Abstract
Being born small (SGA) or large for gestational age (LGA) is associated with adverse birth outcomes and metabolic diseases in later life of the offspring. It is known that aberrations in growth during gestation are related to altered placental function. Placental function is regulated by epigenetic mechanisms such as DNA methylation. Several studies in recent years have demonstrated associations between altered patterns of DNA methylation and adverse birth outcomes. However, larger studies that reliably investigated global DNA methylation are lacking. The aim of this study was to characterize global placental DNA methylation in relationship to size for gestational age. Global DNA methylation was assessed in 1023 placental samples by LC-MS/MS. LGA offspring displayed significantly higher global placental DNA methylation compared to appropriate for gestational age (AGA; p < 0.001). ANCOVA analyses adjusted for known factors impacting on DNA methylation demonstrated an independent association between placental global DNA methylation and LGA births (p < 0.001). Tertile stratification according to global placental DNA methylation levels revealed a significantly higher frequency of LGA births in the third tertile. Furthermore, a multiple logistic regression analysis corrected for known factors influencing birth weight highlighted an independent positive association between global placental DNA methylation and the frequency of LGA births (p = 0.001).
Subject terms: Epidemiology, Molecular medicine
Introduction
Birth size is an important clinical parameter that can be used for determining the health of the newborn and is highly correlated with postpartum morbidity and complications during labour. Increases in birth weight parallel the gestational age1. Relating birth weight to gestational age has the advantage of evaluating the potential growth of the fetus according to the length of pregnancy2. Weight for gestational age is usually classified into small for gestational age (SGA), appropriate for gestational age (AGA), and large for gestational age (LGA)3. Abnormal fetal growth is defined as being born smaller than the 10th percentile (SGA) or larger than the 90th percentile (LGA)3. Both SGA and LGA births display an increased peri- and postnatal morbidity and mortality. Regarding maternal health, SGA and LGA births are associated with maternal disease such as hypertension, preeclampsia, and preexisting or gestational diabetes mellitus. Moreover, SGA and especially LGA births are associated with adverse obstetrical outcomes such as emergency cesarean deliveries and postpartum hemorrhages4. SGA newborns are at an increased risk for stillbirth, seizures, intraventricular hemorrhages, hypoxic ischemic encephalopathy, necrotizing enterocolitis, sepsis, and neonatal mortality5. LGA newborns display an increased risk for stillbirth, traumatic delivery, brachial plexus palsy, mechanical ventilation, and neonatal mortality5. Furthermore, numerous studies have demonstrated that both SGA and LGA newborns are at an increased risk for disease in later life, highlighting the importance of these anthropometric measures as surrogate parameters of fetal programming6–9.
Adaptive processes of the fetus to adverse environmental cues during gestation can have an impact on the physiology and metabolism in later life10,11. An adverse intrauterine environment can be translated into being born small for gestational age (SGA) which is an independent risk factor for metabolic diseases12,13. Interestingly, similar associations have been demonstrated for LGA newborns, displaying an increased susceptibility for obesity, hypertension and diabetes mellitus compared to appropriate for gestational age (AGA) newborns6,14. Overall, studies show an U-shaped relation between birth weight and type-2 diabetes, hypertension or risk of overweight in later life15,16. However, it has been proposed that the underlying mechanisms leading to the same metabolic diseases in adult life differ between SGA and LGA newborns17,18.
The placenta is the primary means of communication between mother and fetus during pregnancy. Placental function has been reported to be altered in pregnancies with SGA or LGA outcomes19. Changes in placental function may occur due to changes in placental structure. However, the molecular pathways responsible for changes in placental structure and function in association with pregnancy outcomes have not been widely studied yet19. More recent studies highlight the importance of epigenetic mechanisms, primarily DNA methylation, in placental performance during pregnancy20–22.
Birth weight for gestational age is a complex phenotype that results from various processes and the expression of numerous genes in the genome. This might explain why association analyses between DNA methylation of certain genes and birth weight for gestational age remained elusive despite a direct connection between the gene of interest and fetal growth processes23,24. The whole human genome contains only about 1.5% of protein encoding genes, while the remaining majority comprises of introns, repetitive elements, and other non-coding sequences25. Initially regarded as “junk DNA” more recent research has revealed important functions of non-coding genomic regions and their epigenetic modification, including DNA methylation26,27. Thus, investigating DNA methylation levels on a global scale might facilitate getting first insights into epigenetic mechanisms of complex phenotypes28,29. However, studies investigating correlations between global DNA methylation and birth weight so far were only performed using rather small sample sizes30–33.
Currently, various approaches to quantify global DNA methylation exist. Several studies applied genome wide array approaches34–36. Although such assays can measure CpG island methylation of about 99% of all RefSeq genes, they only cover about 1.5% of genomic CpGs, are more specifically designed to analyze promotor methylation and neglect other genomic regions where DNA methylation might be of importance37,38. Also newer variants of genome wide array approaches like the Illumina MethylationEPIC bead chip still exclude regions of potentially meaningful biological variation38. More comprehensive methods such as whole genome bisulfite sequencing are not feasible yet for utilization with large sample sizes. Other novel sequencing based approaches that indeed are applicable for large sample sizes do so at the expense of genomic coverage and are further limited by their still quite extensive costs38. Other commonly used methods measure DNA methylation in repetitive elements such as LINE-1 or Alu elements that serve as surrogate parameters for global DNA methylation39. Futhermore, the absolute methyl cytosine content can be measured by using HPLC or LC-MS/MS. LC-MS/MS is reported to be the most sensitive method for absolute methyl cytosine quantification and is considered the current gold standard37.
The aim of this study was to analyze placental global DNA methylation using the current gold standard method, LC-MS/MS, and to investigate whether alterations in birth weight for gestational age are associated with differences in global placental DNA methylation.
Material and Methods
Clinical study
The study was conducted in 1023 mothers with singleton pregnancies delivering at the Charité obstetrics department, Berlin, Germany from 2000 to 2008. The local ethics committee approved the study and all clinical investigations were performed according to the principles expressed in the Declaration of Helsinki. Written informed consent was obtained from all participants. Data on history of diabetes in the family, the status of diabetes before or during pregnancy, smoking status before pregnancy and the ethnic background were obtained from the “Mutterpass” (German pregnancy record booklet). According to the practice guideline of the German Diabetes Assosiation (DGG) and the German Association for Gynaecology and Obstetrics (DGGG), all mothers were screened for gestational diabetes40. Variables describing diabetes in the dataset were diabetes before pregnancy (preexisting diabetes mellitus according to the pregnancy record booklet) and diabetes during pregnancy (composite of diabetes before pregnancy and gestational diabetes). Gestational age was estimated from the date of the last menstrual period. Data on blood pressure during pregnancy were obtained from routine antenatal examinations. Placental samples were collected directly after delivery. One complete placental cotyledon taken from the same location was collected from all participating mothers. Placental samples were stored at −20 °C until further use for DNA isolation. Additionally, data regarding the sex of the newborn, birth weight, birth length, and head circumference were extracted into our database.
