Abstract
Soil-inhabiting streptomycetes are nature’s medicine makers, producing over half of all known antibiotics and many other bioactive natural products. However, these bacteria also produce many volatiles, molecules that disperse through the soil matrix and may impact other (micro)organisms from a distance. Here, we show that soil- and surface-grown streptomycetes have the ability to kill bacteria over long distances via air-borne antibiosis. Our research shows that streptomycetes do so by producing surprisingly high amounts of the low-cost volatile ammonia, dispersing over long distances to inhibit the growth of Gram-positive and Gram-negative bacteria. Glycine is required as precursor to produce ammonia, and inactivation of the glycine cleavage system nullified ammonia biosynthesis and concomitantly air-borne antibiosis. Reduced expression of the porin master regulator OmpR and its cognate kinase EnvZ is used as a resistance strategy by E. coli cells to survive ammonia-mediated antibiosis. Finally, ammonia was shown to enhance the activity of canonical antibiotics, suggesting that streptomycetes adopt a low-cost strategy to sensitize competitors for antibiosis from a distance.
Subject terms: Antibiotics, Soil microbiology
Introduction
Volatile compounds (VCs) are small molecules with high vapor pressure and low molecular weight that easily diffuse through air, water, or soil [1, 2]. VCs have a broad activity spectrum, acting as infochemicals, growth-promoting or inhibiting agents, modulators of quorum sensing and drug resistance or as a carbon-release valve, influencing their neighbor’s behavior and phenotypes such as stress response, colony morphology, biofilm, virulence, and pigmentation [3–7]. In soil, VCs play important roles in inter- and intra-species interactions [8].
Actinobacteria are one of the largest bacterial phyla present in soil [9, 10]. They are known as Nature’s medicine makers [11], with the ability to produce bioactive secondary metabolites that work as antibiotics, anticancer, antifungal, anthelmintic, and immunosuppressant agents [10, 12, 13]. Streptomycetes alone produce half of all known antibiotics used in the clinic. They are also prolific producers of VCs, although their ecological functions remain largely elusive [14, 15]. In terms of antimicrobial activity of VCs, information is primarily available on their activity as antifungals [9, 16]. A rare example of a VC with antibacterial activity is the sesquiterpene albaflavenone produced by Streptomyces albidoflavus [17].
The natural role of antibiotics is subject to intensive debate. It has been argued that their main function lies in cell to cell communication [18]. However, antibiotics may well act as weapons, and bioactivity is influenced by social and competitive interactions between strains [19]. Interestingly, it is becoming more evident that the small inorganic VCs such as hydrogen sulfide (H2S) and nitric oxide (NO) play a major role in modulating antibiotic activity and resistance [20]. For example, H2S production protects bacteria against antibiotics targeting DNA, RNA, protein, and cell wall biosynthesis [21]. Furthermore, Bacillus anthracis produces NO to protect itself against oxidative stress and helps the bacterium to survive in macrophages, thus playing a key role in escaping the host’s defense [22, 23]. Production of NO also directly protects bacteria against a broad spectrum of antibiotics [24, 25]. Ammonia induced resistance to tetracycline, by increasing the level of polyamines which leads to a modification of membrane permeability [26]. There is also some experimental evidence that suggests that VCs may affect membrane integrity [27, 28], which in turn may make the cells more susceptible to other cell-damaging compounds, such as antibiotics. To date, however, more conclusive experimental evidence for the ecological roles of VCs produced by bacteria and for the mechanisms underlying resistance to VCs is limited.
In this study, we show for the first time that Streptomyces can produce surprisingly high levels of ammonia that affect growth of Gram-positive and Gram-negative bacteria over long distances. We also show that the production of ammonia in Streptomyces is affected by the growth conditions and that its biosynthesis depends on the glycine cleavage system. The same is observed in an in vivo assay where we show that the ammonia-producing wild type S. griseus inoculated into soil inhibits the growth of B. subtilis whereas the S. griseus glycine riboswitch-null mutant (gcvT) did not. Finally, E. coli cells gain resistance against ammonia by reducing the expression of the two-component system OmpR-EnvZ, thereby counteracting passage through the outer membrane porins.
Materials and methods
Strains, media, culture conditions, and antimicrobial assays
Strains used in this study are listed in Table S1. The Streptomyces strains were grown on Soy Flour Mannitol (SFM) agar plates to prepare spore stocks. Escherichia coli strain AS19-RlmA− [29] and B. subtilis 168 [30] were used as test microorganisms and grown on Luria-Bertani (LB) agar plates.
Volatile antimicrobial assays were performed using a petri dish with two compartments, one filled with SFM media for Streptomyces growth and the second one with LB +/− TES buffer 50–100 mM. Streptomyces strains were streaked on the SFM side and allowed to grow for 5 days after which, E. coli or B. subtilis were inoculated on the LB side using a concentration of 104 and 103 cfu/mL respectively.
Collection and analysis of VCs
VCs produced by Streptomyces monocultures grown on SFM agar were collected using a glass Petri dish designed for trapping of the volatile headspace [31]. The lid of the glass Petri dish contains an outlet specially designed to hold a stainless-steel column packed with 200 mg Tenax® TA 60/80 material (CAMSCO, Houston, TX, USA). Samples were taken in triplicates from day 3 to day 5 of growth; after that, the Tenax steel traps were sealed and stored at 4 °C until GC-Q-TOF analysis.
Trapped volatiles were desorbed using an automated thermodesorption unit (model UnityTD-100, Markes International Ltd., United Kingdom) at 210 °C for 12 min (Helium flow 50 mL/min) and trapped on a cold trap at −10 °C. The trapped volatiles were introduced into the GC-QTOF (model Agilent 7890B GC and the Agilent 7200A QTOF, USA) by heating the cold trap for 3 min to 280 °C. A 30 × 0.25 mm ID RXI-5MS column with a film thickness of 0.25 μm was used (Restek 13424–6850, USA). Temperature program used was as follows: 39 °C for 2 min, from 39 to 95 °C at 3,5 °C/min, then to 165 °C at 6 °C/min, to 250 °C at 15 °C/min and finally to 300 °C at 40 °C/min, hold 20 min The VCs were detected by the mass spectrometer (MS) operating at 70 eV in EI mode. MS spectra were extracted with MassHunter Qualitative Analysis Software V B.06.00 Build 6.0.633.0 (Agilent Technologies, USA) using the GC-Q-TOF qualitative analysis module. MS spectra were exported as mzData files for further processing in MZmine. The files were imported to MZmine V2.14.2 [32] and compounds were identified via their mass spectra using deconvolution function (Local-Maximum algorithm) in combination with two mass-spectral-libraries: NIST 2014 V2.20 (National Institute of Standards and Technology, USA http://www.nist.gov) and Wiley 9th edition mass spectral libraries and by their linear retention indexes (LRI). The LRI values were calculated using an alkane calibration mix before the measurements in combination with AMDIS 2.72 (National Institute of Standards and Technology, USA). The calculated LRI were compared with those found in the NIST and in the in-house NIOO-KNAW LRI database. After deconvolution and mass identification peak lists containing the mass features of each treatment (MZ-value/Retention time and the peak intensity) were created and exported as CSV files for statistical processing via MetaboAnalyst V3.0 (www.metaboanalyst.ca; [33]).
pH change, ammonia determination, and toxicity
Change in pH of the growth media was determined by the addition of phenol red indicator (0.002%). Pictures were taken after 0, 3, and 5 days of incubation next to Streptomyces biomass. For the ammonia test, Streptomyces strains were grown for 5 days on SFM agar using the two-compartment petri-dish, whereby the other half of the plate was left empty. After 5 days, the ammonia was determined using the Quantofix® ammonium test kit. Pictures were recorded to obtain a qualitative measurement of ammonia production from each strain.
