Abstract
Shewanella oneidensis, a metal reducer and facultative anaerobe, expresses a large number of c-type cytochromes, many of which function as anaerobic reductases. All of these proteins contain the typical heme-binding motif CXXCH and require the Ccm proteins for maturation. Two c-type cytochrome reductases also possess atypical heme-binding sites, the NrfA nitrite reductase (CXXCK) and the SirA sulfite reductase (CX12NKGCH). S. oneidensis MR-1 encodes two cytochrome c synthetases (CcmF and SirE) and two apocytochrome c chaperones (CcmI and SirG). SirE located in the sir gene cluster is required for the maturation of SirA, but not NrfA. Here we show that maturation of SirA requires the combined function of the two apocytochrome c chaperones CcmI and SirG. Loss of either protein resulted in decreased sulfite reductase. Furthermore, SirA was not detected in a mutant that lacked both chaperones, perhaps due to misfolding or instability. These results suggest that CcmI interacts with SirEFG during SirA maturation, and with CcmF during maturation of NrfA. Additionally, we show that CRP regulates expression of sirA via the newly identified transcriptional regulatory protein, SirR.
Subject terms: Bacteriology, Element cycles, Water microbiology
Introduction
Shewanella species are facultative anaerobes that are abundant in freshwater and marine environments and are best known for their ability to use metals as electron acceptors for anaerobic respiration (see Fredrickson et al. for review1). S. oneidensis MR-1 is one of the best studied members of the Shewanella genus and the most diverse with regard to the electron acceptors it uses for respiration. These include O2, fumarate, NO3−, NO2−, trimethylamine N-oxide (TMAO), dimethylsulfoxide (DMSO), metal oxides, thiosulfate, and sulfite2–10. In addition, S. oneidensis can reduce radionuclides and toxic metals such as Tc, U, and Cr11–15. The ability of S. oneidensis to use this wide range of electron acceptors is partly attributed to the large number of c-type cytochromes encoded by its genome16,17.
Dissimilatory sulfite reduction is a key intermediate step in sulfate reduction and has been extensively studied in many sulfate reducing prokaryotes such as Desulfovibrio vulgaris and Archeoglobus fulgidus (for review see18) and in Salmonella typhimurium. Desulfovibrio, Archeoglobus and Salmonella species all utilize a siroheme-containing enzyme to reduce sulfite to sulfide19–21. In contrast, the sulfite reductase of Shewanella oneidensis MR-1 lacks siroheme. In S. oneidensis, the terminal sulfite reductase consists of SirA, an octaheme c cytochrome, SirC, an Fe-S protein, and SirD, which is predicted to be a membrane-bound quinol oxidase, based on similarity to the quinol oxidase component of the formate-dependent nitrite reductases10. A similar system involved in sulfite reduction has been identified in Wolinella succinogenes22.
c-type cytochromes are characterized by the covalent attachment of heme b vinyl groups to the two cysteines present in the typical CXXCH heme-binding motif23. In α and γ Proteobacteria, the System I cytochrome maturation proteins, CcmABCDEFGH, attach a heme moiety to the CXXCH motif (see23–25 for review). The final step in heme attachment and enzyme maturation requires transfer and covalent linkage of heme by the heme lyase complex. In Escherichia coli, CcmFH form the complex that functions as a heme lyase, whereas in other Gram-negative bacteria such as Rhodobacter capsulatus, the heme lyase complex consists of CcmFHI26. Similar to other Gram-negative bacteria, maturation of c-type cytochromes in S. oneidensis requires proteins encoded by genes in the ccmABCDE and ccmFGH operons. Mutations in the ccm genes lead to complete loss of mature c-type cytochromes27,28. SO_0265 was identified as ccmI, and was initially determined to have a nonessential role in cytochrome c maturation in S. oneidensis28, but subsequently found to have a role in maturation of canonical heme binding motifs and to be required for maturation of the nitrite reductase, NrfA29. However, it remains unknown if or what role CcmI plays in maturation of the sulfite reductase.
In addition to heme attachment to the typical CXXCH motif, specialized heme lyase systems can attach heme b to atypical sites, such as CXXCK and CX15CH30,31. In E. coli, ligation of heme to the CXXCK site of NrfA requires the heme lyase NrfEFG30. In Wolinella succinogenes, two distinct heme lyases, NrfI and CcsA1, are required for heme attachment to the CXXCK binding sites of the nitrite reductase and the CX15CH binding site of the sulfite reductase, respectively32,33. Two atypical heme-binding sites have been identified in S. oneidensis c-type cytochromes. The CXXCK motif is found in the periplasmic nitrite reductase NrfA34, and the CX12NKGCH motif is found in SirA10. As indicated above, the heme lyase CcmI is essential for maturation of the nitrite reductase, NrfA, of Shewanella29. Less is known about the complete maturation of the Shewanella sulfite reductase, SirA. SirA is an octaheme c-type cytochrome that is predicted to be the catalytic subunit of the sulfite reductase SirACD, and appears to represent a siroheme-independent class of sulfite reductases10.
In this paper we describe the role of CcmI, SirG and SirEFG in the maturation of the sulfite reductase. Contrary to what has been shown for other c-cytochromes of S. oneidensis, our results indicated that both the Ccm and Sir systems are essential for proper sulfite reductase maturation. In addition, we identify a novel transcriptional regulator, SirR, required for expression of the sulfite reductase, SirA. It has been shown that sulfite reduction is regulated by the cyclic AMP receptor protein (CRP)35, yet it remains unknown if this occurs via direct transcriptional regulation of the sir operon. Here we show that CRP controls expression of SirR, which in turn regulates expression of the sulfite reductase operon.
Materials and Methods
Bacterial strains and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1. Lysogeny Broth (LB) medium was routinely used for aerobic growth of S. oneidensis MR-1 and E. coli strains. Anaerobic cultures of S. oneidensis MR-1 strains were grown in minimal medium (63 mM HEPES pH 7.4, Trace Metals & Minerals) supplemented with 50 mM lactate and 0.02% casamino acids36. Electron acceptors were used at 10 mM unless noted otherwise. Growth and reduction of sulfite (10 mM), thiosulfate (5 mM), and tetrathionate (3 mM), was performed anaerobically in a Coy anaerobic chamber using biometer flasks with 10 ml of 40% KOH in the sidearm to trap H2S10. Chloramphenicol (20 µg/ml), kanamycin (25 µg/ml), and gentamycin (25 µg/ml), were added as appropriate.
Table 1.
List of strains and plasmids used in this study.
| Strain | Description | Source |
|---|---|---|
| MR-1 | Lake Oneida S. oneidensis isolate | Venkateswaran et al.60 |
| SR1207 | MR-1 ∆SO_0479 (sirA) | Shirodkar et al.10 |
| ∆nrfA | MR-1 ∆nrfA | This Work |
| SR1518 | ∆sirA containing pTC1 | This Work |
| SR1550 | MR-1 ∆SO_0265 (ccmI) | This Work |
| SR1576 | ∆ccmI containing pTC2 | This Work |
| SR1566 | MR-1 ∆SO_0476 (sirH) | This Work |
| SR1570 | ∆sirH containing pTC3 | This Work |
| SR1542 | MR-1 ∆SO_0477-8 (sirEF) | This Work |
| SR1548 | ∆sirEF containing pTC4 | This Work |
| SR1593 | MR-1 ∆SO_0482 (sirG) | This Work |
| SR1592 | ∆sirG containing pTC5 | This Work |
| SR1594 | ∆ccmI ∆SO_0482 (sirG) | This Work |
| SR1595 | ∆ccmI∆sirG containing pTC2 | This Work |
| SR1596 | ∆ccmI∆sirG containing pTC5 | This Work |
| EC100D+ | E. coli EC100 derivative, pir+ | Epicenter Technologies |
| β2155 | pir::RP4, KmR | Dehio et al.38 |
| pER21 | R6K ori, GmR, sacB, lacZ α-fragment | Shirodkar et al.10 |
| pJBC1 | Cloning and sequencing vector, CmR | This Work |
| pTRG | Bacterial two-hybrid target plasmid | Agilent Technologies |
| pBT | Bacterial two-hybrid bait plasmid | Agilent Technologies |
| pTC1 | SO_0479N589C in pJBC1 | This Work |
| pTC2 | SO_0265 (ccmI) in pJBC1 | This Work |
| pTC3 | SO_0476 (sirH) in pJBC1 | This Work |
| pTC4 | SO_0477-8 (sirEF) in pJBC1 | This Work |
| pTC5 | SO_0482 (sirG) in pJBC1 | This Work |
| pBTH1 | SO_0479 (sirA) in pTRG | This Work |
| pBTH2 | SO_0479 (sirA) in pBT | This Work |
| pBTH3 | SO_0482 (sirG) in pTRG | This Work |
| pBTH4 | SO_0482 (sirG) in pBT | This Work |
| pBTH5 | SO_0265 (ccmI) in pTRG | This Work |
| pBTH6 | SO_0265 (ccmI) in pBT | This Work |
Generation and complementation of mutants
Chromosomal deletions of sirH, sirEF, sirG and ccmI were generated using the suicide vector pER2137. Approximately 1 kb DNA fragments that flank these genes were amplified by PCR and Phusion polymerase (New England BioLabs). The internal primers were engineered to include restriction enzyme sites and allow ligation of the amplified fragments (see Table 2 for primer sequences). The ligated fragments were cloned into the SmaI digested pER21, and the recombinant plasmids were transferred into S. oneidensis MR-1 cells by conjugation from E. coli β215538. Mutants were isolated following sucrose selection as described previously39 and chromosomal deletions were confirmed by PCR. To complement the mutants, DNA fragments that carry the respective genes were generated by PCR using Phusion polymerase (New England BioLabs). Amplified fragments were cloned into pJBC137 then transferred from E. coli β215538 to S. oneidensis MR-1 strains by conjugation.
