely due to host metabolic dysregulation and upregulated immune response [4–7]. Our findings support that more frequent monitoring of the subgingival microbiome and/or an emphasis on more diligent biofilm control in T2DM patients may be necessary as small shifts in the microbiome could trigger periodontitis in this population.
In addition to the overall microbiome differences observed between T2DM and ND subjects, we investigated the differences in the individual microbial species linked to periodontitis. It is well known that bacterial colonization on the tooth surface and biofilm development are associated with periodontal disease. Streptococcus species (yellow complex) and Actinomyces species are known as early colonizer species that bind to the complementary salivary receptors in the pellicle coating on the tooth surface. F. nucleatum (orange complex) is the key bridge organism that interacts with late colonizer species including pathogenic red complex species and facilitates the succession of bacterial colonization and biofilm formation, progressing toward periodontitis [39]. Pathogenic red complex species were all present and highly correlated in the periodontitis state (Fig. 4), consistent with the essential role of coordinated interactions of the pathogenic species in periodontitis [15, 18]. Comparing T2DM and ND subjects, we found that the subgingival microbiome in the healthy state in T2DM subjects had higher relative abundances of the orange complex and the red complex species than in ND subjects (Fig. 7). This suggests that T2DM patients are predisposed to a more pathogenic state of the subgingival microbiome and a higher risk of developing periodontitis, which is consistent with clinical observations [40]. On the other hand, in the resolved state, we found that the subgingival microbiome in T2DM patients had lower relative abundances of orange complex and red complex species than in ND, suggesting that T2DM patients are less tolerant to the presence of periodontal pathogens and thus have lower abundances of the pathogens in order to maintain the periodontium in a clinically resolved state.
Fig. 7.
Dynamics of the subgingival microbiome associated with periodontitis in T2DM and ND subjects. A schematic quantitatively illustrates the dynamic changes in the subgingival microbiome composition among the healthy state, periodontitis state, and resolved state in T2DM and ND. The health- and disease-associated species are depicted according to the subgingival complexes designated by Socransky et al. [37] and the knowledge of bacterial interactions from the literature [39]. The proportions of the species are scaled based on their relative abundances in our data from this study
We next revealed microbial functions that may link the subgingival microbiome to the two diseases. We identified 21 pathways that were significantly enriched in different disease states (Fig. 6). They include the four pathways that were enriched with virulence factors and were more prevalent in the periodontitis state in both T2DM and ND. These are pathogenic pathways associated with periodontitis. For example, three identified cell motility-related pathways are involved in chemotaxis-guided bacterial motility that enhances the growth of periodontal pathogens and facilitates their colonization to penetrate through protective epithelial cell layers into the underlying tissues [41–43]. We also identified a two-component system in the periodontitis state that facilitates microorganisms to adapt to host environmental changes and modulates the maturation and transportation of the virulence factors by regulating gene expression of the type IX secretion system components. One example of the virulence factors is protease gingipains, which induce inflammation and tissue destruction in the periodontium [44, 45]. In addition, we found three pathogenic pathways that were more prevalent in the periodontitis state in ND, but not in T2DM. They include two pathways in lipid metabolism and inositol phosphate metabolism, which are linked via lipoprotein-associated phospholipases, a group of inflammatory enzymes associated with oral infections [46, 47]. Compared with ND subjects, our data indicate that periodontal inflammation can be induced in T2DM patients without enrichment of the virulence factors in these above mentioned pathways.
We also identified three pathways that were enriched in T2DM, not in ND, in both periodontitis state and healthy state. These are pathways involved in carbohydrate metabolism—butanoate metabolism, pentose and glucuronate interconversions, and ascorbate and aldarate metabolism, and they are linked via dehydrogenases. The ascorbate and aldarate metabolism pathway has been associated with inflammatory diseases including periodontitis [48, 49] and T2DM [50]. Its downstream pathway, pentose and glucuronate interconversions, may be responsible for regulation of ascorbate levels in the local environment. Furthermore, in our data, we found that butanoate metabolism pathway was enriched in the periodontal healthy state in T2DM, but not in ND. Microbial butanoate metabolism has been indicated as a metabolic signature of periodontal inflammation [51], and butyrate can influence insulin sensitivity [52]. These results are consistent with epidemiological studies, which support that periodontal infection has an adverse effect on glycemic control [40]. Our findings show that these pathways may potentially provide a microbial function link between periodontitis and T2DM.
The limitations of our study include the small group size. We analyzed 47 samples from 15 T2DM and 16 ND subjects. While the numbers of subjects and samples analyzed in this study are comparable with other published studies of the subgingival microbiome [8–15], future studies of larger cohorts are needed. Another potential limitation is the difference in age between the T2DM and ND healthy periodontium groups. In our cohort, the average age of the T2DM patients with healthy periodontium was older than that of ND subjects with healthy periodontium (Table 1). In spite of this, we did not find a significant difference in the microbiome composition between these two groups (p = 0.189) (Fig. 2). Based on the literature, it is unclear whether age affects the composition of the subgingival microbiome in this age range [53]. On the other hand, while there was no significant difference in age between the T2DM and ND periodontitis patients in our cohort (p = 0.29), we identified clear differences in the microbiome between these two diseased groups.
