Abstract
DNA methylation is an important epigenetic mechanism involved in many biological processes, i.e. gametogenesis and embryonic development. However, increased copy numbers of DNA methylation related genes (dnmt, tet and tdg) have been found during chordate evolution due to successive whole genome duplication (WGD) events. Their evolutionary history and phylogenetic relationships remain unclear. The present study is the first to clarify the evolutionary history of DNA methylation genes in chordates. In particular, our results highlight the fixation of several dnmt3-related genes following successive WGD throughout evolution. The rainbow trout genome offered a unique opportunity to study the early evolutionary fates of duplicated genes due to a recent round of WGD at the radiation of salmonids. Differences highlighted in transcriptional patterns of these genes during gametogenesis and ontogenesis in trout indicated that they might be subjected to sub- or neo-functionalisation after WDG. The fixation of multiple dnmt3 genes in genomes after WGD could contribute to the diversification and plastic adaptation of the teleost.
Subject terms: Phylogeny, Phylogeny, Molecular evolution, Molecular evolution, Gene expression
Introduction
DNA methylation is an important epigenetic mechanism involving the covalent binding of a methyl group to the 5th carbon position of cytosine in CpG dinucleotides in vertebrates1. This mechanism is generally considered as a repressive epigenetic mark that inhibits gene expression2. DNA methylation plays a critical role in several biological processes such as embryonic development and gametogenesis1,3,4. The DNA methylation process is mediated by DNA methyltransferases (Dnmts): maintenance methyltransferase Dnmt15 and de novo methyltransferase Dnmt36. The erasure of methylation marks can be achieved either passively through the inhibition of Dnmt1 during DNA replication and cell division7, or actively through the action of ten-eleven translocation (Tet) dioxygenase family mediated iterative oxidation of 5-methylcytosine (5mC) and thymine DNA glycosylase (Tdg)-dependent base excision repair (BER)8–10.
The DNA methylation/demethylation machinery has been well described in mammals. While dnmt1 is generally identified as single-copy gene during evolution11, multiple dnmt3 genes were found in vertebrates with different gains and losses among tetrapod lineages12. The mammalian dnmt3 family consists of four members: dnmt3a, dnmt3b, dnmt3c and dnmt3l13,14. dnmt3l serves as a catalytically inactive cofactor for de novo methylation, and is found to exist only in eutherian mammals and in some marsupials15, whilst dnmt3c, previously annotated as a pseudogene, was recently identified in rodent genomes14. In contrast, the discovery of active demethylation-related genes occurred fairly late, with three tet paralogs (tet1, tet2 and tet3) and a single tdg gene found in mammalian genomes9,10,16. The identification of well-conserved DNA methylation genes in vertebrates, including teleosts, suggests that these regulatory pathways may be conserved across vertebrates17,18. However, due to the additional round of whole genome duplication (WGD) event that occurred before the radiation of the teleost lineage [TGD, teleost-specific genome duplication, 320 Mya (million years ago)], an increase in the number of copies of these genes has been found in teleost species19. For instance, the de novo methyltransferase dnmt3 was shown to be more divergent in teleosts compared with mammals: despite the absence of dnmt3l in zebrafish (Danio rerio) genome, up to 6 dnmt genes were identified as orthologous to mammalian dnmt3a and dnmt3b genes20–22. Similarly, 4 to 9 different dnmt3 paralogs were reported to exist in the genome of other teleost species23–28.
It is generally accepted that a WGD event can provide additional genetic material for selection, and are thus associated with phenotypic diversity and evolutionary innovations29. Alternatively, duplicated genes can also originate from small scale duplications (SSD) which can produce different kinds of adaptations compared to WGD30. Following WGD or SSD events, duplicated genes can either be lost or retained with three distinct outcomes: conservation of the ancestral gene functions, sub-functionalisation, or neo-functionalisation31. Through these processes, the fixation of extra copies of DNA methylation genes in teleost genomes may contribute to the diversification and plastic adaptation of teleosts. However, to characterise these adaptation, it is essential to first understand the evolutionary origin of these additional copies.
The increasing availability of sequenced genomes of teleost species facilitates to establish a comprehensive comparative study of DNA methylation genes among different taxa. However, few studies have been done to clarify the evolutionary history of dnmt, tet and tdg genes in vertebrates22–24,32. Hence, the evolutionary history and the orthologous relationship of DNA methylation genes remain incomplete and unclear, especially when considering genomes with higher complexity, i.e. salmonid species, which experienced a fourth round of WGD (Salmonid specific genome duplication, SaGD, 100 Mya).
The present study aimed to refine the current knowledge concerning the evolutionary history of dnmt genes in vertebrates, and to update the existing story with all DNA methylation genes (dnmt, tet and tdg) for extended taxa within the chordate phylum. To conduct the present study, we selected representative species with sequenced genomes of different taxa from a WGD point of view. Rainbow trout (Oncorhynchus mykiss), a salmonid fish, was included as a model species, which is supposed to have the maximum copies of DNA methylation genes among chordates due to SaGD. To explore if WGD events lead to sub- or neo-functionalisation of DNA methylation genes, estimations of the expression patterns of DNA methylation genes were done during gametogenesis and early development in trout.
Results
Evolutionary history of dnmt genes in chordates
Dnmt1
Through analysing the genomes of representative species in Ensembl (Release 91, December 2017) and in NCBI, we found that the dnmt1 gene was generally identified as a single copy in the chordate phylum (Table 1), except that 2 dnmt1 genes were identified in trout and Atlantic salmon (Salmo salar). Phylogenetic analyses showed that both dnmt1 genes identified in trout and salmon grouped together with other vertebrate dnmt1 orthologs, whereas the dnmt1 genes of amphioxus (Branchiostoma. floridae), ciona (Ciona intestinalis) and lamprey (Petromyzon marinus) rooted as outgroups in the tree (Supplemental Fig. S1A, dnmt1 protein aliment in Supplemental Fig. S2). The subsequent syntenic analysis showed that the 2 dnmt1 genes in trout were included in the same syntenic group conserved among vertebrates (angptl6-eif3g-dnmt1-s1pr2-cdc37-tyk2-raver1, Supplemental Fig. S1B). We thus annotated them as dnmt1a and dnmt1b according to ZFIN Nomenclature. Further analysis of conserved domain search showed that both the two dnmt1 encoding protein sequences in trout have all the conserved domain features as their mammalian DNMT1 orthologs (DMAP1-binding domain, DNMT1-RFD domain, zf-CXXC domain, BAH domain, and Dcm domain). They also share identical conserved motifs in C-terminal catalytic domain (Supplemental Fig. S2).
Table 1.
Copy numbers of DNA methylation genes in target species in chordates.