Table 2.
List of primer used in this study.
| Primer | Sequence |
|---|---|
| Chromosomal Deletions Primers | |
| 265dUF | CTGTTCTGCGATGCTGAC |
| 265dUR | TACGAATTCCATCTACAATAGCGCTAAGTC |
| 265dDF | TACGAATTCCACTTTAGTTTGCATAAAAGCG |
| 265dDR | GTCCAGTTACGGAATGCAC |
| 476dUF | GATTCTTCCCAAGCAATTGC |
| 476dUR | TACGAATTCTCCCGTAAAGCACGGCG |
| 476dDF | TACGAATTCTATTCAAACTGATCGAGTCGC |
| 476dDR | ATCCCGCACCTCAAAACAAC |
| 477dUF | GGCGATAAAGTCAATTTCACG |
| 477dUR | GATCGGATCCGGATTAACCCATGAAATCTTAC |
| 478dDF | GATCGGATCCATTAGTTAACGATTTAC |
| 478dDR | CGTCTTAGAGTGACGTGAACGTC |
| 482dUF | CTGGCGCATGTCAGGAAG |
| 482dUR | GACTGGATCCGAGTGCGATGGTGAACCC |
| 482dDF | GACTGGATCCCGCGATGCCATCAATAACGC |
| 482dDR | GAGCGACTACTGCGACTAACG |
| Complementation Primers | |
| 265cF | TGCGCTAAGCCAAGACTTAG |
| 265dDR | GTCCAGTTACGGAATGCAC |
| 476cF | AAGCATATCACATATGATGAAATCTTACACAGCCAAGC |
| 476cR | CATTTACCGCCGTGCTTTAC |
| 477cF | GTAAGATTTCATGGGTTAATCC |
| 478cR | TCCTAATTTATCATTAGTT |
| 482cF | AAGCATATCACATATGGCATGGCAAGTTTAGGGTTC |
| 482cR | GGCTAGCGACTTGATTAATATC |
| Site Directed Mutagenesis Primers | |
| 479subF | CGGCTTCTGACCACGACGTAACTGAATGCAAAGGTTGTCATAGCCAGTTCCAATC |
| 479subR | GATTGGAACTGGCTATGACAACCTTTGCATTCAGTTACGTCGTGGTCAGAAGCCG |
| Bacterial Two-Hybrid Primers | |
| TRGCMIEF | GCATCAGAATTCCAACATTTAGGTGCCTTTGAAAATATAGGC |
| TRGCMIXR | GATCTTACTCGAGTTGTACTTGAGTATCCAGTACTAAGTTTGCGG |
| BT479BF | CCAGCGGGATCCGCTAAATCGGATGGTAAAGTG |
| BT479XR | CCGATCTCCTCGAGCATTTTAGCGTTGTAGCCATTACC |
| TRG482BF | GCGGCCGGATCCGGACGTTATAGCGATTGG |
| TRG482BR | CTGGCGCTCGAGATATCCACTTTCATTCAATTTTATTTG |
Site directed mutagenesis of SirA
The QuikChange II Site-Directed Mutagenesis Kit from Stratagene (Agilent Technologies, Inc., Santa Clara, CA) was used to generate a mutation in the atypical heme-binding site of SirA. The primers extended 26 bases upstream and downstream of the corresponding N589 codon in SirA (see Table 2 for primer sequences). The N589 codon, AAC, was changed to TGC resulting in an N589C substitution in the translated sequence. The mutagenized fragment was amplified by PCR using PfuUltra HF DNA polymerase and the base substitution was confirmed by DNA sequencing (Eurofins MWG Operon, Huntsville, AL). The mutagenized sirA and its native promoter were cloned into the S. oneidensis expression plasmid pJBC137. The resulting plasmid was transferred from E. coli β215538 into ∆sirA by conjugation.
Protein detection by western blot
Antibodies against SirA peptides (N′- CDGSWGAHGPRYTQKRLD and N′- CHGPQYEKWRRSRHSK) were generated and affinity purified by Biomatik corp. (Cambridge, Ontario). Strains of S. oneidensis were incubated anaerobically in basal medium supplemented with 50 mM lactate, 0.02% casamino acids and 10 mM sodium sulfite for 24 hours, unless otherwise indicated. Cells were pelleted by centrifugation and aliquots were used for determination of protein concentrations or resuspended directly in SDS loading buffer and boiled for 10 min40. Proteins were separated on 10% sodium dodecyl sulfate-polyacrylamide gels and transferred to polyvinylidene difluoride membrane (Thermo Scientific, Rockford, IL). The membranes were incubated with SirA antibodies and developed using Supersignal West Pico Chemiluminescent substrate (Thermo Scientific, Rockford, IL). Blots were imaged using a FOTO/Analyst Luminary FX Workstation (FOTODYNE Inc., Hartland, WI). Protein concentrations were determined using the Coomassie Plus Protein Assay kit (PIERCE, Rockford, IL).
Detection of sulfite and nitrite reductase activities in native polyacrylamide gels
S. oneidensis wild type and mutant strains were grown anaerobically in basal medium supplemented with 50 mM lactate, 0.02% casamino acids and 10 mM sodium sulfite or 0.5 mM potassium nitrite as electron acceptors. Cell extracts were prepared using B-PER lysis reagent (PIERCE, Rockford, IL). Protein concentrations were determined using the Coomassie Plus Protein Assay kit and 50 μg total protein was separated on 10% native polyacrylamide gels. Enzyme activities were assayed essentially as previously described41. Gels were transferred to a Coy anaerobic chamber (Coy Laboratory Products, Grass Lake, MI) and stained for 20 min with reduced methyl viologen [38.9 mM methyl viologen dichloride and 57.4 mM sodium hydrosulfite in 10 mM Tris pH 7]. 10 mM of Na2SO3 or KNO3 was added and the gels were incubated further until bands of clearing, indicating reduction of the electron acceptors, were observed. Gels were imaged using a Kodak DC290 digital imaging system (Eastman Kodak, Rochester, NY).