Quantification of ammonia accumulation inside the LB agar was determined by extracting the liquid from the LB agar by centrifugation. For this, centrifuge tube filters were used (spin-X® 0.22 μm cellulose acetate, Corning Inc. USA), 1 cm2 of agar was taken 1 cm away from the plastic barrier separating the Streptomyces biomass. The plug was put inside the filter tube and centrifuge at 13,000 rpm for 20 min. The eluate (∼200 μL) was used to quantify the ammonium concentration in comparison to a standard curve. The standard curve was made with LB agar containing 0–50 mM concentrations of ammonia. Ammonia solution (25% in H2O, J.T. Baker 6051) was used as source of ammonia. The liquid was extracted from the agar the same way as described before and used together with the Quantofix® ammonium kit to obtain a semi-quantitative measure of ammonia accumulation inside the agar.
To determine the toxicity of ammonia, E. coli and B. subtilis were incubated in the automated Bioscreen C (Lab systems Helsinki, Finland) in the presence of increasing concentrations of ammonia. Each dilution was prepared in LB containing an inoculum of 105 cfu/mL + different volumes of ammonia solution (J.T. Baker) to give the following final concentrations: 1, 5, 10, 15, 16, 17, 18, 20, 25, 30, 40, and 50 mM. The final working volume in each well of the honeycomb was 100 μL. Cultures were incubated at 37 °C overnight with continuous shaking. O.D. measurements (wideband) were taken every 30 min for 20 h. The data and growth curves were calculated from triplicates.
Ammonia production in soil
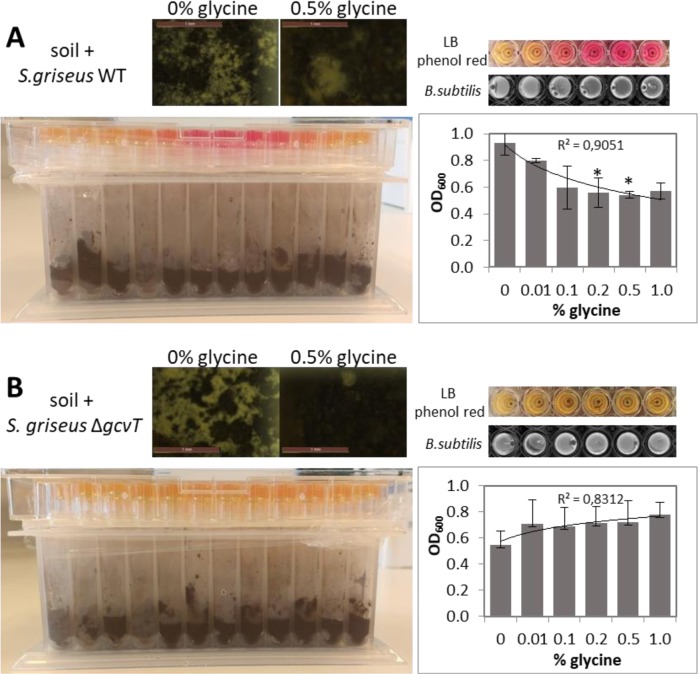
To detect the production of ammonia by Streptomyces inoculated in field soil, a microrespirometry-like system [34] was used. The assembled system was comprised of a 96 deep-well plate with a capacity of 2 mL (Favorgen biotech corp.) interconnected to a top 96 well plate with flat bottom (Greiner bio-one) via a CapMat of 96 naps (Greiner bio-one). Holes were made to the CapMat to allow the passage of volatiles and avoid cross contamination between wells. On the bottom plate 0.5 g of sterile soil were placed in each well and 100 μL of trace elements solution were added (ZnCl2 0.08 mg/mL; FeCl3·6H2O 0.4 mg/mL; CuCl2·2H2O 0.02 mg/mL; MnCl2·4H2O 0.02 mg/mL; Na2B4O7·10H2O; (NH4)6Mo7O24·10H2O 0.02 mg/mL). One hundred microliters of a spore suspension of S. griseus wild-type or S. griseus gcvT [35] were inoculated in order to give a final concentration of 1 × 106 spores/well. The final moisture content was 40% (v/w). In the control treatment, no Streptomyces was inoculated. For the upper plate, each well was filled with 130 μL of LB with 0.002% phenol red as pH indicator. This plate was sealed with the CapMat with holes and the two 96 well plates were sealed together using parafilm. The final setup is shown in Fig. 5. The system was incubated for 10 days at 30 °C. After this, the top plate was inoculated with 2 μL of a suspension of B. subtilis at an OD600 of 0.1, sealed with a new CapMat without holes and incubated at 37 °C. The bottom plate was used to check the growth of Streptomyces. Growth was determined qualitatively by the appearance of Streptomyces colonies on top of the soil and visualized by stereo microscopy using a Zeiss Lumar V12 steremicroscope. Streptomyces growth was determined quantitatively by dilution plating soil suspensions onto SFM.
Fig. 5.
Long-distance inhibition of B. subtilis by volatile ammonia produced by S. griseus in soil. Experiments were done with a wild type S. griseus, and b S. griseus ΔgcvT that lacks the glycine cleavage system. In each panel, the bottom left image shows the growth of Streptomyces in 96 well plates filled with soil with the ammonia detection plate placed on top, and top left stereo micrographs of Streptomyces colonies growing on top of the soil. The top right side shows the color change of phenol red if ammonia is produced and the corresponding B. subtilis inhibition or growth. The graphs on the bottom right side shows the quantitative measurement of the OD600 of each well where B. subtilis was spotted. *indicates statistical significance. Note that growth of B. subtilis was inhibited significantly by wild-type S. griseus but not by its gcvT null mutant
HCN determination
To detect hydrogen cyanide in the headspace of Streptomyces growth we used a method adapted from Castric and Castric [36]. For this, Whatman™ paper was soaked in suspension containing 5 mg/ml of copper(II) ethyl acetoacetate and 4,4′-methylenebis-(N,N-dimethylaniline) (Sigma-Aldrich, USA) dissolved in chloroform and allowed to dry protected from light. The filter paper was placed next to Streptomyces pre-grown for 2 days. Pseudomonas donghuensis P482 was used as positive control. Strains were incubated at 30 °C until blue coloration of the filter paper was evident.
Whole genome sequencing
Genome sequencing of E. coli AS19-RlmA− [37] and its mutant ARM9 was performed using Illumina HiSEQ and PacBio RfavaS at Baseclear BV, Leiden (The Netherlands). Paired-end sequence reads were generated using the Illumina HiSeq 2000 system and mapping the individual reads against the reference genome of E. coli B str. REL606. The contigs were placed into superscaffolds based on the alignment of the PacBio CLC reads. Alignment was performed with BLASR [38]. Genome annotation was performed using the Baseclear annotation pipeline based on the Prokaryotic Genome Annotation System (http://vicbioinformatics.com). Variant detection was performed using the CLC genomics workbench version 6.5. The initial list of variants was filtered using the Phred quality score and false positives were reduced by setting the minimum variant frequency to 70% and the minimum number of reads that should cover a position was set to 10. Relevant mutations were confirmed by PCR analysis. The genome of E. coli AS19-RlmA− has been published, with accession number CP027430 [37].
Genetic complementation of ompR and envZ
E. coli strain AS19-RlmA− suppressor mutant ARM9 was complemented by inserting the ompR or envZ genes in pCA24N from the ASKA collection [39]. Cells of suppressor mutant ARM9 containing the plasmid were inoculated in LB + Chloramphenicol (25 μg/mL) with or without IPTG 0.1 mM for induction of the gene expression.
RNA sequencing
For RNA extraction E. coli cells were grown to an O.D.600 of 0.5, RNA Protect Bacteria Reagent (Qiagen Cat No. 76506) was added according to manufacturer instructions. Cells were pelleted and re-suspended in boiling 2% SDS + 16 mM EDTA followed by extraction with Phenol:chloroform:Isoamyl alcohol (25:24:1) pH 6.6. (VWR Prolabo 436734C). Aqueous phase was precipitated with 3 M sodium acetate pH 5.2 and pure ethanol, washed with 70% Ethanol and re-suspended in RNAse-free water. DNA was removed using five units of DNAseI (Fermentas #EN0521) with further purification using again phenol:chloroform:isoamyl alcohol and precipitation with sodium acetate and ethanol. The final pellet was dissolved in RNase free water.