| dnmt1 | dnmt3 | tet | tdg | |
|---|---|---|---|---|
| Amphioxus | 1 | 1 | 1 | 1 |
| Ciona | 1 | 2 (?) | 1 | 1 |
| Lamprey | 1 | 3 (A/A/A) | 3 (1/2/3?) | 1 |
| Shark | 1 | 2 (A/B) | 3 (1/2/3) | 1 |
| Lizard | 1 | 2 (A/B) | 3 (1/2/3) | 1 |
| Chicken | 1 | 2 (A/B) | 3 (1/2/3) | 1 |
| Mouse | 1 | 3 (A/B/C) | 3 (1/2/3) | 1 |
| Spotted gar | 1 | 3 (a/ba/bb) | 3 (1/2/3) | 1 |
| Zebrafish | 1 | 6 (aa/ab/ba/bba/bbb1/bbb2) | 3 (1/2/3) | 2 (a/b) |
| Medaka | 1 | 4 (aa/ba/bba/bbb) | 3 (1/2/3) | 2 (a/b) |
| Stickleback | 1 | 5 (aa/ab/ba/bba/bbb) | 3 (1/2/3) | 2 (a/b) |
| Fugu | 1 | 5 (aa/ab/ba/bba/bbb) | 3 (1/2/3) | 2 (a/b) |
| Tetraodon | 1 | 5 (aa/ab/ba/bba/bbb) | 3 (1/2/3) | 2 (a/b) |
| Trout | 2 | 8 (aa/ab1/ab2/ba1/ba2/bba1/bba2/bbb) | 7 (1a/1b/2a/2b/2c/3a/3b) | 4 (aa/ab/ba/bb) |
Dnmt3
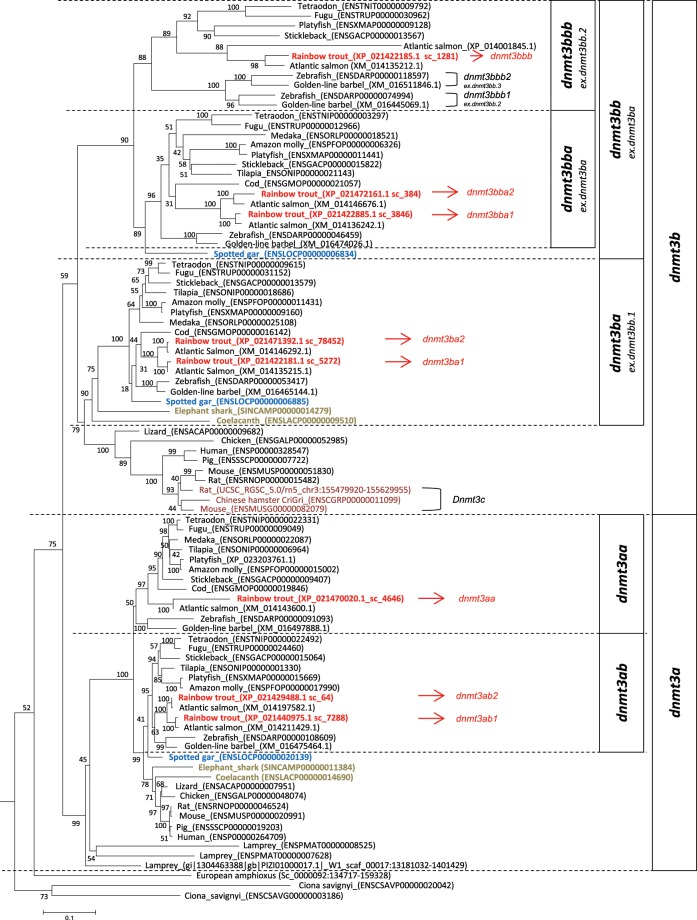
We first evaluated the evolutionary history of dnmt3 in non-vertebrate chordate species. Through a BLAST search against the amphioxus genome, we found that both B. floridae and B. lanceolatum had only one dnmt3 gene located on BRAFLscaffold_185, and sc_0000092, respectively. In ciona (C. savignyi), two putative dnmt3 homologs were identified (Table 1), with 45.5% of their identity shared in their protein sequences. Further analysis of conserved domain search showed that the dnmt3-related protein sequence in B. lanceolatum and one of the dnmt3 in C. savignyi (ENSCSAVP00000020042) have all the domain features specific for dnmt3: PWWP domain, ADD domain and Dcm domain (Supplemental Fig. S3). But the other dnmt3 in C. savignyi (ENSCSAVP00000020042) has only the Dcm domain in its protein sequence. The conserved dnmt3 amino acid sequences (sequences alignment including PWWP, ADD and Dcm domain) of representative species in chordates were used to perform phylogenetic analysis (alignment in Supplemental Fig. S3), results showed that amphioxus and the ciona dnmt3 rooted as outgroup in the tree, whereas vertebrate dnmt3a and dnmt3b related sequences formed two separate groups (Fig. 1). Percentage identity matrix was calculated between this two dnmt3 related sequences in ciona and tetrapods, which showed that ciona sequences seemed no more related to dnmt3a than to dnmt3b (Supplemental Table S1). In order to clarify the specific timing for when dnmt3a and dnmt3b genes arose, we did a dnmt3 syntenic analysis in chordates using Genomicus software v01.01 (www.genomicus.biologie.ens.fr, Supplemental Fig. S4). We showed that the synteny around dnmt3 genes in amphioxus and ciona was not well conserved except for col21a1 and akap7, which were found on reftig_1 and reftig_44 in ciona, respectively. In vertebrates, dnmt3a and dnmt3b genes were included in two distinct syntenic groups: ak7-abhd1-col21a1-akap7-epb41l2-sl3a1-zbtb24 and pcsk2-dusp15-pofut1-b4galt5-zbtb46, respectively.
Figure 1.
Phylogeny of dnmt3 in chordates. The phylogenetic tree was built by the Neighbor-Joining (NJ) method. The reliability of the inferred trees was estimated by the bootstrap method with 1,000 replications. All accession numbers are specified in parentheses.
We then investigate the evolutionary history of dnmt3 in vertebrates. In jawless vertebrate, lamprey, two dnmt3 genes annotated as dnmt3a (ENSPMAG00000007710 and ENSPMAG00000006895) were identified in the Ensembl database. However, we identified a 3rd sequence in the lamprey genome assembly in NCBI (NCBI, GCA_002833325.1). We identified this sequence (W1 scaf_00017) as a novel dnmt3 in lampreys. This new dnmt3 gene and ENSPMAG00000007710 in lamprey have all the conserved domain features for dnmt3, whereas the ENSPMAG00000006895 has no PWWP domain (Supplemental Fig. S3). Percentage identity matrix (Supplemental Table S1) and phylogenetic analysis (Fig. 1) confirmed that all these three dnmt3 related genes in lamprey shared higher conservation with dnmt3a gnathostoma genes than with dnmt3b. Syntenic analysis results also showed a partial syntenic conservation of the first group cited above around dnmt3 genes found in lampreys, confirming that these genes were related to the dnmt3a gene in other vertebrates (Supplemental Fig. S4). We also verified this result in SIMRBASE. Noteworthy, there is another gene PMZ_0040501-RA annotated as dnmt3b in this database. However, we found that this latter sequence contains only a FYVE domain, and clustered with the mammalian DNMT3L orthologs (Data not shown).
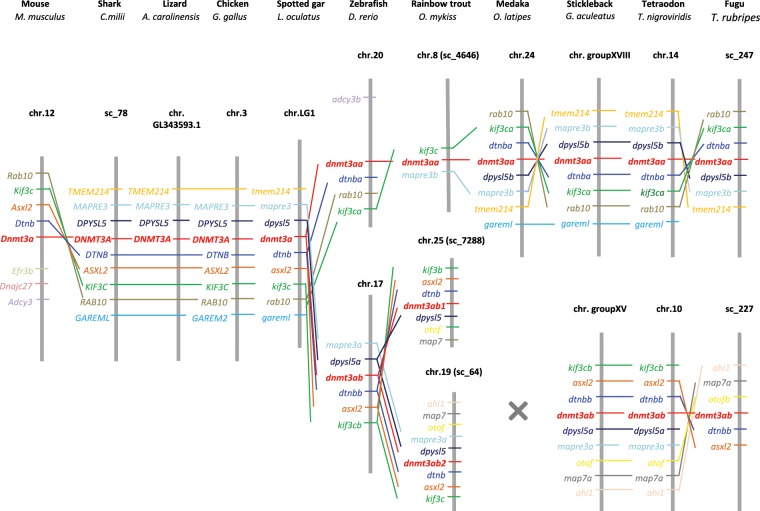
In jawed vertebrates, only one dnmt3a gene was identified in tetrapods, shark, coelacanth and spotted gar, but dnmt3a pseudogenes were identified in humans (ENSG00000224071, chr.2) and mice (ENSMUST00000192011.1, chr.3, Supplemental Fig. S4). By contrast, two dnmt3a genes were found in most of the teleost species (Table 1), which were well grouped as two independent clusters, dnmt3aa and dnmt3ab, in phylogenetic tree (Fig. 1). Both duplicated dnmt3a genes in teleost shared conserved synteny with other vertebrates (Fig. 2). Exceptionally, one dnmt3aa gene and two dnmt3ab genes were found in trout and salmon, whereas dnmt3ab was not found in medaka. Conserved domain analysis showed that no PWWP domain was identified in Dnmt3aa of salmon. Besides, rainbow trout Dnmt3aa lost one of the 6 conserved motifs (I, IV, VI, VIII, IX and X), motif IX, in its catalytic region, and its ADD domain sequence also showed less conserved compared to other DNMT3 related sequences (Supplemental Fig. S3).
Figure 2.
Syntenic analysis of dnmt3a in jawed vertebrates. Data were collected with Genomicus software version 01.01 and NCBI. chr., chromosome; sc., scaffold.