Sulfite reduction assays
Reduction of sulfite was performed anaerobically in a Coy anaerobic chamber using biometer flasks as described previously10. The flasks contained 100 ml of deoxygenated minimal medium supplemented with 50 mM lactate, 0.02% casamino acids and 10 mM sodium sulfite as the sole electron acceptor. Chloramphenicol (20 μg/ml) was added as appropriate. The side arm of the flasks contained 10 ml of 40% KOH to trap H2S42. Overnight cultures of S. oneidensis strains grown in LB were used as inocula. Cultures were sampled to measure H2S and SO3−2 every 12 to 24 hrs for up to 120 hrs. H2S was measured using the mixed diamine assay43 with modification. Briefly, 0.5 ml of the KOH trap was transferred to 25 ml of dH20 and 1 ml of mixed diamine reagent (20 g N, N-dimethyl-p-phenylenediamine sulfate, 30 g ferric chloride (FeCl3·6H20) in 500 ml of 50% hydrochloric acid) was added. The color was allowed to develop for 20 minutes and the absorbance was measured at 670 nm. Hydrogen sulfide concentrations were determined using sodium sulfide as a standard. Sulfite concentrations were determined using the fuchsin assay as described previously44 using sodium sulfite as the standard. Briefly, 100 μl of 1.18 mM fuchsin dye (dissolved in 2.25 N H2SO4) was added to 20 μl of sample mixed with 870 μl of water. Following 10 min incubation at room temperature, 10 µl of formalin was added, and the absorbance was measured at 570 nm.
Nitrite reduction assay
Reduction of nitrite was performed anaerobically in a Coy anaerobic chamber. Serum vials containing deoxygenated basal medium supplemented with 50 mM lactate, 0.02% casamino acids, 0.5 mM potassium nitrite, were inoculated with S. oneidensis cultures grown overnight in LB. Cultures were sampled every hour and nitrite concentrations were measured using N-(1-naphthyl)ethylenediamine dihydrochloride and sulfanilic acid. The mixture was incubated for 10 min, and the absorbance was measured at 540 nm.
β-galactosidase assays
The DNA fragments upstream of sirA and sirR were amplified by PCR and Phusion polymerase (New England Biolabs), digested with HindIII and BamHI, then cloned into pMC1045. Following transformation of E. coli β2155, the resulting plasmids were transferred into S. oneidensis wild type and mutant strains by conjugation. β galactosidase activity was determined using ONPG (9 mg of ONPG in 10 ml of 1 mM MgCl2 and 0.1 mM β-mercaptoethanol) as described previously46.
Quantitative reverse-transcription PCR
Wild-type or mutant cells were grown overnight under aerobic conditions in triplicate. Cultures were pelleted, and RNA was isolated using the TRIzol Plus RNA Purification System (ThermoFisher) according to manufacture instructions. Chromosomal DNA was removed by treatment with DNaseI, cleaned up and total RNA quantified on a Qubit 4 (ThermoFisher). cDNA was generated with the High-Capacity RNA-to-cDNA kit and Realtime-PCR was carried out on a CFX96 (BioRad) with the SsoAdvanced Universal SYBR Green Supermix (Biorad). Gene specific primers were synthesized by IDT Technologies, and primer specificity was confirmed by melt curve analysis followed by agarose gel electrophoresis, no template and no reverse transcriptase were performed as appropriate. Relative expression was determined by the delta-delta-Ct method from three biological replicates assayed in technical triplicates.
Bacterial two-hybrid
Genes SO_479, SO_482 and SO_265 were PCR amplified from S. oneidensis MR-1 chromosomal DNA with Phusion DNA polymerase (New England BioLabs, Ipswich, MA) and purified with the IBI Gel/PCR DNA fragment extraction kit (IBI Scientific, Dubuque, IA) (see Table 2 for primer sequences). The SO_479 (sirA) DNA fragment was digested with BamHI and XhoI and ligated into pTRG. Similarily, SO_482 (sirG) and SO_265 (ccmI) DNA fragments were digested with EcoRI and XhoI and then ligated pBT. The resultant plasmids were transformed into E. coli XL1-Blue MRF´ Kan and screened on LB-chloramphenicol (for pBT transformants) or LB-Tetracycline (for pTRG transformants). Colonies that contained the appropriate vectors were confirmed by PCR. The vector pairs were then co-transformed into the BacterioMatch II two-hybrid reporter strain. Transformed cells were plated on M9 non-selective and selective media that contained 3-amino-1,2,4-triazole (3AT) supplemented with chloramphenicol and tetracycline. Additional vectors and reporter strains were also generated with the fragments switched (i.e. sirA in pBT and 482 and ccmI in pTRG). To confirm interactions between the proteins, reporter strains were grown overnight in M9 + His broth and serially diluted (up to 10-7). 10 µl were spotted on M9, M9 + 3AT and M9 + 3AT agar supplemented with 12.5 µg streptomycin per mL and allowed to grow at 37 °C for up to 48hrs.
Results
CRP regulates sulfite reduction via the transcriptional regulator SirR
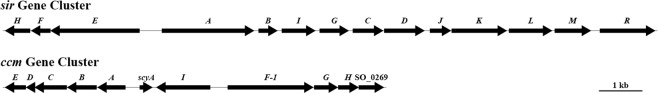
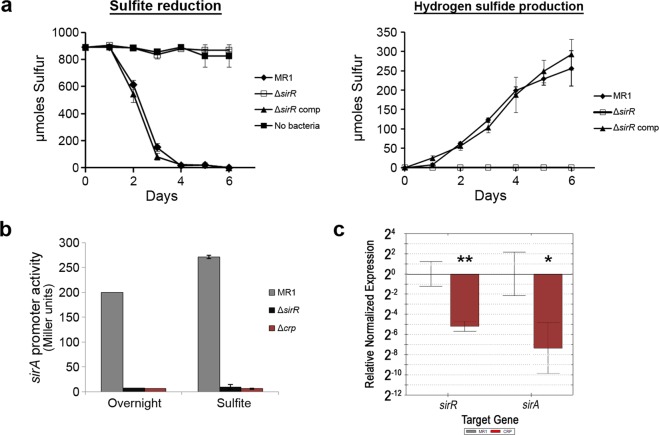
The genes involved in anaerobic sulfite reduction in S. oneidensis MR-1, sirAGCD, are located within a cluster of genes designated the sir locus (Fig. 1). SO_0490, which we designated sirR, lies just downstream of the sir locus and encodes a protein that belongs to the ToxR family of transcriptional activators. To determine if SirR regulates genes required for anaerobic growth, a sirR (∆sirR) mutant was generated and tested for anerobic respiration with diverse electron acceptors utilized by the wild type S. oneidensis MR-1. The ∆sirR mutant was deficient in sulfite reduction but grew with all other electron acceptors tested. Complementation of the ∆sirR mutant restored sulfite reduction and H2S production to wild type levels (Figs. 2a and S1). To demonstrate that SirR regulates the expression of the sirAIGCDJKLM operon, a sirA promoter-lacZ fusion was generated. Expression of the sirA operon was assessed, by way of promoter activity, in the MR-1 parent strain, the ∆sirR mutant, and as a negative control, the ∆crp mutant. Expression of the sirA operon was eliminated in both the ∆sirR and ∆crp mutants (Fig. 2b). These results indicated that both SirR and CRP were required for sulfite reduction, yet it was unknown whether these proteins regulated sirA expression independently or as part of a regulatory cascade.
Figure 1.
Arrangement of the sir and ccm gene clusters on the S. oneidensis chromosome. Letters correspond to the sir or ccm gene names unless indicated otherwise.
Figure 2.
SirR regulates sulfite reduction via regulation of sirA expression. (a) Sulfite reduction is indicated by loss of sulfite (SO3) and accumulation of hydrogen sulfide (H2S). Loss of sirR completely abolished sulfite reduction. Complementation of sirR restored sulfite reduction to wild type levels. (b) Activity of the sirA promoter was measured by lacZ-promoter fusions. Beta-galactosidase activity was measured after overnight aerobic growth or anerobic growth with sulfite as the sole electron acceptor. Both sirR and crp were required for expression of the sulfite reductase operon, as indicated by loss of lacZ expression in the ΔsirR and Δcrp mutants. (c) Expression of sirR and sirA in the wild-type MR-1 and Δcrp mutant. CRP was required for wild-type level expression of sirR and sirA, which indicated that CRP was required for optimal expression of sirR as well as sirA. *p < 0.05, **p < 0.005, ANOVA.
CRP is known to regulate the expression of many genes involved in anaerobic respiration, and sequences that match the E. coli CRP-binding site (TGTGA------TCACA) were identified upstream of sirR (TGAGT------TCACA). To determine if CRP regulates sirR expression, we assessed relative transcripts of sirR and sirA in the MR-1 wild-type and Δcrp mutant. The transcript level of sirR was significantly reduced in the ∆crp mutant compared to the wild type (p = 0.002), and subsequently sirA transcript was further reduced in the Δcrp mutant (p = 0.018) (Fig. 2c). These results indicated that CRP positively regulates the expression of sirR, which correlates with previously published transcriptomic data that indicated reduced expression of sirR in the ∆crp mutant35. These results further suggest that CRP likely regulates sulfite reduction via regulation of the sulfite reductase regulator SirR.