DNA preparation and placental methylation quantification
DNA extraction was performed using a QIAamp DNA Mini Kit (Qiagen). RNase was added during the isolation process to eliminate the influence of RNA in the next step. Quality and quantity of the DNA solution were measured using a spectrophotometer ND-1000 (NanoDrop). Degradase plus DNAse (Zymo Research) was used in the DNA digestion process according to the manufacturer’s instructions. As a control, 200 ng of the digested DNA was analyzed by agarose gel electrophoresis. DNA was prepared by adding 280 µl of 0.1% formic acid to 70 µL of digested DNA to obtain a final concentration of 2 ng/µL. Determination of placental methylation was performed by liquid chromatography-electrospray ionization/multi-stage mass spectrometry (LC-ESI/MS/MS) as described previously41. Analyses were performed with an Agilent 6530 Accurate-Mass Q-TOF instrument with Jet Stream-Interface (Palo Alto, USA). The chromatographic separation process was performed with a X-BridgeTM C18 4.6 mm × 150 mm (3.5 lm particle size) column (Waters). For gradient elution solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in methanol) were used. Flow rate of eluent was maintained constant at 0.5 ml/min. A total of 100 ng digested DNA in a volume of 50 µL was applied in every injection. The parameters used in the ESI-MS/MS step for gas temperature and gas flow rate were 250 °C and 8 L/min respectively, while the sheat nebulizer gas temperature and pressure were maintain at 300 °C and 60 psig. Parameters associated with the voltage were as follows: capillary voltage 4000 V; collision energy for dC, and dG 5mdC row 7 V, 13 V and 10 V. The quantification of placental methylation was calculated based on the formula: Methylation % = [5mdC]/{[5mdC] + [dC]} over m/z signal for dC and mdC at 228.0979/112.0505 and 242.1135/126.0662, respectively.
Statistical analysis
SPSS version 20 was used for the statistical analyses. Descriptive data are given as mean plus/minus standard deviation for continuous variables or percent for dichotomous variables. SGA was defined as birth weight for gestational age below the 10th percentile, while the cutoff for LGA was above the 90th percentile, using a global reference. 2. For the comparison of means between groups, ANOVA was applied according to normal distribution of the data. A Bonferroni post hoc test was used to assess specific significant differences between two groups. Kruskal-Wallis test was applied for non-normally distributed data. Independence of observed associations was investigated using a multivariable ANCOVA model adjusted for suitable confounders. Parameters that were already shown to interact with global DNA methylation and/or variables that were significantly different among SGA, AGA, and LGA births were employed as confounders. For the ANCOVA model these included as diabetes during pregnancy, age of the mother, systolic blood pressure, and smoking before pregnancy42–47. Associations between DNA methylation stratified into tertiles and the frequency for AGA, SGA or LGA was analyzed using Pearson’s Chi-square test. Analysis of the independence of observed associations was performed employing a multiple logistic regression analysis. Variables known to impact on size for gestational age and/or variables that were significantly different among the three ranked global placental DNA methylation groups were included as confounders. These parameters consisted of diabetes during pregnancy, smoking before pregnancy, pre-pregnancy BMI, diastolic blood pressure, ethnicity, and sex of the child48,49. Collinearity between confounders was controlled by using a variance inflation factor <550.
Ethics approval
The study was approved by the ethical comitee of the University Hospital Charité, Berlin-Mitte, Humboldt-University Berlin, Berlin, Germany.
Consent for publication
The final version of this manuscript was approved by all authors for submission to Scientific Reports
Results
Detailed descriptive statistics of the cohort are shown in Table 1. The mean age of the mothers was 30.0 ± 5.9 years. The overall mean gestational age at birth, birth weight, and birth length index were 38.8 ± 2.0 weeks, 3355.4 ± 630.1 g, and 50.7 ± 3.2 cm, respectively. Regarding the distribution of sex, 52.3% were male and 47.7% were female newborns. Placental DNA methylation ranged from 2.01% to 4.83%, with a mean value of 3.01 ± 0.47%. Descriptive statistics of the cohort grouped according to the birth weight for gestational age are displayed in Table 1. There were 127 SGA, 796 AGA and 100 LGA newborns in this study. LGA newborns were heavier (p < 0.001), longer (p < 0.001), and had a bigger head circumference (p < 0.001) compared to AGA and SGA newborns. Gestational age and pre-pregnancy BMI were also significantly different (p = 0.002 and p < 0.001). The distribution of sex was significantly different (p < 0.001), with SGA births showing a higher frequency of female (34.6% males; 65.4% females) and LGA births a higher frequency of male newborns (69.0% males; 31.0% females). Moreover, the distribution of the mothers who suffered from diabetes mellitus during pregnancy was significantly shifted towards LGA newborns (p < 0.001).
Table 1.
Descriptive statistics of all mother-child pairs and group comparison according to birth weight for gestational age; Data are given as mean ± SD or %; SGA = small for gestational age; AGA = appropriate for gestational age; LGA = large for gestational age; BMI = body mass index; SBP = systolic blood pressure; DBP = diastolic blood pressure.