Concerning dnmt3b, only one dnmt3b related gene was found in non-teleost vertebrates except for the spotted gar, whose genome contained 2 dnmt3b related genes in Ensembl annotated as dnmt3ba and dnmt3bb.1. The murine specific Dnmt3c genes were grouped together, and rooted with the Dnmt3b genes of rodents in phylogenetic tree (Fig. 1). In most of the teleost species, there were three dnmt3b genes identified in Ensembl, namely dnmt3ba, dnmt3bb.1 and dnmt3bb.2. Phylogenetic tree showed that the dnmt3bb.1 in spotted gar clustered with the teleost dnmt3bb.1 genes, the elephant shark and the coelacanth dnmt3b orthologs and then rooted together with the tetrapod dnmt3b genes, whereas the gar dnmt3ba rooted together with teleost dnmt3ba and dnmt3bb.2 genes as a separate branch (Fig. 1). We thus proposed to rename dnmt3bb.1 and dnmt3ba in the spotted gar as dnmt3ba and dnmt3bb and the previous dnmt3bb.1, dnmt3ba and dnmt3bb.2 in teleosts as dnmt3ba, dnmt3bba and dnmt3bbb, respectively according to ZFIN Nomenclature guidelines (Table 2). These genes will be referred by their new nomenclature henceforth in this paper.
Table 2.
Proposed nomenclature for dnmt3, tet and tdg genes.
| Species | Name proposed in Ensembl | Name proposed by Simoda et al.(2005) | Name proposed by Campos et al. (2012) | Proposed new nomenclature |
|---|---|---|---|---|
| Spotted gar | dnmt3aa | dnmt3a | ||
| dnmt3ba | dnmt3bb | |||
| dnmt3bb.1 | dnmt3ba | |||
| tdg.1 | tdg | |||
| Zebrafish | dnmt3aa | dnmt8 | dnmt3a2 | dnmt3aa |
| dnmt3ab | dnmt6 | dnmt3a1 | dnmt3ab | |
| dnmt3bb.1 | dnmt4 | dnmt3b1 | dnmt3ba | |
| dnmt3ba | dnmt7 | dnmt3b2 | dnmt3bba | |
| dnmt3bb.2 | dnmt3 | dnmt3b3 | dnmt3bbb1 | |
| dnmt3bb.3 | dnmt5 | dnmt3b4 | dnmt3bbb2 | |
| tdg1 | tdga | |||
| tdg2 | tdgb | |||
| Gene loci in Genomicus | Gene loci in NCBI | Proposed new nomenclature | ||
| Trout | scaffold_644 | chr.12 | dnmt1a | |
| scaffold_1433 | chr.13 | dnmt1b | ||
| scaffold_4646 | chr.8 | dnmt3aa | ||
| scaffold_7288 | chr.25 | dnmt3ab1 | ||
| scaffold_64 | chr.19 | dnmt3ab2 | ||
| scaffold_5272 | chr.16 | dnmt3ba1 | ||
| scaffold_78452 | chr.9 | dnmt3ba2 | ||
| scaffold_3846 | chr.16 | dnmt3bba1 | ||
| scaffold_384 | chr.9 | dnmt3bba2 | ||
| scaffold_1281 | chr.16 | dnmt3bbb | ||
| scaffold_18888 & 2097 | chr.1 | tet1a | ||
| scaffold_189 & 12473 | chr.23 | tet1b | ||
| scaffold_1163 | chr.19: 4604994..4635364 | tet2a | ||
| / | chr.19: 4726729..4738210 | tet2b | ||
| / | chr.10 | tet2c | ||
| scaffold_21498 & 36080 | chr.6 | tet3a | ||
| scaffold_605 | chr.11 | tet3b | ||
| scaffold_2682 | chr.4 | tdgaa | ||
| scaffold_82 | chr.2 | tdgab | ||
| scaffold_2682 | chr.4 | tdgba | ||
| scaffold_82 | chr.2 | tdgbb |
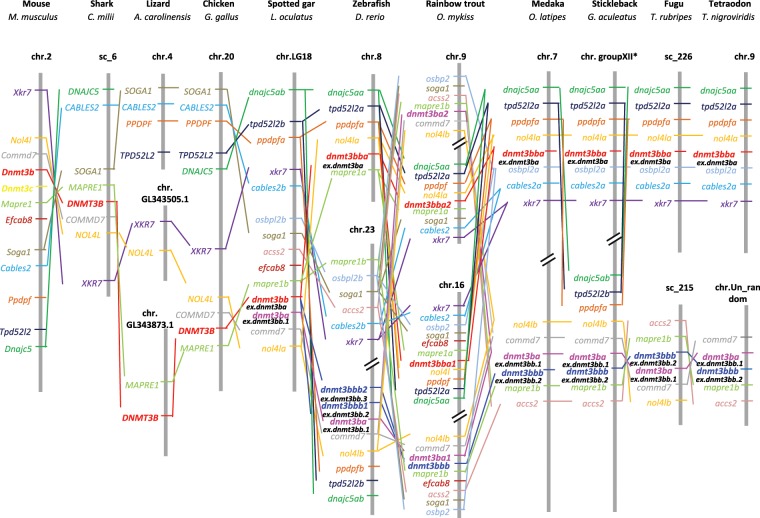
Regarding the dnmt3ba sub-tree, for most of non-salmonid teleost species, only one gene was found grouped together with non-teleost dnmt3b sequences. In salmonids, we identified 2 sequences related to dnmt3ba in our phylogenetic analysis which were located in the conserved and duplicated syntenic group osbp2–soga1-acss2–mapre1b–dnmt3ba-commd –nol4lb (Fig. 3).
Figure 3.
Syntenic analysis of dnmt3b in jawed vertebrates. Data were collected with Genomicus software version 01.01 and NCBI. chr., chromosome; sc., scaffold. *The synteny of stickleback is in reverse direction.
For dnmt3bb sub-tree, the gar dnmt3bb sequence rooted at the basis of the dnmt3bba (ex. dnmt3ba) cluster, whereas dnmt3bbb (ex. dnmt3bb.2) sequences formed a separate cluster (Fig. 1). However, the conserved syntenic group of dnmt3bb in the spotted gar was identified on two distinct chromosomes in teleost species including dnmt3bba and dnmt3bbb. In salmonid species, 3 dnmt3bb paralogs were found in the trout genome, with two of them clustering with dnmt3bba, and the last one clustering with dnmt3bbb sequences (Fig. 1) in the phylogenetic tree. Both dnmt3bba related genes in trout were included in two well-conserved duplicated syntenic groups (dnajc5aa–tpd5212a–ppdpfa-nol4la–dnmt3bba–osbpl2a–cables2a, Fig. 3) across teleosts. dnmt3bbb was also included in the nol4lb–commd7–dnmt3ba–dnmt3bbb–mapre1b syntenic group conserved among teleosts. Compared with trout, salmon were found to have an additional dnmt3bbb paralog (XP_014001845.1) but it is very short in length (150aa). Besides, we found two dnmt3bbb (ex. dnmt3bb.2) paralogs (ex. dnmt3/dnmt3bb.2 and ex. dnmt5/dnmt3bb.3) in zebrafish and another cyprinid fish, golden-line barbel. Further analysis of conserved domain search showed that the protein sequences of Dnmt3 orthologs are highly conserved across chordate phylum. Interestingly, all the Dnmt3bba and Dnmt3bbb related protein sequences have an extra CH domain in their N-terminal (Supplemental Fig. S3).
Evolutionary history of DNA demethylation genes: tet and tdg genes
Tet
We identified one tet related sequence in each genome of amphioxus (BRAFLscaffold_344 in B. floridae; sc_00000095 in B. lanceolatum ill) and ciona (chromosome 12 in C. intestinalis; reftig_54 in C. savignyi) through a BLAST search (Table 1). Our results also showed that, most of the jawed vertebrates have three tet genes, namely tet1, tet2 and tet3 (Table 1) which were included in three distinct syntenic groups conserved among vertebrates (Supplemental Fig. S5). Interestingly, in amphioxus, the only tet gene was included in the wnt8b-scd-tet-rassf-tmem2-cisd2-fam13-cdh-hnrnpd syntenic group. Genes from this syntenic group were also found syntenic with tet1, tet2 and to a lesser extent with tet3 vertebrate genes, suggesting that these 3 genes shared a common ancestor. The phylogenetic analysis showed that tet1/2/3 formed three separate groups in the tree, whereas the amphioxus and ciona tet sequences rooted as an outgroup (Supplemental Fig. S6; protein alignment in Supplemental Fig. S7).