The atypical heme binding motif of SirA is required for sulfite reduction
The S. oneidensis sulfite reductase SirA belongs to the MccA family of c-type cytochromes and in addition to typical CXXCH heme binding sites, contains a CX12NKGCH motif predicted to bind heme47 (Fig. 3a). A CX12AKGCH motif was found to be essential for the activity of the W. succinogenes sulfite reductase22. We hypothesized that this atypical heme-binding motif is essential for the S. oneidensis SirA sulfite reductase activity as well. To test this, we generated a SirA mutant, SirAN589C, where the asparagine in the CX12NKGCH motif was replaced with a cysteine to generate a typical heme-binding site (CX12CKGCH). The ability of SirAN589C to restore sulfite reductase activity to the ∆sirA mutant was tested by measuring H2S production with sulfite as the sole electron acceptor. Our results indicated that SirAN589C was completely deficient in sulfite reduction (Fig. 3b). Furthermore, we did not detect sulfite reductase activity by SirAN589C in native polyacrylamide gels when methyl viologen was used as the electron donor. This deficiency appears to be due to loss of protein as evidenced by the lack of a band that reacted with peptide antibodies generated against SirA (Fig. 3c). These results support the prediction that the atypical CX12NKGCH site is essential for SirA activity, as was observed in its W. succinogenes counterpart.
Figure 3.
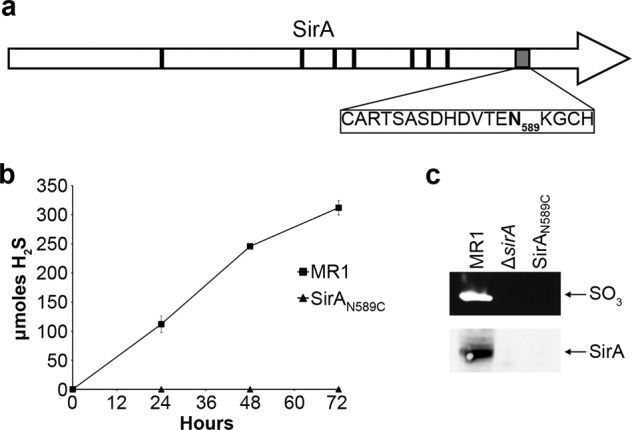
The atypical heme binding site of SirA is required for maturation and stability. (a) Locations of typical and atypical heme binding sites of SirA. Black boxes indicate typical CXXCH binding sites, the gray box indicates the atypical CX12NKGCH binding site. Asparagine 589 (bolded) was changed to a cystine to restore the atypical binding site to the typical CXXCH motif. (b) Shewanella that expressed the wildtype or mutated SirA were grown anaerobically on sulfite. The SirAN589C mutant was unable to reduce sulfite, as indicated by lack of hydrogen sulfide production. (c) Whole cell extracts from the wildtype, ΔsirA and SirAN589C strains were tested for sulfite reductase activity and the SirA protein. Neither ΔsirA nor SirAN589C cell extracts had an active sulfite reductase or a band that reacted with antibodies directed against SirA. Full-length blots/gels are presented in Supplementary Fig. S3.
The Ccm and Sir systems are required for proper maturation of the sulfite reductase SirA
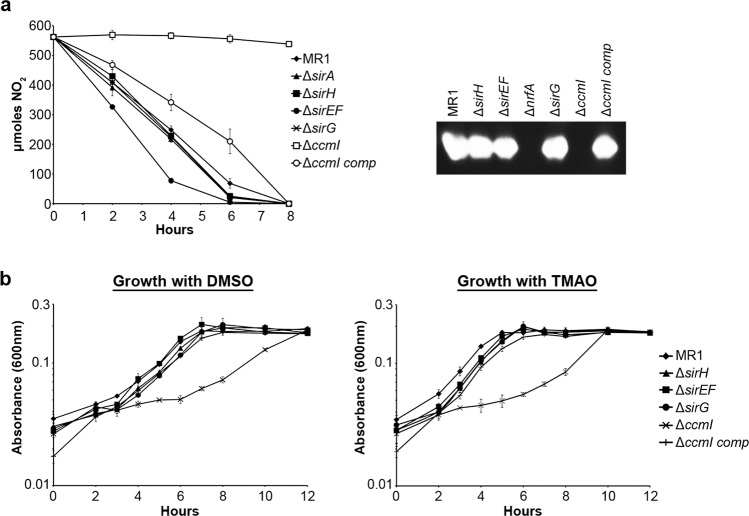
Based on studies of heme maturation systems in other bacteria, such as E. coli and W. succinogenes22,30,31,33,47,48, we hypothesized that heme ligation to the atypical CX12NKGCH motif of SirA would require a specific and dedicated heme lyase complex. To identify the proteins involved in maturation of SirA, we first tested the role of CcmI and SirG in sulfite reduction. Previous studies that investigated the role of CcmI in maturation of the nitrite reductase NrfA, indicated that loss of CcmI may result in a partial, but incomplete, defect in SirA enzyme activity29. Accordingly, we found that mutants that lack either SirG or CcmI were able to reduce sulfite, although a lag phase of 24 hr (SirG) and 48 hr (CcmI), with respect to wildtype, was observed before H2S production was detected (Fig. 4a). However, a mutant that lacked both SirG and CcmI was completely deficient in sulfite reduction, which suggested that SirG and CcmI may be able to partially compensate for loss of one another (Fig. 4a). Introduction of sirG into the ∆sirG∆ccmI mutant partially restored sulfite reduction, with reduction rates similar to that of the single ∆ccmI mutant. Similarly, introduction of ccmI into the ∆sirG∆ccmI mutant resulted in sulfite reduction rates similar to that of the ∆sirG mutant (Fig. 4a,b). These results indicated that, contrary to nitrite reduction where CcmI was essential, neither CcmI nor SirG were essential for sulfite reduction. Both proteins appear to have a partially redundant function, and complete loss of sulfite reduction was observed only when both proteins were absent.
Figure 4.
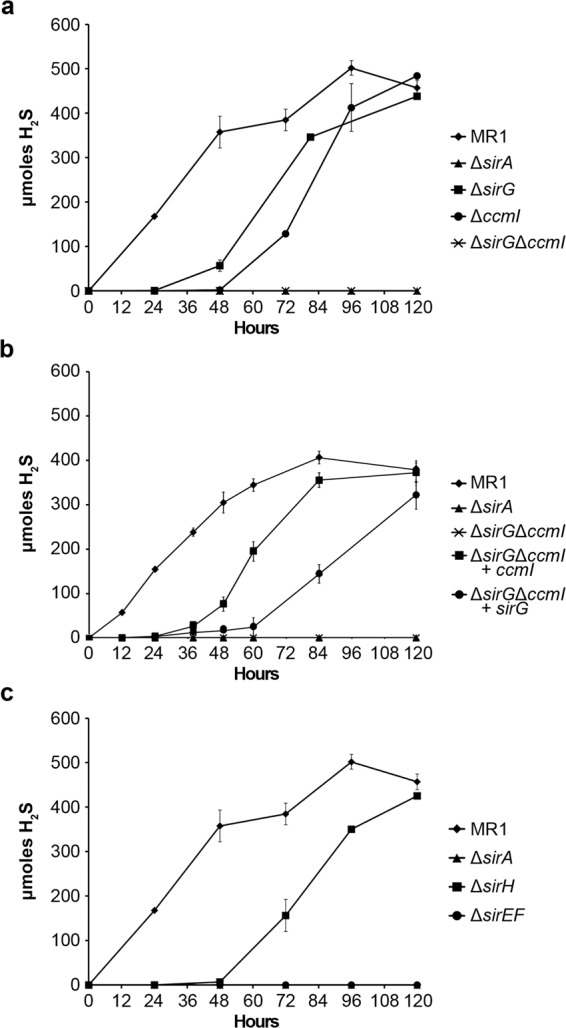
Cytochrome maturation and heme lyase components are required for optimal sulfite reduction. Wildtype and mutant S. oneidensis were grown anaerobically with sulfite as the sole electron acceptor. Sulfite reduction was indicated by the production of hydrogen sulfide (H2S). (a) Loss of either heme chaperone, SirG or CcmI, resulted in a lag in sulfite reduction, likely due to a reduction in functionally active reductase. Sulfite activity was abolished when both sirG and ccmI were deleted. (b) Sulfite reductase activity of the ΔsirGΔccmI double mutant was partially restored, similar to that of the respected single mutant, when complemented with either sirG or ccmI. (c) Loss of the heme lyase components SirH or SirEF resulted in decreased sulfite reduction. The ΔsirH mutant exhibited a 48-hour lag in sulfite reduction compared to wildtype, whereas the ΔsirEF mutant lacked any sulfite reduction. The ΔsirA mutant served as a negative control for sulfite reduction in all assays.