| Parameter | All samples | SGA | AGA | LGA | p |
|---|---|---|---|---|---|
| (N = 1023) | (N = 127) | (N = 796) | (N = 100) | ||
| Placental methylation (%) | 3.01 ± 0.47 | 2.99 ± 0.41 | 2.99 ± 0.47 | 3.19 ± 0.54 | <0.001 |
| Age of the mother (years) | 30.0 ± 5.9 | 29.6 ± 5.9 | 30.0 ± 5.9 | 31.0 ± 5.7 | 0.161 |
| Pre-pregnancy BMI (kg/m2) | 23.2 ± 4.6 | 22.2 ± 4.0 | 23.1 ± 4.5 | 25.1 ± 5.8 | <0.001 |
| SBP 3rd trimester (mmHg) | 116.3 ± 11.2 | 114.8 ± 12.3 | 116.4 ± 10.9 | 117.6 ± 11.7 | 0.162 |
| DBP 3rd trimester (mmHg) | 70.5 ± 7.6 | 70.3 ± 8.9 | 70.5 ± 7.4 | 70.3 ± 7.5 | 0.901 |
| Smoking before pregnancy (%) | 36.3 | 41.3 | 35.9 | 33.0 | 0.393 |
| Diabetes before pregnancy (%) | 1.0 | 0.8 | 0.8 | 3.0 | 0.092 |
| Diabetes during pregnancy (%) | 6.3 | 7.1 | 4.9 | 16.0 | 0.001 |
| Diabetes in family (%) | 36.6 | 43.4 | 35.5 | 36.2 | 0.312 |
| Ethnicity (Caucasian/other; %) | 93.3/6.7 | 89.8/10.1 | 93.6/6.4 | 95.0/5.0 | 0.214 |
| Gestational age at delivery (weeks) | 38.8 ± 2.0 | 38.2 ± 2.3 | 38.9 ± 2.0 | 38.7 ± 2.2 | 0.002 |
| Child birth weight (g) | 3355.4 ± 630.1 | 2515.2 ± 355.3 | 3380.0 ± 493.1 | 4226.9 ± 554.4 | <0.001 |
| Child head circumference (cm) | 34.7 ± 1.7 | 32.9 ± 1.5 | 34.8 ± 1.5 | 36.3 ± 1.3 | <0.001 |
| Child birth length (cm) | 50.7 ± 3.2 | 47.3 ± 3.1 | 50.9 ± 2.8 | 53.5 ± 2.8 | <0.001 |
| Sex of the child (m/f; %) | 52.3/47.7 | 34.6/65.4 | 53.0/47.0 | 69.0/31.0 | <0.001 |
To investigate potential differences in global placental DNA methylation between the three birthweight for gestational age groups, ANOVA analyses followed by a Bonferroni post hoc test were performed. These analyses revealed a significantly higher global placental DNA methylation in LGA compared to AGA (p < 0.001) and SGA (p = 0.004) newborns (Fig. 1). There was no significant difference in global placental DNA methylation between AGA and SGA newborns as shown in Fig. 1. To analyse the potential impact of various confounders on global placental DNA methylation, a multivariable ANCOVA model was calculated including diabetes during pregnancy, age of the mother, systolic blood pressure and smoking before pregnancy (Table 2). This analysis demonstrated that higher levels of global placental DNA methylation are associated with LGA births independent of other confounding factors known to influence DNA methylation (p < 0.001; Partial η2 = 0.013; 95% C.I. = 0.08–0.28; Table 2).
Figure 1.
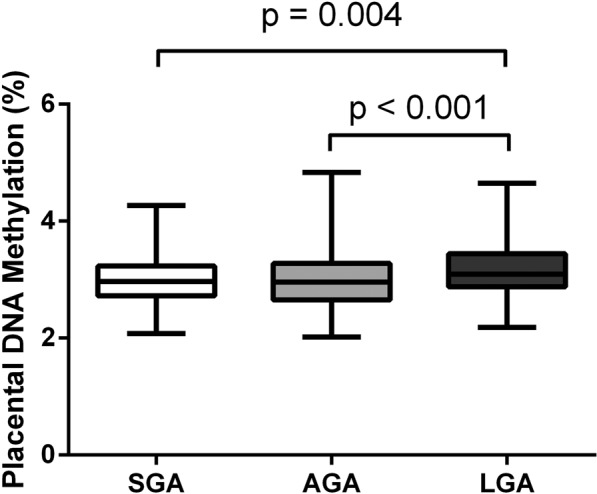
ANOVA analysis followed by a Bonferroni post-hoc test comparing global placental DNA methylation among birth weight for gestational age groups. Box and whisker blot showing median, minimum and maximum; SGA = small for gestational age; AGA = appropriate for gestational age; LGA = large for gestational age.
Table 2.
ANCOVA analysis to investigate the association between global placental DNA methylation (dependent variable) and birth weight for gestational age (aAGA was used as a reference); SGA = small for gestational age; AGA = appropriate for gestational age; LGA = large for gestational age; BMI = body mass index; SBP = systolic blood pressure;.
| Dependent variable: Placental DNA methylation; r2 = 0.026 | B | S.E. | Partial η2 | p | 95% C.I. for B | |
|---|---|---|---|---|---|---|
| Min. | Max. | |||||
| Intercept | 3.29 | 0.17 | 0.262 | <0.001 | 2.95 | 3.63 |
| Pre-pregnancy BMI (kg/m2) | 0.00 | 0.00 | 0.000 | 0.487 | −0.01 | 0.00 |
| Diabetes during pregnancy (yes/no) | 0.19 | 0.06 | 0.009 | 0.003 | 0.07 | 0.31 |
| Smoking before pregnancy (yes/no) | −0.06 | 0.03 | 0.002 | 0.038 | −0.13 | 0.00 |
| SBP (mmHg) | 0.00 | 0.00 | 0.004 | 0.157 | 0.00 | 0.00 |
| Age of the mother (years) | 0.00 | 0.00 | 0.000 | 0.966 | 0.00 | 0.01 |
| SGA | 0.00 | 0.05 | 0.000 | 0.963 | −0.09 | 0.09 |
| LGA | 0.18 | 0.05 | 0.013 | <0.001 | 0.08 | 0.28 |
| AGA | 0a | |||||
To further investigate the association between LGA births and increased global placental DNA methylation, tertiles of global placental DNA methylation were created and ranked into low (n = 341), moderate (n = 341), and high (n = 341) DNA methylation. Descriptive statistics of mothers ranked into tertiles according to the degree of global placental DNA methylation are shown in Table 3. Several parameters were significantly different between the three groups, including diastolic blood pressure (p = 0.017), ethnicity (p = 0.030) smoking before pregnancy (p = 0.005) and diabetes during pregnancy (p = 0.006). Moreover, Pearson’s Chi-square test was performed to assess the prevalence of SGA, AGA and LGA births among the three placental DNA methylation groups (Table 4). There was a significant difference (Pearson’s Chi-Square: 19.051; p = 0.001) in the prevalence for SGA, AGA, and LGA births. Mothers in the group of highest global placental DNA also displayed the highest prevalence for LGA offspring (48.0% (LGA) vs. 32.4% (AGA) vs. 27.6% (SGA); Table 4). Moreover, to investigate a potential independent association between birth weight for gestational age and global placental DNA methylation, an adjusted multiple logistic regression model was calculated using birth weight for gestational age as dependent and global placental DNA methylation as independent continuous variable. The model was adjusted for important factors impacting on size for gestational age and variables that were significantly different among three groups of ranked global placental DNA methylation. These included diabetes during pregnancy, pre-pregnancy BMI, ethnicity, sex of the child, diastolic blood pressure and smoking before pregnancy. The multiple logistic regression model highlighted an independent positive association between LGA birth and global placental DNA methylation (p = 0.001; Exp(B) = 2.06; 95% C.I. = 1.35–3.16; Table 5).
Table 3.