In lamprey, 3 hits were found on chromosomes scaf_00044, scaf_03335 and scaf_00172, respectively, in the Genbank assembly GCA_002833325.1.Further research in SIMRBASE confirmed this result. However, because the tet sequences on scaf_00172 and scaf_03335 were aligned to different region of putative tet gene on scaf_00044, thus it was impossible to include 3 of them together in phylogenetic tree. Our phylogenetic analysis showed that, the putative tet gene in lampreys on scaf_03335 clustered with tet3, whereas the other putative tet gene on scaf_00044 rooted together with ciona and amphioxus as an outgroup in the tree but the bootstrap value for this outgroup was very low (i.e. 18, Supplemental Fig. S6). Interestingly, the conserved syntenic group of both tet1 and tet2 in vertebrates were found on scaf_00044.
In trout, a total of seven tet paralogs were identified, with two orthologs of tet1, three orthologs of tet2 and two orthologs of tet3 (Table 1). tet1 and tet3 most probably duplicated before or around the salmonid radiation following the SaGD, giving rise respectively to tet1a and tet1b, and tet3a and tet3b. In the case of tet2, as previously mentioned, 3 duplicates (annotated as tet2a, tet2b and tet2c, respectively; Table 2) were identified in trout genomes. In trout, tet2a (XM_021573348.1) and tet2b (XM_021573351.1) were located close to one another on chromosome 19 (Supplemental Fig. S5) and shared extremely high identity in the cDNA sequence (96.5%), but tet2b was found to be only half the length of tet2a. The tet2b sequence corresponded to exons 6 to 12 of tet2a. Similarly, tet2c (XM_021617281.1) was found to be closely related to tet2a (89.6% identity), but again, the tet2c sequence was shorter than tet2a and corresponded to exons 1 to 5 of tet2a. There was thus no shared region between tet2b and tet2c, making it impossible to include the three tet2 paralogs of trout together in the phylogenetic analysis (Supplemental Fig. S6). However, the high sequence identity shared by these 3 sequences and the conserved synteny around these 3 loci strongly confirmed that these genes were paralogs. Analysis in salmon also highlighted the 3 tet2 related sequences (XP_014050290.1, XP_014065146.1 and XP_014042431.1) within its genome. Further study of conserved domain search confirmed that that Tet2b and Tet2c share only part of the sequence with Tet2a sequence in trout, with Tet2b lacking the cysteine-rich domain whereas Tet2c lacking the DBSH domain in the C-terminal catalytic domain (Supplemental Fig. S7). Moreover, in most of the studied species, all the conserved motifs in catalytic domain of Tet families (i.e. iron and 2-OG binding sites) can be found (Supplemental Fig. S7), except that the Fe binding, 2-OG binding and 5mC binding sites at the end of C-terminal catalytic domain were not found in amphioxus, two putative tet (located on scaf_00044 and scaf_03335) in lamprey and Tet1a of salmon.
Tdg
Finally, with regards to tdg locus, our results showed that amphioxus, ciona, lamprey, shark, tetrapods and spotted gar had one copy of tdg in their genome (Table 1). In most of teleost species, there were two tdg paralogs (Table 1). Our phylogenetic analysis demonstrated that these two genes were co-orthologous to non-teleost tdg genes (Supplemental Fig. S8A, tdg protein aliment in Supplemental Fig. S9). The syntenic analysis (Supplemental Fig. S8B)showed that these 2 genes were side-by-side on the same chromosome and included in a syntenic group (appl2-nuak1b-rps13-pik3c2a-tdgb-tdga-samm50-api5-nt5dc3-hsp90b1) in teleosts, which is similar to the syntenic block of tdg in non-teleost species (i.e. nuak1, appl2, nfyb, hsp90b1, nt5dc3, and samm50). On the other hand, salmonids retained duplicates of tdga and tdgb genes (Table 1), which were therefore annotated as tdgaa, tdgab, tdgba and tdgbb (Table 2). Phylogenetic analysis showed that the duplicated tdga and tdgb genes in salmon and trout were well clustered with their respective orthologs in other teleosts. The syntenic region around tdga/tdgb in trout was similar to those in other teleost species, with tdgaa/tdgba and tdgab/tdgbb arranged in tandem on chromosome 4 and 2, respectively (Supplemental Fig. S8B). Conserved domain search demonstrated that the functional sites were highly conserved among all the analysed tdg-related sequences (Supplementary Fig. S9).
Expression pattern of DNA methylation genes during gametogenesis and embryogenesis in trout
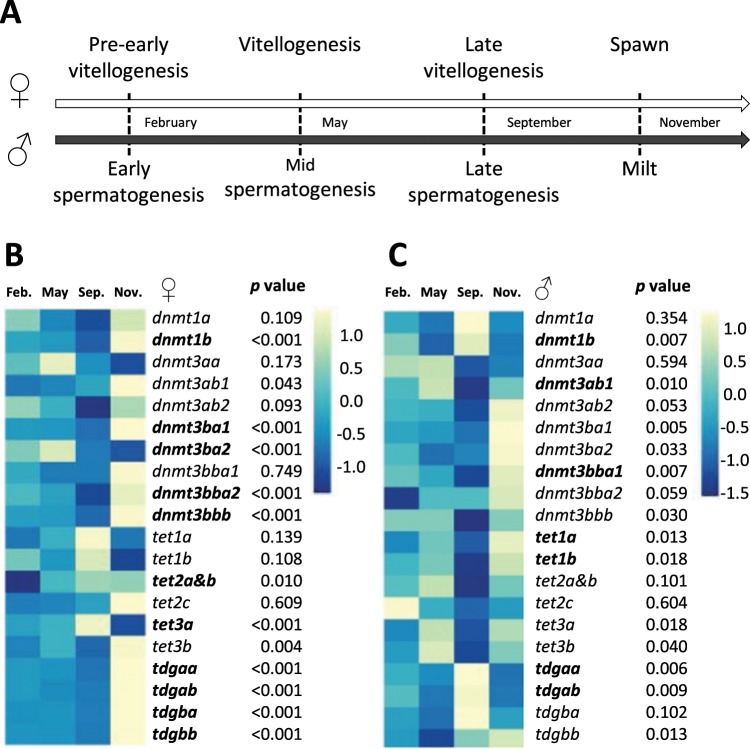
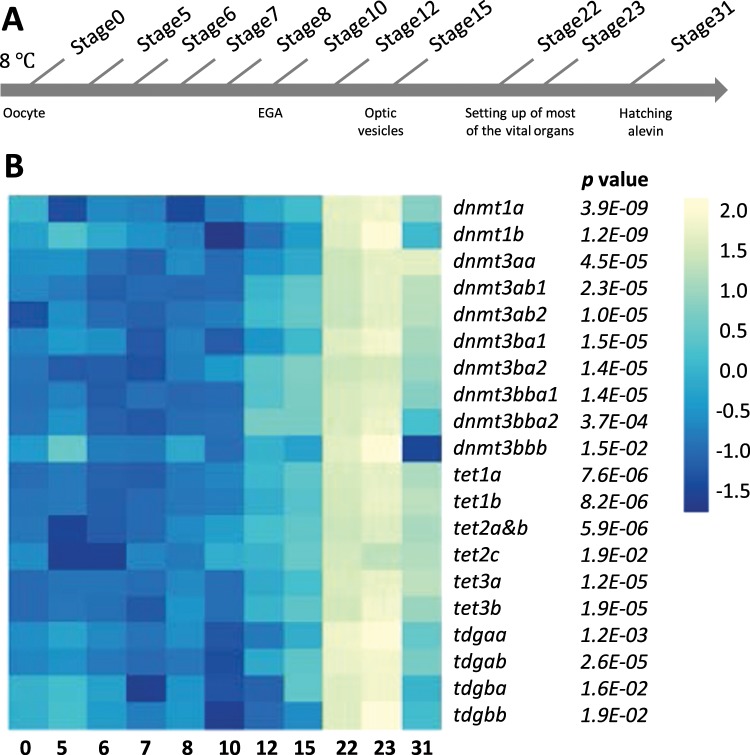
In the Phylofish database33 we found that the high expression of DNA methylation genes in reproduction and development-related tissues seems to have been conserved throughout chordate evolution (data not shown). Therefore, in order to explore the potential different function of duplicated DNA methylation genes after WGD, we investigated their expression patterns during gametogenesis (Fig. 4A) and embryogenesis (Fig. 5A) in trout, which has the maximum DNA methylation genes fixed in its genome.