RNA sequencing and analysis was performed by Baseclear BV (Leiden, The Netherlands). Ribosomal RNA was subsequently removed with a Ribo-Zero kit (Epicenter) and the remaining RNA used as input for the Illumina TruSeq RNA-seq library preparation. Once fragmented and converted into double strand cDNA, the fragments (about 100–200 bp) were ligated with DNA adapters at both ends and amplified via PCR. The resulting library was then sequenced using an Illumina Sequencer. The FASTQ sequence reads were generated using the Illumina Casava pipeline version 1.8.3. Initial quality assessment was based on data passing the Illumina Chastity filtering. Subsequently, reads containing adapters and/or PhiX control signals were removed using an in-house filtering protocol. The second quality assessment was based on the remaining reads using the FASTQC quality control tool version 0.10.0.
For the RNA-Seq analysis the quality of the FASTQ sequences was enhanced by trimming off low-quality bases using the “Trim sequences” option present in CLC Genomics Workbench Version 6.0.4 (QIAGEN, Bioinformatics). The quality-filtered sequence reads were used for further analysis with CLC Genomics Workbench. First an alignment against the reference and calculation of the transcript levels was performed using the “RNA-Seq” option. Subsequent comparison of transcript levels between strains and statistical analysis was done with the “Expression analysis” option, calculating so-called RPKM values. These are defined as the reads per kilobase per million mapped reads [40] and normalizes for the difference in the number of mapped reads between samples and for transcript length. The RNAseq data has been submitted to the Gene Expression Omnibus (GEO) from National Biotechnology Center Information (NCBI) with GEO accession number GSE111370.
Synergism of Streptomyces AB-VCs with soluble antibiotics
Synergistic assays were performed using a petri dish with two compartments, one filled with SFM media for Streptomyces growth and the second one with LB. Streptomyces strains were streaked on the SFM side and allowed to grow for 5 days at 30 °C after which, E. coli or B. subtilis were inoculated on the LB side using a culture grown to an O.D. = 0.5. One hundred microliters were streaked on the LB side. Afterwards, 6 mm filter disks (Whatman™) were placed on top of the LB and 10 μL of each antibiotic spotted on the filter disc. Two different concentrations per antibiotic were tested: ampicillin 500, 31 μg/mL; erythromycin 250, 31 μg/mL; kanamycin 1000, 500 μg/mL; tylosin 500, 62 μg/mL; actinomycin 500, 62 μg/mL; spectinomycin 1000, 500 μg/mL; streptomycin 500, 62 μg/mL. Plates were incubated at 37 °C overnight and pictures recorded after 20 h of growth.
Results and discussion
VCs as bioactive agents in long-distance antibiosis
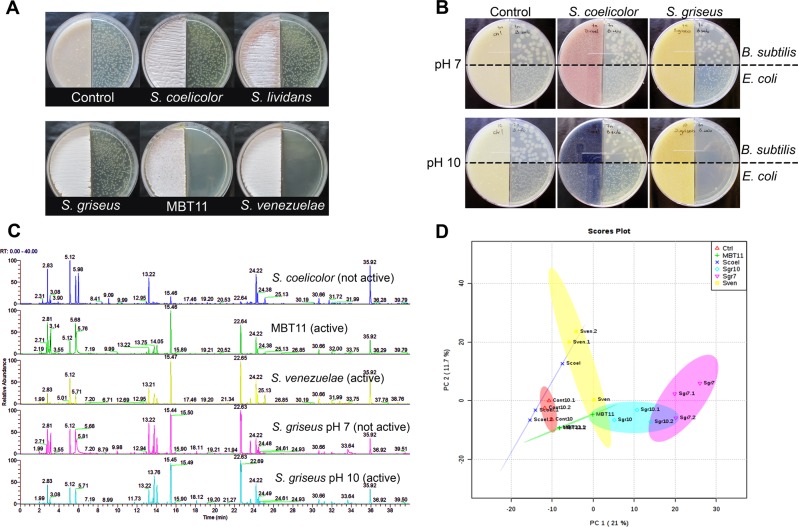
To investigate if streptomycetes produce antibacterial VCs, different Streptomyces strains were grown physically separated from indicator/target bacteria by a polystyrene barrier. Air-borne VCs can pass over the barrier, but it does not allow passage of canonical (soluble) antibiotics. We used Bacillus subtilis and Escherichia coli strain AS19-RlmA− (referred to as E. coli ASD19 from now on) as Gram-positive and Gram-negative indicator strains, respectively. The latter has known antibiotic sensitivity [29]. Interestingly, the streptomycetes showed varying volatile activity against E. coli ASD19; this strain failed to grow adjacent to Streptomyces sp. MBT11 or S. venezuelae, but grew normally next to S. coelicolor, S. lividans, and S. griseus (Fig. 1a). B. subtilis was not inhibited by any of the Streptomyces strains tested.
Fig. 1.
Bioactivity and metabolomic analysis of VCs released by streptomycetes. a Bioactivity of VCs released by selected Streptomyces strains against E. coli strain ASD19. b Volatile antibiotic activity of different Streptomyces strains grown at pH 7 or pH10, the latter by addition of a glycine/NaOH buffer; E. coli strain ASD19 was the indicator strain. c Comparison of GC-chromatogram of VCs from bioactive and non-bioactive strains. d PCA-2D Plot showing the VC-grouping of the different strains. No clear separation between VCs produced by different Streptomyces strains is seen
We then wanted to assess whether the production of antimicrobial volatile compounds (AMVCs) could be elicited by varying the growth conditions. We previously showed that growth at pH 10, N-acetylglucosamine, starch or yeast extract pleiotropically enhanced the production of antibiotics in many Streptomyces species [41]. Interestingly, in contrast to a neutral pH, when glycine/NaOH buffer (25 mM glycine) [42] was added to raise the pH to 10, S. griseus produced VCs that completely inhibited growth of E. coli and B. subtilis (Fig. 1b). The production of AMVCs by Streptomyces sp. MBT11 was also enhanced by growth in the presence of the buffer system (data not shown). In contrast, S. coelicolor failed to produce AMVCs under any of the conditions tested (Table S2). Intriguingly, when grown on 0.8% peptone, S. lividans was able to inhibit B. subtilis but not E. coli. Comparative GC-profiling with S. lividans grown in media with different concentrations of peptone followed by compound identification and bioactivity assays will be needed to establish which volatile compound(s) cause(s) this effect.
The induction of AMVCs by S. griseus when grown on the glycine buffer offered an ideal system to elucidate the nature of the bioactive molecules by statistical methods. Correlation between bioactivity and metabolic profiles allows efficient reduction of candidate molecules [43, 44]. Therefore, GC-Q-TOF-based metabolomics was performed to compare the VC profiles of S. coelicolor, Streptomyces sp. MBT11, S. venezuelae and S. griseus (the latter grown with and without glycine buffer pH 10). Despite the antimicrobial activity, no volatile organic compound (VOC) was detected that correlated statistically to the bioactivity, nor did we see any significant difference between the metabolome profiles of S. griseus grown with or without the glycine buffer (Fig. 1c, d). Some of the mass features suggested differential production of 2-methylisoborneol (2-MIB) and 2-methylenebornane by the active strains compared to the non-active strains. However, mutants of S. griseus that lacked either or even all the terpene cyclases, still retained their antibacterial volatile activity (data not shown).
The main inhibitory molecule is ammonia
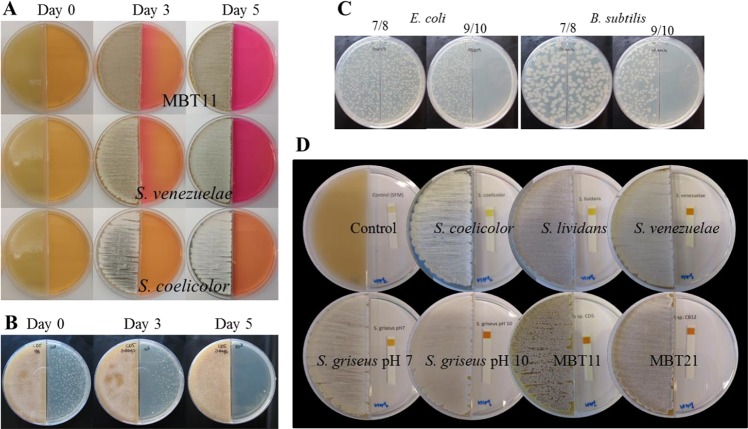
VCs may induce a change in pH away from the producing colonies [45, 46]. To assess this, we used phenol red, which changes from pale orange to bright pink when the media becomes more alkaline. Interestingly, a gradual increase was seen in the pH caused by VCs produced by S. venezuelae or by Streptomyces sp. MBT11 that produce AMVCs, but not by S. coelicolor that lacks the production of AMVCs in our assays. Initially, a pH increase was seen close to the Streptomyces biomass, and after 5 days the receiver side had turned completely pink (Fig. 2a). At that point, the pH in the medium on the receiver side had increased to around 8.5. This pH increase correlated with growth inhibition of E. coli close to the Streptomyces after 3 days, while after 5 days the growth of E. coli was fully inhibited (Fig. 2b).