Descriptive statistics of mother-child pairs stratified into tertiles of global placental DNA methylation (Low; Moderate; High); Data are given as mean ± SD or %; BMI = body mass index; SBP = systolic blood pressure; DBP = diastolic blood pressure.
| Parameter | Methylation level | p | ||
|---|---|---|---|---|
| Low | Moderate | High | ||
| Placental DNA methylation (%) | 2.52 ± 0.18 | 2.98 ± 0.11 | 3.53 ± 0.34 | <0.001 |
| Age of the mother (years) | 29.7 ± 6.0 | 30.1 ± 6.2 | 30.3 ± 5.6 | 0.382 |
| Pre-pregnancy BMI (kg/m2) | 23.3 ± 4.6 | 23.2 ± 4.4 | 23.1 ± 4.8 | 0.819 |
| SBP 3rd trimester (mmHg) | 116.9 ± 10.9 | 116.7 ± 11.3 | 115.2 ± 11.3 | 0.089 |
| DBP 3rd trimester (mmHg) | 71.4 ± 7.5 | 70.1 ± 7.7 | 69.9 ± 7.4 | 0.017 |
| Smoking before pregnancy (%) | 43.2 | 33.5 | 32.2 | 0.005 |
| Diabetes before pregnancy (%) | 0.6 | 1.2 | 1.2 | 0.666 |
| Diabetes during pregnancy (%) | 4.4 | 4.7 | 9.7 | 0.006 |
| Diabetes in family (%) | 37.7 | 35.3 | 36.6 | 0.854 |
| Ethnicity (Caucasian/other; %) | 94.7/5.3 | 94.7/5.3 | 90.3/9.7 | 0.030 |
| Gestational age at delivery (weeks) | 38.8 ± 2.1 | 38.7 ± 2.1 | 38.8 ± 2.0 | 0.573 |
| Child birth weight (g) | 3367.0 ± 585.9 | 3295.9 ± 643.8 | 3403.4 ± 655.4 | 0.077 |
| Child head circumference (cm) | 34.8 ± 1.6 | 34.6 ± 1.8 | 34.8 ± 1.7 | 0.557 |
| Child birth length (cm) | 50.8 ± 2.9 | 50.4 ± 3.3 | 50.9 ± 3.3 | 0.066 |
| Sex of the child (m/f; %) | 50.7/49.3 | 48.1/51.9 | 44.3/55.7 | 0.238 |
Table 4.
Cross-tabulation of global placental DNA methylation ranked in tertiles and the frequency of different birth weight for gestational age groups (SGA; AGA; LGA); SGA = small for gestational age; AGA = appropriate for gestational age; LGA = large for gestational age.
| Pearson’s Chi-Square: 19.051; p = 0.001 | Placental DNA methylation ranked | |||
|---|---|---|---|---|
| Low | Moderate | High | ||
| SGA | Count | 36 | 56 | 35 |
| % within placental DNA methylation rank | 28.3 | 44.1 | 27.6 | |
| AGA | Count | 283 | 255 | 258 |
| % within placental DNA methylation rank | 35.6 | 32.0 | 32.4 | |
| LGA | Count | 22 | 30 | 48 |
| % within placental DNA methylation rank | 22.0 | 30.0 | 48.0 | |
| Total | Count | 341 | 341 | 341 |
Table 5.
Multiple logistic regression analysis of the association between global placental DNA methylation and birth weight for gestational age (dependent variable); aAGA was set as reference for this parameter; AGA = appropriate for gestational age; LGA = large for gestational age; BMI = body mass index; DBP = diastolic blood pressure.
| Parameter | B | S.E | p | Exp(B) | 95% C.I. for Exp(B) | ||
|---|---|---|---|---|---|---|---|
| Min. | Max. | ||||||
| LGAa | Intercept | −4.39 | 1.38 | 0.001 | |||
| Diabetes during pregnancy (yes/no) | 1.05 | 0.34 | 0.002 | 2.86 | 1.47 | 5.57 | |
| Smoking before pregnancy (yes/no) | −0.16 | 0.24 | 0.507 | 0.85 | 0.54 | 1.36 | |
| BMI beginning of pregnancy (kg/m2) | 0.08 | 0.02 | <0.001 | 1.08 | 1.04 | 1.13 | |
| DBP (mmHg) | −0.02 | 0.02 | 0.230 | 0.98 | 0.95 | 1.01 | |
| Ethnicity (Caucasian, other) | 0.40 | 0.51 | 0.430 | 1.49 | 0.55 | 4.01 | |
| Sex of the child (male/female) | −0.65 | 0.23 | 0.005 | 0.52 | 0.33 | 0.83 | |
| Placental methylation | 0.72 | 0.22 | 0.001 | 2.06 | 1.35 | 3.16 | |
Discussion
The current study demonstrated that being born LGA is associated with a significantly higher level of global placental DNA methylation compared to AGA or SGA births. An ANCOVA model furthermore demonstrated that this association is independent of known confounders of placental DNA methylation (p < 0.001; Partial η2 = 0.013; 95% C.I. = 0.08–0.28). To substantiate the finding of an association between LGA and higher global placental DNA methylation, global placental DNA methylation was ranked into three groups comprising of low, moderate and high DNA methylation. In these groups, the frequency of LGA births was compared. These analyses demonstrated a significantly higher frequency of LGA births in the group of mothers with the highest global placental DNA methylation levels. Furthermore, by comparing descriptive statistics of the three methylation groups, potential confounders of the association between higher global placental DNA methylation and LGA births were identified. Diastolic blood pressure, ethnicity, the prevalence of smoking before pregnancy, and the prevalence of diabetes were significantly different among the three methylation groups. Calculating a multiple logistic regression model adjusted for the identified confounders and other well-established confounders of birth weight for gestational age plus using placental methylation as a continuous variable backed results of the univariate analysis, highlighting an independent positive association between global placental DNA methylation and LGA births (p = 0.001; Exp(B) = 2.06; 95% C.I. = 1.35–3.16).
The average global placental methylation assessed by this study was 2.99 ± 0.47% in AGA newborns. This finding is in agreement with previous studies, demonstrating global hypomethylation in comparison to other tissues in the human body51. However, different to previous studies which used HPLC to quantify global placental DNA methylation, this study employed the current gold standard of global DNA methylation quantification, LC-MS/MS, and is by far the largest study of its kind.