In summary, we present the first longitudinal metagenomic analysis of the subgingival microbiome associated with periodontitis in a highly susceptible population T2DM in comparison with ND subjects. We revealed the differences in the microbiome shifts between clinical states in T2DM compared with ND. We also showed that the correlations among bacterial species in relative abundance were different between the two populations. Both suggest that T2DM patients are more susceptible to shifts in the subgingival microbiome toward dysbiosis in developing periodontitis. This study addresses a gap in our knowledge of the composition and dynamics of the subgingival microbiome in the diabetes population, which is at a higher risk for periodontitis. In addition, we identified a set of microbial genes, which were enriched in the pathways associated with periodontitis and in the pathways potentially linking T2DM and periodontitis. Our study sheds new light on the relationship between T2DM and periodontitis from the perspective of the oral microbiome and warrants future investigations of larger cohorts.
Supplementary information
Acknowledgements
We thank Kimberly Mataalii at UCLA and Audrey Simons and her team at the San Fernando Community Health Center for their efforts in subject recruitment and sample collection. This study was supported by NIH/NIDCR grants R01DE021574 and RC1DE020298.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-019-0544-3) contains supplementary material, which is available to authorized users.
References
- 1.Preshaw PM, Alba AL, Herrera D, Jepsen S, Konstantinidis A, Makrilakis K, et al. Periodontitis and diabetes: a two-way relationship. Diabetologia. 2012;55:21–31. doi: 10.1007/s00125-011-2342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res. 2012;91:914–20. doi: 10.1177/0022034512457373. [DOI] [PubMed] [Google Scholar]
- 3.Eke PI, Dye BA, Wei L, Slade GD, Thornton-Evans GO, Borgnakke WS, et al. Update on prevalence of periodontitis in adults in the United States: NHANES 2009 to 2012. J Periodontol. 2015;86:611–22. doi: 10.1902/jop.2015.140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lalla E, Papapanou PN. Diabetes mellitus and periodontitis: a tale of two common interrelated diseases. Nat Rev Endocrinol. 2011;7:738–48. doi: 10.1038/nrendo.2011.106. [DOI] [PubMed] [Google Scholar]
- 5.Mealey BL, Oates TW. American academy of periodontology. Diabetes mellitus and periodontal diseases. J Periodontol. 2006;77:1289–303. doi: 10.1902/jop.2006.050459. [DOI] [PubMed] [Google Scholar]
- 6.Taylor JJ, Preshaw PM, Lalla E. A review of the evidence for pathogenic mechanisms that may link periodontitis and diabetes. J Clin Periodontol. 2013;40:S113–134. doi: 10.1111/jcpe.12059. [DOI] [PubMed] [Google Scholar]
- 7.Xiao E, Mattos M, Vieira GHA, Chen S, Corrê aJD, Wu Y, et al. Diabetes enhances IL-17 expression and alters the oral microbiome to increase its pathogenicity. Cell Host Microbe. 2017;22:120–8. doi: 10.1016/j.chom.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–85. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, Sommer DD, et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS ONE. 2012;7:e37919. doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–74. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7:1016–25. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, He J, He Z, Zhou Y, Yuan M, Xu X, et al. Phylogenetic and functional gene structure shifts of the oral microbiomes in periodontitis patients. ISME J. 2014;8:1879–91. doi: 10.1038/ismej.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jünemann S, Prior K, Szczepanowski R, Harks I, Ehmke B, Goesmann A, et al. Bacterial community shift in treated periodontitis patients revealed by ion torrent 16S rRNA gene amplicon sequencing. PLoS ONE. 2012;7:e41606. doi: 10.1371/journal.pone.0041606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laksmana T, Kittichotirat W, Huang Y, Chen W, Jorgensen M, Bumgarner R, et al. Metagenomic analysis of subgingival microbiota following non-surgical periodontal therapy: a pilot study. Open Dent J. 2012;6:255–61. doi: 10.2174/1874210601206010255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi B, Chang M, Martin J, Mitreva M, Lux R, Klokkevold P, et al. Dynamic changes in the subgingival microbiome and their potential for diagnosis and prognosis of periodontitis. mBio. 2015;6:e01926–14. doi: 10.1128/mBio.01926-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casarin RC, Barbagallo A, Meulman T, Santos VR, Sallum EA, Nociti FH, et al. Subgingival biodiversity in subjects with uncontrolled type-2 diabetes and chronic periodontitis. J Periodontal Res. 2013;48:30–36. doi: 10.1111/j.1600-0765.2012.01498.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhou M, Rong R, Munro D, Zhu C, Gao X, Zhang Q, et al. Investigation of the effect of type 2 diabetes mellitus on subgingival plaque microbiota by high-throughput 16S rDNA pyrosequencing. PLoS ONE. 2013;8:e61516. doi: 10.1371/journal.pone.0061516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganesan SM, Joshi V, Fellows M, Dabdoub SM, Nagaraja HN, O’Donnell B, et al. A tale of two risks: smoking, diabetes and the subgingival microbiome. ISME J. 2017;11:2075–89. doi: 10.1038/ismej.