Fig. 2.
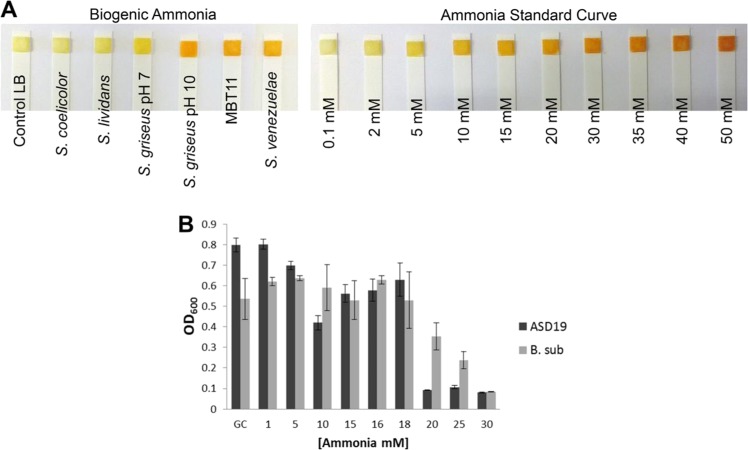
pH increase is caused by high ammonia production. a pH change illustrated by the color change of the indicator (Phenol red 0.002%). LB medium alkalization after 3 and 5 days of growth of Streptomyces sp. MBT11 and S. venezuelae, no alkalization was caused by VCs produced by S. coelicolor. b Streptomyces sp. MBT11 antimicrobial VCs production curve against E. coli strain ASD19. c E. coli strain ASD19 and B. subtilis growth under different pH adjusted with glycine/NaOH buffer. d NH3 emission. Test strips on the right compartment show the production of NH3 by Streptomyces strains. S. coelicolor and S. lividans (∼10 mg/L); S. venezuelae (∼100 mg/L); Streptomyces sp. MBT11, and Streptomyces sp. MBT21 (∼100 mg/L); Control: SFM media (0 mg/L). Concentrations are estimated according to the color chart indicator from the Quantofix® ammonium detection Kit
Further in support of pH-dependent growth inhibition, E. coli grew normally when the media was buffered with 50 mM TES (pH 7; data not shown). However, the pH itself was not the cause of the inhibition, since the E. coli cells grew normal on media adjusted to pH 9 (Fig. 2c). We also previously showed that antibiotic susceptibility of E. coli was similar for pH 10 and pH 7 [41].
Ammonia and trimethylamine (TMA) are VCs known to induce a pH change [26, 45–47]. We also tested production of hydrogen cyanide (HCN), a known AMVC produced by rhizospheric streptomycetes [48]; however, none of the Streptomyces strains produced detectable amounts of HCN (Fig. S1). Furthermore, under our growth conditions, TMA was not detected in the headspace of any of the Streptomyces strains tested, when grown at pH 7 or pH 10. (Fig. S2). Following ammonia detection using the Quantofix® Ammonium detection kit, we found that antibiosis by the streptomycetes strongly correlated with an increase in ammonia production (Fig. 2d). To determine the ammonia concentrations in vitro, the Streptomyces strains were grown for 5 days and the agar on the receiver side extracted. Agar containing different concentrations of ammonia was used to create a standard curve. S. coelicolor, S. lividans, and S. griseus (pH 7) accumulated 2–5 mM ammonia, while the growth-inhibiting S. venezuelae, Streptomyces sp. MBT11, Streptomyces sp. MBT21 as well as S. griseus—the latter only with added Gly/NaOH buffer pH 10—produced 15–30 mM ammonia (Fig. 3a). Subsequent assays showed that E. coli failed to grow on media with 20 mM ammonia or higher, while growth of B. subtilis was inhibited by ammonia concentrations above 30 mM (Fig. 3b). These values are within the range produced by the strains causing volatile-mediated antibiosis, strongly suggesting that ammonia is indeed the AMVC produced by the antagonistic Streptomyces strains.
Fig. 3.
Bioactivity is caused by ammonia. a Ammonia quantification from LB agar extracts exposed to Streptomyces VCs (left). Ammonia standard curve from LB agar extract (right). b Growth of E. coli ASD19 (black) and B. subtilis (gray) under different concentrations of ammonia
Ammonia is derived from glycine cleavage
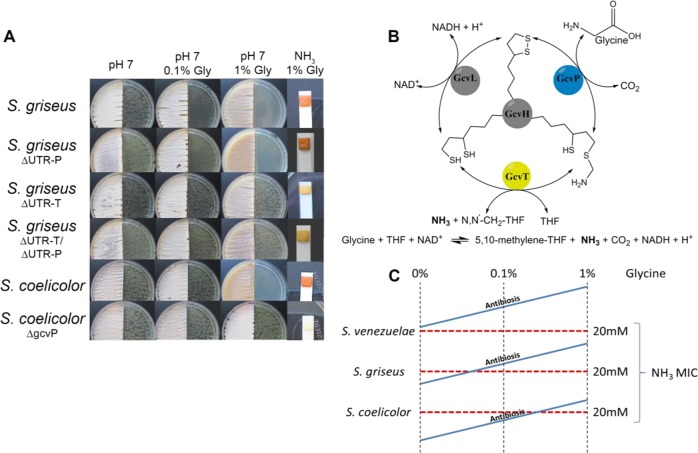
We then wondered if ammonia was generated from glycine metabolism, because a glycine/NaOH buffer was used to set the pH. A major pathway for the catabolism of glycine is the glycine cleavage system (GCV) that converts glycine into CO2, ammonia and a methylene group that is transferred to tetrahydrofolate (THF) to form N5, N10-methylene-THF [35, 49]. Importantly, when S. griseus was grown on SFM agar containing glycine only at concentrations as low as 0.1% (w/v), this time without increasing the pH, the strain still fully inhibited the growth of B. subtilis and E. coli (Fig. 4a). We then also grew S. coelicolor on SFM agar with increasing concentrations of glycine. Interestingly, at concentrations of 1% (w/v) glycine or higher, also S. coelicolor fully inhibited the indicator/target cells. This suggests that at sufficiently high concentrations of glycine, all streptomycetes tested can produce ammonia to a level that inhibits the growth of other bacteria.
Fig. 4.