In a previous study we observed an association between gestational diabetes mellitus and higher levels of global placental DNA methylation in a similar cohort as the one analysed in this study47. As to be expected, ranking global placental DNA methylation into 3 groups demonstrated a higher frequency of diabetes during pregnancy (composite variable encompassing mothers with overt diabetes before and during pregnancy) in the highest global placental DNA methylation tertile. As diabetes during pregnancy is strongly associated with an increased risk for LGA births and, as previously shown, is associated with increased global placental DNA methylation, any multivariable analysis had to be adjusted for maternal diabetes. However, adjusting for diabetes during pregnancy in the multivariable logistic regression model did not affect the association between increased levels of global placental DNA methylation and a higher frequency for LGA births. Furthermore, an evaluation of the study cohort excluding any mothers with diabetes (Supplemental Data) did not weaken but actually slightly strengthen the association between global placental DNA methylation and LGA births. This suggests that the association between gestational diabetes and global placental DNA methylation is an independent phenomenon and does not explain the observed association between global placental DNA methylation and LGA birth.
Several studies have been conducted to investigate the impact of global placental DNA methylation on birth weight. Most of these studies measured LINE-1 methylation as a surrogate for global DNA methylation, were performed in rather small scale settings, and were more focused on intrauterine growth restriction (IUGR) or SGA birth outcomes30–33. Bourque et al., using 35 placenta samples, showed no difference in LINE-1 methylation comparing intrauterine growth restricted newborns to newborns with normal birth weight30. Tzschoppe et al., measuring LINE-1 methylation in 55 placenta samples showed no difference in placental DNA methylation between IUGR, SGA and AGA newborns32. Similarly, Mukhopadhyay et al. did not observe significant associations between placental LINE-1 methylation and birth weight analyzing 109 placenta samples of AGA and SGA births31. Different to just mentioned studies, Xiao et al., using 119 placenta samples, revealed a positive correlation between placental LINE-1 methylation and birth weight standard deviation scores33. Michels et al. who investigated LINE-1 methylation in 319 placenta samples observed a higher degree of placental methylation in the low birth weight group (<2500 g) compared to the normal birth weight group (2500–4000 g)52. Contrary to these results, we did not observe any significant differences in global placental DNA methylation of SGA newborns. Similar to findings of the current study, Wilhelm-Benartzi et al., analyzing DNA methylation of repetitive elements in 380 placenta samples, showed that placental methylation of the Alu element AluYB8 was significantly higher in LGA offspring53. Furthermore, both LINE-1 and Alu methylation levels were significantly positively associated with birth weight percentiles53. Overall, the rather heterogeneous supporting and opposing findings of previous studies can only be carefully used for interpreting our current findings. Major difference of the current study is the usage of a methodology that robustly quantifies global DNA methylation, in contrast to the usage of surrogate parameters of global DNA methylation in previous studies30,32,33,52,53. Furthermore, chosen sample size appears to be an important factor, given the differences in results comparing small scale30–32 to relatively larger scale studies52,53.
The potential biological effect of increased global placental DNA methylation in general and even more so in regards to affecting growth patterns of the offspring remains very poorly understood. The placenta does share certain phenotypic features usually found in solid neoplasms, such as cell invasion, vascular remodelling and suppression of the immune system54. Moreover, the placenta, similar to many neoplasms, is characterized by global DNA hypomethylation in comparison to somatic tissues55. Interestingly, next to global hypomethylation, neoplastic tumors demonstrate local hypermethylation, which is often found in tumor suppressor genes56,57. Hypermethylation of tumor suppressor genes was also demonstrated for the human placenta58. Moreover, as outlined above, increases in methylation of Alu elements, was shown to be positively correlated with fetal growth53. Higher methylation in these evolutionary younger transposons, was found to be relevant for placental function59. Furthermore, it was demonstrated in a study in rats that the administration of the DNA methylation inhibitor 5-Azacitidine lead to significantly smaller placentas, yet again highlighting a connection between the degree of placental DNA methylation and placental growth. Placental weight, which serves as a surrogate parameter of the surface area for materno-fetal nutrient transport, is a very important determinant for fetal growth and offspring birth weight60,61. Hypothetically, above average hypermethylation of growth related regions could induce larger than average placental growth which could translate into an increased risk for LGA births. Furthermore, above average increases in DNA methylation of diverse growth related regions in the placental DNA could result in a slight net increase in global placental DNA methylation that differentiates LGA from AGA births. However, to address these issues future adequately designed studies are necessary before drawing any causal conclusions.
Findings of the current study can be interpreted by findings of studies investigating other aspects of global placental DNA methylation. Nomura et al. showed that obese women display a higher global placental DNA methylation compared to lean women62. It is well established that obesity is a strong maternal risk factor for LGA offspring6. In the current study we did not observe any association between pre-pregnancy BMI and global placental DNA methylation. However, different from the study of Nomura et al. in which obesity was defined as a BMI > 30 kg/m2 and affected 30% of the participating mothers, mean pre-pregnancy BMI in the current study was 23.2 ± 4.6 kg/m2. It is possible that these differences in the makeup of the investigated cohorts accounted for the conflicting observation. Although increases in BMI are associated with dyslipidemia63, studies demonstrated BMI independent associations between the maternal blood lipid profile and LGA births64–66. Global DNA methylation has been reported to be associated with lipid metabolism and plasma glucose concentration67. It was demonstrated in a rat model that maternal dyslipidemia, induced by feeding a high fat diet, increases global placental DNA methylation68. Interestingly, an intervention with dietary n-3 fatty acids reversed these alterations68. Current literature suggests that the link between dietary lipids and DNA methylation might be due to their involvement in one carbon metabolism68,69. Thus, the maternal genetic setup or consumed diet70, key factors that affect blood lipid profiles, might also impact on placental lipid metabolism and DNA methylation during pregnancy. Effects of changes in the placental DNA methylation status on placental lipid metabolism or, vice versa, effects of an altered placental lipid metabolism on the placental DNA methylation status theoretically could be one factor for the positive association between DNA methylation and LGA offspring and warrant further studying.
Study strengths and limitations
One limitation of this and the majority of previous studies is that no cell type separation prior to DNA methylation analyses was performed. The placenta is composed of several different cell types ranging from fibroblast, trophoblasts to mesenchymal cells. Epigenetic patterns are cell specific and different cell types in the placenta are known to have a different DNA methylation profile71. Thus, it is not possible to rule out that the obtained results were influenced by cell type heterogeneity in between samples. However, since this study used more than 1000 placenta samples, isolation and analysis of specific placental cells was not feasible. There are studies that compared the effect of different placental sampling areas and the isolation of specific placental cell types on global placental DNA methylation72,73. Non et al. showed that sampling from different locations in the human placenta still resulted in strongly correlated degrees of LINE-1 methylation73. Schroeder et al. compared global DNA methylation, measured by MethylC-Seq in isolated rhesus trophoblasts to whole rhesus placenta methylation by MethylC-Seq and demonstrated a strong correlation between the two72. However, studies that focus on elucidating the contribution of individual cell types to whole placental DNA methylation are still lacking. This is especially problematic for the analysis of array data, as until now accounting for mixed cell type composition, an important consideration in heterogeneous tissues such as the placenta, is still not feasible74. Thus, future studies that focus on cell type specific differences in placental DNA methylation and their contribution to whole placenta DNA methylation are urgently needed.