To confirm that both SirG and CcmI directly interact with the apo-SirA during maturation, bacterial two-hybrid analysis was performed. Constructs were generated to assess the interaction between SirA and SirG, and between SirA and CcmI. The bacterial two-hybrid assay indicated that both SirG and CcmI interact with SirA independently of one another and in the absence of other components of the S. oneidensis Ccm and Sir maturation systems (Supplementary Fig. S2). Complimentary assays were performed in which SirA served as both the bait (pBT) or the target (pTRG) protein. The results obtained were the same regardless of the orientation of the two-hybrid assay, and further supported the observation that both SirG and CcmI are involved in direct maturation of the SirA sulfite reductase.
Located within the sir gene cluster, are sirEFH. SirEF are similar to the NrfEF cytochrome c synthetase and thiol-oxidoreductase that are required for heme ligation to the CXXCK site in NrfA (31). Interestingly, previous work found that SirE is not required for nitrite reduction in S. oneidensis29. The third gene in the sirEFH operon, sirH, encodes a protein similar to thioredoxin-like proteins of the TlpA- DsbE- ResA family49. It is predicted to be a periplasmic protein with 63% similarity to the protein encoded by SO_0269, which lies downstream of ccmH (Fig. 1). SirH also shares similarity with P. aeruginosa thioredoxins (NP_251167.1 and NP_249644.1) with 54% and 49% similarity, respectively. To determine if SirEFH were required for maturation of a functional sulfite reductase, a ∆sirEF double mutant and a ∆sirH single mutant were generated and tested for the ability to reduce sulfite. Deletion of sirEF resulted in complete loss of sulfite reduction (Fig. 4c). Complementation of the ∆sirEF mutant restored sulfite reduction to wild type levels. In contrast, a mutant that lacked sirH, which appears to form an operon with sirEF, was able to reduce sulfite at a rate similar to wild type after a lag period of 48 hr (Fig. 4c). This delay in reduction may be due to inefficient maturation of the sulfite reductase, suggesting that SirH is important, but is not essential, for maturation of SirA.
Sulfite reductase activity and protein stability
To further elucidate the role of the heme lyase proteins on maturation of the terminal sulfite reductase, sulfite reductase activity was measured in wildtype and mutant strains. S. oneidensis strains were grown for 24 hr with SO3−2 and then tested for sulfite reductase activity on native polyacrylamide gels using methyl viologen as the sole electron donor. Reductase activity was indicated as a band of clearing in the gel. As expected, ∆sirA was completely deficient in sulfite reductase activity (Fig. 5). Cell extracts from the ∆sirH and ∆sirG mutants exhibited decreased sulfite reductase activity, which was restored to wild type levels by complementation of the mutation. Cell extracts from the ∆sirEF mutant completely lacked sulfite reductase activity (Fig. 5), which further supported the hypothesis that SirEF are essential for maturation of the sulfite reductase. Sulfite reductase activity was barely detectable in ∆sirG and completely absent in ∆ccmI grown for 24 hr, further substantiating the sulfite reduction results presented in Fig. 4 and indicating that both SirG and CcmI have a role in sulfite reducatase activity at this time point. Cell extracts of the ∆ccmI∆sirG double mutant completely lacked sulfite reductase activity (Fig. 5). Moreover, introduction of ccmI, but not sirG, into the ∆ccmI∆sirG double mutant restored sulfite reductase activity (Fig. 5). These results support our findings that CcmI has a more prominent role in sulfite reductase maturation than SirG, although neither CcmI or SirG can fully support wild type levels of SirA maturation. Together these observations indicated that sulfite reduction by intact cells is proportional to sulfite reductase activity of cell extracts, and further suggested that the observed sulfite reduction deficiency in the mutants was not due to loss of other c-type cytochromes that are part of the electron transport chain that leads to sulfite reduction.
Figure 5.
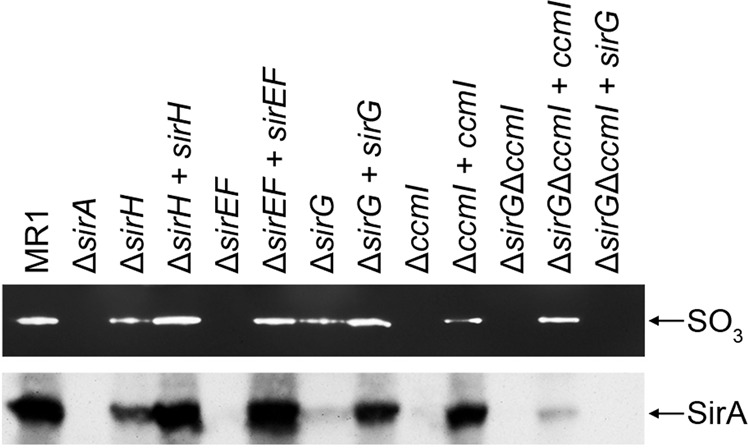
Sulfite reductase activity and protein levels of wild-type, mutant, and complemented mutant cell extracts. Upper panel. Sulfite reductase activity was indicated by bands of clearing. No activity was observed in extracts from ΔsirA, ΔsirEF, ΔccmI or ΔccmIΔsirG. Reduced activity was observed in cell extracts from ΔsirH and ΔsirG. Complementation restored reductase activity to wildtype levels, except with ΔccmI, which partially restored activity. Lower panel. Western blot analysis of the cell extracts was performed using antibodies against SirA. Reactive bands that corresponded to SirA are indicated. Reduced SirA was detected in extracts from ΔsirH and ΔsirG compared to wildtype and was absent in extracts from ΔsirA, ΔsirEF, ΔccmI and ΔccmIΔsirG. Complementation restored SirA levels similar to that of the wildtype or corresponding single mutants, in the case of the ΔsirGΔccmI mutant. Full-length blots/gels are presented in Supplementary Fig. S5.
The observed decrease or absence of sulfite reductase activity in cell extracts of the ccmI and sirEF, sirG, and sirH mutants may be due to decreased catalytic activity or reduced protein levels. To distinguish between these two possibilities, we assessed SirA levels in cell extracts from the wild type strain and mutants using peptide antibodies generated against SirA. A reactive band of 72 kDa that corresponded to the predicted size of SirA was detected in the wild type and to a lesser extent in the ∆sirH and ∆sirG cell extracts (Fig. 5). This band was absent in cell extracts from the ∆sirEF and ∆ccmI mutants and was restored by complementation. Similar to the results obtained for enzyme activity, cell extracts from the ∆sirG∆ccmI double mutant lacked SirA, presumably as a result of protein degradation. Cell extracts of the ∆sirG∆ccmI mutant complemented with ccmI, had a faint band that reacted with SirA antibodies; however, the band was absent in cell extracts when the double mutant was complemented with only sirG (Fig. 5). These results indicated that the sulfite reduction deficiency observed in the cytochrome c maturation mutants was due to decreased protein levels, most likely due to instability and degradation of the unmatured apoprotein.