Bioactivity is caused by ammonia in a glycine cleavage-dependent manner. a Volatile activity and ammonia production by S. griseus, S. griseus glycine cleavage mutant gcvP (ΔUTR-P), S. griseus glycine cleavage mutant gcvT (ΔUTR-T), S. griseus glycine cleavage double mutant gcvT-gcvP (ΔUTR-T/ΔUTR-P), S. coelicolor, and the S. coelicolor glycine cleavage system mutant ΔgcvP. b Scheme representation of the reactions carried by the glycine cleavage (GCV) system, consisting of pyridoxal phosphate-containing glycine decarboxylase GcvP (blue); THF-dependent aminomethyltransferase GcvT (yellow); dihydrolipoamide dehydrogenase GcvL; and lipoic acid-containing carrier protein GcvH. c Illustration of the induction of volatile antibiosis in different Streptomyces strains when increasing concentrations of glycine are added
To investigate the involvement of the GCV system [35], we tested mutants deficient in the glycine cleavage system which consists of three enzymes (GcvL, GcvP, GcvT) and a carrier protein: GcvH (Fig. 4b). The gcvP and gcvT mutants of S. griseus have deletions in the 5′ UTR, and therefore lack a glycine riboswitch that controls the expression of the genes. Conversely, the gcvP mutant in S. coelicolor has the entire gcvP replaced by an apramycin resistance cassette. The gcvP mutant of S. coelicolor [50] and the gcvT mutant of S. griseus [35] were unable to produce inhibiting amounts of ammonia, even when grown on high amounts of glycine. A mutant of S. griseus lacking the 5′UTR of gcvP [35] still produced sufficient ammonia to inhibit the growth of E. coli cells, but this was annihilated by the additional deletion of gcvT (Fig. 4a). Taken together, this strongly suggests that in both S. coelicolor and S. griseus, volatile ammonia is primarily derived from the GCV system, and—as expected—that GcvT is the key enzyme responsible for the production of ammonia from glycine. Inactivation of gcvP is also sufficient to block volatile ammonia production in S. coelicolor, which is in line with the idea that the key system is GCV. The silencing rather than deleting the gcv genes in S. griseus, may allow some residual expression of the genes, and this could explain why the S. griseus gcv mutants still produce sufficient amounts of ammonia to exert antibacterial activity.
A major difference between S. coelicolor and S. griseus on the one hand, and S. venezuelae and Streptomyces sp. MBT11 on the other, is that the latter two strains do not require any added glycine to produce levels of ammonia above the minimal inhibitory concentration (MIC). In this case, ammonia may be derived from the catabolism of other amino acids or from other metabolic enzymes, such as ammonia lyases, deaminases, deiminases, and pyridoxamine phosphate oxidases. We now aim to identify the gene(s) and pathways responsible for the overproduction of ammonia in these strains.
Ammonia production in situ
In order to investigate if volatile-mediated antagonism from a distance could also occur in a more ecologically relevant environment, we introduced S. griseus and the gcvT mutant in soil and assessed the antibacterial activity of the volatiles emitted from the soil in the headspace both in absence and with increasing concentrations of glycine. The production of ammonia was detected using LB with phenol red as pH indicator in a 96 well plate assembled on top of the deep wells with soil inoculated with the Streptomyces strain or mutant.
S. griseus and the gcvT mutant were both able to grow in soil, with and without added glycine. When glycine was added, the growth of S. griseus was boosted indicating that it was able to metabolize the amino acid, whereby 0.1% of glycine was optimal for growth. S. griseus gcvT mutant is unable to metabolize glycine, and expectedly, its growth was not enhanced by glycine; as a result, growth of the gcvT mutant was slightly lower in soil containing glycine than that of the wild type strain, but enough to be comparable (Figs. 5 and S3). Ammonia was produced by S. griseus but not by the gcvT mutant as observed by the change of phenol red from yellow to pink in the top indicator plate. A gradual change in color was observed, which was proportional to the glycine concentration. When B. subtilis was spotted inside the top wells, it grew well when exposed to the VCs from the gcvT mutant, while strong growth inhibition was observed when spotted in the wells exposed to the VCs from wild-type S. griseus (Fig. 5 right). The antagonistic effect correlated with the ammonia levels released into the headspace and with an increasing amount of glycine added to the soil. These data show that long-distance killing by ammonia produced by streptomycetes also occurs in a soil environment, provided that glycine is available. Glycine is an abundant amino acid found in root exudates [51, 52] making its presence in soil feasible. Furthermore, microbial interactions in soil and rhizosphere environments typically occur in microsites where sufficiently high substrate (i.e., glycine) concentrations can be reached to allow for VC-mediated antibiosis.
OmpR is key to ammonia resistance
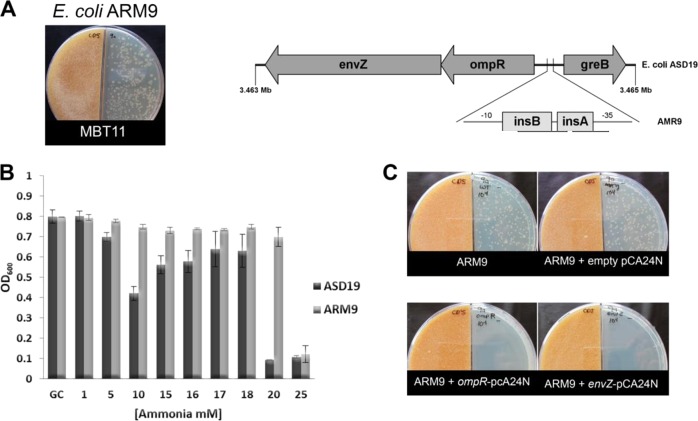
To obtain more insights into the resistance mechanisms to ammonia-mediated antibiosis, we selected for spontaneous ammonia-resistant mutants in E. coli cells exposed to ammonia produced by Streptomyces sp. MBT11. After two days of growth, several E. coli colonies appeared that were able to withstand the accumulated ammonia and likely had sustained one or more suppressor mutations. Four of these colonies were analyzed further, which showed different levels of resistance based on the colony size (Fig. S4A). Of these, suppressor mutant ARM9 was selected for its high resistance (Fig. 6a left). Strain ARM9 was reproducibly more resistant to ammonia than its parent, with MICs of 25 and 20 mM, respectively. This is a biologically significant difference, as we previously showed that 20 mM is precisely the tipping point for ammonia sensitivity of E. coli (Fig. 6b).
Fig. 6.
Insertion sequences in the E. coli ompR-envZ promoter govern resistance to ammonia. a (left) spontaneous suppressor mutant ARM9 derived from E. coli strain ASD19 growing under the presence of volatile compounds produced by Streptomyces sp. MBT11; (right) visualization of insertion sequences in-between the −10 and −35 sequences of the ompR/envZ promoter. b Growth of E. coli strain ASD19 (black) and E. coli strain ASD19 suppressor mutant ARM9 (gray) under the presence of different concentrations of ammonia. c Growth of suppressor mutant ARM9 and transformants harboring either empty plasmid pCA24N, plasmid ompR-pCA24N (expressing ompR), or plasmid envZ-pCA24N (expressing envZ). Note that introduction of a plasmid expressing either envZ or ompR makes ARM9 sensitive again to ammonia
Under low availability of nitrogen, the AmtB transporter facilitates the intake of ammonium inside the cell [53, 54]. Our conditions include high concentrations of ammonia; therefore, we hypothesized that a mechanism other than the AmtB channel would be involved in the resistance towards ammonia. To identify the nature of the mutation(s) sustained by ARM9, its genome sequence was compared to that of its parent E. coli ASD19 (Table S3). In total 658 mutations were found by single nucleotide permutation analysis, of which 198 gave rise to amino acid changes or insertions or deletions. However, one change immediately stood out, namely the introduction of two insertion elements (insA_31 and insB_31) in-between the −35 and −10 consensus sequences of the promoter for ompR-envZ, which encode the two-component system (TCS) consisting of response regulator OmpR and sensory kinase EnvZ (Fig. 6a right). This TCS is involved in osmoregulation in response to environmental signals [55] and regulates the expression of outer membrane porins OmpF and OmpC. Importantly, these are known to be involved in antibiotic resistance regulated by osmotic pressure and pH [56], and to reduce the responsiveness of E. coli cells to VCs [5].
Reduced transcription of the ompR-envZ operon is the cause of ammonia resistance
Considering the location right in the middle of the promoter, we expected that the IS elements in the ompR-envZ promoter reduced the transcription of these crucial TCS genes. To establish the transcriptional consequences of the IS insertion into the ompR-envZ promoter region, RNAseq was performed on E. coli ASD19 and its suppressor mutant ARM9 grown in LB media until mid-exponential phase (OD600 0.5), and the global transcription profiles compared (see Table S4 for the full dataset). Table 1 shows genes highly up/down regulated as a result of a clustering analysis using a cutoff value of a fold change ±2.0. These data confirm the downregulation of ompR and envZ genes and other related genetic elements like omrA, a small noncoding RNA that negatively regulates ompR expression. Additionally, genes involved in amino-acid metabolism were down regulated, including the astABCE gene cluster involved in the ammonia-producing arginine catabolic pathway, aspA that is involved in the conversion of L-aspartate into fumarate and ammonia, and tnaC for catabolism of tryptophan, which again releases ammonia.