Figure 4.
Expression patterns of DNA methylation genes during gametogenesis in trout. Experimental design. (A) Heatmaps were used to depict the mRNA levels of DNA methylation genes during oogenesis (B) and spermatogenesis (C) with the mean value of each stage (n = 6). When there is a significantly difference among groups (p < 0.05), the gene’s name is in bold.
Figure 5.
Expression patterns of DNA methylation genes during ontogenesis in trout. (A) Experimental design: Stages numbers referred to the Vernier developmental table 46. (B) The mRNA levels of DNA methylation genes during ontogenesis were shown in heatmap with the mean value of each stage. Data were subjected to log transformation. EGA, embryonic genome activation.
During oogenesis, we observed that dnmt1b, dnmt3ba1, dnmt3bba2, dnmt3bbb exhibited a similar expression pattern (Fig. 4B, mean ± SD data in Supplemental Table S4), which remained steady from February to May (pre-early vitellogenesis and vitellogenesis stages), was halved in September (late vitellogenesis stage), and increased several fold in November (spawn stage). In contrast, the mRNA level of dnmt3ba2 was significantly lower in September and November compared to February and May in trout ovaries (Fig. 4B). No significant difference was observed in the mRNA levels of dnmt1a, all dnmt3a paralogs and dnmt3bba1 during oogenesis (Fig. 4B). Regarding tet and tdg genes, mRNA levels of both tet1 ohnologs, tet2c and tet3b were not significantly different among different stages. As for tet2a&b (we were not able to design discriminating primers to separately amplify tet2a and tet2b), the mRNA level increased slightly but significantly during oogenesis (Fig. 4B). The same pattern was observed for tet3a except in spawned oocytes (November) in which tet3a mRNA level was lower than the other stages. All tdg paralogs displayed a similar pattern with a strong increase in mRNA levels at the spawn stage (November) (Fig. 4B).
In males, we observed that the expression profile of DNA methylation genes fluctuated during spermatogenesis (Fig. 4C, mean ± SD data in Supplemental Table S5). Increased mRNA level of dnmt1b was found in testes sampled in September (late spermatogenesis stage), whereas the expression of dnmt3ab1 and dnmt3bba1 decreased to its lowest level in September, and increased again to a high level in November (milt stage, Fig. 4C). No significant difference was found in mRNA levels for other dnmt genes during spermatogenesis (Fig. 4C). There was no significant difference in all tet genes in testes whatever the gonadal developmental stages, except for tet1 ohnologs, which displayed a significantly lower mRNA abundance in September than in earlier stages, but increased to a higher level in November (Fig. 4C). For tdg genes, tdga ohnologs displayed similar expression patterns and remarkably reached a maximum in September and then decreased to a low level in November. No significant difference was found in the mRNA level of tdgb ohnologs across all stages (Fig. 4C).
During trout ontogenesis (Fig. 5A), our results showed that mRNA levels for most DNA methylation genes displayed a similar expression pattern, with no or low mRNA levels from stages 0 to 15, followed by a remarkable increase to reach the maximum expression at stage 22/23, and then a decrease after hatching (Fig. 5B, mean ± SD data in Table S6). Exceptionally, relatively high mRNA abundances of dnmt1b, dnmt3bbb and tdgb ohnologs were observed at the beginning of ontogenesis. Besides, significantly high mRNA level of dnmt3aa and low mRNA level of dnmt3bbb were found at stage 31.
Discussion
In silico analysis of DNA methylation genes in the present study showed that only one dnmt1 gene was fixed in the genomes of most species in chordates, which was in accordance with previous studies34,35. A recent study concerning the molecular evolution of dnmt1 in vertebrates demonstrated that an additional SSD event occurred before the radiation of major marsupial groups, giving rise to the fixation of two dnmt1 copies in marsupials11. Here, we also identified two dnmt1 genes in trout and salmon, which were supposed to arise from the SaGD thus being ohnologous genes.
Similar results were found for tet1, tet2, and tet3, which represent as single-copy gene across chordate evolution, except that amphioxus and ciona have only one tet related gene, whereas trout has 7 tet related genes. Previous studies did by Zhang et al.36 failed to identify any tet related genes in lamprey genome, probably due to the fact that their analyses were done with lamprey somatic genome, which contains only a part of the germline genome due to the programmed genome rearrangement event that happened specifically in lamprey37. In the present study, we identified 3 putative tet genes in lamprey, 1 of them are likely to be ortholog of tet3, but with a remaining question about the identity of the other two genes. So it still remains elusive to identify the origin of tet1/2/3 during evolution. In terms of the fixation of multiple tet genes, especially the 3 tet2 genes in salmonid species, it could be hypothesised that a lineage-specific duplication of tet2 and synthenic genes may have occurred following the SaGD event.
Concerning tdg genes, our results showed an increase in the copy number of this gene during chordate evolution. Results in this study suggested that, a single copy of tdg was present at least in the gnathostomata ancestor, and a duplication of the tdg gene occurred before or around the teleost radiation, which resulted in the presence of the duplicated genes tdga and tdgb in the ancestral teleost. These results were in contrast with the previous hypothesis proposed by Best et al. (2018), which stated that the two tdg paralogs derived from a lineage-specific duplication event28. This is the reason why in the present study we renamed tdg1 and tdg2 as tdga and tdgb, respectively according to ZFIN Nomenclature guidelines. On the other hand, salmonids possess duplicates of tdga and tdgb genes, which most probably duplicated before or around the salmonid radiation and were retained following the SaGD.
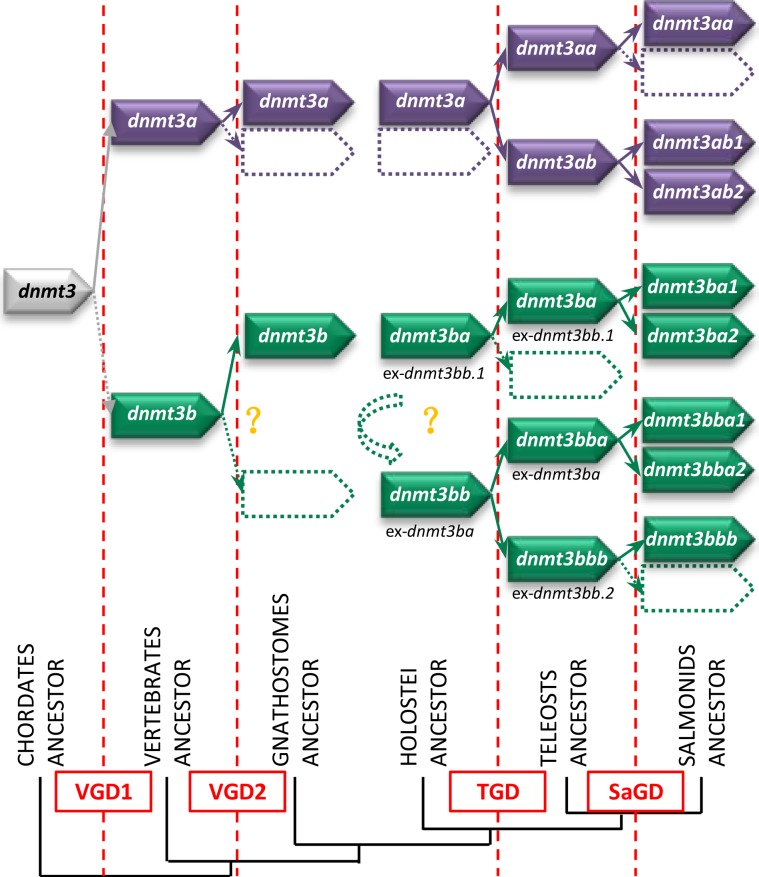
Finally, the most important work in the present study should be the clarification of the evolutionary history of dnmt3 genes in chordates. First, our investigation of the origin timing of dnmt3a and dnmt3b takes advantage of the inclusion of cephalochordate, tunicate and agnatha data. Campos et al. suggested that the ancestral dnmt3a and dnmt3b genes seem to have arisen during the VGD2 (second vertebrate whole genome duplication) event22, but the identification of multiple dnmt3 genes in ciona and more specifically the confirmation of several dnmt3a genes in lamprey in the present study challenged this view. Several authors are now in support of the idea that one round of genome duplication (VDG1) occurred during the evolution of jawless vertebrates from chordate invertebrates, but whether the VGD2 occurred before or after the cyclostome-gnathostome split remains a controversy38,39. It can be thus hypothesised that dnmt3a and dnmt3b arose before the radiation of the lamprey ancestor, probably during VGD1 event. The dnmt3b seems to be lost in lamprey, whereas the 3 dnmt3a exist in its genome suggested that lamprey dnmt3a experienced several duplication events either through VGD2 followed by another SSD event, or through other possible duplication scenarios i.e. triplication, chromosome-scale duplications37.