The Sir system is exclusive to the maturation of the SirA reductase
Previous reports indicated that the SirEFH complex is not required for maturation of c-cytochromes that possess only typical CXXCH heme-binding sites28 and that SirE is not required for nitrite reductase activity29. However, it remained unclear what role, if any, SirG plays in the maturation of any of these other c-cytochrome reductases. As both CcmI and SirG were involved in maturation of the sulfite reductase, which possesses both typical and atypical heme-binding sites, we first determined if SirG was involved in maturation of the nitrite reductase, NrfA, which also possesses an atypical, albeit unique, heme-binding site. Nitrite reduction assays and direct measurement of enzymatic activity suggest that SirG is not required for maturation of NrfA and that SirG is unable to compensate for loss of CcmI, with respect to NrfA maturation (Fig. 6a). We also assessed the role of SirG in maturation of two c-cytochromes that only have typical CXXCH sites, the DMSO and TMAO reductases. Growth with DMSO or TMAO as the sole electron acceptor was unaffected by loss of any of the sir genes. As expected, loss of CcmI resulted in decreased growth with either DMSO or TMAO. Interestingly, following a long lag phase, compared to wild type, the ΔccmI mutant was able to grow to wild type levels with both DMSO or TMAO as the sole electron acceptor (Fig. 6b). These results confirmed that SirEFGH are required for optimal maturation of the sulfite reductase, but do not have an appreciable role in the maturation of other c-type cytochromes.
Figure 6.
The sir cytochrome maturation genes are not involved in maturation of the NrfA nitrite reductase. (a) Nitrite reduction was not significantly decreased in any of the sir mutants, however the ΔsirEF mutants reduced nitrite faster than the wildtype. As anticipated, ΔccmI was unable to reduce nitrite and reduction was restored by complementation. Enzymatic activity of the nitrate reductase was not affected in any of the sir mutants. There was no nitrite reductase activity in the cell lysates of ΔnrfA or ΔccmI. (b) ΔsirH, ΔsirEF and ΔsirG were all able to grow on DMSO or TMAO similar to wildtype. The ΔccmI mutant exhibited a lag in growth with either DMSO or TMAO, which was restored by complementation. Full-length gel is presented in Supplementary Fig. S4.
Discussion
Maturation of c-type cytochromes requires specialized systems for the covalent attachment of heme b to CXXCH motifs of apocytochromes. In bacteria, System I (Ccm) or System II (Ccs) is required for holocytochrome c maturation and involves an apocytochrome c chaperone and a cytochrome c synthetase. In bacteria such as Rhodobacter capsulatus and Pseudomonas aeruginosa, CcmI, CcmH, and CcmF fulfill these roles, while in Escherichia coli CcmH appears to be a fusion protein that carries out the functions of both CcmI and CcmH23,25,50,51. For c-type cytochromes with atypical heme-binding motifs, such as CXXCK found in the nitrite reductase of E. coli and other bacteria, specific heme lyases are required for heme attachment to these non-conventional sites. The genome of the metal reducer S. oneidensis MR-1 encodes 42 c-type cytochromes, many of which serve as terminal reductases during anaerobic respiration and all contain the typical CXXCH motif16,17. The System I proteins CcmABCDEFGH were found to be essential for maturation of all c-type cytochromes in S. oneidensis28,29, but none of these proteins were predicted to act as the apocytochrome c chaperone component of the heme lyase. Recently, CcmISo (SO_0265) was identified as a cytochrome c maturation protein required for maturation of the nitrite reductase NrfA28,29 and that we predicted functions as an apocytochrome c chaperone for maturation of the sulfite reductase SirA. In contrast to an R. capsulatus ccmI null mutant that is completely deficient in mature c-type cytochromes52, an S. oneidensis ccmI mutant was able to grow anaerobically similar to the wild type with some electron acceptors, but failed to grow with others28,29.
S. oneidensis MR-1 possess two c-cytochromes, NrfA and SirA, that in addition to typical CXXCH heme-binding motifs, contain atypical heme-binding sites. The S. oneidensis nitrite reductase, NrfA, is a pentaheme c-type cytochrome with 4 CXXCH and one CXXCK heme-binding sites, and is similar to the NrfA proteins of E. coli and W. succinogenes30,53–55. Maturation of NrfA in W. succinogenes and E. coli requires the dedicated heme lyases NrfI and NrfEFG, respectively30,32,48,56. Unlike these bacteria, S. oneidensis does not appear to have a dedicated heme lyase system for the maturation of its NrfA protein. Although CcmI is essential for the maturation of NrfA, it also participates in the maturation of other c-type cytochromes. Furthermore, CcmF is required for maturation of NrfA, yet loss of SirEF has no effect on nitrite reduction. The lack of a dedicated cytochrome c synthetase system for maturation of the nitrite reductase in S. oneidensis, suggests that CcmFHI may form the heme lyase complex responsible for heme attachment to the CXXCK site. If this is the case, then it further suggests that the same heme synthetase CcmF of S. oneidensis is able to ligate heme to apocytochromes c with both CXXCH and CXXCK heme-binding motifs.
The S. oneidensis sulfite reductase subunit, SirA, is an octaheme c-type cytochrome with 7 CXXCH motifs and an atypical CX12NKGCH site10. Similar atypical sequences have also been identified in the sulfite reductase of Wolinella succinogenes and in the MccA family of c-type cytochromes22,47. In S. oneidensis, this site appears to be important for the activity and stability of SirA, and may be the catalytic site of the enzyme. Substitution of the asparagine residue in CX12NKGCH with a cysteine, to mimic the typical CXXCH heme-binding motif, completely abolished sulfite reductase activity. Furthermore, we did not detect SirAN589C in Western blots suggesting that the mutation led to instability and degradation of the protein. These findings are similar to previous reports where amino acid substitutions in the heme-binding site or deletion of ccm genes lead to instability of the apocytochromes30,31,57,58 and ineffective maturation of the NrfA reductase leads to rapid degradation29.
To date, maturation of the MccA family of c type cytochromes has been studied only in W. succinogenes. In this bacterium, a specific heme lyase system is responsible for heme attachment to the CX15CH motif22,33. Our results described above show that in S. oneidensis, ccmISo is involved in the maturation of the sulfite reductase SirA, which contains an atypical CX12NGKCH site. A ccmISo null mutant produced mature sulfite reductase more slowly than the wild type as indicated by reduction and enzyme assays. We consistently observed a lag phase of 48 hr in sulfite reduction by ∆ccmISo, which suggested that unlike its essential role in the maturation of the nitrite reductase NrfA, CcmI was involved in, but not required for, maturation of the sulfite reductase SirA. ∆ccmISo was also deficient in DMSO and TMAO respiration, as has been shown previously28, but was able to grow to wild type levels after an extended lag phase. The ∆ccmISo phenotypes were surprising for several reasons. First, all c-type cytochromes involved in TMAO, DMSO, and fumarate reduction contain only typical CXXCH heme-binding motifs and a role for CcmI in the maturation of these proteins is unexpected. Second, the non-conventional heme-binding sites in the nitrite and sulfite reductases (CXXCK and CX12NGKCH respectively) are different, yet CcmI appears to be involved in the maturation of both proteins, but is only essential for the maturation of NrfA29.
In S. oneidensis, we have identified SirEFG as a heme lyase that appears to be specific for the maturation of SirA. SirEFG are similar to the E. coli proteins NrfEFG that constitute the heme lyase required for maturation of NrfA30,53. SirEF, which are similar to cytochrome c synthetases (CcmF) and thiol oxidoreductases (CcmH)59, appear to be essential for sulfite reductase activity and stability. Our results suggest that SirA requires a dedicated cytochrome c synthetase for its maturation, but unlike other c-type cytochromes studied to date, it appears to require two apocytochrome c chaperones for this process. Single mutants that lack either SirG or CcmI were able to express an active SirA enzyme, albeit at lower levels than the wild type, whereas the double mutant ∆ccmI∆sirG mutant completely lacked sulfite reductase activity and the SirA protein. Based on these results, we propose that SirEFG along with CcmI constitute a 4-subunit heme-lyase complex that functions in maturation of SirA.
Our results described above, in combination with previously published findings, indicate an unusual cytochrome c maturation system in S. oneidensis. This bacterium has two predicted heme synthetases, CcmF and SirE and two apocytochromes c chaperones, CcmI and SirG. We predict that multiple combinations of these proteins in S. oneidensis can form heme lyase complexes depending on growth conditions. For example, CcmI may interact with CcmF and CcmH when nitrite and DMSO are used as electron acceptors. CcmI may also interact with SirEFG when cells are grown with sulfite. Unlike other bacteria studied to date, S. oneidensis does not appear to have a dedicated and specific heme lyase complex for each of the heme-binding motifs found in its c-type cytochromes. These findings add another twist to the already complex mechanism of cytochrome c maturation in bacteria.