Table 1.
Clustering of the differentially down/upregulated genes in ARM9 compared with ASD19. Only fold changes >2.0 or ≤2.0 are shown
| Cluster | Gene | Function | Fold change |
|---|---|---|---|
| Downregulated | |||
| Membrane function/transport | envZ | Sensory histidine kinase in two-component regulatory system with OmpR | −16.43 |
| omrA | Small regulatory RNA | −15.86 | |
| yhdV | Putative outer membrane protein | −15.86 | |
| ompR | Response regulator in two-component regulatory system with EnvZ | −14.84 | |
| dacD | D-alanyl-D-alanine carboxypeptidase, penicillin-binding protein 6b | −6.47 | |
| yqhH | Outer membrane lipoprotein, Lpp paralog | −6.20 | |
| ydiM | Putative MFS transporter, membrane protein | −4.83 | |
| yiaD | Multicopy suppressor of BamB, outer membrane lipoprotein | −4.54 | |
| yajR | Putative transporter | −4.37 | |
| yhfL | Small lipoprotein | −3.22 | |
| Domain: EAL | bluF | Anti-repressor for YcgE, blue light-responsive, FAD-binding, inactive c-di-GMP phosphodiesterase-like EAL domain protein | −2.55 |
| yhjH | Cyclic-di-GMP phosphodiesterase, FlhDC-regulated | −2.13 | |
| ycgG | Putative membrane-anchored cyclic-di-GMP phosphodiesterase | −2.36 | |
| yliE | Putative membrane-anchored cyclic-di-GMP phosphodiesterase | −2.04 | |
| Aminoacyl-tRNA biosynthesis | alaW | tRNA-Ala | −2.02 |
| alaX | tRNA-Ala | −2.30 | |
| argX | tRNA-Arg | −2.19 | |
| asnV | tRNA-Asn | −2.76 | |
| asnW | tRNA-Asn | −2.12 | |
| glnV | tRNA-Gln | −2.18 | |
| glnX | tRNA-Gln | −2.35 | |
| hisR | tRNA-His | −2.03 | |
| leuP | tRNA-Leu | −2.37 | |
| leuT | tRNA-Leu | −3.34 | |
| leuU | tRNA-Leu | −2.43 | |
| pheV | tRNA-Phe | −3.56 | |
| proK | tRNA-Pro | −2.24 | |
| proL | tRNA-Pro | −2.94 | |
| proM | tRNA-Pro | −2.76 | |
| selC | tRNA-Sec | −2.07 | |
| valV | tRNA-Val | −3.52 | |
| Aminoacid metabolism | astA | Arginine succinyltransferase | −2.17 |
| astB | Succinylarginine dihydrolase | −2.09 | |
| astC | Succinylornithine transaminase, PLP-dependent | −2.13 | |
| astE | Succinylglutamate desuccinylase | −2.14 | |
| feaR | Transcriptional activator for tynA and feaB | −2.02 | |
| tnaC | Tryptophanase leader peptide | −2.21 | |
| Fatty acid oxidation | fadA | 3-ketoacyl-CoA thiolase (thiolase I) | −2.20 |
| fadB | Fatty acid oxidation complex, α component | −2.19 | |
| fadH | 2,4-dienoyl-CoA reductase, NADH and FMN-linked | −2.14 | |
| prpB | 2-methylisocitrate lyase | −2.07 | |
| Upregulated | |||
| Prophage | xisD | Pseudogene, exisionase in defective prophage DLP12 | 7.252 |
| ylcI | DUF3950 family protein, DLP12 prophage | 5.318 | |
| Ribosome | rrsC | 16S ribosomal RNA | 3.93 |
| rrfB | 5S ribosomal RNA | 3.32 | |
| rrfC | 2.18 | ||
| rrfD | 5.08 | ||
| rrfG | 3.55 | ||
| rrfH | 3.38 | ||
| Pilus | fimC | Periplasmic chaperone | 2.42 |
| fimF | Minor component of type 1 fimbriae | 2.05 | |
| ppdD | Putative prepilin peptidase-dependent pilin | 4.35 | |
| ydeR | Putative fimbrial-like adhesin protein | 2.18 |
To confirm that indeed the reduced transcription of ompR-envZ was the major cause for the acquired ammonia resistance, E. coli mutant ARM9 was genetically complemented by the introduction of constructs from the ASKA collection [39] expressing either ompR or envZ. Introduction of constructs expressing either ompR or envZ restored ammonia sensitivity, while transformants harboring the empty plasmid continued to be resistant (Fig. 6c). This strongly suggests that the reduced expression of ompR and envZ was the sole cause of the acquired ammonia resistance. It is important to note that mutant ARM9 had also become resistant to AMVCs produced by S. venezuelae and by S. griseus (the latter grown on glycine), again providing evidence that all strains act by producing ammonia as the AMVC (Fig. S4B).
Taken together, these data show that E. coli responds to exposure to ammonia by reducing ompR-envZ transcription, down regulating the expression of OMPs to minimize the passage of small molecules, and by the reduction of ammonia biosynthesis. Both responses are aimed at defense against the accumulation of toxic levels of ammonia. When exposed to ammonia, E. coli ompR mutants were shown to be significantly more sensitive to tetracycline than the parental strain [26]. Our results show that reducing the expression of OMPs is a defense mechanism against ammonia toxicity extending also earlier observations that ompF mutants show impaired response to VOCs that affect the motility of E. coli [5].
Ammonia released by Streptomyces modifies sensitivity to canonical antibiotics
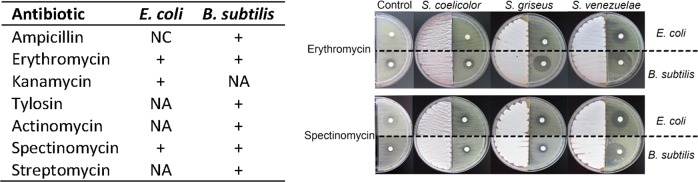
Since ammonia is an AMVC that can reach far from the colony, we hypothesized that the molecule may play a role in long-distance competition with other microbes in the soil, e.g., by modifying the effect of other antibiotics produced by actinomycetes, such as erythromycin, kanamycin, actinomycin, spectinomycin, and streptomycin. This could be an interesting synergistic effect whereby weapons produced by the strain itself are potentiated via ammonia. To have an indication, the streptomycetes were grown on the left side of the agar plates for 4 days to allow accumulation of compounds on the receiver side containing LB. After that, B. subtilis and E. coli BREL606 (more resistant to AMVCs) were plated next to Streptomyces strains and a filter disk placed on the agar containing different canonical antibiotics. Interestingly, we noticed a significant increase in the sensitivity of B. subtilis and E. coli to most antibiotics when ammonia-producing streptomycetes were grown adjacent to the receiver cells (Fig. 7). Thus, bioactive VCs from Streptomyces modulate the activity of soluble antibiotics at longer distances, thereby allowing them to exploit antibiotics produced by other bacteria, or to enhance the activity of its own antibiotics. This novel concept should be worked out further to consider antibiotic-related susceptibility/resistance but also tested in ecological settings in situ, such as competition experiments in controlled microcosms with microbial consortia.
Fig. 7.