On the other hand, we refined the evolutionary history of dnmt3 in vertebrates, especially for dnmt3b. A previous study proposed that dnmt3bba (ex. dnmt3b2/dnmt3ba) was derived from TDG, whereas dnmt3ba (ex. dnmt3b1/dnmt3bb.1) and dnmt3bbb (ex. dnmt3b3/dnmt3bb.2) arose from a common ancestor through lineage-specific duplication after TDG22. This conclusion, however, took only tetrapod and teleost sequences into account. Thus through the inclusion of the missing link which was previously described as the bridge from tetrapods to teleosts40, namely the spotted gar, as well as shark and coelacanth, we clarified and reviewed the phylogenetic relationship between dnmt3b paralogs in teleosts. Our results showed that 2 dnmt3b genes exist in the genome of gar; one of them (ex. dnmt3bb.1) clustered together on one hand with dnmt3b orthologs in shark, coelacanth and tetrapods and one the other hand with teleost orthologs, while the other dnmt3b gene in gar clustered with ex- dnmt3ba and ex- dnmt3bb.2 in teleosts. We thus hypothesised that the ancestral dnmt3b duplicated at VGD2, both duplicates were fixed at least in holeostei, whereas one copy was lost in gnathostomes. Although we cannot exclude the possibility that the dnmt3ba/bb might arise from a punctual duplication occurred in ancestral holeostei, we are much in favour of the former hypothesis. In addition, we demonstrated a closer phylogenetic relationship between ex- dnmt3ba and ex- dnmt3bb.2 than between these genes and ex- dnmt3bb.1, which is in accordance with the previous studies22–24,41. This is the reason why we proposed new nomenclatures for these genes. Besides, we found two dnmt3bbb (ex. dnmt3bb.2) paralogs (ex. dnmt3/dnmt3bb.2 and ex. dnmt5/dnmt3bb.3) in zebrafish and another cyprinid fish, indicating that these two genes may arose from a lineage-specific duplication in cyprinid ancestor, as was reported before22.
In summary, we shed new light onto the evolutionary history of complex dnmt3 among chordates. Based on our findings, we proposed a model for the evolutionary history of these genes in chordates depicted in Fig. 6.
Figure 6.
Proposed evolutionary history model of dnmt3 in chordates.
Our findings prove that DNA methylation genes seemed to have conserved their importance during gametogenesis in teleosts as in mammals. However, the increasing number of DNA methylation related genes in teleosts raises the question of their divergence in term of expressional territories and functions.
In the present study, we demonstrated that most of the identified ohnologous/paralogous genes displayed divergent expression pattern during oogenesis or spermatogenesis (i.e among the dnmt3b encoding genes), suggesting that after duplication these genes encountered a sub- or neo-functionalisation. This hypothesis might be supported by our analyses of the branch test of dN/dS ratio for duplicated dnmt, tet, and tdg genes in trout (Supplemental Fig. S10), which demonstrated that these genes evolved differently. For instance, among the 6 dnmt3 paralogs in trout, the dnmt3aa and dnmt3bba2 exhibited higher dN/dS ratio compared to the other paralogs, indicating that these genes accumulated more non-synonymous substitutions and evolved faster (Supplemental Fig. S10B).
In addition, results obtained from the present study showed that these duplications also led to new expression patterns compared to what was previously described in other teleosts or mammals. During oogenesis, dnmt1b was found maternally expressed in trout oocytes whereas its zebrafish ortholog dnmt1 was not found in mature oocytes42. Moreover trout tet3a expression pattern strongly differed from those previously described for the mammalian tet343. To date, little is known concerning methylome modification during gametogenesis in fish. Nonetheless, low mRNA levels of dnmt1b, dnmt3ba1, dnmt3bba2, dnmt3bbb together with the high mRNA level of tet3a in September (late vitellogenesis stage) may possibly coincide with a demethylation event. Additionally, our results highlighted a significant increase in mRNA levels for genes involved in both methylation and demethylation (all tdg paralogs) pathways of mature oocytes, indicating that they may be important for early embryonic development before zygotic genome activation.
In terms of spermatogenesis, a previous study in zebrafish reported that the testicular transcription of dnmt1 and dnmt3 was correlated with global DNA methylation level44. In trout, the higher mRNA levels of dnmt3ab1 and dnmt3bba1 observed at the end of spermatogenesis may indicate that a de novo methylation event occurred before the release of sperm. Moreover, tet1 ohnologs were the only tet encoding genes that differentially regulated during spermatogenesis in trout, suggesting an important role of tet1 genes as was previously proposed in mammals45. Overall, our results suggest that late spermatogenesis stage may be as a possible “active phase” for the transcriptional regulation of DNA methylation genes, as most of the affected DNA methylation genes exhibited either their highest (dnmt1b, tdgaa, and tdgab) or lowest (dnmt3ab1, dnmt3bba1 and tet1b) mRNA level at this stage.
Concerning early development, we demonstrated that all DNA methylation genes have similar expression pattern with an increase of mRNA levels after the embryonic genome activation (stage 10) and strongly increased from stage 15, which corresponds to the developmental stage of visible optic vesicles46. This observation could possibly be explained by the indispensable function of DNA methylation in lens development as was previously described in zebrafish47. The peak of expression was observed occurred during the setting up of vital organs (stage 22, 23). This result is in accordance with previous studies, which showed that the expression of DNA methylation genes was largely increased during the organogenesis period in teleosts18,48–50. The high expression of DNA methylation related genes at these specific stages maybe explained by the phylotic stage related widespread DNA demethylation of enhancers of regulatory genes during embryogenesis50. However this latter publication was conducted on zebrafish, and comparative embryology of 2 different ectotherm species bred at different temperature remained elusive without performing histological analysis.
Looking at the results in detail we showed that some ohnologous/paralogous genes displayed divergent expression patterns at specific stages, suggesting a neo- or sub-functionalisation after duplication. For some of these genes, expression patterns diverged from what was previously described in other species, which is in accordance with previous studies, indicating that DNA methylation related genes could be differentially regulated from species to species22–24,48. Indeed, in contrast to what was previously described in zebrafish and sole, we did not observe a peak in the expression of dnmt3b paralogs during the blastula or gastrula stages22,23,51, which were in favour of neo-functionalisation of these genes in trout during development.
On the other hand, early embryonic expression of dnmt1 was in accordance with previous studies in other vertebrate species23,52–54, suggesting that the vital function of methylation maintenance during this period was conserved during evolution. Besides, the high mRNA level of dnmt3aa in hatched alevins as was previously showed in several teleosts22,23,48 may indicate a special role of this gene in alevins compared to the other DNA methylation genes.
Conclusion
The present study is the first to analyse the evolutionary history of DNA methylation genes (dnmt, tet and tdg) in chordates. Results showed that while dnmt1, tet1, tet2, and tet3 remain as a single copy gene in the genomes of most species of chordates, dnmt3 and tdg duplicates were preferentially fixed in genomes after successive WGD events, increasing their copy numbers during evolution. The case study for trout revealed that these duplicated genes were regulated independently during gametogenesis and embryogenesis, suggesting that they may have suffered sub- or neo-functionalisation after WDG.
Methods
Ethical issues and approval
Investigations were conducted according to the guiding principles for the use and care of laboratory animals and in compliance with French and European regulations on animal welfare (Decree 2001-464, 29 May 2001 and Directive 2010/63/EU, respectively). This protocol and the project as a whole were approved by the French National Consultative Ethics Committee (201610061056842).