Supplementary information
Acknowledgements
This work was supported by U. S. Department of Energy (DOE) Grant No. DE-FG02-07ER64382 to D.A.S. and by a University of Wisconsin-Milwaukee Office of Undergraduate Research award to T.J.C. and a Distinguished Dissertator award to K.L.B. We thank Mark McBride for helpful comments and critical reading of the manuscript.
Author contributions
K.L.B., S.S. and D.A.S. conceived and planned the experiments. K.L.B., S.S., T.J.C., R.B. and D.A.S. performed experiments. K.L.B. and D.A.S. wrote the manuscript with support from S.S. All authors reviewed and contributed to the final version of the manuscript.
Data availability
No datasets were generated or analysed during the current study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-57587-6.
References
- 1.Fredrickson JK, et al. Towards environmental systems biology of Shewanella. Nature reviews. Microbiology. 2008;6:592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 2.Beliaev AS, Saffarini DA. Shewanella putrefaciens mtrB encodes an outer membrane protein required for Fe(III) and Mn(IV) reduction. Journal of bacteriology. 1998;180:6292–6297. doi: 10.1128/jb.180.23.6292-6297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beliaev AS, Saffarini DA, McLaughlin JL, Hunnicutt D. MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR-1. Molecular microbiology. 2001;39:722–730. doi: 10.1046/j.1365-2958.2001.02257.x. [DOI] [PubMed] [Google Scholar]
- 4.Cruz-Garcia C, Murray AE, Klappenbach JA, Stewart V, Tiedje JM. Respiratory nitrate ammonification by Shewanella oneidensis MR-1. Journal of bacteriology. 2007;189:656–662. doi: 10.1128/jb.01194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dos Santos JP, Iobbi-Nivol C, Couillault C, Giordano G, Mejean V. Molecular analysis of the trimethylamine N-oxide (TMAO) reductase respiratory system from a Shewanella species. Journal of molecular biology. 1998;284:421–433. doi: 10.1006/jmbi.1998.2155. [DOI] [PubMed] [Google Scholar]
- 6.Gao H, et al. Reduction of nitrate in Shewanella oneidensis depends on atypical NAP and NRF systems with NapB as a preferred electron transport protein from CymA to NapA. The ISME journal. 2009;3:966–976. doi: 10.1038/ismej.2009.40. [DOI] [PubMed] [Google Scholar]
- 7.Gralnick JA, Vali H, Lies DP, Newman DK. Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4669–4674. doi: 10.1073/pnas.0505959103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moser DP, Nealson KH. Growth of the facultative anaerobe Shewanella putrefaciens by elemental sulfur reduction. Applied and environmental microbiology. 1996;62:2100–2105. doi: 10.1128/AEM.62.6.2100-2105.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myers CR, Nealson KH. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science (New York, N.Y.) 1988;240:1319–1321. doi: 10.1126/science.240.4857.1319. [DOI] [PubMed] [Google Scholar]
- 10.Shirodkar S, Reed S, Romine M, Saffarini D. The octahaem SirA catalyses dissimilatory sulfite reduction in Shewanella oneidensis MR-1. Environmental microbiology. 2011;13:108–115. doi: 10.1111/j.1462-2920.2010.02313.x. [DOI] [PubMed] [Google Scholar]
- 11.Myers CR, Carstens BP, Antholine WE, Myers JM. Chromium(VI) reductase activity is associated with the cytoplasmic membrane of anaerobically grown Shewanella putrefaciens MR-1. Journal of applied microbiology. 2000;88:98–106. doi: 10.1046/j.1365-2672.2000.00910.x. [DOI] [PubMed] [Google Scholar]
- 12.Bencheikh-Latmani R, et al. Global transcriptional profiling of Shewanella oneidensis MR-1 during Cr(VI) and U(VI) reduction. Applied and environmental microbiology. 2005;71:7453–7460. doi: 10.1128/aem.71.11.7453-7460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C, Gorby YA, Zachara JM, Fredrickson JK, Brown CF. Reduction kinetics of Fe(III), Co(III), U(VI), Cr(VI), and Tc(VII) in cultures of dissimilatory metal-reducing bacteria. Biotechnology and bioengineering. 2002;80:637–649. doi: 10.1002/bit.10430. [DOI] [PubMed] [Google Scholar]
- 14.Marshall MJ, et al. c-Type cytochrome-dependent formation of U(IV) nanoparticles by Shewanella oneidensis. PLoS biology. 2006;4:e268. doi: 10.1371/journal.pbio.0040268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall MJ, et al. Hydrogenase- and outer membrane c-type cytochrome-facilitated reduction of technetium(VII) by Shewanella oneidensis MR-1. Environmental microbiology. 2008;10:125–136. doi: 10.1111/j.1462-2920.2007.01438.x. [DOI] [PubMed] [Google Scholar]
- 16.Heidelberg JF, et al. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nature biotechnology. 2002;20:1118–1123. doi: 10.1038/nbt749. [DOI] [PubMed] [Google Scholar]
- 17.Meyer TE, et al. Identification of 42 possible cytochrome C genes in the Shewanella oneidensis genome and characterization of six soluble cytochromes. Omics: a journal of integrative biology. 2004;8:57–77. doi: 10.1089/153623104773547499. [DOI] [PubMed] [Google Scholar]
- 18.Rabus, R., A. Hansen, T. & Widdel, F. Vol. 2 659–768 (2006).
- 19.Dahl C, Kredich NM, Deutzmann R, Truper HG. Dissimilatory sulphite reductase from Archaeoglobus fulgidus: physico-chemical properties of the enzyme and cloning, sequencing and analysis of the reductase genes. Journal of general microbiology. 1993;139:1817–1828. doi: 10.1099/00221287-139-8-1817. [DOI] [PubMed] [Google Scholar]
- 20.Huang CJ, Barrett EL. Identification and cloning of genes involved in anaerobic sulfite reduction by Salmonella typhimurium. Journal of bacteriology. 1990;172:4100–4102. doi: 10.1128/JB.172.7.4100-4102.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang CJ, Barrett EL. Sequence analysis and expression of the Salmonella typhimurium asr operon encoding production of hydrogen sulfide from sulfite. Journal of bacteriology. 1991;173:1544–1553. doi: 10.1128/JB.173.4.1544-1553.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kern M, Klotz MG, Simon J. The Wolinella succinogenes mcc gene cluster encodes an unconventional respiratory sulphite reduction system. Molecular microbiology. 2011;82:1515–1530. doi: 10.1111/j.1365-2958.2011.07906.x. [DOI] [PubMed] [Google Scholar]
- 23.Sanders C, Turkarslan S, Lee DW, Daldal F. Cytochrome c biogenesis: the Ccm system. Trends in microbiology. 2010;18:266–274. doi: 10.1016/j.tim.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kranz, R. G., Richard-Fogal, C., Taylor, J. S. & Frawley, E. R. Cytochrome c biogenesis: mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiology and molecular biology reviews: MMBR73, 510–528, Table of Contents, 10.1128/mmbr.00001-09 (2009). [DOI] [PMC free article] [PubMed]
- 25.Travaglini-Allocatelli C. Protein Machineries Involved in the Attachment of Heme to Cytochrome c: Protein Structures and Molecular Mechanisms. Scientifica. 2013;2013:505714. doi: 10.1155/2013/505714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanders C, et al. The cytochrome c maturation components CcmF, CcmH, and CcmI form a membrane-integral multisubunit heme ligation complex. The Journal of biological chemistry. 2008;283:29715–29722. doi: 10.1074/jbc.M805413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouhenni R, Gehrke A, Saffarini D. Identification of genes involved in cytochrome c biogenesis in Shewanella oneidensis, using a modified mariner transposon. Applied and environmental microbiology. 2005;71:4935–4937. doi: 10.1128/aem.71.8.4935-4937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin M, et al. Unique organizational and functional features of the cytochrome c maturation system in Shewanella oneidensis. PloS one. 2013;8:e75610. doi: 10.1371/journal.pone.0075610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu H, Jin M, Wan F, Gao H. Shewanella oneidensis cytochrome c maturation component CcmI is essential for heme attachment at the non-canonical motif of nitrite reductase NrfA. Molecular microbiology. 2015;95:410–425. doi: 10.1111/mmi.12865. [DOI] [PubMed] [Google Scholar]