Cooperativity between VCs and soluble antibiotics produced by streptomycetes. Left: changes in antibiotic sensitivity caused by the presence of Streptomyces VCs. values indicated increase (+) in halo size. NC, no changes; NA, not active. Right: representative images showing changes in halo size. Streptomycetes were grown on SFM agar, allowed to growth for 4 days, prior to streaking of the indicator strains. Filter disks containing the antibiotic were placed immediately after plating the indicator strains
Implications on ecology and antibiotic activity
Streptomyces VCs have a perceivable impact on the pH of their surroundings. Research has shown that richness and diversity of bacterial soil microbiomes is largely explained by the soil pH [57] with acidic soils having the lowest diversity. Basic environments favor bacterial growth while acidic environments do it for fungi [58, 59]. Studies have shown that VCs are more strongly adsorbed in alkaline soils, especially those containing high organic carbon content (2.9%) [60]. High concentrations of ammonia are achieved in densely populated environments, like the human intestine where ammonia concentrations range from 12 to 30 mM [61]. Besides, its volatile characteristic allows ammonia to travel far from the producer, enabling it to mediate long-distance interactions. To the best of our knowledge, this is the first report showing that abundant soil bacteria such as Streptomyces actively kill other bacteria through the air, via the production of ammonia. Additional ecological impact is provided by work of others that have shown that ammonia is a trait that correlates positively and significantly to some plant characteristics such as elongation of roots and shoots [62]. For some plants like A. thaliana, however, exposure to biogenic ammonia produced by Serratia odorifera reduced its growth [63]. With the knowledge we obtained in our controlled experiments, we now have generated tools (e.g., mutants) to begin to address the ecological importance of ammonia and other volatiles produced by streptomycetes in situ. In this context, our results suggest that the release of ammonia could help the solubility and diffusion of other types of secondary metabolites, while sensitizing competing bacteria. Therefore, the production of a small low-cost ammonia is also a logical strategy for bacteria to enhance the activity of more complex and costly antibiotics, such as polyketides, non-ribosomal peptides or β-lactams. After all, synthesis of these compounds requires expensive high-energy precursors like ATP, NADPH and acyl-CoAs. This is applicable both to antibiotics produced by the organism itself, and to those produced by bacteria further away from the colony. The validity of this concept of “antibiotic piracy” requires further experimental testing.
In conclusion, our work shows that several streptomycetes can use ammonia as a low-cost airborne weapon to change their surrounding environment, thereby making their own, more costly defense mechanisms more effective. In this microbial warfare, the surrounding bacteria then respond by reducing the permeability of their outer membrane and by switching off ammonia production. The field of AMVCs should continue to be studied as it may offer new opportunities for agricultural and/or medical applications, such as for crop protection and natural product discovery.
Supplementary information
Acknowledgements
This work was supported by Grant No. 313599 from The Mexican National Council of Science and Technology (CONACYT) to MA, by VIDI grant 864.11.015 from the Netherlands Organization for Scientific Research (NWO) to PG and by grant 14221 from the Netherlands Organization for Scientific Research (NWO) to GPvW. We thank Hans Zweer for technical help with GC/Q-TOF analysis and Lisanne Storm for the help with the volatile antimicrobial screening, Yasuo Ohnishi and Le Zhang for sharing the glycine cleavage system mutants from S. griseus and S. coelicolor respectively, and Stephen Douthwaite for providing E. coli AS19-RlmA−.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-019-0537-2) contains supplementary material, which is available to authorized users.
References
- 1.Schmidt R, Cordovez V, de Boer W, Raaijmakers J, Garbeva P. Volatile affairs in microbial interactions. ISME J. 2015;9:2329–35. doi: 10.1038/ismej.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schulz S, Dickschat JS. Bacterial volatiles: the smell of small organisms. Nat Prod Rep. 2007;24:814–42. doi: 10.1039/b507392h. [DOI] [PubMed] [Google Scholar]
- 3.Audrain B, Farag MA, Ryu CM, Ghigo JM. Role of bacterial volatile compounds in bacterial biology. FEMS Microbiol Rev. 2015;39:222–33. doi: 10.1093/femsre/fuu013. [DOI] [PubMed] [Google Scholar]
- 4.Kai M, Haustein M, Molina F, Petri A, Scholz B, Piechulla B. Bacterial volatiles and their action potential. Appl Microbiol Biotechnol. 2009;81:1001–12. doi: 10.1007/s00253-008-1760-3. [DOI] [PubMed] [Google Scholar]
- 5.Kim KS, Lee S, Ryu CM. Interspecific bacterial sensing through airborne signals modulates locomotion and drug resistance. Nat Commun. 2013;4:1809. doi: 10.1038/ncomms2789. [DOI] [PubMed] [Google Scholar]
- 6.Nijland R, Burgess JG. Bacterial olfaction. Biotechnol J. 2010;5:974–7. doi: 10.1002/biot.201000174. [DOI] [PubMed] [Google Scholar]
- 7.Que YA, Hazan R, Strobel B, Maura D, He J, Kesarwani M, et al. A quorum sensing small volatile molecule promotes antibiotic tolerance in bacteria. PLoS One. 2013;8:e80140. doi: 10.1371/journal.pone.0080140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulz-Bohm K, Martín-Sánchez L, Garbeva P. Microbial volatiles: small molecules with an important role in intra- and inter-kingdom interactions. Front Microbiol. 2017;8:2484. doi: 10.3389/fmicb.2017.02484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordovez V, Carrion VJ, Etalo DW, Mumm R, Zhu H, van Wezel GP, et al. Diversity and functions of volatile organic compounds produced by Streptomyces from a disease-suppressive soil. Front Microbiol. 2015;6:1081. doi: 10.3389/fmicb.2015.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barka EA, Vatsa P, Sanchez L, Gaveau-Vaillant N, Jacquard C, Klenk HP, et al. Taxonomy, physiology, and natural products of actinobacteria. Microbiol Mol Biol Rev. 2016;80:1–43. doi: 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopwood DA. Streptomyces in nature and medicine: the antibiotic makers. New York: Oxford University Press; 2007. [Google Scholar]
- 12.Hopwood DA. Streptomyces in nature and medicine. New York, NY: The Antibiotic Makers. Oxford University Press Inc; 2007. [Google Scholar]
- 13.Berdy J. Thoughts and facts about antibiotics: where we are now and where we are heading. J Antibiot. 2012;65:385–95. doi: 10.1038/ja.2012.27. [DOI] [PubMed] [Google Scholar]
- 14.Citron CA, Barra L, Wink J, Dickschat JS. Volatiles from nineteen recently genome sequenced actinomycetes. Org Biomol Chem. 2015;13:2673–83. doi: 10.1039/c4ob02609h. [DOI] [PubMed] [Google Scholar]
- 15.Schöller CEG, Gürtler H, Pedersen R, Molin S, Wilkins K. Volatile metabolites from actinomycetes. J Agric Food Chem. 2002;50:2615–21. doi: 10.1021/jf0116754. [DOI] [PubMed] [Google Scholar]
- 16.Wang C, Wang Z, Qiao X, Li Z, Li F, Chen M, et al. Antifungal activity of volatile organic compounds from Streptomyces alboflavus TD-1. FEMS Microbiol Lett. 2013;341:45–51. doi: 10.1111/1574-6968.12088. [DOI] [PubMed] [Google Scholar]
- 17.Gürtler H, Pedersen R, Anthoni U, Christophersen C, Nielsen PH, Wellington EM, et al. Albaflavenone, a sesquiterpene ketone with a zizaene skeleton produced by a streptomycete with a new rope morphology. J Antibiot. 1994;47:434–9. doi: 10.7164/antibiotics.47.434. [DOI] [PubMed] [Google Scholar]
- 18.Davies J. Are antibiotics naturally antibiotics? J Ind Microbiol Biotechnol. 2006;33:496–9. doi: 10.1007/s10295-006-0112-5. [DOI] [PubMed] [Google Scholar]