In silico analysis
Paralogs for DNA methylation genes (dnmt, tet and tdg) in trout were identified in in NCBI assembly database (GCF_002163495.1) using the BLAST tool. Orthologous genes of DNA methylation genes and deduced amino acid sequences for other species were identified from the Ensembl genome browser (Ensembl release 91, December 2017, http://www.ensembl.org) or NCBI database with BLAST tool. The corresponding genes in amphioxus, elephant shark were identified in their respective genome database (https://genome.ucsc.edu/) or NCBI database through BLAST searches. SIMRBASE (https://genomes.stowers.org/search/loc), a database which provides preliminary annotations to characterise the latest lamprey assembly (GCA_002833325.1) was also used to verify the results in lamprey. All the BLAST searches were performed using TBLASTN method with default algorithm parameters: expect threshold was set as 10; word size was set as 6; and low complexity regions were filtered. All the identified sequences were subsequently subjected to conserved domain search in NCBI (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) to further confirm their identities and check if there is any alteration in conserved functional sites.
Phylogenetic analyses were carried out using MEGA package version 7 software55 with the deduced amino acid sequences of DNA methylation genes. All the protein sequences were aligned with the MUSCLE method in MEGA software before constructing phylogenetic tree. Phylogenetic trees of Dnmt1, Tet and Tdg were built using Maximum Likelihood method and the whole protein sequences, whereas the phylogenetic tree of Dnmt3 was constructed using Neighbour-Joining (NJ) method with the protein alignment containing only the conserved functional domains (PWWP, ADD and Dcm). The reliability of the inferred trees was estimated using the bootstrap method with 500 or 1000 replications in Maximum likelihood method and NJ method, respectively. New gene annotations were allocated according to ZFIN Nomenclature guidelines (http://zfin.org/).
Syntenic analyses were performed with the Genomicus software, version 01.01 (www.genomicus.biologie.ens.fr) to confirm identities of the target genes in various vertebrates. The gene loci of DNA methylation genes in lampreys, elephant sharks and trout and their syntenic genes were identified using the BLAST tool against their corresponding assembly databases in NCBI.
In order to have a preliminary idea about the prediction of sub- or neo-functionalisation among multiple paralogues, the ratios between non-synonymous and synonymous differences (dN/dS) were also calculated with the codeml program in the PAML package version 4.9i56. The aligned amino acids sequences of dnmt1, dnmt3, tet1, tet2, tet3 and tdg genes in selected representative species were used to guide the alignment of the nucleotides sequences by PAL2NAL to get codon alignments of these genes57. We further built phylogenetic trees with their respective codon alignment of these genes using Maximum likelihood method. The codon alignments and phylogenetic trees were used as the input file for codeml program. Branch tests with the free-ratios model (Alternative hypothesis: heterogeneity of dN/dS among branches) and model 0 (Null hypothesis: homogeneous dN/dS across all sites and branches) were performed to calculate the dN/dS for each of the branches of the gene tree. The comparison between two models was conducted using a likelihood ratio test (LRT) followed by a chi-squared test of significance56.
Fish and experimental design
For gametogenesis experiment: two year old genitors were obtained and reared at 8 °C in INRA experimental facilities. Ovaries and testes were sampled (6 individuals per time point) during gametogenesis process in February, May, September and November, corresponding to pre-early vitellogenesis/early spermatogenesis, vitellogenesis/mid spermatogenesis, late vitellogenesis/late spermatogenesis, and spawn stage, respectively (Fig. 4A). For ontogenesis experiment: embryos were collected on triplicate spawns intermittently before the fertilisation stage (oocyte) and during embryonic development: stages 5, 6, 7, 8, 10, 12, 15, 22, 23 and 31 (hatching alevins, 384°D (degree days)) according to Vernier46. Embryos were directly snap-frozen whereas alevins and fish were sacrificed by terminal anaesthetisation with a benzocaine bath (60 mg·L−1) prior to sampling and subsequently frozen in liquid nitrogen. Samples were stored at −80 °C until further analysis (Fig. 5A).
Total RNA extraction, cDNA synthesis and RT-qPCR analysis
Total RNA, cDNA synthesis and RT-qPCR analysis on whole embryos (30 embryos extracted together per spawn) and whole alevins (6 alevins extracted individually per spawn) was performed as previously described58. Data were normalised to the exogenous luciferase transcript abundance in samples diluted at 1:2559.
For gonads, total RNA and cDNA synthesis were performed as previously described59. However, samples were diluted at 1:40, and the geometric mean of several reference genes: β-actin, elongation factor 1α (ef1α), 18S ribosomal RNA (18S rRNA), 40 S ribosomal protein S6 (rps16), and 60 S Ribosomal Protein L27 (rpl27) was used as a normalisation for qPCR data during gametogenesis using the E method (Light Cycler software) as previously described60–62. The primer sets used for analysis are listed in Supplemental Table S2 (target genes) and Supplemental Table S3 (reference genes).
Statistical analysis
The qPCR data were presented as heatmap with mean value of each condition using ClustVis (https://biit.cs.ut.ee/clustvis/). Normality of distributions was assessed using the Shapiro-Wilk test using R (v3.4.0)/Rcmdr Package. Data were analysed by a Kruskal-Wallis non-parametric test followed by a Tukey test as post hoc analysis.
Supplementary information
Acknowledgements
We warmly thank F. Vallée, P. Maunas and N. Turonnet for animal care at INRA Lees-Athas facilities and for their indispensable help with sampling. We also thank F. Terrier for the preparation of the diets. J.L. received a doctoral fellowship from the China Scholarship Council (File No. 201506330063).
Author contributions
L.M. designed the study. L.M. and J.L. managed the study and performed in silico analysis. J.L. and H.H. performed qPCR analysis. J.L. did statistical analysis and wrote the manuscript. L.M. and S.P. contributed to the manuscript corrections.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-57753-w.
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 4.Dean W. DNA methylation and demethylation: a pathway to gametogenesis and development. Mol. Reprod. Dev. 2014;81:113–125. doi: 10.1002/mrd.22280. [DOI] [PubMed] [Google Scholar]
- 5.Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 2004;279:48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- 6.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998;19:219. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 7.Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M. Replication‐coupled passive DNA demethylation for the erasure of genome imprints in mice. The EMBO journal. 2013;32:340–353. doi: 10.1038/emboj.2012.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Y-F, et al. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science (New York, N.Y.) 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alvarez-Ponce D, Torres-Sánchez M, Feyertag F, Kulkarni A, Nappi T. Molecular evolution of DNMT1 in vertebrates: Duplications in marsupials followed by positive selection. PLoS One. 2018;13:e0195162. doi: 10.1371/journal.pone.0195162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018;19:81–92. doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- 13.Bestor TH. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 14.Barau J, et al. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science. 2016;354:909–912. doi: 10.1126/science.aah5143. [DOI] [PubMed] [Google Scholar]
- 15.Yokomine T, Hata K, Tsudzuki M, Sasaki H. Evolution of the vertebrate DNMT3 gene family: a possible link between existence of DNMT3L and genomic imprinting. Cytogenet. Genome Res. 2006;113:75–80. doi: 10.1159/000090817. [DOI] [PubMed] [Google Scholar]
- 16.Krokan HE, Nilsen H, Skorpen F, Otterlei M, Slupphaug G. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000;476:73–77. doi: 10.1016/S0014-5793(00)01674-4. [DOI] [PubMed] [Google Scholar]
- 17.Goll, M. G. & Halpern, M. E. In Prog. Mol. Biol. Transl. Sci. Vol. 101 (eds Xiaodong Cheng & Robert M. Blumenthal) 193–218 (Academic Press, 2011).