- 30.Eaves DJ, et al. Involvement of products of the nrfEFG genes in the covalent attachment of haem c to a novel cysteine-lysine motif in the cytochrome c552 nitrite reductase from Escherichia coli. Molecular microbiology. 1998;28:205–216. doi: 10.1046/j.1365-2958.1998.00792.x. [DOI] [PubMed] [Google Scholar]
- 31.Kern M, Simon J. Production of recombinant multiheme cytochromes c in Wolinella succinogenes. Methods in enzymology. 2011;486:429–446. doi: 10.1016/b978-0-12-381294-0.00019-5. [DOI] [PubMed] [Google Scholar]
- 32.Kern M, Simon J. Electron transport chains and bioenergetics of respiratory nitrogen metabolism in Wolinella succinogenes and other Epsilonproteobacteria. Biochimica et biophysica acta. 2009;1787:646–656. doi: 10.1016/j.bbabio.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 33.Kern M, Eisel F, Scheithauer J, Kranz RG, Simon J. Substrate specificity of three cytochrome c haem lyase isoenzymes from Wolinella succinogenes: unconventional haem c binding motifs are not sufficient for haem c attachment by NrfI and CcsA1. Molecular microbiology. 2010;75:122–137. doi: 10.1111/j.1365-2958.2009.06965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Youngblut M, et al. Laue crystal structure of Shewanella oneidensis cytochrome c nitrite reductase from a high-yield expression system. Journal of biological inorganic chemistry: JBIC: a publication of the Society of Biological Inorganic Chemistry. 2012;17:647–662. doi: 10.1007/s00775-012-0885-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charania MA, et al. Involvement of a membrane-bound class III adenylate cyclase in regulation of anaerobic respiration in Shewanella oneidensis MR-1. Journal of bacteriology. 2009;191:4298–4306. doi: 10.1128/jb.01829-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saffarini DA, Schultz R, Beliaev A. Involvement of cyclic AMP (cAMP) and cAMP receptor protein in anaerobic respiration of Shewanella oneidensis. Journal of bacteriology. 2003;185:3668–3671. doi: 10.1128/JB.185.12.3668-3671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouhenni RA, et al. The Role of Shewanella oneidensis MR-1 Outer Surface Structures in Extracellular Electron Transfer. Electroanalysis. 2010;22:856–864. doi: 10.1002/elan.200880006. [DOI] [Google Scholar]
- 38.Dehio C, Meyer M. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. Journal of bacteriology. 1997;179:538–540. doi: 10.1128/JB.179.2.538-540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wan XF, et al. Transcriptomic and proteomic characterization of the Fur modulon in the metal-reducing bacterium Shewanella oneidensis. Journal of bacteriology. 2004;186:8385–8400. doi: 10.1128/jb.186.24.8385-8400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 41.Lund K, DeMoss JA. Association-dissociation behavior and subunit structure of heat-released nitrate reductase from Escherichia coli. The Journal of biological chemistry. 1976;251:2207–2216. [PubMed] [Google Scholar]
- 42.Haschke RH, Campbell LL. Thiosulfate reductase of Desulfovibrio vulgaris. Journal of bacteriology. 1971;106:603–607. doi: 10.1128/JB.106.2.603-607.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cline JD. Spectrophotometric Determination of Hydrogen Sulfide in Natural Waters. Limnology and Oceanography. 1969;14:454–458. doi: 10.4319/lo.1969.14.3.0454. [DOI] [Google Scholar]
- 44.Kletzin A. Coupled enzymatic production of sulfite, thiosulfate, and hydrogen sulfide from sulfur: purification and properties of a sulfur oxygenase reductase from the facultatively anaerobic archaebacterium Desulfurolobus ambivalens. Journal of bacteriology. 1989;171:1638–1643. doi: 10.1128/JB.171.3.1638-1643.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shroff NP, Charania MA, Saffarini DA. ArcB1, a homolog of Escherichia coli ArcB, regulates dimethyl sulfoxide reduction in Shewanella oneidensis MR-1. Journal of bacteriology. 2010;192:3227–3230. doi: 10.1128/jb.01695-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller, J. H. Experiments in Molecular Genetics. (Cold Spring Harbor Laboratory, 1972).
- 47.Hartshorne S, Richardson DJ, Simon J. Multiple haem lyase genes indicate substrate specificity in cytochrome c biogenesis. Biochemical Society transactions. 2006;34:146–149. doi: 10.1042/bst0340146. [DOI] [PubMed] [Google Scholar]
- 48.Pisa R, Stein T, Eichler R, Gross R, Simon J. The nrfI gene is essential for the attachment of the active site haem group of Wolinella succinogenes cytochrome c nitrite reductase. Molecular microbiology. 2002;43:763–770. doi: 10.1046/j.1365-2958.2002.02784.x. [DOI] [PubMed] [Google Scholar]
- 49.Crow A, Acheson RM, Le Brun NE, Oubrie A. Structural basis of Redox-coupled protein substrate selection by the cytochrome c biosynthesis protein ResA. The Journal of biological chemistry. 2004;279:23654–23660. doi: 10.1074/jbc.M402823200. [DOI] [PubMed] [Google Scholar]
- 50.Di Silvio E, Di Matteo A, Malatesta F, Travaglini-Allocatelli C. Recognition and binding of apocytochrome c to P. aeruginosa CcmI, a component of cytochrome c maturation machinery. Biochimica et biophysica acta. 2013;1834:1554–1561. doi: 10.1016/j.bbapap.2013.04.027. [DOI] [PubMed] [Google Scholar]
- 51.Verissimo AF, Yang H, Wu X, Sanders C, Daldal F. CcmI subunit of CcmFHI heme ligation complex functions as an apocytochrome c chaperone during c-type cytochrome maturation. The Journal of biological chemistry. 2011;286:40452–40463. doi: 10.1074/jbc.M111.277764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanders C, Boulay C, Daldal F. Membrane-spanning and periplasmic segments of CcmI have distinct functions during cytochrome c Biogenesis in Rhodobacter capsulatus. Journal of bacteriology. 2007;189:789–800. doi: 10.1128/jb.01441-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hussain H, Grove J, Griffiths L, Busby S, Cole J. A seven-gene operon essential for formate-dependent nitrite reduction to ammonia by enteric bacteria. Molecular microbiology. 1994;12:153–163. doi: 10.1111/j.1365-2958.1994.tb01004.x. [DOI] [PubMed] [Google Scholar]
- 54.Bamford VA, et al. Structure and spectroscopy of the periplasmic cytochrome c nitrite reductase from Escherichia coli. Biochemistry. 2002;41:2921–2931. doi: 10.1021/bi015765d. [DOI] [PubMed] [Google Scholar]
- 55.Einsle O, et al. Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs. The Journal of biological chemistry. 2000;275:39608–39616. doi: 10.1074/jbc.M006188200. [DOI] [PubMed] [Google Scholar]
- 56.Han D, Kim K, Oh J, Park J, Kim Y. TPR domain of NrfG mediates complex formation between heme lyase and formate-dependent nitrite reductase in Escherichia coli O157:H7. Proteins. 2008;70:900–914. doi: 10.1002/prot.21597. [DOI] [PubMed] [Google Scholar]
- 57.Thony-Meyer L, James P, Hennecke H. From one gene to two proteins: the biogenesis of cytochromes b and c1 in Bradyrhizobium japonicum. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:5001–5005. doi: 10.1073/pnas.88.11.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao T, O’Brian MR. Iron-dependent cytochrome c1 expression is mediated by the status of heme in Bradyrhizobium japonicum. Journal of bacteriology. 2005;187:5084–5089. doi: 10.1128/jb.187.15.5084-5089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richard-Fogal CL, et al. A conserved haem redox and trafficking pathway for cofactor attachment. The EMBO journal. 2009;28:2349–2359. doi: 10.1038/emboj.2009.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Venkateswaran K, et al. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. International journal of systematic bacteriology. 1999;49(Pt 2):705–724. doi: 10.1099/00207713-49-2-705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.