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcel Martin. Cutadapt removes adapter sequences high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 22.Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The human oral microbiome database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database. 2010;2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2006;34:D16–20. doi: 10.1093/nar/gkj157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol. 2008;11:472–7. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Tomida S, Nguyen L, Chiu BH, Liu J, Sodergren E, Weinstock GM, et al. Pan-genome and comparative genome analyses of Propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. mBio. 2013;4:e00003–13. doi: 10.1128/mBio.00003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen LH, Yang J, Yu J, Yao ZJ, Sun LL, Shen Y, et al. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33(Database issue):D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 31.Bleharski JR, Li H, Meinken C, Graeber TG, Ochoa MT, Yamamura M, et al. Use of genetic profiling in leprosy to discriminate clinical forms of the disease. Science. 2003;301:1527–30. doi: 10.1126/science.1087785. [DOI] [PubMed] [Google Scholar]
- 32.Barnard E, Shi B, Kang D, Craft N, Li H. The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci Rep. 2016;6:39491. doi: 10.1038/srep39491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18:117–43. [Google Scholar]
- 35.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 37.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 38.Aruni AW, Mishra A, Dou Y, Chioma O, Hamilton BN, Fletcher HM. Filifactor alocis-a new emerging periodontal pathogen. Microbes Infect. 2015;17:517–30. doi: 10.1016/j.micinf.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolenbrander PE, Palmer RJ, Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8:471–80. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 40.Taylor GW. Bidirectional interrelationships between diabetes and periodontal diseases: an epidemiologic perspective. Ann Periodontol. 2001;6:99–112. doi: 10.1902/annals.2001.6.1.99. [DOI] [PubMed] [Google Scholar]
- 41.Charon NW, Goldstein SF. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu Rev Genet. 2002;36:47–73. doi: 10.1146/annurev.genet.36.041602.134359. [DOI] [PubMed] [Google Scholar]
- 42.Dashper SG, Seers CA, Tan KH, Reynolds EC. Virulence factors of the oral spirochete Treponema denticola. J Dent Res. 2011;90:691–703. doi: 10.1177/0022034510385242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lux R, Shi W. Chemotaxis-guided movements in bacteria. Crit Rev Oral Biol Med. 2004;15:207–20. doi: 10.1177/154411130401500404. [DOI] [PubMed] [Google Scholar]
- 44.Frederick JR, Rogers EA, Marconi RT. Analysis of a growth-phase-regulated two-component regulatory system in the periodontal pathogen Treponema denticola. J Bacteriol. 2008;190:6162–9. doi: 10.1128/JB.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kadowaki T, Yukitake H, Naito M, Sato K, Kikuchi Y, Kondo Y, et al. A two-component system regulates gene expression of the type IX secretion component proteins via an ECF sigma factor. Sci Rep. 2016;6:23288. doi: 10.1038/srep23288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wyss C, Choi BK, Schupbach P, Moter A, Guggenheim B, Gobel UB. Treponema lecithinolyticum sp. Nov., a small saccharolytic spirochaete with phospholipase A and C activities associated with periodontal diseases. Int J Syst Bacteriol. 1999;4:1329–39. doi: 10.1099/00207713-49-4-1329. [DOI] [PubMed] [Google Scholar]
- 47.Ishida H, Shinohara H, Nagata T, Nishikawa S, Wakano Y. Phospholipase A2 activity in gingival crevicular fluid from patients with periodontal disease: a possible marker of disease activity. Mediators Inflamm. 1994;3:17–21. doi: 10.1155/S0962935194000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishida M, Grossi SG, Dunford RG, Ho AW, Trevisan M, Genco RJ. Dietary vitamin C and the risk for periodontal disease. J Periodontol. 2000;71:1215–23. doi: 10.1902/jop.2000.71.8.1215. [DOI] [PubMed] [Google Scholar]
- 49.Iwasaki M, Manz MC, Taylor GW, Yoshihara A, Miyazaki H. Relations of serum ascorbic acid and α-tocopherol to periodontal disease. J Dent Res. 2012;91:167–72. doi: 10.1177/0022034511431702. [DOI] [PubMed] [Google Scholar]
- 50.Zhou C, Na L, Shan R, Cheng Y, Li Y, Wu X, et al. Dietary vitamin C intake reduces the risk of type 2 diabetes in chinese adults: HOMA-IR and T-AOC as potential mediators. PLoS ONE. 2016;11:e0163571. doi: 10.1371/journal.pone.0163571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakanaka A, Kuboniwa M, Hashino E, Bamba T, Fukusaki E, Amano A. Distinct signatures of dental plaque metabolic byproducts dictated by periodontal inflammatory status. Sci Rep. 2017;7:42818. doi: 10.1038/srep42818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58:1509–17. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feres M, Teles F, Teles R, Figueiredo LC, Faveri M. The subgingival periodontal microbiota of the aging mouth. Periodontology. 2016;72:30–53. doi: 10.1111/prd.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.