- 19.Abrudan MI, Smakman F, Grimbergen AJ, Westhoff S, Miller EL, van Wezel GP, et al. Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc Natl Acad Sci USA. 2015;112:11054–9. doi: 10.1073/pnas.1504076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avalos M, van Wezel GP, Raaijmakers JM, Garbeva P. Healthy scents: microbial volatiles as new frontier in antibiotic research? Curr Opin Microbiol. 2018;45:84–91. doi: 10.1016/j.mib.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 21.Shatalin K, Shatalina E, Mironov A, Nudler E. H2S: a universal defense against antibiotics in bacteria. Science. 2011;334:986–90. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- 22.Gusarov I, Nudler E. NO-mediated cytoprotection: instant adaptation to oxidative stress in bacteria. Proc Natl Acad Sci USA. 2005;102:13855–60. doi: 10.1073/pnas.0504307102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shatalin K, Gusarov I, Avetissova E, Shatalina Y, McQuade LE, Lippard SJ, et al. Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proc Natl Acad Sci USA. 2008;105:1009–13. doi: 10.1073/pnas.0710950105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gusarov I, Shatalin K, Starodubtseva M, Nudler E. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science. 2009;325:1380–4. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Sorge NM, Beasley FC, Gusarov I, Gonzalez DJ, von Kockritz-Blickwede M, Anik S, et al. Methicillin-resistant Staphylococcus aureus bacterial nitric-oxide synthase affects antibiotic sensitivity and skin abscess development. J Biol Chem. 2013;288:6417–26. doi: 10.1074/jbc.M112.448738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernier SP, Letoffe S, Delepierre M, Ghigo JM. Biogenic ammonia modifies antibiotic resistance at a distance in physically separated bacteria. Mol Microbiol. 2011;81:705–16. doi: 10.1111/j.1365-2958.2011.07724.x. [DOI] [PubMed] [Google Scholar]
- 27.Fadli M, Chevalier J, Hassani L, Mezrioui NE, Pages JM. Natural extracts stimulate membrane-associated mechanisms of resistance in Gram-negative bacteria. Lett Appl Microbiol. 2014;58:472–7. doi: 10.1111/lam.12216. [DOI] [PubMed] [Google Scholar]
- 28.Yung PY, Grasso LL, Mohidin AF, Acerbi E, Hinks J, Seviour T, et al. Global transcriptomic responses of Escherichia coli K-12 to volatile organic compounds. Sci Rep. 2016;6:19899. doi: 10.1038/srep19899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu M, Douthwaite S. Activity of the ketolide telithromycin is refractory to Erm monomethylation of bacterial rRNA. Antimicrob Agents Chemother. 2002;46:1629–33. doi: 10.1128/AAC.46.6.1629-1633.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barbe V, Cruveiller S, Kunst F, Lenoble P, Meurice G, Sekowska A, et al. From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology. 2009;155:1758–75. doi: 10.1099/mic.0.027839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garbeva P, Hordijk C, Gerards S, de Boer W. Volatile-mediated interactions between phylogenetically different soil bacteria. Front Microbiol. 2014;5:289. doi: 10.3389/fmicb.2014.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010;11:11. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell CD, Chapman SJ, Cameron CM, Davidson MS, Potts JM. A rapid microtiter plate method to measure carbon dioxide evolved from carbon substrate amendments so as to determine the physiological profiles of soil microbial communities by using whole soil. Appl Environ Microbiol. 2003;69:3593–9. doi: 10.1128/AEM.69.6.3593-3599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tezuka T, Ohnishi Y. Two glycine riboswitches activate the glycine cleavage system essential for glycine detoxification in Streptomyces griseus. J Bacteriol. 2014;196:1369–76. doi: 10.1128/JB.01480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castric KF, Castric PA. Method for rapid detection of cyanogenic bacteria. Appl Environ Microbiol. 1983;45:701–2. doi: 10.1128/aem.45.2.701-702.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avalos M, Boetzer M, Pirovano W, Arenas NE, Douthwaite S, van Wezel GP. Complete genome sequence of Escherichia coli AS19, an antibiotic-sensitive variant of E. coli strain B REL606. Genome Announc. 2018;6:e00385–00318. doi: 10.1128/genomeA.00385-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinforma. 2012;13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–9. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 40.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 41.Zhu H, Swierstra J, Wu C, Girard G, Choi YH, van Wamel W, et al. Eliciting antibiotics active against the ESKAPE pathogens in a collection of actinomycetes isolated from mountain soils. Microbiology. 2014;160:1714–25. doi: 10.1099/mic.0.078295-0. [DOI] [PubMed] [Google Scholar]
- 42.Mohan C. Buffers. A guide for the preparation and use of buffers in biological systems, 2006.
- 43.Gubbens J, Zhu H, Girard G, Song L, Florea BI, Aston P, et al. Natural product proteomining, a quantitative proteomics platform, allows rapid discovery of biosynthetic gene clusters for different classes of natural products. Chem Biol. 2014;21:707–18. doi: 10.1016/j.chembiol.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 44.Wu C, Kim HK, van Wezel GP, Choi YH. Metabolomics in the natural products field—a gateway to novel antibiotics. Drug Discov Today Technol. 2015;13:11–7. doi: 10.1016/j.ddtec.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 45.Jones SE, Ho L, Rees CA, Hill JE, Nodwell JR, Elliot MA. Streptomyces exploration is triggered by fungal interactions and volatile signals. Elife. 2017;6:e21738. doi: 10.7554/eLife.21738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Letoffe S, Audrain B, Bernier SP, Delepierre M, Ghigo JM. Aerial exposure to the bacterial volatile compound trimethylamine modifies antibiotic resistance of physically separated bacteria by raising culture medium pH. MBio. 2014;5:e00944–00913. doi: 10.1128/mBio.00944-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Čepl JJPI, Blahůšková A, Cvrčková F, Markoš A. Patterning of mutually interacting bacterial bodies: close contacts and airborne signals.pdf. BMC Microbiol. 2010;10:139. doi: 10.1186/1471-2180-10-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anwar S, Ali B, Sajid I. Screening of rhizospheric actinomycetes for various in-vitro and in-vivo plant growth promoting (PGP) traits and for agroactive compounds. Front Microbiol. 2016;7:1334. doi: 10.3389/fmicb.2016.01334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kikuchi G, Motokawa Y, Yoshida T, Hiraga K. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad Ser B. 2008;84:246–63. doi: 10.2183/pjab/84.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L. Identification and characterization of developmental genes in Streptomyces (PhD thesis) Leiden: Leiden University; 2015. [Google Scholar]
- 51.Bobille H, Limami AM, Robins RJ, Cukier C, Le Floch G, Fustec J. Evolution of the amino acid fingerprint in the unsterilized rhizosphere of a legume in relation to plant maturity. Soil Biol Biochem. 2016;101:226–36. [Google Scholar]
- 52.Zhalnina K, Louie KB, Hao Z, Mansoori N, da Rocha UN, Shi S, et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol. 2018;3:470–80. doi: 10.1038/s41564-018-0129-3. [DOI] [PubMed] [Google Scholar]
- 53.Conroy MJ, Durand A, Lupo D, Li X-D, Bullough PA, Winkler FK, et al. The crystal structure of the Escherichia coli AmtB–GlnK complex reveals how GlnK regulates the ammonia channel. Proc Natl Acad Sci USA. 2007;104:1213. doi: 10.1073/pnas.0610348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wirén Nicolaus von, Merrick Mike. Molecular Mechanisms Controlling Transmembrane Transport. Berlin, Heidelberg: Springer Berlin Heidelberg; 2004. Regulation and function of ammonium carriers in bacteria, fungi, and plants; pp. 95–120. [Google Scholar]
- 55.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fernandez L, Hancock RE. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev. 2012;25:661–81. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–31. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rousk J, Brookes PC, Bååth E. Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization. Appl Environ Microbiol. 2009;75:1589–96. doi: 10.1128/AEM.02775-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bárcenas-Moreno G, Rousk J, Bååth E. Fungal and bacterial recolonisation of acid and alkaline forest soils following artificial heat treatments. Soil Biol Biochem. 2011;43:1023–33. [Google Scholar]
- 60.Serrano A, Gallego M. Sorption study of 25 volatile organic compounds in several Mediterranean soils using headspace–gas chromatography–mass spectrometry. J Chromatogr A. 2006;1118:261–70. doi: 10.1016/j.chroma.2006.03.095. [DOI] [PubMed] [Google Scholar]
- 61.Hughes R, Magee EA, Bingham S. Protein degradation in the large intestine: relevance to colorectal cancer. Curr Issues Intest Microbiol. 2000;1:51–8. [PubMed] [Google Scholar]
- 62.Marques APGC, Pires C, Moreira H, Rangel AOSS, Castro PML. Assessment of the plant growth promotion abilities of six bacterial isolates using Zea mays as indicator plant. Soil Biol Biochem. 2010;42:1229–35. [Google Scholar]
- 63.Weise T, Kai M, Piechulla B. Bacterial ammonia causes significant plant growth inhibition. PLoS One. 2013;8:e63538. doi: 10.1371/journal.pone.0063538. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.