Next to the mentioned limitations, this study has several strengths. To the best of our knowledge, it is by far the largest study of its type, investigating global placental DNA methylation in over a thousand placenta samples. More so, the current gold standard for the quantification of global DNA methylation, LC-MS/MS, was used75.
Conclusions
The current study demonstrated a significant positive association between the degree of global placental DNA methylation and being born LGA, which was independent of maternal diabetes during pregnancy, a very important risk factor for LGA births. Taken together, evidence in literature is existing that supports the observed positive association between global placental DNA methylation and being born LGA. So far the underlying mechanisms remain elusive. Hypothetically, hypermethylation of growth related DNA regions, such as tumor suppressor genes and certain transposable elements could affect placental size and function which could impact on fetal growth. Above average increases of DNA methylation in growth related DNA regions could result in pathologically increased growth. Furthermore, this might be reflected by significantly elevated levels of global placental DNA methylation. Findings of previous studies suggest a possible connection between anthropometric measurements, lipid metabolism, one-carbon metabolism, and DNA methylation. Taken together, it remains largely unknown how changes in global placental DNA methylation can affect fetal growth. Moreover, the cause of altered global placental DNA methylation levels in LGA births is still highly elusive. Thus, future studies are needed to be able to draw any more causal conclusions.
Supplementary information
Acknowledgements
We acknowledge the cooperation of the participating mother/child pairs. The study was supported by the Deutsche Forschungsgemeinschaft to Dr. B. Hocher
Author contributions
Dwi Putra, Reichetzeder, Kleuser, Hasan: laboratory measurements. Dwi Putra, Reichetzeder, Hasan, Hocher: writing of the manuscript. Dwi Putra, Reichetzeder, Hasan, Chu, Slowinski, Krämer, Kleuser: data collection. Slowinski, Dwi Putra, Reichetzeder, Hocher: statistical analysis. Hocher: design of the study.
Data availability
Data will be made available upon request to the corresponding author
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: S. E. Dwi Putra and C. Reichetzeder
Contributor Information
C. Reichetzeder, Email: reichetz@uni-potsdam.de
B. Hocher, Email: berthold.hocher@medma.uni-heidelberg.de
Supplementary information
is available for this paper at 10.1038/s41598-020-57725-0.
References
- 1.Morisaki N, Esplin MS, Varner MW, Henry E, Oken E. Declines in birth weight and fetal growth independent of gestational length. Obstet. Gynecol. 2013;121:51–58. doi: 10.1097/AOG.0b013e318278d014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mikolajczyk RT, et al. A global reference for fetal-weight and birthweight percentiles. Lancet Lond. Engl. 2011;377:1855–1861. doi: 10.1016/S0140-6736(11)60364-4. [DOI] [PubMed] [Google Scholar]
- 3.Ding G, et al. Application of a global reference for fetal-weight and birthweight percentiles in predicting infant mortality. BJOG Int. J. Obstet. Gynaecol. 2013;120:1613–1621. doi: 10.1111/1471-0528.12381. [DOI] [PubMed] [Google Scholar]
- 4.Chavkin U, Wainstock T, Sheiner E, Sergienko R, Walfisch A. Perinatal outcome of pregnancies complicated with extreme birth weights at term. J. Matern.-Fetal Neonatal Med. Off. J. Eur. Assoc. Perinat. Med. Fed. Asia Ocean. Perinat. Soc. Int. Soc. Perinat. Obstet. 2019;32:198–202. doi: 10.1080/14767058.2017.1376048. [DOI] [PubMed] [Google Scholar]
- 5.Chauhan SP, et al. Neonatal Morbidity of Small- and Large-for-Gestational-Age Neonates Born at Term in Uncomplicated Pregnancies. Obstet. Gynecol. 2017;130:511–519. doi: 10.1097/AOG.0000000000002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiavaroli V, Derraik JGB, Hofman PL, Cutfield WS. Born Large for Gestational Age: Bigger Is Not Always Better. J. Pediatr. 2016;170:307–311. doi: 10.1016/j.jpeds.2015.11.043. [DOI] [PubMed] [Google Scholar]
- 7.Chiavaroli V, et al. Progression of cardio-metabolic risk factors in subjects born small and large for gestational age. PloS One. 2014;9:e104278. doi: 10.1371/journal.pone.0104278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffman DJ, Reynolds RM, Hardy DB. Developmental origins of health and disease: current knowledge and potential mechanisms. Nutr. Rev. 2017;75:951–970. doi: 10.1093/nutrit/nux053. [DOI] [PubMed] [Google Scholar]
- 9.Padmanabhan V, Cardoso RC, Puttabyatappa M. Developmental Programming, a Pathway to Disease. Endocrinology. 2016;157:1328–1340. doi: 10.1210/en.2016-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanson MA, Gluckman PD. Early Developmental Conditioning of Later Health and Disease: Physiology or Pathophysiology? Physiol. Rev. 2014;94:1027–1076. doi: 10.1152/physrev.00029.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henriksen T, Clausen T. The fetal origins hypothesis: placental insufficiency and inheritance versus maternal malnutrition in well-nourished populations. Acta Obstet. Gynecol. Scand. 2002;81:112–114. doi: 10.1034/j.1600-0412.2002.810204.x. [DOI] [PubMed] [Google Scholar]
- 12.Hong YH, Chung S. Small for gestational age and obesity related comorbidities. Ann. Pediatr. Endocrinol. Metab. 2018;23:4–8. doi: 10.6065/apem.2018.23.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ratnasingham A, Eiby YA, Dekker Nitert M, Donovan T, Lingwood BE. Review: Is rapid fat accumulation in early life associated with adverse later health outcomes? Placenta. 2017;54:125–130. doi: 10.1016/j.placenta.2017.01.101. [DOI] [PubMed] [Google Scholar]
- 14.Calkins K, Devaskar SU. Fetal origins of adult disease. Curr. Probl. Pediatr. Adolesc. Health Care. 2011;41:158–176. doi: 10.1016/j.cppeds.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harder T, Rodekamp E, Schellong K, Dudenhausen JW, Plagemann A. Birth weight and subsequent risk of type 2 diabetes: a meta-analysis. Am. J. Epidemiol. 2007;165:849–857. doi: 10.1093/aje/kwk071. [DOI] [PubMed] [Google Scholar]
- 16.Huxley R, Neil A, Collins R. Unravelling the fetal origins hypothesis: is there really an inverse association between birthweight and subsequent blood pressure? Lancet Lond. Engl. 2002;360:659–665. doi: 10.1016/S0140-6736(02)09834-3. [DOI] [PubMed] [Google Scholar]