- 18.Ge L, et al. TET2 plays an essential role in erythropoiesis by regulating lineage-specific genes via DNA oxidative demethylation in a zebrafish model. Mol. Cell. Biol. 2014;34:989–1002. doi: 10.1128/MCB.01061-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ravi V, Venkatesh B. The Divergent Genomes of Teleosts. Annu Rev Anim Biosci. 2018;6:47–68. doi: 10.1146/annurev-animal-030117-014821. [DOI] [PubMed] [Google Scholar]
- 20.Shimoda N, Yamakoshi K, Miyake A, Takeda H. Identification of a gene required for de novo DNA methylation of the zebrafish no tail gene. Dev. Dyn. 2005;233:1509–1516. doi: 10.1002/dvdy.20455. [DOI] [PubMed] [Google Scholar]
- 21.Smith TH, Collins TM, McGowan RA. Expression of the dnmt3 genes in zebrafish development: similarity to Dnmt3a and Dnmt3b. Dev. Genes Evol. 2011;220:347–353. doi: 10.1007/s00427-010-0347-z. [DOI] [PubMed] [Google Scholar]
- 22.Campos C, Valente LM, Fernandes JM. Molecular evolution of zebrafish dnmt3 genes and thermal plasticity of their expression during embryonic development. Gene. 2012;500:93–100. doi: 10.1016/j.gene.2012.03.041. [DOI] [PubMed] [Google Scholar]
- 23.Firmino J, et al. Phylogeny, expression patterns and regulation of DNA Methyltransferases in early development of the flatfish, Solea senegalensis. BMC Dev. Biol. 2017;17:11. doi: 10.1186/s12861-017-0154-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skjærven KH, Hamre K, Penglase S, Finn RN, Olsvik PA. Thermal stress alters expression of genes involved in one carbon and DNA methylation pathways in Atlantic cod embryos. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology. 2014;173:17–27. doi: 10.1016/j.cbpa.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Dasmahapatra AK, Khan IA. DNA methyltransferase expressions in Japanese rice fish (Oryzias latipes) embryogenesis is developmentally regulated and modulated by ethanol and 5-azacytidine. Comparative Biochemistry and Physiology Part C: Toxicology &. Pharmacology. 2015;176–177:1–9. doi: 10.1016/j.cbpc.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Sun X, Zhang L, Zhang W. Testicular Dnmt3 expression and global DNA methylation are down-regulated by gonadotropin releasing hormones in the ricefield eel Monopterus albus. Sci. Rep. 2017;7:43158. doi: 10.1038/srep43158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood RK, Crowley E, Martyniuk CJ. Developmental profiles and expression of the DNA methyltransferase genes in the fathead minnow (Pimephales promelas) following exposure to di-2-ethylhexyl phthalate. Fish Physiol. Biochem. 2016;42:7–18. doi: 10.1007/s10695-015-0112-3. [DOI] [PubMed] [Google Scholar]
- 28.Best, C. et al. Epigenetics in teleost fish: From molecular mechanisms to physiological phenotypes. Comp. Biochem. Physiol. B Biochem. Mol. Biol., 10.1016/j.cbpb.2018.01.006 (2018). [DOI] [PubMed]
- 29.Zhang J. Evolution by gene duplication: an update. Trends Ecol. Evol. 2003;18:292–298. doi: 10.1016/S0169-5347(03)00033-8. [DOI] [Google Scholar]
- 30.Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nature Reviews Genetics. 2008;9:938. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 31.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 32.Akahori H, Guindon S, Yoshizaki S, Muto Y. Molecular Evolution of the TET Gene Family in Mammals. International journal of molecular sciences. 2015;16:28472–28485. doi: 10.3390/ijms161226110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasquier J, et al. Gene evolution and gene expression after whole genome duplication in fish: the PhyloFish database. BMC Genomics. 2016;17:368. doi: 10.1186/s12864-016-2709-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponger L, Li W-H. Evolutionary diversification of DNA methyltransferases in eukaryotic genomes. Mol. Biol. Evol. 2005;22:1119–1128. doi: 10.1093/molbev/msi098. [DOI] [PubMed] [Google Scholar]
- 35.Albalat R. Evolution of DNA-methylation machinery: DNA methyltransferases and methyl-DNA binding proteins in the amphioxus Branchiostoma floridae. Dev. Genes Evol. 2008;218:691–701. doi: 10.1007/s00427-008-0247-7. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, Z. et al. Genome‐wide and single‐base resolution DNA methylomes of the Sea Lamprey (Petromyzon marinus) Reveal Gradual Transition of the Genomic Methylation Pattern in Early Vertebrates, 10.1101/033233 (2015).
- 37.Smith JJ, et al. The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat. Genet. 2018;50:270. doi: 10.1038/s41588-017-0036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, J. J. & Keinath, M. C. The sea lamprey meiotic map improves resolution of ancient vertebrate genome duplications. Genome Res. (2015). [DOI] [PMC free article] [PubMed]
- 39.Kasahara M. The 2R hypothesis: an update. Curr. Opin. Immunol. 2007;19:547–552. doi: 10.1016/j.coi.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 40.Braasch I, et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 2015;47:427. doi: 10.1038/ng.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang F-L, et al. Genome-wide identification, evolution of DNA methyltransferases and their expression during gonadal development in Nile tilapia. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 2018;226:73–84. doi: 10.1016/j.cbpb.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Mhanni AA, McGowan RA. Variations in DNA (cytosine-5)-methyltransferase-1 expression during oogenesis and early development of the zebrafish. Dev. Genes Evol. 2002;212:530–533. doi: 10.1007/s00427-002-0275-7. [DOI] [PubMed] [Google Scholar]
- 43.Masala L, et al. Methylation dynamics during folliculogenesis and early embryo development in sheep. Reproduction. 2017;153:605–619. doi: 10.1530/REP-16-0644. [DOI] [PubMed] [Google Scholar]
- 44.Laing L, et al. Sex-specific transcription and DNA methylation profiles of reproductive and epigenetic associated genes in the gonads and livers of breeding zebrafish. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology. 2018;222:16–25. doi: 10.1016/j.cbpa.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 45.Nettersheim D, et al. Analysis of TET Expression/Activity and 5mC Oxidation during Normal and Malignant Germ Cell Development. PLoS One. 2013;8:e82881. doi: 10.1371/journal.pone.0082881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vernier J. Table chronogique du development embryonnaire de la truite arc-en-ciel, Salmo gairdneri Rich. Ann. Embryol. Morphogenet. 1969;2:495–520. [Google Scholar]
- 47.Seritrakul P, Gross JM. Expression of the de novo DNA methyltransferases (dnmt3–dnmt8) during zebrafish lens development. Dev. Dyn. 2014;243:350–356. doi: 10.1002/dvdy.24077. [DOI] [PubMed] [Google Scholar]
- 48.Fellous, A. et al. DNA methylation in adults and during development of the self‐fertilizing mangrove rivulus, Kryptolebias marmoratus. Ecol. Evol. (2018). [DOI] [PMC free article] [PubMed]
- 49.Takayama K, Shimoda N, Takanaga S, Hozumi S, Kikuchi Y. Expression patterns of dnmt3aa, dnmt3ab, and dnmt4 during development and fin regeneration in zebrafish. Gene Expression Patterns. 2014;14:105–110. doi: 10.1016/j.gep.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 50.Bogdanović O, et al. Active DNA demethylation at enhancers during the vertebrate phylotypic period. Nat. Genet. 2015;47:417. doi: 10.1038/ng.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aluru N, et al. Developmental exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin alters DNA methyltransferase (dnmt) expression in zebrafish (Danio rerio) Toxicol. Appl. Pharmacol. 2015;284:142–151. doi: 10.1016/j.taap.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nature Reviews Genetics. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 53.Ko Y-G, et al. Stage-by-stage change in DNA methylation status of Dnmt1 locus during mouse early development. J. Biol. Chem. 2005;280:9627–9634. doi: 10.1074/jbc.M413822200. [DOI] [PubMed] [Google Scholar]
- 54.Vassena R, Dee Schramm R, Latham KE. Species‐dependent expression patterns of DNA methyltransferase genes in mammalian oocytes and preimplantation embryos. Mol. Reprod. Dev. 2005;72:430–436. doi: 10.1002/mrd.20375. [DOI] [PubMed] [Google Scholar]
- 55.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 57.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marandel, L., Véron, V., Surget, A., Plagnes-Juan, É. & Panserat, S. Glucose metabolism ontogenesis in rainbow trout (Oncorhynchus mykiss) in the light of the recently sequenced genome: new tools for intermediary metabolism programming. J. Exp. Biol., jeb. 134304 (2016). [DOI] [PubMed]
- 59.Marandel L, Labbe C, Bobe J, Le Bail P-Y. nanog 5′-upstream sequence, DNA methylation, and expression in gametes and early embryo reveal striking differences between teleosts and mammals. Gene. 2012;492:130–137. doi: 10.1016/j.gene.2011.10.037. [DOI] [PubMed] [Google Scholar]
- 60.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034.0031–research0034.0011. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deloffre LA, Andrade A, Filipe AI, Canario AV. Reference genes to quantify gene expression during oogenesis in a teleost fish. Gene. 2012;506:69–75. doi: 10.1016/j.gene.2012.06.047. [DOI] [PubMed] [Google Scholar]
- 62.Nicol B, Guiguen Y. Expression profiling of Wnt signaling genes during gonadal differentiation and gametogenesis in rainbow trout. Sex. Dev. 2011;5:318–329. doi: 10.1159/000334515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.