- 17.Dessì A, Puddu M, Ottonello G, Fanos V. Metabolomics and fetal-neonatal nutrition: between ‘not enough’ and ‘too much’. Mol. Basel Switz. 2013;18:11724–11732. doi: 10.3390/molecules181011724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sookoian S, Gianotti TF, Burgueño AL, Pirola CJ. Fetal metabolic programming and epigenetic modifications: a systems biology approach. Pediatr. Res. 2013;73:531–542. doi: 10.1038/pr.2013.2. [DOI] [PubMed] [Google Scholar]
- 19.Roland MCP, et al. Fetal growth versus birthweight: the role of placenta versus other determinants. PloS One. 2012;7:e39324. doi: 10.1371/journal.pone.0039324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burris HH, et al. Association between birth weight and DNA methylation of IGF2, glucocorticoid receptor and repetitive elements LINE-1 and Alu. Epigenomics. 2013;5:271–281. doi: 10.2217/epi.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filiberto AC, et al. Birthweight is associated with DNA promoter methylation of the glucocorticoid receptor in human placenta. Epigenetics. 2011;6:566–572. doi: 10.4161/epi.6.5.15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maccani JZJ, Koestler DC, Houseman EA, Marsit CJ, Kelsey KT. Placental DNA methylation alterations associated with maternal tobacco smoking at the RUNX3 gene are also associated with gestational age. Epigenomics. 2013;5:619–630. doi: 10.2217/epi.13.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubois L, et al. Genetic and environmental contributions to weight, height, and BMI from birth to 19 years of age: an international study of over 12,000 twin pairs. PloS One. 2012;7:e30153. doi: 10.1371/journal.pone.0030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, et al. Genome wide screening of candidate genes for improving piglet birth weight using high and low estimated breeding value populations. Int. J. Biol. Sci. 2014;10:236–244. doi: 10.7150/ijbs.7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kellis M, et al. Defining functional DNA elements in the human genome. Proc. Natl. Acad. Sci. USA. 2014;111:6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng H, Gifford DK. Predicting the impact of non-coding variants on DNA methylation. Nucleic Acids Res. 2017;45:e99. doi: 10.1093/nar/gkx177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barroso I, McCarthy MI. The Genetic Basis of Metabolic Disease. Cell. 2019;177:146–161. doi: 10.1016/j.cell.2019.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joyce BT, et al. Prospective changes in global DNA methylation and cancer incidence and mortality. Br. J. Cancer. 2016;115:465–472. doi: 10.1038/bjc.2016.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung M, Pfeifer GP. Aging and DNA methylation. BMC Biol. 2015;13:7. doi: 10.1186/s12915-015-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bourque DK, Avila L, Peñaherrera M, von Dadelszen P, Robinson WP. Decreased placental methylation at the H19/IGF2 imprinting control region is associated with normotensive intrauterine growth restriction but not preeclampsia. Placenta. 2010;31:197–202. doi: 10.1016/j.placenta.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Mukhopadhyay A, et al. Placental expression of DNA methyltransferase 1 (DNMT1): Gender-specific relation with human placental growth. Placenta. 2016;48:119–125. doi: 10.1016/j.placenta.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 32.Tzschoppe A, et al. DNA methylation of the p66Shc promoter is decreased in placental tissue from women delivering intrauterine growth restricted neonates. Prenat. Diagn. 2013;33:484–491. doi: 10.1002/pd.4096. [DOI] [PubMed] [Google Scholar]
- 33.Xiao X, et al. Fetal growth restriction and methylation of growth-related genes in the placenta. Epigenomics. 2016;8:33–42. doi: 10.2217/epi.15.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boers R, et al. Genome-wide DNA methylation profiling using the methylation-dependent restriction enzyme LpnPI. Genome Res. 2018;28:88–99. doi: 10.1101/gr.222885.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerring ZF, McRae AF, Montgomery GW, Nyholt DR. Genome-wide DNA methylation profiling in whole blood reveals epigenetic signatures associated with migraine. BMC Genomics. 2018;19:69. doi: 10.1186/s12864-018-4450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yong W-S, Hsu F-M, Chen P-Y. Profiling genome-wide DNA methylation. Epigenetics Chromatin. 2016;9:26. doi: 10.1186/s13072-016-0075-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurdyukov, S. & Bullock, M. DNA Methylation Analysis: Choosing the Right Method. Biology5 (2016). [DOI] [PMC free article] [PubMed]
- 38.Carmona JJ, et al. Empirical comparison of reduced representation bisulfite sequencing and Infinium BeadChip reproducibility and coverage of DNA methylation in humans. Npj Genomic Med. 2017;2:1–10. doi: 10.1038/s41525-017-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 2011;13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kleinwechter H, et al. Gestational Diabetes Mellitus (GDM) Diagnosis, Therapy and Follow-Up Care. Exp. Clin. Endocrinol. Diabetes. 2014;122:395–405. doi: 10.1055/s-0034-1366412. [DOI] [PubMed] [Google Scholar]
- 41.Dwi Putra SE, Neuber C, Reichetzeder C, Hocher B, Kleuser B. Analysis of genomic DNA methylation levels in human placenta using liquid chromatography-electrospray ionization tandem mass spectrometry. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2014;33:945–952. doi: 10.1159/000358666. [DOI] [PubMed] [Google Scholar]
- 42.Dick KJ, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet Lond. Engl. 2014;383:1990–1998. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 43.Dwi Putra SE, et al. DNA methylation of the glucocorticoid receptor gene promoter in the placenta is associated with blood pressure regulation in human pregnancy. J. Hypertens. 2017;35:2276–2286. doi: 10.1097/HJH.0000000000001450. [DOI] [PubMed] [Google Scholar]
- 44.Fuke C, et al. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann. Hum. Genet. 2004;68:196–204. doi: 10.1046/j.1529-8817.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- 45.Kulkarni A, Chavan-Gautam P, Mehendale S, Yadav H, Joshi S. Global DNA methylation patterns in placenta and its association with maternal hypertension in pre-eclampsia. DNA Cell Biol. 2011;30:79–84. doi: 10.1089/dna.2010.1084. [DOI] [PubMed] [Google Scholar]
- 46.Maccani JZ, Maccani MA. Altered placental DNA methylation patterns associated with maternal smoking: current perspectives. Adv. Genomics Genet. 2015;2015:205–214. doi: 10.2147/AGG.S61518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reichetzeder C, et al. Increased global placental DNA methylation levels are associated with gestational diabetes. Clin. Epigenetics. 2016;8:82. doi: 10.1186/s13148-016-0247-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cogswell ME, Yip R. The influence of fetal and maternal factors on the distribution of birthweight. Semin. Perinatol. 1995;19:222–240. doi: 10.1016/S0146-0005(05)80028-X. [DOI] [PubMed] [Google Scholar]
- 49.Steer PJ, Little MP, Kold-Jensen T, Chapple J, Elliott P. Maternal blood pressure in pregnancy, birth weight, and perinatal mortality in first births: prospective study. BMJ. 2004;329:1312. doi: 10.1136/bmj.38258.566262.7C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hossain MG, Saw A, Alam R, Ohtsuki F, Kamarul T. Multiple regression analysis of anthropometric measurements influencing the cephalic index of male Japanese university students. Singapore Med. J. 2013;54:516–520. doi: 10.11622/smedj.2013175. [DOI] [PubMed] [Google Scholar]
- 51.Chatterjee A, et al. Placental Hypomethylation Is More Pronounced in Genomic Loci Devoid of Retroelements. G3 Bethesda Md. 2016;6:1911–1921. doi: 10.1534/g3.116.030379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michels KB, Harris HR, Barault L. Birthweight, maternal weight trajectories and global DNA methylation of LINE-1 repetitive elements. PloS One. 2011;6:e25254. doi: 10.1371/journal.pone.0025254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilhelm-Benartzi CS, et al. In utero exposures, infant growth, and DNA methylation of repetitive elements and developmentally related genes in human placenta. Environ. Health Perspect. 2012;120:296–302. doi: 10.1289/ehp.1103927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lorincz MC, Schübeler D. Evidence for Converging DNA Methylation Pathways in Placenta and Cancer. Dev. Cell. 2017;43:257–258. doi: 10.1016/j.devcel.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 55.Ehrlich M, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues or cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ehrlich M. DNA hypermethylation in disease: mechanisms and clinical relevance. Epigenetics. 2019;14:1141–1163. doi: 10.1080/15592294.2019.1638701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nordor AV, et al. The early pregnancy placenta foreshadows DNA methylation alterations of solid tumors. Epigenetics. 2017;12:793–803. doi: 10.1080/15592294.2017.1342912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chatterjee A, et al. Placental Hypomethylation Is More Pronounced in Genomic Loci Devoid of Retroelements. G3 GenesGenomesGenetics. 2016;6:1911–1921. doi: 10.1534/g3.116.030379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fowden AL, Ward JW, Wooding FPB, Forhead AJ, Constancia M. Programming placental nutrient transport capacity. J. Physiol. 2006;572:5–15. doi: 10.1113/jphysiol.2005.104141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brett KE, Ferraro ZM, Yockell-Lelievre J, Gruslin A, Adamo KB. Maternal–Fetal Nutrient Transport in Pregnancy Pathologies: The Role of the Placenta. Int. J. Mol. Sci. 2014;15:16153–16185. doi: 10.3390/ijms150916153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nomura Y, et al. Global methylation in the placenta and umbilical cord blood from pregnancies with maternal gestational diabetes, preeclampsia, and obesity. Reprod. Sci. Thousand Oaks Calif. 2014;21:131–137. doi: 10.1177/1933719113492206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Denke MA, Sempos CT, Grundy SM. Excess Body Weight: An Under-recognized Contributor to Dyslipidemia in White American Women. Arch. Intern. Med. 1994;154:401–410. doi: 10.1001/archinte.1994.00420040061010. [DOI] [PubMed] [Google Scholar]
- 64.Hou R-L, et al. Effect of maternal lipid profile, C-peptide, insulin, and HBA1c levels during late pregnancy on large-for-gestational age newborns. World J. Pediatr. WJP. 2014;10:175–181. doi: 10.1007/s12519-014-0488-7. [DOI] [PubMed] [Google Scholar]
- 65.Jin W-Y, et al. Associations between maternal lipid profile and pregnancy complications and perinatal outcomes: a population-based study from China. BMC Pregnancy Childbirth. 2016;16:60. doi: 10.1186/s12884-016-0852-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liang N, et al. The high maternal TG level at early trimester was associated with the increased risk of LGA newborn in non-obesity pregnant women. Lipids Health Dis. 2018;17:294. doi: 10.1186/s12944-018-0936-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delgado-Cruzata L, et al. Dietary modifications, weight loss, and changes in metabolic markers affect global DNA methylation in Hispanic, African American, and Afro-Caribbean breast cancer survivors. J. Nutr. 2015;145:783–790. doi: 10.3945/jn.114.202853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ramaiyan B, Talahalli RR. Dietary Unsaturated Fatty Acids Modulate Maternal Dyslipidemia-Induced DNA Methylation and Histone Acetylation in Placenta and Fetal Liver in Rats. Lipids. 2018;53:581–588. doi: 10.1002/lipd.12074. [DOI] [PubMed] [Google Scholar]
- 69.Voisin S, et al. Dietary fat quality impacts genome-wide DNA methylation patterns in a cross-sectional study of Greek preadolescents. Eur. J. Hum. Genet. EJHG. 2015;23:654–662. doi: 10.1038/ejhg.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pollin TI, Quartuccio M. What We Know About Diet, Genes, and Dyslipidemia: Is There Potential for Translation? Curr. Nutr. Rep. 2013;2:236–242. doi: 10.1007/s13668-013-0065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grigoriu A, et al. Cell specific patterns of methylation in the human placenta. Epigenetics. 2011;6:368–379. doi: 10.4161/epi.6.3.14196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schroeder DI, et al. Early Developmental and Evolutionary Origins of Gene Body DNA Methylation Patterns in Mammalian Placentas. PLoS Genet. 2015;11:e1005442. doi: 10.1371/journal.pgen.1005442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Non AL, et al. DNA methylation of stress-related genes and LINE-1 repetitive elements across the healthy human placenta. Placenta. 2012;33:183–187. doi: 10.1016/j.placenta.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deyssenroth Maya A., Marsit Carmen J., Chen Jia, Lambertini Luca. In-depth characterization of the placental imprintome reveals novel differentially methylated regions across birth weight categories. Epigenetics. 2019;15(1-2):47–60. doi: 10.1080/15592294.2019.1647945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quinlivan EP, Gregory JF. DNA methylation determination by liquid chromatography-tandem mass spectrometry using novel biosynthetic [U-15N]deoxycytidine and [U-15N]methyldeoxycytidine internal standards. Nucleic Acids Res. 2008;36:e119. doi: 10.1093/nar/gkn534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available upon request to the corresponding author


